Abstract
Aim:
To characterize inter- and intra-strain variability of variable-number tandem repeats (VNTRs) in Mycoplasma pneumoniae to determine the optimal multilocus VNTR analysis scheme for improved strain typing.
Methods:
Whole genome assemblies and next-generation sequencing data from diverse M. pneumoniae isolates were used to characterize VNTRs and their variability, and to compare the strain discriminability of new VNTR and existing markers.
Results:
We identified 13 VNTRs including five reported previously. These VNTRs displayed different levels of inter- and intra-strain copy number variations. All new markers showed similar or higher discriminability compared with existing VNTR markers and the P1 typing system.
Conclusion:
Our study provides novel insights into VNTR variations and potential new multilocus VNTR analysis schemes for improved genotyping of M. pneumoniae.
KEYWORDS : Mycoplasma pneumoniae, next-generation sequencing, tandem repeats
DNA tandem repeats (TRs), also known as satellite DNA, are widely distributed in genomes across all domains of life. TRs are inherently unstable in length due to slipped strand mispairing during DNA replication, and as a result mutate at 10–100,000-times higher frequencies than nonrepetitive sequences in the genome [1]. Variations in TRs within genes and promoters can alter gene expression and function, resulting in harmful as well as beneficial consequences. While TRs are less abundant in prokaryotes including mycoplasmas compared with eukaryotes, TR variations play an important role in genome plasticity and bacterial adaptation strategies, including diversification of surface proteins to avoid host immune attacks and mediation of response to environmental stresses [2–4]. Aside from their physiologic and pathogenic role, TRs have been widely used for genome mapping and molecular typing due to their hypervariability in repeat copy number between individuals of the same species. To increase the discriminatory power, usually multiple variable TR loci are simultaneously used in the typing scheme, termed multilocus variable-number tandem repeat (VNTR) analysis (MLVA).
Mycoplasma pneumoniae belongs to the Mollicutes class, a group of wall-less bacteria with the smallest cell and genome size known to date for any self-replicating organism [5–8]. The comparison of the genome between M. pneumoniae and its closest relative Mycoplasma genitalium has helped define the essential gene set of a self-replicating minimal cell [9,10]. Both species are human pathogens, with M. pneumoniae being one of the most common causes of community-acquired pneumonia [11] and M. genitalium being an important cause of sexually transmitted urogenital infection [12]. Despite their extremely small genome size, approximately 5–8% of the genome sequence in both species is composed of repetitive DNA, mostly present in the P1 adhesion operon in M. pneumoniae or its counterpart MgPa operon in M. genitalium, and their chromosomal repetitive elements, known as RepMPs and MgPars, respectively [5,6]. These loci have been frequently used as the targets for molecular typing to help understand the epidemiology and pathogenesis of both pathogens [13–18]. Of note, numerous studies of VNTRs in M. genitalium have demonstrated extensive intrastrain repeat number variations despite its genome size being the smallest among all sequenced mycoplasma species [14,18–24], whereas there has been a scarcity of reports of such variations in M. pneumoniae [25] and other mycoplasma species. It is unknown whether this difference reflects intrinsic genomic divergence between different species or technical limitations of the detection methods used.
While P1 typing is the gold standard for M. pneumoniae typing [26–30], MLVA based on five VNTR loci has been increasingly used worldwide due largely to its greater discriminatory power [15,31–32]. However, there have been rising concerns of instability at one of these five loci (Mpn1) [25,33] though the extent of instability in this locus and other loci is not well understood. In addition, both P1 and MLVA typing have been shown to be insufficient for meaningful differentiation and lack of epidemiological and clinical relevance [25,32]. There is a clear need to characterize in more detail the variability of VNTRs and identify new VNTR markers with higher stability and discriminatory power [31,34]. Traditionally, identification of VNTRs have depended on an initial computational screening of TR loci from a single reference genome, and a subsequent experimental verification of the variability of selected TRs by Sanger sequencing or fragment sizing analysis of PCR amplified TR targets [14–15,18]. This approach is very tedious and time consuming, and has low throughput and low sensitivity to detect low abundant alleles in heterozygous samples. In addition, the analysis of TRs is prone to errors caused by slipped-strand mispairing during PCR [35].
With the advent of high-throughput next-generation sequencing (NGS) technologies, there has been a rapid expansion of publicly available genome sequences. Many complete and fragmentary genome assemblies as well as high volumes of raw sequence reads are available for M. pneumoniae isolates from diverse geographic origins, time periods and clinical background [7–8,34,36–39]. The wealth of genome data provides an excellent opportunity to characterize VNTRs in M. pneumoniae. The objectives of this study are: to identify VNTRs based on published complete genome assemblies for 20 M. pneumoniae isolates; to characterize the inter- and intrastrain variability of VNTRs using high-throughput NGS reads for 26 M. pneumoniae isolates; to compare the performance of new VNTR markers with that of the existing five VNTR markers as well as the P1 typing system using archived genome sequence data for a panel of 41 M. pneumoniae isolates originated from nine countries, which have been previously typed by the P1 and MLVA typing.
Materials & methods
• Genome assemblies & NGS data sources
To identify VNTR loci, we retrieved complete genome sequences of 20 geographically diverse M. pneumoniae isolates from the NCBI database (Supplementary Table 1). To examine intrastrain copy number variation, we used archived NGS raw reads for 26 M. pneumoniae isolates, including 454 reads (average length 509 bp) for one isolate 309 reported by Kenri et al. [36]; Illumina reads (251 bp each) for 15 isolates reported by Xiao et al. [8]; Illumina reads (116 bp each) for two isolates reported by Li et al. [37,38]; Illumina reads (95 bp each) for seven isolates reported by Lluch-Senar et al. [7]; Illumina reads (36 bp each) for the strain FH reported by Touati et al. [39]. We selected only seven of the 23 isolates reported by Lluch-Senar et al. [7] that represented different genome size and geographic regions, instead of using all of them because their read lengths were too short (95 bp) to cover the long repeats in Mpn14–16. To evaluate the performance of VNTRs for strain typing, we used whole genome sequence assemblies or NGS raw reads from 41 M. pneumoniae isolates that have been previously typed by MLVA and P1 typing. The background information for all isolates, including country of origin and GenBank accession code, is shown in Supplementary Table 1.
