Abstract
Pancreatic cancer is a devastating malignancy that ranks as the fourth leading cause of cancer-related deaths worldwide. Dismal prognosis is mainly attributable to limited knowledge of the molecular pathogenesis of the disease. miRNAs have been found to be deregulated in pancreatic cancer, affecting several steps of initiation and aggressiveness of the disease by regulating important signaling pathways, such as the KRAS and Notch pathways. Moreover, the effect of miRNAs on regulating cell cycle events and expression of transcription factors has gained a lot of attention. Recent studies have highlighted the application of miRNAs as biomarkers and therapeutic tools. The current review focuses on latest advances with respect to the roles of miRNAs in pancreatic ductal adenocarcinoma associated signaling pathways and miRNA-based therapeutics.
KEYWORDS : antisense oligonucleotides, miRNAs, pancreatic cancer
Pancreatic cancer overview
Pancreatic ductal adenocarcinoma (PDAC) is the predominant form of pancreatic neoplasms and accounts for greater than 85% of the clinical cases [1]. The disease develops via acinar-ductal metaplasia and neoplastic precursor lesions [2]. In the USA alone, 48,960 new cases of pancreatic cancer were expected to occur in 2015, with an estimated 40,560 deaths, about the same number in women (19,850) as in men (20,710). Over 96% are cancers arising from exocrine pancreas. Endocrine carcinomas are often diagnosed at a younger age and exhibit a better prognosis. From 2007 to 2011, mortality rates increased slightly by 0.3% per year. For all stages combined, the 1- and 5-year relative survival rates are 28 and 7%, respectively. For the small percentage of people diagnosed with local disease (9%), the 5-year survival is 26%, while more than half of patients (53%) are diagnosed at a late stage for which 1- and 5-year survival rates reach 15 and 2%, respectively [3]. These numbers are a staggering example of the poor prognosis associated with PDAC.
While new therapeutic options are emerging for nonhematologic malignancies, molecular-targeted therapies for pancreatic cancer have failed to make any positive impact on patient survival. In 1993, the Nobel Prize-winning discovery of small interference RNAs (siRNAs) led to an outburst of knowledge on RNA interference and gene regulation [4]. Since then, thousands of research studies have described the functional role of small noncoding RNAs, named miRNAs, in a vast panel of human pathologies. In 2002, a small genomic region in chromosome 13q14 comprising miR-15a and miR-16-1 genes was found to be commonly deleted in chronic lymphocytic leukemia, suggesting a link between miRNAs and human tumors [5]. Typically acting as negative regulators of gene expression, miRNAs have provided new directions for research on mechanisms underlying disease, serving as potential targets for therapeutic intervention.
In the current review, we discuss the role(s) of miRNAs in major signaling pathways related to pancreatic oncogenesis, with emphasis to KRAS and Notch signaling pathways that often intersect. We consequentially highlight the important functional relationship of miRNAs with the regulation of cell cycle events and expression of transcription factors. Furthermore, we provide an overview of the clinical aspects of miRNAs as potential biomarkers and discuss the therapeutic potential of manipulating these pathways via miRNAs intervention.
• Biogenesis & function of miRNAs
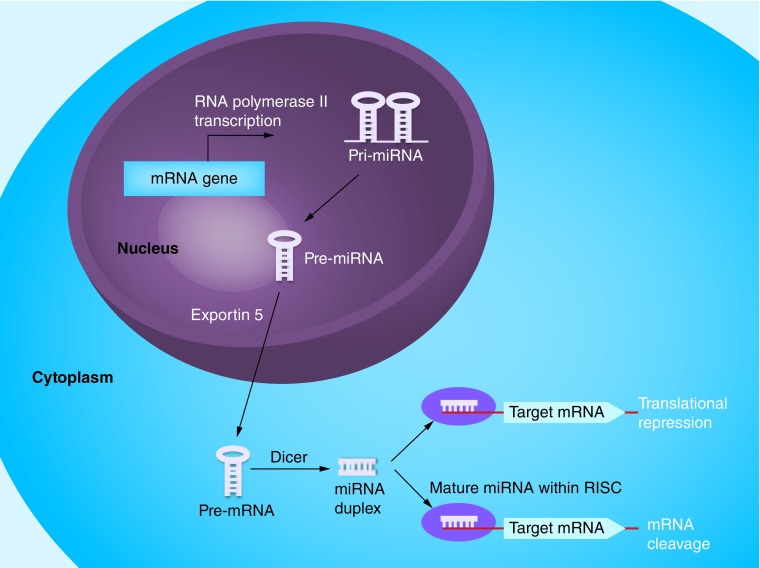
miRNAs pair to the messages of protein-coding genes, ultimately regulating gene expression through mRNA cleavage or translational inhibition [6,7]. The miRNA biogenesis pathway (Figure 1) has an important role in gene regulatory networks, considering that: more than 1000 individual miRNA genes have been identified; an individual miRNA can target hundreds or thousands of different mRNAs; an individual mRNA can be coordinately suppressed by multiple different miRNAs. Numerous studies have revealed that miRNAs mediate essential processes such as cell proliferation, apoptosis and inflammation [8–14]. Importantly, miRNAs have emerged as critical players in cancer initiation and progression by modulating pathological aspects related to tumor development, growth, metastasis and drug resistance [15,16]. Pointing to this direction, overexpression of tumor suppressor miRNAs or inhibition of oncogenic miRNAs has shown therapeutic potential in model systems [17]. Mounting evidence highlights the aberrant expression of miRNAs in various types of cancer [18–21] supporting the notion that miRNAs potentially serve as better indicators of tumor prognosis than conventional protein-coding gene arrays.
Figure 1. . Schematic presentation of miRNA biogenesis and mechanism of action.
RNA precursors called pri-miRNAs are processed by the microprocessor complex into pre-miRNA hairpins, transported into the cytoplasm and further processed by Dicer into miRNA duplexes. Following strand separation, the mature miRNAs are loaded into the RISCs to guide the repression of protein synthesis or mRNA degradation [22]. Direct binding in the 3′untranslated region of genes and specifically through sequence complementarity between nucleotides 2 and 8 of miRNAs (called seed sequence) represents their major post-transcriptional mechanism of action [7,23]. A particular miRNA recognition sequence could be found in a number of genes, which allows a single miRNA to regulate multiple functionally connected genes simultaneously and/or chronologically [16].
miRNA-mediated pathways implicated in pancreatic oncogenesis
Due to the devastating natural course of pancreatic adenocarcinoma, research has focused on the identification, classification and biology of this cancer [24,25]. Recent advances in pancreatic cancer biology have emerged important roles for miRNA families in regulating tumor responses. The following data support the strong interplay between miRNAs and pancreatic cancer initiation and development. Li and colleagues have described the implication of the entire miR-200 family of miRNAs in gemcitabine resistance of PDAC cells through reversal of epithelial–mesenchymal transition (EMT) [26]. Among the target proteins regulated by miR-34 are Notch pathway proteins and Bcl-2, suggesting a role for miR-34 in the maintenance and survival of cancer stem cells. It has also been suggested that miR-34 upregulation may restore, at least in part, the tumor suppressing function of p53 in p53-deficient human pancreatic cancer cells [27]. Importantly, oncogenic KRAS activation leads to repression of the miR-143/145 cluster in pancreatic cancer and is dependent on RREB1. KRAS and RREB1 are targets of miR-143/miR-145, revealing a feed-forward mechanism that potentiates Ras signaling [28]. However, regulation of the molecular pathways underlying pancreatic tumor initiation and progression, as well as miRNAs function in mediating signals within the tumor microenvironment still remain poorly explored. Here, we provide an updated view on the interplay between miRNAs function and signaling pathways associated to pancreatic oncogenesis, as well as miRNA-based PDAC therapeutics.
• KRAS signaling pathway
Over two decades ago, the KRAS gene was identified. Activating mutations of KRAS constitute one of the most frequently activated oncogenes, with 25% of all human tumors harboring these mutations [29]. KRAS encodes an ˜21 kDa small GTPase, which cycles between GTP-bound active and GDP-bound inactive states. GTPase-activating proteins (GAPs) induce hydrolysis of GTP resulting in KRAS inactivation. Activating mutations of KRAS found in human PDAC impair intrinsic GTPase activity of the KRAS protein and prevent its interaction with GAPs. This leads to constitutive activation of KRAS and persistent stimulation of downstream signaling pathways including sustained proliferation, metabolic reprogramming, anti-apoptosis, remodeling of the tumor microenvironment, evasion of the immune response, cell migration and metastasis [30]. KRAS is conceived as a principal initiator of pancreatic cancer, based on the early occurrence of KRAS genetic alterations (mostly point mutations at codon G12) in PDAC formation [31], as well as their prevalence at the time of diagnosis (>90% of the clinical cases) [32]. In the same direction, recently developed genetically engineered mouse models expressing mutated KRAS in the pancreas, sufficiently initiate development of neoplastic precursor lesions that are histologically identical to those in humans [33]. A recent clinical trial, describes a clinically applicable siRNA delivery method that seems to overcome the major obstacles of toxicity and organ accessibility. The miniature biodegradable implant, siG12D-LODER™ in combination with chemotherapy displayed potential efficacy in patients with locally advanced pancreatic cancer. Although, the prolonged clinical benefit warrants further evaluation of this agent in combination with chemotherapy [34]. However, KRAS is still widely considered undruggable [35] and attention is now focused on the therapeutic exploitation of its effector pathways that contribute to PDAC initiation, progression and maintenance.
