Abstract
In medical practices, doctors detail patients’ care plan via discharge summaries written in the form of unstructured free texts, which among the others contain medication names and prescription information. Extracting prescriptions from discharge summaries is challenging due to the way these documents are written. Handwritten rules and medical gazetteers have proven to be useful for this purpose but come with limitations on performance, scalability, and generalizability. We instead present a machine learning approach to extract and organize medication names and prescription information into individual entries. Our approach utilizes word embeddings and tackles the task in two extraction steps, both of which are treated as sequence labeling problems. When evaluated on the 2009 i2b2 Challenge official benchmark set, the proposed approach achieves a horizontal phrase-level F1-measure of 0.864, which to the best of our knowledge represents an improvement over the current state-of-the-art.
Keywords: NLP, Machine Learning, Word Embeddings, CRFs, Prescription Extraction
Graphical Abstract
1. Introduction
In medical practices, doctors detail patients’ care plan in unstructured free texts. These documents contain medication names and prescription information, which are important components of patients’ overall care.
Extracting medication names and other prescription information from discharge summaries is challenging due to the way these documents are written. While highly readable to those with medical background, such documents are not intended to be digested by computers. The presence of different expressions conveying the same information, medical acronyms, misspellings, and ambiguous terminologies makes the automatic analysis of these documents difficult.
For example, consider the following excerpts: (1) The doctor prescribed him 325 mg Aspirin p.o. 4x/day for 2 weeks as needed for inflammation. (2) We gave her a seven-day course of 200 mg Cefpodoxime q.h.s. for bronchitis, which was taken through mouth. From both excerpts, we want to extract mentioned medication names along with information related to their dosage, mode of administration, frequency, duration, and medical reason for prescription. We refer to this task as medication information extraction, where medication names, dosages, modes, frequencies, durations, and reasons are medication entities. Medication entities corresponding to the above examples are demonstrated in Table 1 below.
Table 1.
Medication entries extracted from the prescription excerpts (1) and (2)
| Medication Name | Dosage | Mode | Frequency | Duration | Reason |
|---|---|---|---|---|---|
| Aspirin | 325 mg | p.o. | 4x/day | for 2 weeks | inflammation |
| Cefpodoxime | 200 mg | through mouth | q.h.s. | a seven-day course | bronchitis |
We then group medication entities together through relation extraction to create medication entries, which link medications to their signature information and constitute the final output. Medication information extraction and relation extraction collectively make up what we refer to as prescription extraction.
In this paper, we present a system for automatic prescription extraction from unstructured discharge summaries. We treat prescription extraction as a two-step sequence labeling task: we first apply Conditional Random Fields (CRFs) with word embeddings to extract medication information; we then tackle relation extraction as a second sequence labeling task. We evaluated our system against the i2b2 2009 official benchmark set on medication entries, achieving a horizontal phrase-level F1-measure of 0.864 (see section 4.4 for a description of the evaluation metrics). The proposed system achieves a significantly higher overall performance than the current state-of-the-art system.
2. Related Work
2.1 Medication Extraction Systems
MedLEE[1] is one of the earliest medication extraction systems, built with the purpose of extracting, structuring, and encoding clinical information within free text patient reports. MedLEE extracts medication information using hand-written rules. Similar to MedLEE, MetaMap[2] is a rule-based system that extracts medical concepts (which includes medications) by querying the Unified Medical Language System (UMLS) Metathesaurus[3]. Both systems are unable to extract medication entries since they cannot interpret the relations of medication entities.
Research in automatic prescription extraction has been fostered by the Third i2b2 Challenge on NLP for Clinical Records[4]. The best performing system[5] in this challenge used a hybrid of machine learning classifiers and handwritten rules. It utilized Conditional Random Fields (CRFs) for medication information extraction and applied Support Vector Machines (SVMs) for relation extraction, reaching a horizontal phrase-level F1-measure of 0.857 on the i2b2 official benchmark set. Similarly, Li et al.[6] from the University of Wisconsin-Milwaukee trained CRFs with rules for medication information extraction but reached a relatively low performance (horizontal phrase-level F1-measure of 0.764). The significant performance differences using CRFs indicate the importance of system architecture, feature extraction, and parameter optimization. Besides, seven out of the top 10 systems, ranked from 2nd to 8th, were purely rule-based[7–13]. They utilized pattern matching rules with existing knowledge bases such as medication gazetteers.