• Sequence analysis
To identify VNTR loci, we made an alignment of published complete genome sequences of 20 M. pneumoniae isolates using the MAUVE algorithm of the MegAlign Pro software in the Lasergene version 12 package (DNAStar, WI, USA). We did not include the incomplete genome assemblies (with 19–45 scaffolds) reported by Lluch-Senar et al. [7] since they caused alignment problems and interfered with the visualization of the VNTR regions. A VNTR locus was defined as a region containing a tandem repeat unit of 2 bp or longer that was variable in the repeat copy number between any two or more of the 20 genomes analyzed. Excluded were loci related to the interspersed repetitive elements RepMPs of the MPN141 and MPN142 genes in the P1 operon due to their high sequence identify and potential to recombine with homologous sequences to generate intrastrain variation [40,41]. Also excluded were all but one (Mpn1) loci related to the multicopy hsdS gene family encoding the S subunit of type I restriction enzyme due to the high sequence identity among them [8], which prevents accurate alignment with short NGS reads. The Mpn1 locus was included since it is one of the five VNTR loci in the current MLVA scheme [15]. While mononucleotide TRs are dominant in bacterial genomes they were excluded in this study since their role is less understood and their identification is complicated by a high rate of homopolymer indel errors in NGS data [42].
Analysis of repeat copy number variation in NGS reads involved two major steps. In the first step, a region of approximately 500 bp flanking each end of the VNTR repeat region was retrieved from published M. pneumoniae genome sequences, and then aligned to the NGS reads from each isolate by SeqMan NGen software in the Lasergene version 12 package (DNAStar, WI, USA) using the templated genome assembly module. In the second step, NGS reads aligned to both flanking regions of each VNTR locus in the first step were retrieved and realigned using Sequencher software (version 5.3; Gene Codes Corporation, MI, USA), resulting in contig(s) containing the VNTR locus and its flanking regions. The contigs were trimmed by removing reads containing any ambiguous sequences in the repeat unit and removing reads containing only one flanking region (to avoid overcounting reads containing lower repeat copy numbers). Repeat copy numbers were determined from qualified NGS reads, which were defined as reads that completely span the entire repeat region plus at least one unique flanking base on both ends. The repeat copy numbers for the five loci in the current MLVA scheme were counted following previous reports [15,31].
To compare the relative rates of deletion and insertion of tandem repeats, we included all VNTR loci containing two or more different repeat copy numbers within isolates (heterozygous repeats). Deletion and insertion were defined as loss or gain of one or more repeat copies, respectively, by comparing the second most predominant allele to the most predominant allele while ignoring the remaining minor alleles if there were three or more alleles. The difference between the deletion and insertion rates was assessed by binomial test.
• Comparison of new genetic markers with P1 & MLVA typing systems
A panel of 41 isolates, including 38 clinical isolates and 3 laboratory strains (FH, M129 and MAC), were used to compare the performance of various known and new genetic markers for strain typing. These isolates represent a broad geographical distribution including North America, Europe, Asia and North Africa. All these isolates have been previously typed by the P1 and/or MLVA systems (Supplementary Table 1). In this study we determined the repeat copy numbers for all previously and newly identified VNTR markers based on NGS reads as described above for 18 isolates sequenced by 454 or Illumina with read lengths >115 bp. For 23 isolates with short Illumina reads (95 bp each), the repeat copy numbers in 16 isolates were determined based primarily on assembled contigs [7] followed by confirmation by aligning the contigs with raw reads, and in seven isolates based on Illumina reads alone for all VNTR loci except for the three loci (Mpn14–16) containing long repeat units, which were determined based on contigs. As expected, in isolates containing heterozygous repeats, the repeat copy numbers identified from contigs represented the dominant population in raw reads. While there was no or only a very small number of qualified raw reads for Mpn14–16 in these 23 isolates, the repeat copy numbers determined as described above were all consistent with previous results obtained by MLVA typing [7]. For all VNTR loci showing heterozygous repeats, only the dominant types were used to assess the discriminatory power. To compare the discriminatory power of individual markers and various selected combinations, we calculated Hunter-Gaston discriminatory indices (HGDIs) according to the method described previously [43].
Results
• Identification of VNTR loci in Mycoplasma pneumoniae genomes
We identified a total of 13 VNTR loci, including the five loci in the current MLVA scheme [15], the AGT triplet repeat in the P1 adhesin gene reported previously [8,30], termed herein as P1-AGT, and seven new loci (Table 1). Based on comparison of the 20 complete M. pneumoniae genome sequences, seven VNTR loci (Mpn13–16, Mpn003, Mpn103 and Mpn651) showed only two different copy numbers while all the remaining six loci showed four to seven different copy numbers among different isolates.
Table 1. . Variable-number tandem repeats in Mycoplasma pneumoniae.
| Repeat name | Location on genome† | Repeat unit (consensus) | Repeat unit size (bp) | Copy number variation‡ | Affected gene ID and its function | HGDI§ |
|---|---|---|---|---|---|---|
| Mpn003 |
2849–2878 |
ATTAAATGGAAGACA |
15 |
1–2 |
MPN003, gyrB |
0.5098 |
| Mpn1¶ |
111920–111991 |
CCGAGCTAAGCG |
12 |
2–6 |
MPN089, hsdS |
0.7890 |
| Mpn103 |
134124–134131 |
TTTA |
4 |
2–3 |
MPN103, unknown |
0.5098 |
| Mpn108a |
141296–141317 |
AG |
2 |
6–11 |
MPN108, 3′-UTR |
0.8207 |
| Mpn108b |
141349–141362 |
AC/AG |
2 |
7–20 |
MPN108, 3′-UTR |
0.6963 |
| Mpn111 |
146255–146274 |
AC/AG |
2 |
10–17 |
MPN111, YtxK |
0.5598 |
| P1-AGT# |
182793–182813 |
AGT |
3 |
5–16 |
MPN141, P1 adhesin |
0.8268 |
| Mpn444 |
538724–53835 |
TAC |
3 |
3–6 |
MPN444, lipoprotein |
0.5988 |
| Mpn13¶ |
596741–596804 |
TATTAATAACTATTCT |
16 |
3–4 |
MPN490, 3′-UTR |
0.5098 |
| Mpn14¶ |
608651–608755 |
TGGACAAAATGGAAGTAAAAA |
21 |
5–6 |
MPN501, unknown |
0.4024 |
| Mpn15¶ |
645561–645707 |
TTGTCCATTCTTTCTTCCATC |
21 |
6–7 |
MPN524, unknown |
0.5098 |
| Mpn16¶ |
736123–736216 |
ATTTTTTAAAAGTTTTTATTTATCCGTTTTGACAACTGCTTTTTGTT |
47 |
1–2 |
MPN613, unknown |
0 |
| Mpn651 | 775850–775852 | AAT | 3 | 1–2 | MPN651, 5′-UTR | 0.5098 |
†Position on the M. pneumoniae M129 genome sequence (GenBank accession code U00089).