Over the last several years, a handful of miRNAs have been identified as regulators of the KRAS signaling pathway in pancreatic oncogenesis. Data obtained by locked nucleic acid in situ hybridization (LNA-ISH) and real-time quantitative PCR showed that miR-217 is frequently downregulated in PDAC tissues. Manipulation of miR-217 expression provides evidence for its role as a tumor suppressor that exerts its function by specifically targeting the KRAS oncogene [36]. Another study identified miR-96 as a potent regulator of KRAS signaling. As evidenced by a subset of in vitro and in vivo assays, miR-96 directly targets KRAS, negatively regulates the phosphorylated AKT signaling pathway downstream of KRAS and exerts antiproliferative, proapoptotic and antimetastatic effects [37]. Mechanistic studies also demonstrated that downregulation of miR-126 and let-7d contributes to PDAC transformation by post-transcriptional upregulation of KRAS [38]. LNA-ISH analysis was used to confirm increased production of miR-21 in human PDAC. miR-21 has recently been linked to the activation of the AP-1 transcription factor in response to RAS [39] and thus not surprisingly, precursor lesions originating from the KRAS(G12D) model display high concentrations of this miRNA. Accordingly, it has been shown that activated KRAS(G12D) stimulates the miR-21 promoter in human pancreatic cells.
Moreover, EGFR production, which is intensified in PDAC, promotes miR-21 expression in PDAC-derived cells [40]. It has been recently reported that miR-206 is abrogated in PDAC specimens and cell lines and exerts tumor suppressive effects through combinatorial targeting of the KRAS and ANXA2 oncogenes. This study revealed the role of miR-206 as a negative regulator of oncogenic KRAS-induced NF-κB transcriptional activity, resulting in a concomitant reduction of the expression and secretion of proangiogenic and proinflammatory factors. Suppression of the potent prolymphangiogenic factor VEGF-C takes place through an NF-κB-independent mechanism. Notably, re-expression of miR-206 in PDAC cells seems to be sufficient to inhibit tumor blood and lymphatic vessel formation, thus leading to a significant delay of tumor growth and progression [41].
• Cell cycle signaling pathway
Synthesis of cyclin D and activation of CDK4/CDK6 constitute an early response to growth factor stimulation [42,43]. Subsequently, cyclin D-CDK4/6 phosphorylates Rb, a well-known tumor suppressor that binds and inhibits E2F [44]. As a result, E2F is released and mediates transcription of genes whose products are required for cell cycle progression and DNA replication [45]. Cyclin D1 overexpression and p16 inactivation are very common events in pancreatic cancer, highlighting the importance of disrupting G1 progression to disease development [46,47]. The inactivation of multiple regulatory pathways of the cell cycle may account for the aggressiveness of pancreatic cancer.
Extensive studies indicate that miRNAs play critical roles in pancreatic cancer development and progression. Zhao and colleagues revealed that miR-192 promotes cell proliferation and facilitates cell cycle progression through the G1 to S-phase in pancreatic cancer cell lines. The growth promoting effects of miR-192 enforced expression were indicated in the in vitro colony formation assay and in a xenograft tumor model in vivo. Interestingly, reduced expression of CDK inhibitors and RB family members occurred upon miR-192 overexpression, while increased expression of positive cell cycle regulators such as cyclin D1, cyclin D2, CDK4, CDC2 and SKP2 was observed [48]. Another study demonstrated that miR-301a promotes pancreatic cancer cell proliferation, at least in part, by directly targeting the 3′UTR of Bim gene [49]. Bim serves as an apoptotic stimuli sensor and initiates apoptosis through activation of multidomain proapoptotic proteins such as Bak and Bax [50].
Activation of ERK pathway is known to protect pancreatic tumor cells from apoptosis, as well as to regulate their progression in the cell cycle [51]. miR-424-5p was found to be frequently upregulated in pancreatic cancer specimens and modulate ERK1/2 signaling pathway by negatively regulating SOCS6. Data further supporting this view, show that downregulation of miR-424-5p inhibits the expression of SOCS6 downstream targets, such as Bcl-2 and MCL1, thus attenuating the ERK pathway activity [52]. A recent study unveiled the involvement of miR-193b-mediated deregulation of the KRAS axis in pancreatic carcinogenesis. Specifically, miR-193b was found to function as a cell cycle brake in PDAC cells by inducing G1-phase arrest and reducing the fraction of cells in S phase, thereby leading to decreased cell proliferation. Malignant transformation phenotype of PDAC cells was also modulated by miR-193b via anchorage-independent growth suppression. Mechanistically, KRAS was verified as a direct effector of miR-193b, through which the AKT and ERK pathways were modulated and cell growth of PDAC cells was suppressed [53].
• Notch signaling pathway
Molecular knowledge of the Notch signaling pathway with respect to pancreatic cancer is considered important for discovering new drugs and the design of novel therapeutic strategies against pancreatic cancer. Notch signals are known to affect stem cell self-renewal and differentiation, and have been suggested to play a role during pancreatic carcinogenesis. It is conceived that activation of Notch and KRAS pathways displays synergistic effects in the initiation of pancreatic carcinogenesis [54]. Interestingly, the concurrent inhibition of the EGF and Notch pathways results in decreased cell proliferation, with concomitant increase in cell apoptosis. Blockage of Notch signaling cascade has also been shown to result in attenuation of NF-κB activity and upregulation of the p21 and p27 [55].
A growing number of miRNAs has been shown to crosstalk with the Notch pathway in human PDAC. Preclinical studies have shown that TP53 directly regulates miR-34, which further downstream targets Notch, indicating a role in the maintenance and survival of PDAC initiating cells. Markedly, treatment of pancreatic cancer stem cells (CSCs) with chromatin-modulating agents resulted in suppression of the miR-34a putative targets Bcl-2, CDK6 and SIRT1 [56]. Moreover, restoration of miR-34 expression in pancreatic CSCs downregulates Notch-1 and -2, while tumor initiating cells display high levels of Notch-1 and -2, consistent with the loss of miR-34 expression. These results imply that self-renewal of pancreatic tumor initiating cells relates to direct modulation of Notch by miR-34 [27]. As illustrated by Brabletz and colleagues, miR-200 members target Notch pathway components, such as Jagged1 and the mastermind-like coactivators Maml2 and Maml3, thereby mediating enhanced Notch activation by the activator of EMT, ZEB1. These results provide a link for the latter and its cancer promoting properties to Notch activation [57]. Furthermore, examination of mouse xenografts lacking expression of the pancreatic stem cell marker, DCLK1, revealed increased miR-145, let-7a and miR-200 levels resulting in significant reduction of pluripotency and EMT-related transcription factors, along with inhibition of Notch1 via miR-144 [58].
• Crosstalk between miRNAs & transcription factors in pancreatic cancer
Transcription factors affect downstream gene transcription of signal transduction pathways triggered by genetic and epigenetic changes linked to the aggressive nature of cancer [59]. Accumulating data suggest that miRNAs exert a widespread impact on regulating either directly or indirectly the expression of transcription factors in pancreatic cancer. Inhibiting miR-22 with antisense oligonucleotides (ASOs) enhances SP1 and ESR1 expression levels in pancreatic cancer cell lines [60]. The constitutive activation of NF-κB, which regulates important genes and thereby affects many cellular processes, is also known to contribute to the aggressive behavior of pancreatic cancer [61]. miR-146a overexpression has been shown to result in inhibition of the invasive capacity of pancreatic cancer cells with concomitant downregulation of NF-κB [62]. A positive feedback loop as a mechanism for persistent NF-κB activation has been recently proposed. As suggested, miR-301a represses Nkrf to elevate NF-κB activity, which in turn promotes miR-301a transcription. Accordingly, miR-301a inhibition or Nkrf upregulation in pancreatic cancer cells led to reduced NF-κB target gene expression and attenuated xenograft tumor growth [63]. STAT3 is activated in primary pancreatic cancer and is involved in various physiologic functions [64], including apoptosis, cell cycle regulation, angiogenesis and metastasis. Mechanistic studies showed that miR-20a regulates STAT3 at the post-transcriptional level, leading to attenuation of cell proliferation and invasion of pancreatic carcinoma [65]. Dual luciferase assays revealed that STAT3 is directly targeted by miR-130b, which was further confirmed by the inverse expression of miR-130b and STAT3 in pancreatic cancer specimens [66]. Recently, another transcription factor named FOXM1 is perceived to play an important role in pancreatic cancer progression [67]. Notably, FOXM1 overexpression is responsible for acquisition of the EMT and CSC phenotypes, which is mediated in part by the regulation of miR-200b expression [68]. In addition, FOXC1 transcription factor has been recently proposed as a target of miR-138-5p, the latter exerting antiproliferative effects both in vitro and in vivo [69]. Reporter expression and chromatin immunoprecipitation assays shed insights into a novel mechanism where increased zinc, mediated by the zinc importer ZIP4, transcriptionally induces miR-373 in pancreatic cancer to promote tumor growth. Further analysis of miR-373 in vivo oncogenic function revealed that it is mediated through its negative regulation of TP53INP1, LATS2 and CD44 [70]. Moreover, TP53 has been described to be downregulated by miR-155, accelerating pancreatic tumor development [71]. miR-222 and miR-203 are also able to target p53 and affect its function as a crucial regulator of the cell cycle [72].