2.2 Word Embeddings in Named-Entity Recognition (NER)
Word embeddings[14] have been used in several NER tasks[15–17] to capture meaningful syntactic and semantic regularities using unsupervised learning from selected training corpora. In clinical NER, there are two prior studies that included word embeddings in their experiments.
First, De Vine et al.[18] analyzed the effectiveness of word embeddings in clinical concept extraction and studied the influence of various corpora used to generate embeddings. During feature extraction, they clustered word vectors into categories and used categorical labels as features for the classifier. They found that real-valued vectors did not show advantages when applied as feature set as the reason to use nominal categories via clustering. In our study, we want to evaluate the efficacy of real-valued word vectors instead of categorical labels, when directly used as classifier features. Second, Wu et al.[19] explored two neural word embedding algorithms (i.e., word2vec[20] and ranking-based[21]) in two clinical NER tasks. Both algorithms use a local context window model that do not explicitly consider global word-word co-occurrence statistics[22], which may contain important linguistic properties. In contrast, GloVe[23], introduced after word2vec, specifically focused on the ratio of co-occurrence probabilities between different set of words when extracting word vectors. To the best of our knowledge, it is still unclear whether GloVe with real-valued vectors positively contribute to the clinical NER tasks.
3. Data
The Third i2b2 Challenge on NLP for Clinical Records[4] provided a corpus of 696 unannotated clinical records for development and 252 manually annotated records for testing. When participating into this challenge, Patrick et al.[5] manually annotated 145 developmental records and used them as training set. This training set contained 250,436 tokens, 21,077 medication entities, and 8,516 medication entries. For testing, we used the official benchmark set from the i2b2 2009 challenge. See Table 2 for the per-category statistics.
Table 2.
Number of entities per category in the training and test data
| Dataset | Record | Token | Medication | Dosage | Mode | Frequency | Duration | Reason | Entry |
|---|---|---|---|---|---|---|---|---|---|
| Training | 145 | 250,436 | 7,988 | 4,132 | 3,052 | 3,881 | 508 | 1,516 | 8,516 |
| Testing | 252 | 284,311 | 8,440 | 4,371 | 3,299 | 3,925 | 499 | 1,325 | 9,193 |
4. Methods
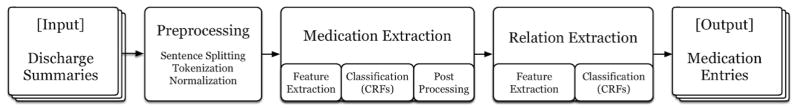
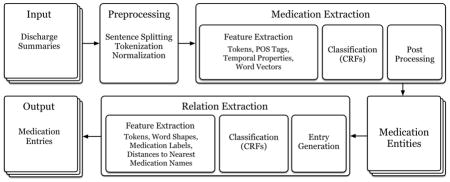
We tackled prescription extraction in two consecutive steps: (1) medication information extraction, and (2) relation extraction. Figure 1 depicts the workflow for our system. We present the details of each component below.
Figure 1.
Prescription extraction system workflow
4.1 Pre-processing
We first pre-processed the data. We split the documents into sentences and then into tokens by simply splitting periods (excluding periods in acronyms, lists, and numbers) and whitespaces. We lowercased tokens and assigned part-of-speech (POS) tags using Natural Language Toolkit (NLTK)[24]. We replaced numbers, including literal and numerical forms, by placeholders (e.g., five days → D days, 10am → DDam, .95 → .DD).
4.2 Medication Information Extraction
We experimented with four different classifiers: Multinomial Naïve Bayes, SVMs, Decision Trees, and CRFs. We tuned the parameters of our classifiers using 5-fold cross validation on the training set. We then experimented with various feature sets and complemented our approach with post-processing rules. The results from each experiment were evaluated using phrase-level F1-measure (exact match, see section 4.4).