‡Based on comparison of 20 complete genome sequences except for Mpn16, which is based on the previous report [15].
§Based on typing of 41 isolates and excluding minor variants in heterogeneous loci (Supplementary Table 10).
¶Reported by Degrange et al. [15].
gyrB: DNA gyrase subunit B; hsdS: Type 1 restriction enzyme S subunit; UTR: Untranslated region;YtxK: Adenine-specific methyltransferase.
Of the 13 VNTR loci, seven were located within coding regions, four were located exclusively in noncoding regions, and the remaining two spanned from the coding region to either the 5′-untranslated region (UTR in MPN003) or 3′-UTR (in MPN613). Repeat copy number variation affects the promoter region of one gene (MPN003) and the coding region of seven genes, resulting in reading frame shift and translational stop codon in two genes (MPN103 and MPN111), and a variable number of short peptide repeats in five genes (MPN089, MPN141, MPN444, MPN501 and MPN524).
• Different levels of intrastrain copy number variation
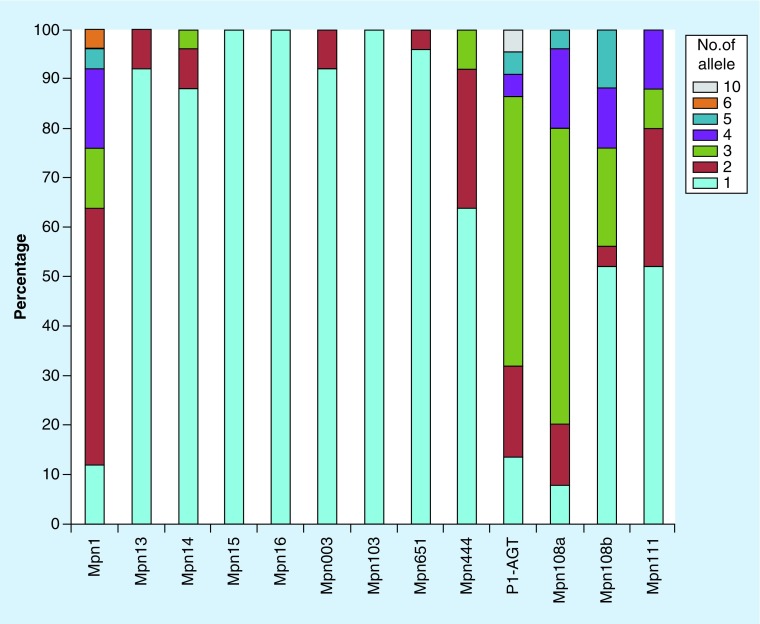
To examine repeat copy number variation within individual genomes (intrastrain variation), we mapped NGS sequence reads from 26 M. pneumoniae isolates to the VNTR loci. In average, each VNTR locus was fully covered by 135 to 301 qualified reads (Supplementary Table 2). Three loci (Mpn15, Mpn16 and Mpn103) displayed homogeneous reads, without variation in the repeat copy number within any isolates (homozygous repeats) while the remaining ten loci showed different degrees of heterogeneity (heterozygous repeats), including five loci (Mpn13, Mpn14, Mpn003, Mpn444 and Mpn651) containing a mixture of two or three different repeat copy numbers in no more than five isolates, and five loci (Mpn1, P1-AGT, Mpn108a, Mpn108b and Mpn111) containing a mixture of two to ten different copy numbers in nine or more isolates (Supplementary Tables 3–8). In all loci showing heterozygous repeats, the most predominant copy number in each VNTR was consistent with the sequence in the corresponding region of the published genome assembly for each isolate surveyed; all minor copy numbers differed mostly by only one or two from the dominant copy number and also had a much lower read coverage than the dominant copy number. Two loci (Mpn108b and Mpn111) contained a mixture of two different repeat units (AC and AG); alleles containing the same repeat copy number may have different distribution patterns of these repeat units as exemplified in Supplementary Figure 1 for Mpn108b. The Mpn108b and Mpn108a loci were located in the 3′ UTR of the gene MPN108 and separated by 31 bp. These two loci as well as P1-AGT, Mpn1 and Mpn111 showed much greater intrastrain copy number variation than other loci (Figure 1). Almost all isolates surveyed contained heterozygous repeats in Mpn1, Mpn108a and P1-AGT (Table 2).
Figure 1. . Distribution of homozygous and heterozygous variable-number tandem repeats in Mycoplasma pneumoniae.
Homozygous repeats contain only allele with the same repeat copy number in each isolate, which is dominant (52–100%) among all the 13 variable-number tandem repeat loci analyzed, except for 3 loci (Mpn1, P1-AGT and Mpn108a). Heterozygous repeats contain two to ten different alleles with different repeat copy numbers with each isolate (see details in Table 2).
Table 2. . Tandem repeat copy number variation in Mycoplasma pneumoniae genomes based on next-generation sequencing data.