miRNAs as potential biomarkers in pancreatic cancer
To date, high-throughput screening strategies are used to discover relevant clinical biomarkers. High stability of miRNAs in tissues and high detection levels in very low amounts of material, represent key advantages over protein and mRNA. Historically, Schmittgen's group was the first to report the expression profiles of miRNAs in PDAC. miR-376a and miR-301 were found to be significantly overexpressed in PDAC tissues [73]. miR-132 [74,75], miR-96 [37], miR-34a [27] and miR-21 [76] have been reported to be altered in PDAC as compared with normal adjacent tissue. Since discriminating PDAC from chronic pancreatitis may prevent from unnecessary surgery, several studies focus on this issue. Initially, 21 miRNAs with increased expression and four underexpressed miRNAs that differentiate PDAC from pancreatitis with exceptional accuracy were reported [77]. Twenty additional miRNAs were discovered to discriminate between PDAC, chronic pancreatic and normal pancreas [78]. Later, expression of miR-203 [78], miR-148a, miR-196b, miR-196a and miR-205 [79] were demonstrated to be altered in PDAC versus chronic pancreatitis. In addition, miRNA expression profiles have been recently used to distinguish PDAC from cholangiocarcinoma, two practically indistinguishable cancers using conventional histopathological and clinical characteristics [80].
While the actual diagnosis of pancreatic cancer is most often done by traditional methods, such as biopsies of tissues and ultrasound-guided fine-needle aspiration, the detection of miRNAs in the plasma/serum could represent a noninvasive diagnostic method. In 2008, remarkable insights into circulating miRNAs as biomarkers for cancer classification and prognostication were reported [81]. One of the most important advantages of using circulating miRNAs as biomarkers is their stability in plasma and serum, where they are most likely protected from RNase degradation by binding to Argonaute proteins [82]. Wang and colleagues were the first to report detection of miRNAs in the blood of PDAC patients [83]. Eighty seven differentially expressed miRNAs have been identified as potential valuable markers for the assessment of cancer recurrence in patients and for people with a familial risk of PDAC [84]. In addition, miR-221 has been proposed as a useful biomarker for predicting malignant outcomes [85], while miR-486-5p and miR-938 seem to discriminate PDAC patients from healthy controls and those with chronic pancreatitis. Remarkably, the diagnostic ability of miR-486-5p was comparable to that of CA19-9, which stands for the most reliable diagnostic serum marker [86]. Recently, Goggin's group demonstrated that miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls [87]. Besides, a recent report shows that salivary miRNAs may serve as discriminatory factors in pancreatic cancer patients that are not eligible for surgery. Specifically, miR-21, miR-23a, miR-23b and miR-29c have been found significantly upregulated in the saliva of pancreatic cancer patients compared with control, showing high sensitivities and excellent specificity. Noticeably, miR-21 detection precedes detection of cancer cell specific tumor markers in the experimental model used by Humeau and colleagues. This study strongly suggests that salivary miR-21 is more sensitive than systemic-based protein markers for the diagnosis of PDAC [88]. Furthermore, the correlation between miRNA profiles and the responses of specific therapies is of great clinical significance. In support of this view, high miR-21 expression involved in gemcitabine chemoresistance, is able to predict significantly shorter overall survival in pancreatic cancer patients treated with gemcitabine [89]. So, miRNA signatures could also potentially predict responses to various cancer treatments, apart from their prognostic value. However, to establish serum/salivary miRNAs as novel biomarkers for pancreatic cancer patient's management, possibly along with additional clinical variables, validation in extended cohorts of patients is mandatory.
miRNAs relate to chemotherapy treatment in PDAC
The poor prognosis of PDAC is mainly due to its propensity to acquire resistance to chemotherapeutic agents and metastasize [90]. The combination of chemotherapy and radiation is typically not curative and provides only minor increases in survival rates in most cases. Chemotherapy drugs such as gemcitabine, albumin-bound paclitaxel nanoparticles (Abraxane) or the combination of four chemotherapy drugs, known as FOLFIRINOX (folinic acid, fluorouracil, irinotecan hydrochloride, oxaliplatin) are the standards of care for metastatic disease [91]. These treatments have limited efficacy and significant side effects, often only improving the quality of life of patients for a period of time rather than curing the disease itself. Consequently, there is an urgent need to develop a better understanding of the molecular drivers of PDAC progression and how they may relate to chemotherapy responses.
• miRNA-targeting alters chemosensitivity or radiosensitivity of PDAC cells
It is of note that, miRNA-targeting approaches have been shown to induce changes in the chemosensitivity or radiosensitivity of PDAC cells in a variety of settings. Several studies reported that antisense targeting of miR-21 and miR-221 could improve the chemosensitivity of gemcitabine resulting in significant cell killing under various conditions [92]. Consistently, anti-miR-21 transfection rendered the pancreatic cancer cells more susceptible to the cytotoxic effects of gemcitabine treatment and increased the expression of FasL. The latter proved to serve as a direct target of miR-21. Importantly, an inverse correlation between the expression of miR-21 and FasL during gemcitabine treatment was observed [93]. MiR-181b transfection has been shown to sensitize PDAC cells to gemcitabine treatment in vivo and in vitro, as evidenced by higher levels of apoptosis [94]. Nagano and colleagues suggested that activation of the Wnt/beta-catenin signaling pathway mediates the miR-29a-induced resistance to gemcitabine in PDAC cells [95]. In another study, transfection of PANC-1 and BxPC-3 cells with an miR-17-5p inhibitor showed growth inhibition, spontaneous apoptosis, higher caspase-3 activation and increased chemosensitivity to gemcitabine [96]. Additionally, miR-205 was found to be consistently downregulated across pancreatic cancer specimens, making it a suitable therapeutic target. Treatment of PDAC cells with miR-205 mimic resulted in the restoration of chemosensitivity to gemcitabine, with decreased expression of stem cell markers OCT3/4 and CD44 and chemoresistance marker class III b-tubulin [97].
Furthermore, it has been demonstrated that miR-23b overexpression inhibits radiation-induced autophagy and sensitizes PDAC cells to radiation. The authors proposed that reduced miR-23b levels increase levels of its target AGT12 and autophagy to promote radioresistance [98]. Treatment of cells with a combination of the tumor suppressor miR-205 and gemcitabine micelles reduced cell invasion and restored the gemcitabine chemosensitivity, while intratumoral injection of the combinatorial treatment in mice bearing gemcitabine-resistant xenografts potently arrested tumor growth [99].
• Combination of anticancer agents & miRNA modulators effectively inhibits tumor growth
The desmoplastic microenvironment promoting tumor growth and metastasis forms a barrier to chemotherapy. Hedgehog (Hh) signaling, that is implicated in the initiation and progression of PDAC, also contributes to desmoplasia [100]. A combination therapy comprising GDC-0449 (inhibitor of Hh pathway) and the miR-let7b mimic, effectively inhibited tumor growth when injected to athymic nude mice bearing ectopic tumors, compared with micelles carrying GDC-0449 or miR-let7b alone. Immunohistochemical analysis revealed attenuated tumor cell proliferation with augmented apoptosis in the animals treated with miR-let7b and GDC-0449 combination [101]. Based upon this evidence, rational design of combined therapeutic approaches comprising anticancer agents and miRNA modulators that act synergistically, will potentially improve therapeutic response.
Manipulation of miRNA expression levels as a therapeutic strategy in PDAC
After being widely demonstrated to be deregulated in cancer cells, researchers are now exploring therapeutic strategies based on modulation of miRNA activity. Molecular approaches are being applied that reverse the aberrant miRNA expression levels. The therapeutic application of miRNAs involves direct administration of miRNA formulations (miRNA mimics) naked, coupled to a carrier or delivered via a viral vector in order to boost miRNA endogenous levels or single-stranded oligonucleotides with miRNA complementary sequences (antisense miRNAs) either small molecules so as to diminish miRNA functions.
• miRNA replacement therapy
miRNA mimics are double-stranded synthetic miRNA oligonucleotides, used to restore miRNAs that show a loss of function. When transfected into cells, miRNA mimics are processed into a single-strand form and regulate protein-coding genes in a miRNA-like manner. This approach, also known as miRNA replacement therapy, has gained a lot of attention as it enables therapeutic exploitation of tumor suppressors [102]. Using in vitro and in vivo models based on pre-miRNA precursors (synthetic miRNA mimics), lentiviral-mediated stable miRNA overexpression or vector-mediated miRNA transient overexpression in pancreatic carcinoma cell lines, the oncogenic or tumor-suppressive role of certain miRNAs has been validated in PDAC (summarized in Table 1).