4.2.1 Feature Extraction
Tokens and POS tags are the base features for our models. For durations and frequencies, we found that most phrases are introduced with and/or closed by specific signals. We captured this information by using two binary features representing whether the current token is a starting signal (e.g., for, before, after) or an ending signal (e.g., weeks, days, hours). We collected a list of these signals by harvesting the training data. Starting signals are mostly temporal prepositions, whereas ending signals tend to be names of time periods or clinical events (see Figure 2 for additional examples).
Figure 2.
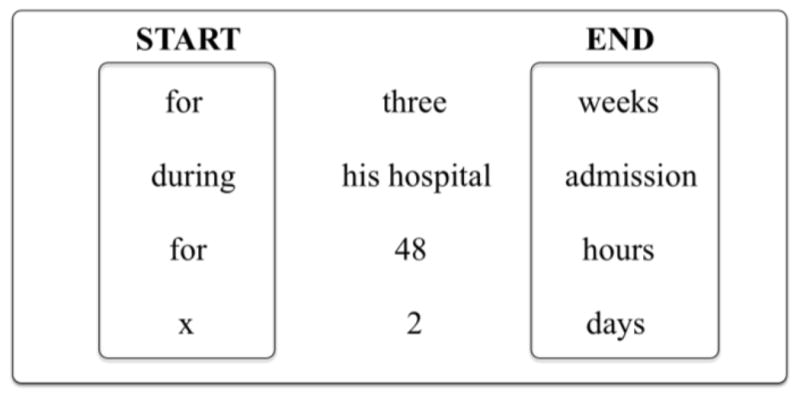
Duration example phrases
Similarly, we extracted five more temporal binary features derived from the ones mostly used in the literature[25]. These features indicate whether a token represents a time (e.g., 8am, 7-pm), temporal period (e.g., decades, weekends), part of the day (e.g., morning, afternoon), temporal reference (e.g., today, yesterday), and numbers (e.g., 0.25, 700). Duration is one of the challenging medication categories. These signal features add characteristics to the tokens that belong to temporal expressions, which helps the classifier better identifying the beginning and the end of duration phrases.
Finally, we concluded feature extraction with the addition of word embeddings, which have been shown to capture meaningful syntactic and semantic regularities in NER tasks. In particular, we used GloVe to extract word vectors from MIMIC III[26]: a large critical care database, which among the others, contains about 2 million clinical notes for about 46 thousand patients. In contrast to other related studies, we preprocessed this dataset using the same normalizer applied on our own medication dataset to create a specific training corpus for GloVe that is more suitable for our task. Besides, we took a balanced approach on computational resource and performance to use 100 dimensions when representing each word. When using word vectors, if a token in our dataset is not included in our pre-calculated vector set, we used the vector from token <unknown> instead.
To compare real-valued vectors with categorical labels, we used k-means algorithm to cluster word vectors extracted from MIMIC III into 1024 distinct categories, which is the optimal cluster size suggested by De Vine et al.[18]. We converted all vectors for each token in the training set into a categorical label by mapping the token to one of the 1024 categories. If a token was not found in all clusters, we used category <unknown> instead.
4.2.2 Supervised classification
We experimented with four different supervised classifiers, i.e., Multinomial Naïve Bayes, Decision Trees, SVMs, and CRFs. We used Scikit-Learn[27] for the first three classifiers and CRFSuite[28] for latter. With the introduction of neighboring tokens in the feature set, we made the first three classifiers capable of understanding token sequences, whereas CRFs is naturally suitable when extracting sequences.
4.2.3 Post-processing
The post-processing step is mainly devoted to boost recall for medication names, which play a strategic role in the sequences since medication names often start a prescription sequence. We looked up tokens in the Drug category of Wikipedia and in the online database from Drugs.com1. If one of those two external resources contained the current token, we marked it as a medication name. In addition, we used Google Search API 2 and its automatic spelling correction module, which we found particularly useful for correcting misspelled medication names. We marked a token as a medication name when the top retrieved web pages were from the domains Drugs.com or RxList.com.