|
Isolate† |
P1 type‡ |
Repeat copy number at VNTR loci§ |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mpn1 | Mpn13 | Mpn14 | Mpn15 | Mpn16 | P1-AGT | Mpn003 | Mpn103 | Mpn444 | Mpn651 | Mpn108a | Mpn108b | Mpn111 | ||
| 85138 |
1 |
6, 4, 5, 3 |
4 |
5 |
7 |
2 |
7, 6, 8 |
2 |
2 |
6, 5 |
1 |
9, 10, 8 |
7 |
10, 9 |
| 85084 |
1 |
7, 6, 4, 5, 8 |
4 |
5 |
7 |
2 |
7, 8, 6 |
2 |
2 |
5, 4, 6 |
1 |
9, 10, 8, 11 |
7 |
10 |
| M129-a |
1 |
4, 5 |
4 |
5 |
7 |
2 |
7, 8, 6 |
2 |
2 |
4, 5 |
1 |
8, 9, 7, 6 |
7 |
10 |
| 54524 |
1 |
4, 5 |
4 |
5 |
7 |
2 |
8, 7, 9 |
2 |
2 |
4, 3 |
1 |
9, 10, 8, 11 |
7 |
10 |
| 54089 |
1 |
3, 4 |
4 |
5 |
7 |
2 |
5 |
2 |
2 |
4 |
1 |
10, 9, 11, 8 |
7 |
10, 9 |
| 51494 |
1 |
4, 3, 5 |
4 |
5 |
7 |
2 |
8, 7, 9, 6, 5 |
2 |
2 |
4, 3, 5 |
1 |
6, 7, 5 |
7 |
10 |
| 1428 |
1 |
4, 3 |
4 |
5 |
7 |
2 |
7, 6, 8 |
2, 1 |
2 |
4 |
1 |
10, 11, 9 |
7 |
10 |
| S355 |
1 |
6, 4 |
4 |
5 |
7 |
2 |
9, 8, 10 |
2 |
2 |
5, 4 |
1 |
6 |
7 |
10 |
| C267 |
1 |
5, 6, 3 |
4 |
5 |
7 |
2 |
7, 8, 6 |
2, 1 |
2 |
5 |
1 |
7, 8, 6 |
7 |
10, 9 |
| 1145 |
1 |
5, 4 |
4, 3 |
5 |
7 |
2 |
12, 11, 13 |
2 |
2 |
4 |
1 |
8, 7, 10 |
7 |
10, 9 |
| 6250 |
1 |
4, 5, 6, 2 |
4 |
5 |
7 |
2 |
9, 8, 10 |
2 |
2 |
4, 5 |
1 |
8, 9, 7 |
7 |
10 |
| 4802 |
1 |
4 |
4 |
5 |
7 |
2 |
6, 7 |
2 |
2 |
4, 3 |
1, 2 |
9, 8, 7 |
7 |
10 |
| 5392 |
1a |
3 |
4 |
5 |
7 |
2 |
10 |
2 |
2 |
4, 3 |
1 |
9, 8, 10 |
7 |
10 |
| 2882 |
2 |
4 |
3 |
6 |
6 |
2 |
6, 5 |
1 |
3 |
3 |
2 |
7 |
14, 15 |
12 |
| 4358 |
2 |
2, 3 |
2 |
6 |
6 |
2 |
6 |
1 |
3 |
3 |
2 |
8, 9, 7 |
16, 15, 17 |
18, 17, 19, 16 |
| 309 |
2a |
7, 8, 5, 6, 4, 3 |
3 |
5 |
6 |
2 |
7, 6 |
1 |
3 |
3 |
2 |
7, 6 |
17, 15, 16, 18, 14 |
10 |
| M2592 |
2 |
3, 4 |
33 |
5 |
6 |
2 |
7, 8, 6, 9 |
1 |
3 |
3 |
2 |
7, 6, 8 |
20, 19, 21, 18 |
13, 12 |
| M2192 |
2 |
4, 5 |
3 |
6 |
6 |
2 |
7, 6 |
1 |
3 |
3 |
2 |
9, 10, 8 |
15, 14, 16, 13 |
17, 16, 18, 15 |
| M1139 |
2 |
5, 6 |
3 |
6 |
6 |
2 |
7, 8, 6 |
1 |
3 |
3 |
2 |
9, 10, 8 |
12, 11, 13 |
15, 14, 16 |
| PO1 |
2 |
4, 5 |
3, 4 |
6, 5, 7 |
6 |
2 |
6, 7, 5 |
1 |
3 |
3 |
2 |
7, 8 |
15, 14, 16 |
12 |
| MAC |
2 |
5, 4, 3, 6 |
3 |
5 |
6 |
2 |
6, 8, 7 |
1 |
3 |
3 |
2 |
8, 7, 9 |
17, 16, 18, 15, 19 |
12, 11, 10 |
| FH-a |
2 |
5, 7, 6 |
3 |
6, 7 |
6 |
2 |
6, 7, 5 |
1 |
3 |
3 |
2 |
7, 6 |
15, 14, 16 |
12, 11 |
| 39443 |
2 |
5, 4 |
3 |
6 |
6 |
2 |
8, 7 |
1 |
3 |
3 |
2 |
9, 10, 8 |
19, 18, 20 |
10, 9 |
| 19294 |
2 |
4, 5, 3, 6 |
3 |
5, 6 |
6 |
2 |
16, 17, 15, 18, 14, 8, 12, 13, 19, 11 |
1 |
3 |
3 |
2 |
11, 10, 12 |
19, 18, 20, 17 |
14, 13, 15, 12 |
| 3896 | 2d | 5, 4 | 3 | 5 | 8 | 2 | 10, 9, 11 | 1 | 3 | 3 | 2 | 11, 10, 9, 12, 8 | 17, 16, 15, 18, 19 | 10 |
†Based on references as detailed in Supplementary Table 1.
‡Determined based on NGS reads, except for those underlined which were determined using assembled contigs [7]. For loci containing variable numbers of repeats, the copy number is listed in the order from predominant to minor.
§Isolate FH-b is shown in the Supplementary Table 11 but not here since its NGS reads are too short to cover the repeat regions in most of the VNTR loci.
NGS: Next-generation sequencing; VNTR: Variable-number tandem repeat.
Including all loci showing heterogeneous repeat copy numbers, the overall rate of repeat deletions was significantly higher than that of repeat insertions (60.6 vs 39.4%; p = 0.0276; Supplementary Table 9), in agreement with previous reports [44]. In addition, in two loci (Mpn108b and Mpn111) almost all alleles with >10 repeats showed extensive intrastrain heterogeneity in the repeat copy number in sharp contrast to the high homogeneity in alleles with <10 repeats (Table 2), consistent with the notion that higher copy number alleles are more unstable than lower copy number alleles [45].
• Comparative analysis of the previously & newly identified markers
Based on typing 41 M. pneumoniae isolates, distinct genotypes in individual markers varied from 1 to 9 (Supplementary Table 10). The P1 typing system and seven VNTR markers (Mpn13–15, Mpn003, Mpn103, Mpn651 and Mpn444) each yielded two predominant types; the two types in each of these VNTR markers showed good correlation with the two P1 types. For example, almost all P1 type 1 isolates contained two repeats in Mpn003 while almost all P1 type 2 isolates contained only one repeat in Mpn003. In two VNTR markers (Mpn108b and Mpn111), the dominant type (seven and ten repeats, respectively) was primarily present in P1 type 1 isolates while the other seven or eight minor types in Mpn108b and Mpn111 were present in P1 type 2 isolates. The genotypes in three VNTR markers (Mpn1, Mpn108a and P1-AGT) appeared to be randomly distributed between the P1 type 1 and 2 isolates. Some isolates with identical P1 and MLVA types could be clearly distinguished by the copy number variation in P1-AGT, Mpn108a or Mpn108b. Based on clustering analysis, the genotypes were not associated with geographic location of the isolates (data not shown).