Table 1. . miRNA-based therapeutic approaches in pancreatic cancer.
| miRNA target | Role | Targeting approach | Functional outcome of miRNA-based targeting | Ref. |
|---|---|---|---|---|
| miR-483-3p |
Oncogenic |
Anti-miRNA |
Reduces cell proliferation and colony formation |
[103] |
| miR-155 |
Oncogenic |
Anti-miRNA |
Significant increase in cell apoptosis |
[71] |
| miR-21/mir-221 |
Oncogenic |
ASO |
Increases cell apoptosis/cell cycle arrest |
[92] |
| miR-21 |
Oncogenic |
HIV-1-based lentiviral vectors |
Inhibits pancreatic cancer tumor growth both in vitro and in vivo |
[104] |
| miR-27a |
Oncogenic |
Anti-miRNA |
Suppresses growth, colony formation and migration |
[105] |
| miR-371-5p |
Oncogenic |
Anti-miRNA |
Proliferative inhibition |
[106] |
| miR-21/miR-23a/miR-27a |
Oncogenic |
Anti-miRNA |
Synergistic effects in reducing cell proliferation both in vivo and in vitro |
[107] |
| miR-200 and let-7 families |
Downregulated in gemcitabine-resistant cells |
miRNA mimic or isoflavone |
Reversal of EMT phenotype leading to epithelial morphology |
[26] |
| ↑ miR-22 and ↓ miR-199a |
– |
miRNA mimic or curcumin |
Suppressed expression SP1 transcription factor and ESR1 |
[60] |
| Let-7 |
Tumor suppressor |
Plasmid-based synthetic miRNAs or by lentiviral transduction |
Strongly diminishes cell proliferation |
[108] |
| miR-34a |
Tumor suppressor |
Lentiviral system |
Inhibits clonogenic cell growth and invasion and induces apoptosis and cell cycle arrest at G1 and G2/M phase |
[27] |
| miR-217 |
Tumor suppressor |
miRNA-expressing plasmids |
Suppresses tumor cell growth in vivo |
[36] |
| miR-20a |
Tumor suppressor |
Lentiviral system |
Inhibits proliferation and metastasis |
[65] |
| miR-96 |
Tumor suppressor |
miRNA mimic and miRNA-expressing plasmid |
Reduces proliferation and invasion capacity in vitro and in vivo |
[37] |
| miR-148a |
Tumor suppressor |
miRNA mimic |
Drastic inhibition of the invasive properties of HCC |
[109] |
| miR-148a |
Tumor suppressor |
miRNA mimic and miRNA-expressing vector |
Suppresses the EMT and metastasis of HCC |
[110] |
| miR-148a |
Tumor suppressor |
Lentiviral system |
Inhibits tumor cell growth and colony formation |
[111] |
| miR-126 |
Tumor suppressor |
miRNA mimic |
Inhibition of invasive growth |
[112] |
| miR-148b |
Tumor suppressor |
miRNA mimic and lentiviral system |
Remarkably suppresses growth and invasion and enhances chemosensitivity/inhibits tumorigenicity in nude mice |
[113] |
| miR-204 |
Tumor suppressor |
miRNA mimic or triptolide treatment |
Decrease in cell viability and cell death |
[114] |
| miR-137 |
Tumor suppressor |
Lentiviral system |
Inhibits cell invasion/increases sensitivity to Fluorouracil/suppresses tumor growth in vivo |
[115] |
| miR-216a |
Tumor suppressor |
miRNA mimic |
Inhibition of the JAK2/STAT3 signaling pathway and xenograft tumor growth in vivo |
[116] |
| miR-135a |
Tumor suppressor |
Lentiviral system |
Reduced proliferation and clonogenicity/induced G1 arrest and apoptosis |
[117] |
| miR-218 |
Tumor suppressor |
miRNA mimic and lentiviral system |
Attenuation of cell migration/invasion |
[118] |
| miR-663 | Tumor suppressor | Lentiviral system | Antiproliferative, anti-invasive and pro-apoptotic effects in vivo and in vitro | [119] |
The oncogenic or tumor-suppressive effect of miRNA-based targeting is denoted in several pancreatic adenocarcinoma settings.
ASO: Antisense oligonucleotide; EMT: Epithelial–mesenchymal transition; HCC: Hepatocellular carcinoma cell.
Restoration of let-7 levels in cancer-derived cell lines strongly inhibits cell proliferation, KRAS expression and MAPK activation, but fails to impede tumor growth progression after intratumoral gene transfer or after implantation of Capan-1 cells stably overexpressing let-7 miRNA [108]. A preclinical in vivo experiment has been described, according to which PDAC cells were directly injected with miR-217-expressing plasmids using in vivo jet PEI. The results from these assays indicated that miR-217 overexpression sufficiently suppresses tumor cell growth [36]. In another study, stable lentivirus-mediated overexpression of miR-148a in IMIM-PC2 cells was shown to inhibit tumor growth and colony formation possibly via targeting of CDC25B [111]. As evidenced by miR-126 overexpression and silencing of its downstream target ADAM9, the miR-126/ADAM9 axis controls pancreatic cancer cell invasive growth [112]. Overexpression of miR-148b dramatically suppresses growth, which is attributable to induction of apoptosis and cell-cycle arrest at S phase, remarkably inhibits invasion and enhances chemosensitivity of pancreatic cancer cells. Moreover, ectopic expression of miR-148b was able to inhibit tumorigenicity in nude mice [113].
As previously described, overexpression of miR-204 either by an miR-204 mimic or by triptolide treatment, downregulates MCL1 by directly binding to the 3′UTR of the gene and causes a subsequent decrease in cell viability and pancreatic cancer cell death [114]. In addition, transfection of lentivirus containing miR-137 mimic inhibits cancer cell invasion, increases sensitivity to Fluorouracil and suppresses tumor growth in vivo [115]. miR-216a overexpression markedly inhibits the JAK2/STAT3 signaling pathway and xenograft tumor growth in vivo [116]. Furthermore, stably miR-135a overexpressing cells display reduced proliferation and clonogenicity, at least in part via the regulation of BMI1 [117]. Restoring the expression of miR-218 in pancreatic cancer resulted in downregulation of ROBO1 and efficient attenuation of cell migration and invasion [118]. A recent study refers to stably miR-663 overexpression exerting antiproliferative, anti-invasive and proapoptotic effects on pancreatic cancer cells by targeting EEF1A1 [119].
It should be highlighted that discrepancies related to assaying miRNAs as therapeutic compounds have been reported. For example, restoration of miR-148a has been shown to suppress invasiveness of hepatoma cells by targeting MET indirectly [109], as well as directly [110]. Such divergence mainly results from different cell lines and experimental strategies used.
• Novel miRNA delivery approaches
More recently, novel miRNA delivery approaches have been described. Systemic administration of an miR-34a delivery system comprising nanocomplexes with a tumor-targeting and -penetrating bifunctional CC9 peptide in a pancreatic xenograft cancer model seems to significantly inhibit tumor growth and induce cancer cell apoptosis [120]. In the same line, a lipid-based nanoparticle has been synthesized for systemic delivery of miRNA expression vectors to cancer cells. Pramanic and colleagues, demonstrated the significant therapeutic efficacy of restituting either miR-34a or miR-143/145 expression in subcutaneous and orthotropic pancreatic cancer xenograft models [121]. These approaches affirm the usefulness of miRNA uptake delivery systems in therapeutic intervention for the disease.
Furthermore, small vehicles (40–100 nm) derived from early endosomes, which fuse to multivesicular bodies called exosomes [122,123], are being discussed as possibly the most potent gene delivery system. Exosomes are secreted by many cells and abundantly by tumor cells [124] and are found in all body fluids [125]. Due to their ubiquitous presence, their particular protein profile, their equipment with mRNA and miRNA, as well as their most efficient transfer in target cells, the use of exosomes in noninvasive diagnostics and therapeutics holds great promise [126–128]. Ongoing studies confirm recovery of exosomes carrying pancreatic cancer stem cell markers [129]. Most importantly, it has been recently shown that circulating exosomes positive for the cell surface proteoglycan GPC1 may serve as a potential noninvasive diagnostic and screening tool to detect early stages of pancreatic cancer and thus facilitate possible curative surgical therapy. By using flow cytometry, circulating exosomes were detected in the serum of cancer patients with absolute specificity and sensitivity, distinguishing healthy subjects and patients with a benign pancreatic disease from patients with early- and late-stage pancreatic cancer [130]. Focalizing exosomal matrix degrading enzymes, a process that facilitates metastasizing CSCs toward the premetastatic niche, has been confirmed for a rat metastasizing pancreatic adenocarcinoma [131,132]. Furthermore, a detailed analysis has recently been presented showing that exosomes derived from malignant pancreatic lesions play a key role in liver premetastatic niche initiation [133].
Interestingly, the tumor exosome miRNA uptake from a metastasizing rat pancreatic adenocarcinoma affected premetastatic organ stroma cells toward supporting tumor cell hosting. Analysis of the exosomal miRNA-modulated gene expression in target cells showed increased protease activity, pronounced adhesion molecule and chemokine ligand expression and upregulation of cell cycle- and angiogenesis-promoting genes, as well as of genes engaged in oxidative stress response, all fitting the demands of metastasizing tumor cells for settlement and growth [134]. These findings potentially offer a means to interfere with tumor exosome promoted metastasis, a major target in pancreatic cancer therapy. Issues of mechanism elucidation, selective uptake and side effects need to be addressed and further experimental studies are necessary in order to possibly place exosomes into therapeutic settings.