4.3 Relation Extraction
After medication information extraction, medication entities are grouped together through relation extraction to create medication entries. Each entry contains a medication name, and when available, links the medication to its dosage, mode, frequency, duration, and reason. This task is a very special case of relation extraction, since the relation holding between medication entities (e.g., “Tylenol” and “325 mg”) is simply binary: the entities belong to the same medication entry or not.
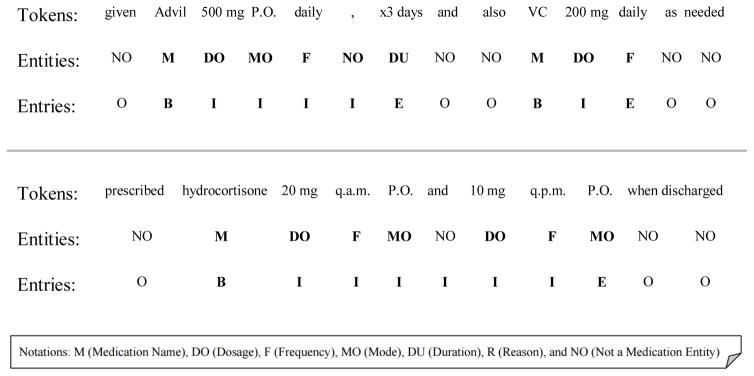
We tackled the task as a sequence labeling problem from the observation that medication names are mostly related to their neighboring entities. Seeing each medication entry as a sequence of tokens allows us to predict its boundaries and relate the entities inside to the same medication. We used tokens and the previously predicted medication information labels as base features to predict spans of textual excerpts containing medication entries. We added to these the absolute distances between the current token and its nearest medication name (both to the left and right), and the collapsed word shape (e.g., “Tylenol” → “Aa”, “325mg” → “0a”). We applied CRFs with window sizes ranging from ±1 to ±3 in CRFSuite[29]. Figure 3 shows two examples of textual excerpts with their entries represented with the BIOE (Begin, Inside, End, and Outside) format. In particular, the second excerpt in Figure 3 shows an example of a medication mentioned with two dosages, modes, and frequencies. In this case, we need to extract two medication entries with different entities sharing the same medication name.
Figure 3.
Example excerpts of medication entry representation
At the end of relation extraction, the extracted sequences are textual excerpts that contain medication entities. We then grouped medication entities inside each excerpt into individual entries.
4.4 Evaluation Metrics and Significance Tests
The results from each experiment on medication information extraction were evaluated using phrase-level F1-measure (i.e., exact match). As for the final output of our prescription extraction system, in order to preserve the comparability of this research with related work, we utilized the same evaluation metrics used in the i2b2 2009 Challenge: horizontal phrase-level F1-measure, which evaluates our system’s final output on the entry level. Therefore, every entity in the entry needs to match exactly in accordance with the gold standard.
In terms of significance test, we used z-test (95% confidence level, population equals to total number of entities, proportion equals to the correctly extracted entities over population) to benchmark the significance of our experiments internally on various settings as well as externally against other existing systems.
5. Results and Discussion
For medication information extraction, we experimented with four multi-label classifiers and generated five sets of results (i.e., window sizes ranging from ±1 to ±5) for each classifier. Cross-validation on the training set indicated that window size of ±1 performed lower than the remaining four window sizes on all four experimented classifiers. However, we did not find any notable differences among window sizes ranging from ±2 to ±5. Therefore, we selected the shortest and simplest window size from this range, i.e., ±2. In terms of classifier selection, Table 3 shows the performance of the four experimented classifiers, when the same feature set mentioned in section 4.2.1 was applied with window size of ±2. We observed that SVMs and CRFs performed significantly better than Naive Bayes and Decision Trees on all types of entities. Furthermore, CRFs performed significantly better than SVMs on medication durations and reasons due to its strength in identifying entities with longer spans. Therefore, multi-label CRFs is selected as the optimal classifier for medication information extraction.
Table 3.