We further compared the discriminatory power of all markers. When evaluating individually, the five VNTR markers in the current MLVA scheme showed HGDIs ranging from 0 to 0.7890 while the seven new VNTR markers showed HGDIs ranging from 0.5098 to 0.8207 (Table 1). The P1 typing system showed an HGDI of 0.6037. Among all 13 VNTR markers, 2 previously identified VNTR markers (Mpn14 and Mpn16) showed the lowest HGDIs (0.4024 and 0, respectively) while 2 new VNTR markers (Mpn108a and P1-AGT) showed the highest HGDIs (0.8207 and 0.8268, respectively). To identify new MLVA schemes with improved performance, we evaluated combinations of various VNTR markers (Table 3). Given that Mpn16 showed no discriminatory ability (HGDI = 0), replacement of this marker in the current MLVA scheme (HGDI = 0.8967) with any of the seven new VNTR markers could improve the HGDI to 0.9024–0.9841 (new schemes MLVA-2 to MLVA-7). Nevertheless, one unstable marker Mpn1 remained in all these new schemes and an additional unstable marker (Mpn108b, Mpn111, P1-AGT or Mpn108a) was included in four of them. The scheme MVLA-8 included five stable markers, which is expected to provide the highest stability though with a relatively low discriminatory power (HGDI = 0.8366). The scheme MLVA-9, including five unstable markers, showed the highest HGDI (0.9988). The scheme MLVA-10, including the three most variable (unstable) markers, showed the second highest HGDI (0.9927).
Table 3. . Performance of different multilocus variable-number tandem repeat analysis schemes for Mycoplasma pneumoniae strain typing.
| MLVA schemes | VNTR loci included | HGDI† |
|---|---|---|
| MLVA original‡ |
Mpn1, Mpn13, Mpn14, Mpn15 and Mpn16 |
0.8967 |
| MLVA-2 |
Mpn1, Mpn13, Mpn14, Mpn15 and Mpn003§ |
0.9024 |
| MLVA-3 |
Mpn1, Mpn13, Mpn14, Mpn15 and Mpn444 |
0.9183 |
| MLVA-4 |
Mpn1, Mpn13, Mpn14, Mpn15 and Mpn108b |
0.9183 |
| MLVA-5 |
Mpn1, Mpn13, Mpn14, Mpn15 and Mpn111 |
0.9244 |
| MLVA-6 |
Mpn1, Mpn13, Mpn14, Mpn15 and P1-AGT |
0.9817 |
| MLVA-7 |
Mpn1, Mpn13, Mpn14, Mpn15 and Mpn108a |
0.9841 |
| MLVA-8 |
Mpn13, Mpn14, Mpn15, Mpn003§ and Mpn444 |
0.8366 |
| MLVA-9 |
Mpn1, Mpn108a, Mpn108b, Mpn111 and P1-AGT |
0.9988 |
| MLVA-10 | Mpn1, Mpn108a and P1-AGT | 0.9927 |
Bold indicates relatively unstable loci.
†Based on analysis of 41 isolates as shown in Supplementary Table 10.
‡As described by Degrange et al. [15].
§Same to Mpn103 and Mpn651 in variability and stability but containing a longer repeat unit than the latter two. Longer repeats could be more easily detected than shorter repeats by fragment sizing.
MLVA: Multilocus variable-number tandem repeat analysis; VNTR: Variable-number tandem repeat.
Discussion
Following the first report of its use in M. pneumoniae in 2009 [15], many studies over the world have consistently shown that MLVA typing is clearly more powerful than the standard P1 typing system in differentiating clinical strains [25,32–33,46–48], and meanwhile there have been rising concerns about its stability, discriminability and correlation to clinical and epidemiological parameters [25,31–33]. These concerns motivated us to seek deeper understanding of the variability of VNTRs in the M. pneumoniae genome and to identify new VNTR markers, by taking the advantage of the availability of abundant NGS data for M. pneumoniae isolates from a variety of continental sources [6–8,36–38,49]. We identified a total of 13 VNTRs in M. pneumoniae, very similar to M. genitalium, in which a total of 12 VNTRs have been reported (excluding those in MgPar repeats) [18]. The detection of a limited number of VNTRs is consistent with the high compaction and conservation of the M. pneumoniae genomes [6–8]. It is noteworthy that variations in eight VNTR loci of M. pneumoniae affect the promoter region, interrupt the reading frame or generate variable-length peptide repeats (Table 1), implying a potential role in modulating gene expression and function.
The high sequence coverage of NGS data from 26 isolates has allowed us to gain deeper insights into the VNTR variation in M. pneumoniae. We have demonstrated that intrastrain variation occurred in different degrees in all 13 VNTR loci surveyed except for 3 loci (Mpn15, Mpn16 and Mpn103) (Table 2). Strikingly, five loci (Mpn1, Mpn108a, Mpn108b, Mpn111 and P1-AGT) showed a very high-level intrastrain variation (Figure 1). These findings suggest that, despite an extremely reduced and conserved genome in M. pneumoniae, these VNTR loci are highly dynamic and rapidly evolving during cell replication and that the repeat copy number may undergo expansion or contraction from one generation to the next. In support of this hypothesis, we found a clear change in the repeat copy number in these loci for two reference strains M129 and FH maintained and sequenced by different laboratories (Supplementary Table 11). The detection of extensive intrastrain copy number variation in Mpn1 (Supplementary Table 3) supports the instability of Mpn1 as reported previously [25,33], whereas the absence or presence of low-level intrastrain variation in other four loci in the current MVLA scheme suggests a relatively high stability of these loci. Of note, despite the absence of report of instability of Mpn13 and Mpn14, we detected two or more repeat copy numbers in both loci in a few isolates (Table 2).
Based on our study of NGS data, P1-AGT showed the greatest inter- and intra-strain variation among all VNTRs analyzed (Supplementary Table 8). Of special note, one strain (no. 19294) showed ten alleles at P1-AGT with repeat copy number varying from 8 to 19 (Supplementary Table 12), suggesting an extreme instability of this locus. Interestingly, the P1-AGT repeat of M. pneumoniae has a counterpart in M. genitalium, which is located in the MPN141 homolog MG191 and which is also hypervariable among and within M. genitalium strains [19–20,22]. The conservation of this repeat across species and its hypervariability within each species implies an important role in functional consequences, presumably in adaptation and pathogen–host interactions [8,22].