• Strategies blocking miRNA functions
On the other hand, for strategies that block miRNA functions, both oligonucleotide-based and small molecule-based approaches are being explored. It has been shown that ASOs can competitively inhibit upregulated oncogenic miRNAs in tumors in a specific, efficient and long-lasting manner [135]. Preclinical studies indicate potent activity of AEG35156 (targets X-linked inhibitor of apoptosis mRNA) in combination with gemcitabine in PDAC [136]. These data imply that targeting miRNAs with ASOs could be a potential new therapeutic strategy for PDAC (summarized in Table 1). Chemical modifications to the backbones of these nucleotides led to the development of miRNA antagonists (also known as anti-miRs), including antagomiRs and LNAs. AntagomiRs represent often cholesterol-conjugated, single-stranded RNA molecules about 21–23 nucleotides in length and complementary to mature target miRNAs. The introduction of 2′-O-methyl groups or 2′-O-methoxyethyl groups contributes to nuclease resistance, as well as improved affinity and specificity toward the endogenous miRNA that is further unable to be processed by RISC [137,138]. LNAs are a class of nucleic acid analogs in which the ribose ring is ‘locked’ by a methylene bridge connecting the 2′-O atom and the 4′-C atom. By ‘locking’ the molecule with the methylene bridge, LNA oligonucleotides display remarkable hybridization affinity toward complementary single-stranded RNA [139]. miRNA antagonists specifically silence the expression of miRNA leading to upregulation of miRNA's downstream gene targets [140].
Multiple studies indicate the therapeutic potential of anti-miRs in pancreatic cancer. Specifically, transfection of Capan-2 cells with an oligonucleotide able to inhibit the miR-155 activity caused TP53INP1 re-expression, as well as a significant increase in apoptosis [71]. Inhibition of miR-27a was shown to inhibit growth, colony formation and migration of pancreatic cancer cells [105]. Likewise, anti-miR-483-3p transfection in SW1990 and PANC-1 cells significantly reduced cell proliferation and colony formation [103]. Another study showed that anti-miR-371-5p treatment causes proliferative inhibition of pancreatic cancer cells, which is partially due to a G1-phase arrest [106]. By analyzing the combined effects of altered activities of miRNAs in PDAC cell lines and in PDAC samples from patients, a combination of three miRNAs (miR-21, miR-23a and miR-27a) that cooperatively promote tumor growth, was identified. Importantly, inhibition of miR-21, miR-23a and miR-27a has synergistic effects in reducing proliferation of PDAC cells in culture and growth of xenograft tumors in mice. These findings provide evidence that coinhibition of multiple miRNAs holds promise for the development of novel cancer therapies [107]. Notably, failure of LNA-antagomiRs to successfully inhibit miR-21 function in exponentially growing tumors in vivo, while being efficient in vitro, suggests that the lack of stability and the incapacity of these molecules to overcome the tumor microenvironment comprise major hurdles for direct miRNA-based therapy of pancreatic tumors. On the other hand, transfection with HIV-1-based lentiviral vectors expressing anti-miR-21 strongly inhibits pancreatic cancer tumor growth both in vitro and in vivo. The latter results, point to the reliance of pancreatic cancer miRNA-based therapy on the delivery vector. However, the possible genotoxicity associated with the use of lentiviral vectors requires the design of novel integrase-deficient lentiviral vectors for the safe and efficient gene delivery of miRNA inhibitors in PDAC-derived cells [104].
The formation of stem loops in pre-miRNAs and bulges in miRNAs found in their secondary structure reinforced the idea of small-molecule targeting. Such structural features provide specificity basis for structure-based drug design. Small molecule inhibitors strongly interact with the surface of miRNAs, thereby interfering with the processing and the biological functions of the latter [141]. Several compounds, including isoflavone, have been shown to significantly downregulate miR-200b, miR-200c, Let-7b, Let-7c, Let-7d and Let-7e in gemcitabine-resistant cancer cells [26]. A naturally occurring flavonoid, curcumin (diferuloylmethane) has been shown to have dramatic effects on the expression profiles of miRNAs in pancreatic cancer models. Curcumin causes upregulation of miR-22 and downregulation of miR-199a, concomitantly regulating SP1 and ESR1, which are involved in cell growth, metastasis and apoptosis [60].
Future perspective
The number of studies on miRNAs in a PDAC setting is increasing at an exponential rate in recent years. Multiple deregulated miRNAs and genes in different steps of pancreatic oncogenesis have been described, supporting the link of miRNAs expression with cancer pathogenesis. Importantly, several studies provide evidence of miRNA's functional significance as mediators of important molecular drivers of PDAC. Identification of the role of miRNAs in the regulation of cell signaling pathways involved in pancreatic cancer development and progression is needed in order to broaden our understanding of the molecular background of the disease. Notably, combinatorial treatments comprising miRNA-intervening approaches and chemotherapeutic agents regulate tumor responses more efficiently.
From a clinical point of view, the perspective that introduction of miRNA mimics may contribute to pancreatic cancer control provides a novel method for pancreatic cancer therapy. Given that several reports about restoring miRNA function based on in vitro but also in vivo models have yielded significant results in repressing pancreatic cancer development, efficient reconstitution of miRNAs through in vivo delivery of pre-miRNA precursors is a crucial factor for the development of successful miRNA-based treatment modalities. However, the in vivo delivery to target sites, its penetration into tumor tissues, stability and pharmacokinetics, remain great challenges. Establishment of effective delivery systems is necessary to improve the stability and uptake of miRNAs. Toward this direction, novel miRNA delivery systems such as nanocomplexes or exosomes should gain further attention as potential means of miRNA-based PDAC therapeutics.
Silencing of miRNAs using intravenously administered chemically engineered oligonucleotides has been successfully performed in many solid organs and in vivo models [142]. So far, efficient delivery of antagomiRs via systemic administration to the pancreas and hypovascular PDACs has not been demonstrated. A major disadvantage of miRNA antagonists is that they possibly exert side effects due to nonselective distribution to nontarget organs. Conjugating the anti-miRNA oligonucleotide with ligands for target organ specific cell surface receptors could contribute in overcoming the latter issue. Furthermore, development of small-molecule drugs seems to be a promising prospect on the miRNA-targeted drug discovery.
Conclusion
Taken together, there is increasing evidence that modulating miRNA levels and functions in pancreatic cancer could be potentially exploited for therapeutic gain. As discussed above, each miRNA can have different targets and could also possibly display bivalent behavior depending on the cellular environment. Therefore, therapeutic strategies directed toward targeting of a single miRNA may yield dramatic alterations in several cellular processes and also in a cell-type specific manner. These features of miRNA activity could be utilized for cancer therapy where targeting of multiple pathways might be desirable. Since the miRNA mimics and antagonists may also affect nontarget tissue when administered, be susceptible to nuclease degradation and be targeted by the innate immune system, the aforementioned methods have certain limitations with regard to their therapeutic applications. Despite these impediments, miRNA-based approaches remain a promising option for effectively targeting endogenous miRNAs. Rapid progress in clinical translation may be anticipated based on the applicability of novel miRNA-delivery systems such as nanocomplexes, exosomes, lentiviral vectors and ligand-conjugated anti-miR oligonucleotides, as well as the discovery of small-molecule drugs. Over and above, combinations of anticancer agents and miRNA modulators will potentially improve therapeutic response.
EXECUTIVE SUMMARY.
Pancreatic cancer overview
Pancreatic ductal adenocarcinoma (PDAC) is the predominant form of pancreatic neoplasms, characterized by poor prognosis.
Discovery of miRNAs has provided new directions for research on mechanisms underlying PDAC.
miRNA-mediated pathways implicated in pancreatic oncogenesis
Interplay between miRNAs function and the KRAS signaling pathway.
mRNAs play critical roles in the regulation of cell cycle events.
Crosstalk between miRNAs and the Notch signaling pathway.
miRNAs exert a widespread impact on regulating either directly or indirectly the expression of transcription factors.
miRNAs relate to chemotherapy responses in PDAC
miRNA targeting approaches induce changes in the chemosensitivity or radiosensitivity in a variety of settings.
Rational design of combinatorial treatments comprising anticancer agents and miRNA modulators will potentially improve therapeutic response.
miRNAs as potential biomarkers in pancreatic cancer
Certain miRNAs have been discovered to discriminate between PDAC, chronic pancreatic and normal pancreas tissue.
Differentially expressed miRNAs detected in serum/saliva have been identified as potential valuable markers for the assessment of pancreatic cancer.
Manipulation of miRNA expression levels as a therapeutic strategy in PDAC
miRNA replacement therapy enables therapeutic exploitation of tumor suppressors.
Nanocomplexes and exosomes could serve as more efficient miRNA-delivery systems.
Targeting single or multiple miRNAs with antisense oligonucleotides or small molecule inhibitors holds promise for the development of novel pancreatic cancer therapies.