Phrase-level F1-measure from four different classifiers without post-processing3
| Classifier | Word Embeddings | Medication | Dosage | Mode | Frequency | Duration | Reason |
|---|---|---|---|---|---|---|---|
| CRFs | Yes | 0.885 | 0.915 | 0.935 | 0.924 | 0.618 | 0.407 |
| No | 0.838 | 0.908 | 0.912 | 0.917 | 0.543 | 0.405 | |
|
| |||||||
| SVMs | Yes | 0.889 | 0.890 | 0.941 | 0.911 | 0.316 | 0.340 |
| No | 0.818 | 0.866 | 0.888 | 0.836 | 0.294 | 0.280 | |
|
| |||||||
| Naive Bayes | No | 0.742 | 0.817 | 0.844 | 0.802 | 0.008 | 0.138 |
|
| |||||||
| Decision Trees | No | 0.735 | 0.816 | 0.887 | 0.756 | 0.123 | 0.165 |
Before we proceed to relation extraction, we checked the contribution of word embeddings on both CRFs and SVMs. The addition of word embeddings was not implemented with Naive Bayes and Decision Trees because these classifiers performed significantly worse in prior experiments. Table 3 shows significant performance improvement after the introduction of word embeddings in both CRFs and SVMs, especially on medication names and durations (all results are significantly higher except reason when using CRFs).
We also verified the performance differences between real-valued and categorical implementation of word vectors. We ran our system twice using the training set with real-valued embeddings and categorical-labeled embeddings each, in a 5-fold cross-validation setting. Results show insignificant performance differences on medication name, dosage, frequency, and reason. However, performance improvement on medication mode and duration were significant, when real-valued embeddings were used (see Table 4).
Table 4.
Phrase-level F1-measure using CRFs using word embeddings in real-valued vectors and categorical labels without post-processing
| Classifier | Word Embeddings | Medication | Dosage | Mode | Frequency | Duration | Reason |
|---|---|---|---|---|---|---|---|
| CRFs | Real-valued | 0.885 | 0.915 | 0.935 | 0.924 | 0.618 | 0.407 |
| Categorical | 0.882 | 0.917 | 0.921 | 0.924 | 0.579 | 0.409 |
To evaluate the generalizability of our approach, we also tested the contribution of word embeddings on two separate tasks. We ran our system applying the same parameters used in the prescription extraction, but swapping the training data with the ones corresponding to these two tasks. First task is the medical concept extraction from the 2010 i2b2/VA Challenge[29]. The objective of this task is to extract medical problems, lab tests, and treatments from discharge summaries. We used the official training set from this challenge as the training data for our system, which contains 349 annotated records.
The second task is the de-identification of protected health information (PHI) from the 2016 CEGS N-GRID Challenge[30]. The objective of this task is to identify PHI (e.g., patient names, phone numbers, hospitals) from psychiatric evaluation records. We applied our system using the official training set released for this challenge, which contains 600 annotated with 30 predefined PHI categories. We tested our system on 11 categories, which are the ones having more than 50 training samples.
We applied our system on both tasks with and without word embeddings. Table 5 and 6 show significant performance improvement on both tasks after the introduction of word embeddings. All results are phrase-level F1-measure, 5-fold cross-validated.
Table 5.
Phrase-level F1-measure on the medical concept extraction task
| Word Embeddings | Problem | Test | Treatment |
|---|---|---|---|
| Yes | 0.759 | 0.793 | 0.744 |
| No | 0.627 | 0.719 | 0.628 |
Table 6.
Phrase-level F1-measure on the de-identification of PHI task
| Word Embeddings | Age | City | Country | Date | Doctor | Hospital |
|---|---|---|---|---|---|---|
| Yes | 0.951 | 0.718 | 0.493 | 0.948 | 0.889 | 0.724 |
| No | 0.940 | 0.664 | 0.439 | 0.931 | 0.874 | 0.705 |
|
| ||||||
| Word Embeddings | Organization | Patient | Phone | Profession | State | |
|
| ||||||
| Yes | 0.567 | 0.637 | 0.851 | 0.648 | 0.794 | |
| No | 0.551 | 0.569 | 0.800 | 0.554 | 0.672 | |
In terms of relation extraction, we conducted a manual analysis of the medication entries from the training set. We found that in more than 90% of the cases, medication names appear prior to their dosages, modes, frequencies, durations, and reasons. Given this observation, as a baseline for relation extraction, we assigned all entities found between two medication names to the first medication name. This approach returned a horizontal phrase-level F1-measure of 0.893 on gold standard medication entities in the training set.