While the presence of a high-level intrastrain variation in the repeat copy number in some loci seemingly complicates the interpretation of the genotypes, this variation reflects the inherent instability of VNTRs as a result of slipped-strand mispairing, as has been well documented in M. genitalium by analysis of PCR products by plasmid cloning [18,20] and fragment sizing [14]. It is likely that intrastrain variation always occurs in all VNTRs, though in different degrees, but the minor variants (<5–10%) usually cannot be detected by conventional low-throughput methods including Sanger sequencing [30] or fragment sizing analysis of the PCR products [15,31–32]. In case two or more variants are detectable (usually differing by only one repeat copy), one can consider reporting only the dominant type or all types as has been described for M. genitalium [14,18].
By comparative analysis of all VNTR markers, we found that the current MLVA scheme could be improved by selected combinations of the existing and new markers (Table 3). The Mpn16 marker in the current MLVA scheme showed no or little discriminatory ability in this study (Tables 1 & 2) and previous reports [15,25,32,46], and can be replaced by any of the seven new VNTR markers. Of the nine new potential MLVA schemes, MVLA-8 had the highest stability though with a relatively low discriminatory power and MLVA-9 showed the highest discriminatory power though with a low stability. Clearly, further studies are needed of how to balance the stability and discriminability using a larger number of clinical isolates. Given the limited availability of VNTR markers for M. pneumoniae, it would be very challenging to develop a typing scheme with both high stability and discriminability. The choice of the MLVA schemes would depend on the purpose of strain typing. In general, a scheme with high stability is the preferred choice for long-term or global epidemiological studies (macroepidemiology) while a scheme with high discriminability is the preferred choice for short-term epidemiological studies (microepidemiology) [50]. Of note, the MLVA-10 scheme showed very high discriminability (HGDI = 0.9927) despite having only three VNTR loci, which could reduce the cost and simplify the detection process, thus making it an attractive choice for microepidemiological studies.
In this study we determined the repeat copy numbers for all new VNTR markers and some of the existing VNTR markers based on nucleotide sequences obtained by whole genome sequencing, which should be more accurate than any indirect sequence detection methods, including fragment sizing analysis of PCR products involved in the current MLVA system [15]. As documented in the recent report [31], fragment sizing analysis may produce inconsistent results among different laboratories using the same set of clinical isolates. In addition, it may be technically challenging for fragment sizing analysis to differentiate short repeat units of 2–3 bp present in most of the new VNTRs identified in this study though there are reports of successful resolution of such short repeats by this method in other organisms [14]. If these short repeats cannot be accurately quantified by other methods, direct sequencing of the VNTR PCR products from both directions [18] would be an option. For isolates with very high-level intrastrain variation, sequencing of the VNTR PCR products directly by NGS or after cloning into plasmids [18] may be helpful. Sequencing should be the best approach to differentiate the distribution patterns of the two different repeat units in Mpn108b and Mpn111 (Table 1 & Supplementary Figure 1), which have the potential to further improve the discriminatory power but are not accessible by fragment sizing.
Conclusion & future perspective
We have characterized the inter- and intra-strain repeat copy number variations of previously and newly identified VNTR markers in M. pneumoniae, and provided, for the first time, direct evidence of extensive intrastrain variation of some VNTRs despite a highly reduced and conserved genome. These studies expand our understanding of the VNTR variations and provide potential new MLVA schemes for M. pneumoniae genotyping. Further studies are needed of the performance of these new MLVA schemes in comparison with multilocus sequence typing [34], multiplex single nucleotide polymorphisms [39] and genome-wide single nucleotide polymorphisms and indels [7] using whole genome sequence data currently available and/or from new clinical isolates with diverse clinical and epidemiological parameters. This report presents the first successful attempt of using primarily the wealth of publicly available NGS sequences to address the fundamental questions on M. pneumoniae biology. We believe that such data are also important resources to investigate other aspects of M. pneumoniae, such as strain variation, recombination, evolution, and population structure and dynamics.
EXECUTIVE SUMMARY.
A total of 13 variable-number tandem repeats (VNTR) loci were identified by comparison of published complete genome assemblies for 20 Mycoplasma pneumoniae isolates.
VNTRs displayed inter- and intra-strain copy number variations at different levels from zero to exceptionally high.
Variations in eight VNTR loci affect the promoter region, interrupt the reading frame or generate variable-length peptide repeats, implying potential impacts on gene function.
All new markers showed similar or higher discriminability compared with existing VNTR markers and the P1 typing system.
Selected combinations of the existing and new markers could provide multilocus variable-number tandem repeat analysis schemes with efficient discriminatory power for either long-term or short-term epidemiological studies.
Supplementary Material
Acknowledgements
The authors thank Dr Myers L for statistical analysis, Drs Chalker V and Pereyre S for information about MLVA copy number assignment, and Drs Cozzuto L, Pubul L S and Senar M L for discussion about their genome data.
Footnotes
Financial & competing interests disclosure
This study was supported in part by the Intramural Research Program of the US NIH Clinical Center; the NIH Clinical and Translational Science Award (grant UL1TR001417 to T Ptacek); the NIH grant (R01AI63909 to K Dybvig); the Ministry of Health, Labor, and Welfare of Japan (grant (H26-Kokui-Shitei-001 to K Shibayama); the Grant-in Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (grants 20018029 and 15H01337 to T Kenri); and the Natural Science Foundation of Beijing Municipality, Beijing, China (grant 7152025 to H Sun). J Zhang was supported by a scholarship from Chongqing Medical University (Chongqing, China). X Song was supported by the China Scholarship Council. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 2010;44:445–477. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- 2.Mrazek J. Analysis of distribution indicates diverse functions of simple sequence repeats in Mycoplasma genomes. Mol. Biol. Evol. 2006;23(7):1370–1385. doi: 10.1093/molbev/msk023. [DOI] [PubMed] [Google Scholar]
- 3.Mrazek J, Guo X, Shah A. Simple sequence repeats in prokaryotic genomes. Proc. Natl Acad. Sci. USA. 2007;104(20):8472–8477. doi: 10.1073/pnas.0702412104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou K, Aertsen A, Michiels CW. The role of variable DNA tandem repeats in bacterial adaptation. FEMS Microbiol. Rev. 2014;38(1):119–141. doi: 10.1111/1574-6976.12036. [DOI] [PubMed] [Google Scholar]
- 5.Fraser CM, Gocayne JD, White O, et al. The minimal gene complement of Mycoplasma genitalium . Science. 1995;270(5235):397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 6.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li BC, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae . Nucleic Acids Res. 1996;24(22):4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lluch-Senar M, Cozzuto L, Cano J, et al. Comparative -‘omics’ in Mycoplasma pneumoniae clinical isolates reveals key virulence factors. PLoS ONE. 2015;10(9):e0137354. doi: 10.1371/journal.pone.0137354. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Comparative genomic and transcriptomic analysis of 23 Mycoplasma pneumoniae clinical solates provides novel insights into strain variations and virulence factors.