Lentiviral vectors efficiently deliver therapeutic miRNAs into PDAC cells, both in vitro and in vivo.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363(9414):1049–1057. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- 2.Morris JPT, Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat. Rev. Cancer. 2010;10(10):683–695. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Facts & Figures 2015. American Cancer Society; Atlanta, GA, USA: 2015. [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Presents evidence for the involvement of miRNA genes in human tumors.
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33(4):1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanno B, Cesi V, Vitali R, et al. Silencing of endogenous IGFBP-5 by micro RNA interference affects proliferation, apoptosis and differentiation of neuroblastoma cells. Cell Death Differ. 2005;12(3):213–223. doi: 10.1038/sj.cdd.4401546. [DOI] [PubMed] [Google Scholar]
- 10.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139(4):693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell. 2010;39(4):493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatziapostolou M, Iliopoulos D. Epigenetic aberrations during oncogenesis. Cell Mol. Life Sci. 2011;68(10):1681–1702. doi: 10.1007/s00018-010-0624-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatziapostolou M, Polytarchou C, Aggelidou E, et al. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147(6):1233–1247. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bakirtzi K, Hatziapostolou M, Karagiannides I, et al. Neurotensin signaling activates microRNAs-21 and -155 and Akt, promotes tumor growth in mice, and is increased in human colon tumors. Gastroenterology. 2011;141(5):1749–1761.e1741. doi: 10.1053/j.gastro.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Highlights the dependence of malignant pathologies on the deregulated expression of miRNA genes.
- 16.Xue J, Niu J, Wu J, Wu ZH. MicroRNAs in cancer therapeutic response: friend and foe. World J. Clin. Oncol. 2014;5(4):730–743. doi: 10.5306/wjco.v5.i4.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat. Rev. Cancer. 2011;11(12):849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 19.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jamieson NB, Morran DC, Morton JP, et al. MicroRNA molecular profiles associated with diagnosis, clinicopathologic criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinoma. Clin. Cancer Res. 2012;18(2):534–545. doi: 10.1158/1078-0432.CCR-11-0679. [DOI] [PubMed] [Google Scholar]
- 21.Frampton AE, Giovannetti E, Jamieson NB, et al. A microRNA meta-signature for pancreatic ductal adenocarcinoma. Expert Rev. Mol. Diagn. 2014;14(3):267–271. doi: 10.1586/14737159.2014.893192. [DOI] [PubMed] [Google Scholar]
- 22.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 23.Drakaki A, Iliopoulos D. MicroRNA gene networks in oncogenesis. Curr. Genomics. 2009;10(1):35–41. doi: 10.2174/138920209787581299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bardeesy N, Depinho RA. Pancreatic cancer biology and genetics. Nat. Rev. Cancer. 2002;2(12):897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- 25.Iacobuzio-Donahue CA, Hruban RH. Gene expression in neoplasms of the pancreas: applications to diagnostic pathology. Adv. Anat. Pathol. 2003;10(3):125–134. doi: 10.1097/00125480-200305000-00002. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Vandenboom TG, 2nd, Kong D, et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69(16):6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji Q, Hao X, Zhang M, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS ONE. 2009;4(8):e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kent OA, Fox-Talbot K, Halushka MK. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene. 2013;32(20):2576–2585. doi: 10.1038/onc.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer. 2011;11(11):761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanda M, Matthaei H, Wu J, et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology. 2012;142(4):730–733 e739. doi: 10.1053/j.gastro.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laghi L, Orbetegli O, Bianchi P, et al. Common occurrence of multiple K-RAS mutations in pancreatic cancers with associated precursor lesions and in biliary cancers. Oncogene. 2002;21(27):4301–4306. doi: 10.1038/sj.onc.1205533. [DOI] [PubMed] [Google Scholar]
- 33.Hingorani SR, Petricoin EF, Maitra A, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4(6):437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 34.Golan T, Khvalevsky EZ, Hubert A, et al. RNAi therapy targeting KRAS in combination with chemotherapy for locally advanced pancreatic cancer patients. Oncotarget. 2015;6(27):24560–24570. doi: 10.18632/oncotarget.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat. Rev. Cancer. 2011;11(11):775–791. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM, Chen J. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS . Carcinogenesis. 2010;31(10):1726–1733. doi: 10.1093/carcin/bgq160. [DOI] [PubMed] [Google Scholar]
- 37.Yu S, Lu Z, Liu C, et al. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res. 2010;70(14):6015–6025. doi: 10.1158/0008-5472.CAN-09-4531. [DOI] [PubMed] [Google Scholar]; • Demonstrating potent regulation of the KRAS signaling pathway by miR-96, which could potentially provide a novel therapeutic strategy for treatment of pancreatic cancer and other KRAS-driven cancers.
- 38.Jiao LR, Frampton AE, Jacob J, et al. MicroRNAs targeting oncogenes are down-regulated in pancreatic malignant transformation from benign tumors. PLoS ONE. 2012;7(2):e32068. doi: 10.1371/journal.pone.0032068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Talotta F, Cimmino A, Matarazzo MR, et al. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene. 2009;28(1):73–84. doi: 10.1038/onc.2008.370. [DOI] [PubMed] [Google Scholar]
- 40.Du Rieu MC, Torrisani J, Selves J, et al. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin. Chem. 2010;56(4):603–612. doi: 10.1373/clinchem.2009.137364. [DOI] [PubMed] [Google Scholar]
- 41.Keklikoglou I, Hosaka K, Bender C, et al. MicroRNA-206 functions as a pleiotropic modulator of cell proliferation, invasion and lymphangiogenesis in pancreatic adenocarcinoma by targeting ANXA2 and KRAS genes. Oncogene. 2015;34(37):4867–4878. doi: 10.1038/onc.2014.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ladha MH, Lee KY, Upton TM, Reed MF, Ewen ME. Regulation of exit from quiescence by p27 and cyclin D1-CDK4. Mol. Cell. Biol. 1998;18(11):6605–6615. doi: 10.1128/mcb.18.11.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohtsubo M, Chibazakura T. [G1 phase regulation] Tanpakushitsu Kakusan Koso. 1996;41(12 Suppl.):1712–1718. [PubMed] [Google Scholar]
- 44.Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nat. Rev. Cancer. 2008;8(9):714–724. doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer. 2009;9(11):785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Li D, Killary AM, et al. Polymorphisms of p16, p27, p73, and MDM2 modulate response and survival of pancreatic cancer patients treated with preoperative chemoradiation. Ann. Surg. Oncol. 2009;16(2):431–439. doi: 10.1245/s10434-008-0220-8. [DOI] [PubMed] [Google Scholar]
- 47.Fry LC, Monkemuller K, Malfertheiner P. Molecular markers of pancreatic cancer: development and clinical relevance. Langenbecks Arch. Surg. 2008;393(6):883–890. doi: 10.1007/s00423-007-0276-0. [DOI] [PubMed] [Google Scholar]
- 48.Zhao C, Zhang J, Zhang S, et al. Diagnostic and biological significance of microRNA-192 in pancreatic ductal adenocarcinoma. Oncol. Rep. 2013;30(1):276–284. doi: 10.3892/or.2013.2420. [DOI] [PubMed] [Google Scholar]
- 49.Chen Z, Chen LY, Dai HY, Wang P, Gao S, Wang K. miR-301a promotes pancreatic cancer cell proliferation by directly inhibiting Bim expression. J. Cell. Biochem. 2012;113(10):3229–3235. doi: 10.1002/jcb.24200. [DOI] [PubMed] [Google Scholar]
- 50.Willis SN, Chen L, Dewson G, et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005;19(11):1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boucher MJ, Morisset J, Vachon PH, Reed JC, Laine J, Rivard N. MEK/ERK signaling pathway regulates the expression of Bcl-2, Bcl-X(L), and Mcl-1 and promotes survival of human pancreatic cancer cells. J. Cell. Biochem. 2000;79(3):355–369. [PubMed] [Google Scholar]
- 52.Wu K, Hu G, He X, et al. MicroRNA-424-5p suppresses the expression of SOCS6 in pancreatic cancer. Pathol. Oncol. Res. 2013;19(4):739–748. doi: 10.1007/s12253-013-9637-x. [DOI] [PubMed] [Google Scholar]
- 53.Jin X, Sun Y, Yang H, et al. Deregulation of the miR-193b–KRAS axis contributes to impaired cell growth in pancreatic cancer. PLoS ONE. 2015;10(4):e0125515. doi: 10.1371/journal.pone.0125515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De La OJ, Murtaugh LC. Notch and Kras in pancreatic cancer: at the crossroads of mutation, differentiation and signaling. Cell Cycle. 2009;8(12):1860–1864. doi: 10.4161/cc.8.12.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z, Zhang Y, Li Y, Banerjee S, Liao J, Sarkar FH. Down-regulation of Notch-1 contributes to cell growth inhibition and apoptosis in pancreatic cancer cells. Mol. Cancer. Ther. 2006;5(3):483–493. doi: 10.1158/1535-7163.MCT-05-0299. [DOI] [PubMed] [Google Scholar]
- 56.Nalls D, Tang SN, Rodova M, Srivastava RK, Shankar S. Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS ONE. 2011;6(8):e24099. doi: 10.1371/journal.pone.0024099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brabletz S, Bajdak K, Meidhof S, et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 2011;30(4):770–782. doi: 10.1038/emboj.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Show the importance of the ZEB1/miR-200 feedback loop controlling Notch signaling in pancreatic cancer cells.