To check the contribution of our sequence labeling approach on relation extraction, we 5-fold cross-validated the same dataset (i.e., training set with gold standard annotation) using CRFs on the feature set described in section 4.3 with window size ±2. This approach obtained a horizontal phrase-level F1-measure of 0.945, which is significantly higher than the baseline. We also experimented with window sizes ±1 and ±3, but window size ±2 performed the highest among them.
Finally, we evaluated our prescription extraction system on the i2b2 official benchmark set. We first ran the medication information extraction component to find medication entities, then ran the relation extraction component on the predicted entities to create entries. On a laptop with an Intel i5–4300u processor and 8 GB RAM, the training process for medication and relation extraction takes about 12 and 2 minutes each to complete. Once the CRFs models are generated, the end to end prescription extraction from one discharge summary to medication entries is almost instantaneous.
The system proposed here achieved a horizontal phrase-level F1-measure of 0.864. Table 7 shows the performance of our method, when compared to the top 6 systems submitted during the challenge. Results from the test set (Table 7) is consistent with our cross validated results on the training set (Table 3). We also observed significant performance improvement after the introduction of word embeddings (see Table 8), and the application of post-processing rules (see Table 9). We then analyzed post-processed results to determine the contribution of each knowledge base. From 652 corrected medication names, 221 were corrected by Drugs.com, 307 were corrected by Wikipedia, and 78 were corrected by both. The remaining instances were corrected by querying the Google search engine. Post-processing boosted the recall of medication names, resulting into more entities being related to these medications. However, the main contribution is still word embeddings, with a 5.4% improvement with respect to the horizontal score.
Table 7.
Horizontal Phrase-level F1-measure on the benchmark set using i2b2 official evaluation scripts
| Method | Medication | Dosage | Mode | Frequency | Duration | Reason | Horizontal Score |
|---|---|---|---|---|---|---|---|
| Yang[10] | 0.859 | 0.804 | 0.849 | 0.817 | 0.399 | 0.229 | 0.796 |
| Solt et al.[7] | 0.848 | 0.827 | 0.880 | 0.813 | 0.393 | 0.303 | 0.797 |
| Mork et al.[9] | 0.845 | 0.882 | 0.884 | 0.866 | 0.400 | 0.275 | 0.803 |
| Spasic et al.[12] | 0.839 | 0.841 | 0.846 | 0.854 | 0.525 | 0.459 | 0.812 |
| Doan et al.[13] | 0.858 | 0.853 | 0.888 | 0.868 | 0.363 | 0.361 | 0.821 |
| Patrick et al.[5] | 0.884 | 0.893 | 0.899 | 0.897 | 0.446 | 0.444 | 0.857 |
| Our Approach | 0.896 | 0.894 | 0.909 | 0.902 | 0.483 | 0.368 | 0.864 |
Table 8.
Phrase-level F1-measure on the benchmark set with and without word embeddings, not postprocessed
| Embeddings | Medication | Dosage | Mode | Frequency | Duration | Reason | Horizontal Score |
|---|---|---|---|---|---|---|---|
| No | 0.800 | 0.831 | 0.827 | 0.853 | 0.254 | 0.257 | 0.778 |
| Yes | 0.857 | 0.860 | 0.885 | 0.893 | 0.473 | 0.320 | 0.832 |
Table 9.