- 8.Xiao L, Ptacek T, Osborne JD, et al. Comparative genome analysis of Mycoplasma pneumoniae . BMC Genomics. 2015;16:610. doi: 10.1186/s12864-015-1801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Comparative whole genome analysis of 15 M. pneumoniae isolates reveals an overall high-degree conservation with about 1500 single nucleotide polymorphisms and indels between type 1 and type 2 isolates.
- 9.Hutchison CA, 3rd, Chuang RY, Noskov VN, et al. Design and synthesis of a minimal bacterial genome. Science. 2016;351(6280):aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 10.Hutchison CA, Peterson SN, Gill SR, et al. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286(5447):2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- 11.Jain S, Williams DJ, Arnold SR, et al. Community-acquired pneumonia requiring hospitalization among U.S. children. N. Engl. J. Med. 2015;372(9):835–845. doi: 10.1056/NEJMoa1405870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blanchard A, Bebear C. The evolution of Mycoplasma genitalium . Ann. NY Acad. Sci. 2011;1230:E61–E64. doi: 10.1111/j.1749-6632.2011.06418.x. [DOI] [PubMed] [Google Scholar]
- 13.Brown RJ, Spiller BO, Chalker VJ. Molecular typing of Mycoplasma pneumoniae: where do we stand? Future Microbiol. 2015;10:1793–1795. doi: 10.2217/fmb.15.96. [DOI] [PubMed] [Google Scholar]; • A thorough and up-to-date review on M. pneumoniae strain typing.
- 14.Cazanave C, Charron A, Renaudin H, Bebear C. Method comparison for molecular typing of French and Tunisian Mycoplasma genitalium-positive specimens. J. Med. Microbiol. 2012;61(Pt 4):500–506. doi: 10.1099/jmm.0.037721-0. [DOI] [PubMed] [Google Scholar]
- 15.Degrange S, Cazanave C, Charron A, Renaudin H, Bebear C, Bebear CM. Development of multiple-locus variable-number tandem-repeat analysis for molecular typing of Mycoplasma pneumoniae . J. Clin. Microbiol. 2009;47(4):914–923. doi: 10.1128/JCM.01935-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The first report of multilocus variable-number tandem repeat analysis (MLVA) for M. pneumoniae strain typing.
- 16.Diaz MH, Winchell JM. The evolution of advanced molecular diagnostics for the detection and characterization of Mycoplasma pneumoniae . Front. Microbiol. 2016;7:232. doi: 10.3389/fmicb.2016.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]; • A detailed and up-to-date review on molecular detection and characterization of M. pneumoniae.
- 17.Hjorth SV, Bjornelius E, Lidbrink P, et al. Sequence-based typing of Mycoplasma genitalium reveals sexual transmission. J. Clin. Microbiol. 2006;44(6):2078–2083. doi: 10.1128/JCM.00003-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma L, Taylor S, Jensen JS, Myers L, Lillis R, Martin DH. Short tandem repeat sequences in the Mycoplasma genitalium genome and their use in a multilocus genotyping system. BMC Microbiol. 2008;8:130. doi: 10.1186/1471-2180-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma L, Jensen JS, Mancuso M, et al. Genetic variation in the complete MgPa operon and its repetitive chromosomal elements in clinical strains of Mycoplasma genitalium . PLoS ONE. 2010;5(12):e15660. doi: 10.1371/journal.pone.0015660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma L, Jensen JS, Mancuso M, et al. Variability of trinucleotide tandem repeats in the MgPa operon and its repetitive chromosomal elements in Mycoplasma genitalium . J. Med. Microbiol. 2012;61(Pt 2):191–197. doi: 10.1099/jmm.0.030858-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma L, Jensen JS, Mancuso M, Myers L, Martin DH. Kinetics of genetic variation of the Mycoplasma genitalium MG192 gene in experimentally infected chimpanzees. Infect. Immun. 2015;84(3):747–753. doi: 10.1128/IAI.01162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma L, Jensen JS, Myers L, et al. Mycoplasma genitalium: an efficient strategy to generate genetic variation from a minimal genome. Mol. Microbiol. 2007;66(1):220–236. doi: 10.1111/j.1365-2958.2007.05911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma L, Mancuso M, Williams JA, et al. Extensive variation and rapid shift of the MG192 sequence in Mycoplasma genitalium strains from patients with chronic infection. Infect. Immun. 2014;82(3):1326–1334. doi: 10.1128/IAI.01526-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma L, Martin DH. Single-nucleotide polymorphisms in the rRNA operon and variable numbers of tandem repeats in the lipoprotein gene among Mycoplasma genitalium strains from clinical specimens. J. Clin. Microbiol. 2004;42(10):4876–4878. doi: 10.1128/JCM.42.10.4876-4878.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benitez AJ, Diaz MH, Wolff BJ, et al. Multilocus variable-number tandem-repeat analysis of Mycoplasma pneumoniae clinical isolates from 1962 to the present: a retrospective study. J. Clin. Microbiol. 2012;50(11):3620–3626. doi: 10.1128/JCM.01755-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atkinson TP, Balish MF, Waites KB. Epidemiology, clinical manifestations, pathogenesis and laboratory detection of Mycoplasma pneumoniae infections. FEMS Microbiol. Rev. 2008;32(6):956–973. doi: 10.1111/j.1574-6976.2008.00129.x. [DOI] [PubMed] [Google Scholar]
- 27.Cousin-Allery A, Charron A, De Barbeyrac B, et al. Molecular typing of Mycoplasma pneumoniae strains by PCR-based methods and pulsed-field gel electrophoresis. Application to French and Danish isolates. Epidemiol. Infect. 2000;124(1):103–111. doi: 10.1017/s0950268899003313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorigo-Zetsma JW, Dankert J, Zaat SA. Genotyping of Mycoplasma pneumoniae clinical isolates reveals eight P1 subtypes within two genomic groups. J. Clin. Microbiol. 2000;38(3):965–970. doi: 10.1128/jcm.38.3.965-970.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su CJ, Dallo SF, Baseman JB. Molecular distinctions among clinical isolates of Mycoplasma pneumoniae . J. Clin. Microbiol. 1990;28(7):1538–1540. doi: 10.1128/jcm.28.7.1538-1540.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao F, Cao B, Li J, et al. Sequence analysis of the p1 adhesin gene of Mycoplasma pneumoniae in clinical isolates collected in Beijing in 2008–2009. J. Clin. Microbiol. 2011;49(8):3000–3003. doi: 10.1128/JCM.00105-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chalker VJ, Pereyre S, Dumke R, et al. International Mycoplasma pneumoniae typing study: interpretation of M. pneumoniae multilocus variable-number tandem-repeat analysis. New Microbes New Infect. 2015;7:37–40. doi: 10.1016/j.nmni.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Comparision of MLVA typing of 30 predefined M. pneumoniae DNA samples by six international laboratories provided new guidelines for performing and interpreting MLVA typing.