- 58.Sureban SM, May R, Qu D, et al. DCLK1 regulates pluripotency and angiogenic factors via microRNA-dependent mechanisms in pancreatic cancer. PLoS ONE. 2013;8(9):e73940. doi: 10.1371/journal.pone.0073940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nebert DW. Transcription factors and cancer: an overview. Toxicology. 2002;181–182:131–141. doi: 10.1016/s0300-483x(02)00269-x. [DOI] [PubMed] [Google Scholar]
- 60.Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol. Cancer Ther. 2008;7(3):464–473. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- 61.Fujioka S, Sclabas GM, Schmidt C, et al. Function of nuclear factor kappaB in pancreatic cancer metastasis. Clin. Cancer Res. 2003;9(1):346–354. [PubMed] [Google Scholar]
- 62.Li Y, Vandenboom TG, 2nd, Wang Z, et al. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010;70(4):1486–1495. doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu Z, Li Y, Takwi A, et al. miR-301a as an NF-kappaB activator in pancreatic cancer cells. EMBO J. 2011;30(1):57–67. doi: 10.1038/emboj.2010.296. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Reveals a novel feedback loop mechanism of NF-κB activation by a miRNA.
- 64.Corcoran RB, Contino G, Deshpande V, et al. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res. 2011;71(14):5020–5029. doi: 10.1158/0008-5472.CAN-11-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan H, Wu J, Liu W, et al. MicroRNA-20a overexpression inhibited proliferation and metastasis of pancreatic carcinoma cells. Hum. Gene Ther. 2010;21(12):1723–1734. doi: 10.1089/hum.2010.061. [DOI] [PubMed] [Google Scholar]
- 66.Zhao G, Zhang JG, Shi Y, et al. MiR-130b is a prognostic marker and inhibits cell proliferation and invasion in pancreatic cancer through targeting STAT3. PLoS ONE. 2013;8(9):e73803. doi: 10.1371/journal.pone.0073803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xia JT, Wang H, Liang LJ, et al. Overexpression of FOXM1 is associated with poor prognosis and clinicopathologic stage of pancreatic ductal adenocarcinoma. Pancreas. 2012;41(4):629–635. doi: 10.1097/MPA.0b013e31823bcef2. [DOI] [PubMed] [Google Scholar]
- 68.Bao B, Wang Z, Ali S, et al. Over-expression of FoxM1 leads to epithelial–mesenchymal transition and cancer stem cell phenotype in pancreatic cancer cells. J. Cell. Biochem. 2011;112(9):2296–2306. doi: 10.1002/jcb.23150. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Yu C, Wang M, Li Z, et al. MicroRNA-138-5p regulates pancreatic cancer cell growth through targeting FOXC1. Cell. Oncol. (Dordr.) 2015;38(3):173–181. doi: 10.1007/s13402-014-0200-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, Yang J, Cui X, et al. A novel epigenetic CREB–miR-373 axis mediates ZIP4-induced pancreatic cancer growth. EMBO Mol. Med. 2013;5(9):1322–1334. doi: 10.1002/emmm.201302507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gironella M, Seux M, Xie MJ, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc. Natl Acad. Sci. USA. 2007;104(41):16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Greither T, Grochola LF, Udelnow A, Lautenschlager C, Wurl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int. J. Cancer. 2010;126(1):73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 73.Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int. J. Cancer. 2007;120(5):1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Park JK, Henry JC, Jiang J, et al. miR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochem. Biophys. Res. Commun. 2011;406(4):518–523. doi: 10.1016/j.bbrc.2011.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang S, Hao J, Xie F, et al. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis. 2011;32(8):1183–1189. doi: 10.1093/carcin/bgr105. [DOI] [PubMed] [Google Scholar]
- 76.Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J. Gastrointest. Surg. 2008;12(12):2171–2176. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297(17):1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 78.Szafranska AE, Davison TS, John J, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26(30):4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 79.Schultz NA, Werner J, Willenbrock H, et al. MicroRNA expression profiles associated with pancreatic adenocarcinoma and ampullary adenocarcinoma. Mod. Pathol. 2012;25(12):1609–1622. doi: 10.1038/modpathol.2012.122. [DOI] [PubMed] [Google Scholar]
- 80.Collins AL, Wojcik S, Liu J, et al. A differential microRNA profile distinguishes cholangiocarcinoma from pancreatic adenocarcinoma. Ann. Surg. Oncol. 2014;21(1):133–138. doi: 10.1245/s10434-013-3240-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl Acad. Sci. USA. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl Acad. Sci. USA. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang J, Chen J, Chang P, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev. Res. (Phila.) 2009;2(9):807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bauer AS, Keller A, Costello E, et al. Diagnosis of pancreatic ductal adenocarcinoma and chronic pancreatitis by measurement of microRNA abundance in blood and tissue. PLoS ONE. 2012;7(4):e34151. doi: 10.1371/journal.pone.0034151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kawaguchi T, Komatsu S, Ichikawa D, et al. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br. J. Cancer. 2013;108(2):361–369. doi: 10.1038/bjc.2012.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Le Large TY, Meijer LL, Mato Prado M, Kazemier G, Frampton AE, Giovannetti E. Circulating microRNAs as diagnostic biomarkers for pancreatic cancer. Expert Rev. Mol. Diagn. 2015;15(12):1525–1529. doi: 10.1586/14737159.2015.1112273. [DOI] [PubMed] [Google Scholar]
- 87.Li A, Yu J, Kim H, et al. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin. Cancer Res. 2013;19(13):3600–3610. doi: 10.1158/1078-0432.CCR-12-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Humeau M, Vignolle-Vidoni A, Sicard F, et al. Salivary microRNA in pancreatic cancer patients. PLoS ONE. 2015;10(6):e0130996. doi: 10.1371/journal.pone.0130996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Giovannetti E, Funel N, Peters GJ, et al. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70(11):4528–4538. doi: 10.1158/0008-5472.CAN-09-4467. [DOI] [PubMed] [Google Scholar]
- 90.Wang Z, Li Y, Ahmad A, et al. Pancreatic cancer: understanding and overcoming chemoresistance. Nat. Rev. Gastroenterol. Hepatol. 2011;8(1):27–33. doi: 10.1038/nrgastro.2010.188. [DOI] [PubMed] [Google Scholar]
- 91.Thota R, Pauff JM, Berlin JD. Treatment of metastatic pancreatic adenocarcinoma: a review. Oncology (Williston Park) 2014;28(1):70–74. [PubMed] [Google Scholar]
- 92.Park JK, Lee EJ, Esau C, Schmittgen TD. Antisense inhibition of microRNA-21 or -221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas. 2009;38(7):e190–e199. doi: 10.1097/MPA.0b013e3181ba82e1. [DOI] [PubMed] [Google Scholar]
- 93.Wang P, Zhuang L, Zhang J, et al. The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR-21 expression confers chemoresistance by targeting FasL. Mol. Oncol. 2013;7(3):334–345. doi: 10.1016/j.molonc.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cai B, An Y, Lv N, et al. miRNA-181b increases the sensitivity of pancreatic ductal adenocarcinoma cells to gemcitabine in vitro and in nude mice by targeting BCL-2. Oncol. Rep. 2013;29(5):1769–1776. doi: 10.3892/or.2013.2297. [DOI] [PubMed] [Google Scholar]
- 95.Nagano H, Tomimaru Y, Eguchi H, et al. MicroRNA-29a induces resistance to gemcitabine through the Wnt/beta-catenin signaling pathway in pancreatic cancer cells. Int. J. Oncol. 2013;43(4):1066–1072. doi: 10.3892/ijo.2013.2037. [DOI] [PubMed] [Google Scholar]
- 96.Yan HJ, Liu WS, Sun WH, et al. miR-17-5p inhibitor enhances chemosensitivity to gemcitabine via upregulating Bim expression in pancreatic cancer cells. Dig. Dis. Sci. 2012;57(12):3160–3167. doi: 10.1007/s10620-012-2400-4. [DOI] [PubMed] [Google Scholar]
- 97.Singh S, Chitkara D, Kumar V, Behrman SW, Mahato RI. miRNA profiling in pancreatic cancer and restoration of chemosensitivity. Cancer Lett. 2013;334(2):211–220. doi: 10.1016/j.canlet.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 98.Wang P, Zhang J, Zhang L, et al. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology. 2013;145(5):1133–1143 e1112. doi: 10.1053/j.gastro.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 99.Mittal A, Chitkara D, Behrman SW, Mahato RI. Efficacy of gemcitabine conjugated and miRNA-205 complexed micelles for treatment of advanced pancreatic cancer. Biomaterials. 2014;35(25):7077–7087. doi: 10.1016/j.biomaterials.2014.04.053. [DOI] [PubMed] [Google Scholar]; • Emphasizes on the increased therapeutic efficiency of combinatorial treatments comprising anticancer agents and miRNA modulators.