Phrase-level F1-measure on the benchmark set with and without post-processing
| Method | Medication | Dosage | Mode | Frequency | Duration | Reason | Horizontal Score |
|---|---|---|---|---|---|---|---|
| Not-processed | 0.857 | 0.860 | 0.885 | 0.893 | 0.473 | 0.320 | 0.832 |
| Post-processed | 0.896 | 0.894 | 0.909 | 0.902 | 0.483 | 0.368 | 0.864 |
Table 7 shows that our system can extract the most medication names, dosages, modes, and frequencies from the test set, with a horizontal score of 0.864. Comparatively, we also applied the same predicted medication entities on the rule-based relation extraction method (i.e., assigned all entities found between two medication names to the first medication name) and obtained a significantly lower horizontal score of 0.849. Upon further analysis of the system outputs from challenge participants, with the addition of word embeddings and post-processing rules, our system performed particularly well in extracting 365 medications that are not mentioned in the training set, as compared to 259 such medications extracted by the best performing system[5] in the challenge. Our system is also able to extract some misspelled medication names during post-processing using Google Search API (e.g., solsite, clobetizol, lopipd), which are not included in the current knowledge bases. Also, more dosages that contain numbers are properly extracted in our system output due to token normalization in the pre-processing steps. However, our system does not improve the extraction of duration and reason.
5.1 Error Analysis
We performed an error analysis to pinpoint some noteworthy mistakes from system output.
For medication information extraction, the most prominent source of erroneous predictions is related to medication names that are followed by other information, surrounded by or close to brackets. We identified 42 cases in the test set. For example, in “lovenox (50mg)”, 50mg is extracted as part of the medication name instead of being recognized as a dosage. This happened because in many training samples bracketed information was labeled as medication names for their additional properties. Also, we identified 14 prepositions and conjunctions that are incorrectly predicted as part of a medication name (e.g., advil of 500 mg, tylenol and aspirin). This happens more frequently when these tokens are in the middle of two medication entities. Additionally, our system has difficulty distinguishing 63 generic medication names that refer to groups of medications (e.g., home medication, antibiotics). In few cases, tokens are incorrectly categorized because of their misleading lexical context. For example, “alcoholic beverage” and “vomiting” are both predicted as medication names when appearing respectively in “she took 600 ml alcoholic beverage for the day” and “vomiting x 5 hours”. Despite having the best performance in duration extraction among systems that used machine learning, our system misses 78 durations phrases that are longer or lack temporal signals (e.g., while the patient is recovering from his pneumonia, after your cardiac catheterization, possibly lifelong).
5.2 System Limitation
As for post-processing, the use of Google Search API is controversial because some potential PHIs may accidentally have been submitted to the search engine. Both datasets used in this paper are de-identified, however, this could be a limitation when users consider submitting their own discharge summaries.
In terms of relation extraction, some duration and reason entities are not properly linked to their entries due to longer distances to their related medication names. Also, considering every medication entry is treated as an independent sequence, if two medication names share one single entity (e.g., give her tylenol and aspirin, 200 mg each), this shared entity is related to only one of the medications. This is a limitation embedded in the model used to tackle the problem, rather than in the scarcity of the training data.
6. Conclusions
In this paper, we presented a high-performing system to automatically extract and organize prescription information into individual entries. Our system employs CRFs to extract medication information with word embeddings. Significant performance improvement after the integration of real-valued word embeddings extracted using GloVe confirmed their contribution in the clinical NER task. Our system also redefined the problem of relation extraction on medication names and their prescription information as a sequence labeling task by using CRFs. Our approach achieves a horizontal phrase-level F1-measure of 0.864 when evaluated using official scripts, which to the best of our knowledge represents the current state-of-the-art.
Highlights.
A high-performing system to extract and organize prescription information
Evaluating the contribution of real-valued word embeddings in clinical NER
Tackling medication relation extraction as a sequence labeling task using CRFs
Acknowledgments
The authors would like to acknowledge Jon Patrick and Min Li from The University of Sydney for sharing their annotated discharge summaries with us. This work was supported in part by “NIH P50 Neuropsychiatric Genome-Scale and RDoC Individualized Domains (N-GRID)” National Institutes of Health, NIH P50 MH106933, PI: Isaac Kohane, and by Philips Research NA, PI: Peter Szolovits. The content is solely the responsibility of the authors and does not necessarily represent the official views of the sponsors.
Footnotes
The online database is retrieved from https://www.drugs.com/drug_information.html.
Python bindings to Google search engine used in our system is from https://pypi.python.org/pypi/google.