- 32.Diaz MH, Benitez AJ, Cross KE, et al. Molecular detection and characterization of Mycoplasma pneumoniae among patients hospitalized with community-acquired pneumonia in the United States. Open Forum Infect. Dis. 2015;2(3):ofv106. doi: 10.1093/ofid/ofv106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun H, Xue G, Yan C, et al. Multiple-locus variable-number tandem-repeat analysis of Mycoplasma pneumoniae clinical specimens and proposal for amendment of MLVA nomenclature. PLoS ONE. 2013;8(5):e64607. doi: 10.1371/journal.pone.0064607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown RJ, Holden MT, Spiller OB, Chalker VJ. Development of a multilocus sequence typing scheme for molecular typing of Mycoplasma pneumoniae . J. Clin. Microbiol. 2015;53(10):3195–3203. doi: 10.1128/JCM.01301-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fungtammasan A, Ananda G, Hile SE, et al. Accurate typing of short tandem repeats from genome-wide sequencing data and its applications. Genome Res. 2015;25(5):736–749. doi: 10.1101/gr.185892.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kenri T, Horino A, Matsui M, et al. Complete genome sequence of Mycoplasma pneumoniae type 2a strain 309, isolated in Japan. J. Bacteriol. 2012;194(5):1253–1254. doi: 10.1128/JB.06553-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S, Liu F, Sun H, Zhu B, Lv N, Xue G. Whole-genome sequencing of macrolide-resistant Mycoplasma pneumoniae strain S355, isolated in China. Genome Announc. 2016;4(2):e00087–e00116. doi: 10.1128/genomeA.00087-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S, Sun H, Liu F, Zhao H, Zhu B, Na L. Complete genome sequence of the macrolide-resistant Mycoplasma pneumoniae strain C267 in China. Genome Announc. 2016;4:e00236–e00216. doi: 10.1128/genomeA.00236-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Touati A, Blouin Y, Sirand-Pugnet P, et al. Molecular epidemiology of Mycoplasma pneumoniae: genotyping using single nucleotide polymorphisms and SNaPshot technology. J. Clin. Microbiol. 2015;53(10):3182–3194. doi: 10.1128/JCM.01156-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spuesens EB, Oduber M, Hoogenboezem T, et al. Sequence variations in RepMP2/3 and RepMP4 elements reveal intragenomic homologous DNA recombination events in Mycoplasma pneumoniae . Microbiology. 2009;155(Pt 7):2182–2196. doi: 10.1099/mic.0.028506-0. [DOI] [PubMed] [Google Scholar]
- 41.Spuesens EB, Van De Kreeke N, Estevao S, et al. Variation in a surface-exposed region of the Mycoplasma pneumoniae P40 protein as a consequence of homologous DNA recombination between RepMP5 elements. Microbiology. 2011;157(Pt 2):473–483. doi: 10.1099/mic.0.045591-0. [DOI] [PubMed] [Google Scholar]
- 42.Goodwin S, Mcpherson JD, Mccombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016;17(6):333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 1988;26(11):2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Metzgar D, Liu L, Hansen C, Dybvig K, Wills C. Domain-level differences in microsatellite distribution and content result from different relative rates of insertion and deletion mutations. Genome Res. 2002;12(3):408–413. doi: 10.1101/gr.198602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamada NA, Smith GA, Castro A, Roques CN, Boyer JC, Farber RA. Relative rates of insertion and deletion mutations in dinucleotide repeats of various lengths in mismatch repair proficient mouse and mismatch repair deficient human cells. Mutat. Res. 2002;499(2):213–225. doi: 10.1016/s0027-5107(01)00282-2. [DOI] [PubMed] [Google Scholar]
- 46.Dumke R, Jacobs E. Culture-independent multi-locus variable-number tandem-repeat analysis (MLVA) of Mycoplasma pneumoniae . J. Microbiol. Methods. 2011;86(3):393–396. doi: 10.1016/j.mimet.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 47.Yan C, Sun H, Xue G, et al. A single-tube multiple-locus variable-number tandem-repeat analysis of Mycoplasma pneumoniae clinical specimens by use of multiplex PCR-capillary electrophoresis. J. Clin. Microbiol. 2014;52(12):4168–4171. doi: 10.1128/JCM.02178-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao F, Liu G, Cao B, et al. Multiple-locus variable-number tandem-repeat analysis of 201 Mycoplasma pneumoniae isolates from Beijing, China, from 2008 to 2011. J. Clin. Microbiol. 2013;51(2):636–639. doi: 10.1128/JCM.02567-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krishnakumar R, Assad-Garcia N, Benders GA, Phan Q, Montague MG, Glass JI. Targeted chromosomal knockouts in Mycoplasma pneumoniae . Appl. Environ. Microbiol. 2010;76(15):5297–5299. doi: 10.1128/AEM.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unemo M, Dillon JA. Review and international recommendation of methods for typing Neisseria gonorrhoeae isolates and their implications for improved knowledge of gonococcal epidemiology, treatment, and biology. Clin. Microbiol. Rev. 2011;24(3):447–458. doi: 10.1128/CMR.00040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.