- 100.Huang FT, Zhuan-Sun YX, Zhuang YY, et al. Inhibition of hedgehog signaling depresses self-renewal of pancreatic cancer stem cells and reverses chemoresistance. Int. J. Oncol. 2012;41(5):1707–1714. doi: 10.3892/ijo.2012.1597. [DOI] [PubMed] [Google Scholar]
- 101.Kumar V, Mondal G, Slavik P, Rachagani S, Batra SK, Mahato RI. Codelivery of small molecule hedgehog inhibitor and miRNA for treating pancreatic cancer. Mol. Pharm. 2015;12(4):1289–1298. doi: 10.1021/mp500847s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bader AG, Brown D, Stoudemire J, Lammers P. Developing therapeutic microRNAs for cancer. Gene Ther. 2011;18(12):1121–1126. doi: 10.1038/gt.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hao J, Zhang S, Zhou Y, Hu X, Shao C. MicroRNA 483-3p suppresses the expression of DPC4/Smad4 in pancreatic cancer. FEBS Lett. 2011;585(1):207–213. doi: 10.1016/j.febslet.2010.11.039. [DOI] [PubMed] [Google Scholar]
- 104.Sicard F, Gayral M, Lulka H, Buscail L, Cordelier P. Targeting miR-21 for the therapy of pancreatic cancer. Mol. Ther. 2013;21(5):986–994. doi: 10.1038/mt.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ma Y, Yu S, Zhao W, Lu Z, Chen J. miR-27a regulates the growth, colony formation and migration of pancreatic cancer cells by targeting Sprouty2. Cancer Lett. 2010;298(2):150–158. doi: 10.1016/j.canlet.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 106.He D, Miao H, Xu Y, et al. MiR-371-5p facilitates pancreatic cancer cell proliferation and decreases patient survival. PLoS ONE. 2014;9(11):e112930. doi: 10.1371/journal.pone.0112930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Frampton AE, Castellano L, Colombo T, et al. MicroRNAs cooperatively inhibit a network of tumor suppressor genes to promote pancreatic tumor growth and progression. Gastroenterology. 2014;146(1):268–277.e18. doi: 10.1053/j.gastro.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 108.Torrisani J, Bournet B, Du Rieu MC, et al. let-7 microRNA transfer in pancreatic cancer-derived cells inhibits in vitro cell proliferation but fails to alter tumor progression. Hum. Gene Ther. 2009;20(8):831–844. doi: 10.1089/hum.2008.134. [DOI] [PubMed] [Google Scholar]
- 109.Gailhouste L, Gomez-Santos L, Hagiwara K, et al. miR-148a plays a pivotal role in the liver by promoting the hepatospecific phenotype and suppressing the invasiveness of transformed cells. Hepatology. 2013;58(3):1153–1165. doi: 10.1002/hep.26422. [DOI] [PubMed] [Google Scholar]
- 110.Zhang JP, Zeng C, Xu L, Gong J, Fang JH, Zhuang SM. MicroRNA-148a suppresses the epithelial-mesenchymal transition and metastasis of hepatoma cells by targeting Met/Snail signaling. Oncogene. 2014;33(31):4069–4076. doi: 10.1038/onc.2013.369. [DOI] [PubMed] [Google Scholar]
- 111.Liffers ST, Munding JB, Vogt M, et al. MicroRNA-148a is down-regulated in human pancreatic ductal adenocarcinomas and regulates cell survival by targeting CDC25B. Lab. Invest. 2011;91(10):1472–1479. doi: 10.1038/labinvest.2011.99. [DOI] [PubMed] [Google Scholar]
- 112.Hamada S, Satoh K, Fujibuchi W, et al. MiR-126 acts as a tumor suppressor in pancreatic cancer cells via the regulation of ADAM9. Mol. Cancer Res. 2012;10(1):3–10. doi: 10.1158/1541-7786.MCR-11-0272. [DOI] [PubMed] [Google Scholar]
- 113.Zhao G, Zhang JG, Liu Y, et al. miR-148b functions as a tumor suppressor in pancreatic cancer by targeting AMPKalpha1. Mol. Cancer Ther. 2013;12(1):83–93. doi: 10.1158/1535-7163.MCT-12-0534-T. [DOI] [PubMed] [Google Scholar]
- 114.Chen Z, Sangwan V, Banerjee S, et al. miR-204 mediated loss of Myeloid cell leukemia-1 results in pancreatic cancer cell death. Mol. Cancer. 2013;12(1):105. doi: 10.1186/1476-4598-12-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xiao J, Peng F, Yu C, et al. microRNA-137 modulates pancreatic cancer cells tumor growth, invasion and sensitivity to chemotherapy. Int. J. Clin. Exp. Pathol. 2014;7(11):7442–7450. [PMC free article] [PubMed] [Google Scholar]
- 116.Wang S, Chen X, Tang M. MicroRNA-216a inhibits pancreatic cancer by directly targeting Janus kinase 2. Oncol. Rep. 2014;32(6):2824–2830. doi: 10.3892/or.2014.3478. [DOI] [PubMed] [Google Scholar]
- 117.Dang Z, Xu WH, Lu P, et al. MicroRNA-135a inhibits cell proliferation by targeting Bmi1 in pancreatic ductal adenocarcinoma. Int. J. Biol. Sci. 2014;10(7):733–745. doi: 10.7150/ijbs.8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.He H, Hao SJ, Yao L, et al. MicroRNA-218 inhibits cell invasion and migration of pancreatic cancer via regulating ROBO1. Cancer Biol. Ther. 2014;15(10):1333–1339. doi: 10.4161/cbt.29706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zang W, Wang Y, Wang T, et al. miR-663 attenuates tumor growth and invasiveness by targeting eEF1A2 in pancreatic cancer. Mol. Cancer. 2015;14:37. doi: 10.1186/s12943-015-0315-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 120.Hu QL, Jiang QY, Jin X, et al. Cationic microRNA-delivering nanovectors with bifunctional peptides for efficient treatment of PANC-1 xenograft model. Biomaterials. 2013;34(9):2265–2276. doi: 10.1016/j.biomaterials.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 121.Pramanik D, Campbell NR, Karikari C, et al. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol. Cancer. Ther. 2011;10(8):1470–1480. doi: 10.1158/1535-7163.MCT-11-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gyorgy B, Szabo TG, Pasztoi M, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell. Mol. Life Sci. 2011;68(16):2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Simons M, Raposo G. Exosomes–vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009;21(4):575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 124.Kharaziha P, Ceder S, Li Q, Panaretakis T. Tumor cell-derived exosomes: a message in a bottle. Biochim. Biophys. Acta. 2012;1826(1):103–111. doi: 10.1016/j.bbcan.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 125.Lee TH, D'asti E, Magnus N, Al-Nedawi K, Meehan B, Rak J. Microvesicles as mediators of intercellular communication in cancer – the emerging science of cellular ‘debris’. Semin. Immunopathol. 2011;33(5):455–467. doi: 10.1007/s00281-011-0250-3. [DOI] [PubMed] [Google Scholar]
- 126.Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8(19):4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 127.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev. Proteomics. 2009;6(3):267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 128.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell. Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 129.Wang H, Rana S, Giese N, Buchler MW, Zoller M. Tspan8, CD44v6 and alpha6beta4 are biomarkers of migrating pancreatic cancer-initiating cells. Int. J. Cancer. 2013;133(2):416–426. doi: 10.1002/ijc.28044. [DOI] [PubMed] [Google Scholar]
- 130.Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ngora H, Galli UM, Miyazaki K, Zoller M. Membrane-bound and exosomal metastasis-associated C4.4A promotes migration by associating with the alpha(6)beta(4) integrin and MT1-MMP. Neoplasia. 2012;14(2):95–107. doi: 10.1593/neo.111450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mu W, Rana S, Zoller M. Host matrix modulation by tumor exosomes promotes motility and invasiveness. Neoplasia. 2013;15(8):875–887. doi: 10.1593/neo.13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Costa-Silva B, Aiello NM, Ocean AJ, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015;17(6):816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rana S, Malinowska K, Zoller M. Exosomal tumor microRNA modulates premetastatic organ cells. Neoplasia. 2013;15(3):281–295. doi: 10.1593/neo.122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Esau CC. Inhibition of microRNA with antisense oligonucleotides. Methods. 2008;44(1):55–60. doi: 10.1016/j.ymeth.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 136.Mahadevan D, Chalasani P, Rensvold D, et al. Phase I trial of AEG35156 an antisense oligonucleotide to XIAP plus gemcitabine in patients with metastatic pancreatic ductal adenocarcinoma. Am. J. Clin. Oncol. 2013;36(3):239–243. doi: 10.1097/COC.0b013e3182467a13. [DOI] [PubMed] [Google Scholar]
- 137.Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13(6):496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 138.Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2(4):E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43(42):13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 140.Khan S, Ansarullah, Kumar D, Jaggi M, Chauhan SC. Targeting microRNAs in pancreatic cancer: microplayers in the big game. Cancer Res. 2013;73(22):6541–6547. doi: 10.1158/0008-5472.CAN-13-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang S, Chen L, Jung EJ, Calin GA. Targeting microRNAs with small molecules: from dream to reality. Clin. Pharmacol. Ther. 2010;87(6):754–758. doi: 10.1038/clpt.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]