All four classifiers used multi-label classification on window size ±2. All results are 5-fold cross-validated on the training set.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedman C. Proceedings of the AMIA Symposium. American Medical Informatics Association; 2000. A broad-coverage natural language processing system; p. 270. [PMC free article] [PubMed] [Google Scholar]
- 2.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. Journal of the American Medical Informatics Association. 2010 May 1;17(3):229–36. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research. 2004 Jan 1;32(suppl 1):D267–70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. Journal of the American Medical Informatics Association. 2010 Sep 1;17(5):514–8. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patrick J, Li M. A cascade approach to extracting medication events. Australasian Language Technology Association Workshop; 2009 Dec 3; 2009. p. 99. [Google Scholar]
- 6.Li Z, Cao Y, Antieau L, et al. Extracting medication information from patient discharge summaries. Proceedings of the Third i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2009. [Google Scholar]
- 7.Solt I, Tikk D. Yet another rule-based approach for extracting medication information from discharge summaries. Proceedings of the Third i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2009. [Google Scholar]
- 8.Grouin C, Deleger L, Zweigenbaum P. A simple rule-based medication extraction system. Proceedings of the Third i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2009. [Google Scholar]
- 9.Mork JG, Bodenreider O, Demner-Fushman D, et al. NLM’s I2b2 tool system description. Proceedings of the Third i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2009. [Google Scholar]
- 10.Yang HA. Linguistic Approach for medication extraction from medical discharge. Proceedings of the Third i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2009. [Google Scholar]
- 11.Hamon T, Grabar N. Concurrent linguistic annotations for identifying medication names and the related information in discharge summaries. Proceedings of the Third i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2009. [Google Scholar]
- 12.Spasic I, Sarafraz F, Keane JA, et al. Medication information extraction with linguistic pattern matching and semantic rules. Proceedings of the Third i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2009. [Google Scholar]
- 13.Doan S, Bastarache L, Klimkowski S, et al. Vanderbilt’s system for medication extraction. Proceedings of the Third i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2009. [Google Scholar]
- 14.Bengio Y, Ducharme R, Vincent P, Jauvin C. A neural probabilistic language model. journal of machine learning research. 2003;3(Feb):1137–1155. [Google Scholar]
- 15.Tang B, Cao H, Wang X, Chen Q, Xu H. Evaluating word representation features in biomedical named entity recognition tasks. BioMed research international. 2014 Mar;6:2014. doi: 10.1155/2014/240403. [DOI] [PMC free article] [PubMed] [Google Scholar]; Passos Alexandre, Kumar Vineet, McCallum Andrew. Lexicon infused phrase embeddings for named entity resolution. 2014. arXiv preprint arXiv:1404.5367. [Google Scholar]
- 16.Turian J, Ratinov L, Bengio Y, Roth D. A preliminary evaluation of word representations for named-entity recognition. NIPS Workshop on Grammar Induction, Representation of Language and Language Learning; 2009. pp. 1–8. [Google Scholar]
- 17.Mikolov T, Yih WT, Zweig G. Linguistic Regularities in Continuous Space Word Representations. InHLT-NAACL. 2013 Jun 9;13:746–751. [Google Scholar]
- 18.De Vine L, Kholghi M, Zuccon G, Sitbon L, Nguyen A. Analysis of word embeddings and sequence features for clinical information extraction [Google Scholar]
- 19.Wu Y, Xu J, Jiang M, Zhang Y, Xu H. A Study of Neural Word Embeddings for Named Entity Recognition in Clinical Text. AMIA Annual Symposium Proceedings; 2015; American Medical Informatics Association; 2015. p. 1326. [PMC free article] [PubMed] [Google Scholar]
- 20.Mikolov T, Chen K, Corrado G, Dean J. Efficient estimation of word representations in vector space. 2013 Jan 16; arXiv preprint arXiv:1301.3781. [Google Scholar]
- 21.Collobert R, Weston J, Bottou L, Karlen M, Kavukcuoglu K, Kuksa P. Natural language processing (almost) from scratch. Journal of Machine Learning Research. 2011;12(Aug):2493–537. [Google Scholar]
- 22.Zhang Y, Wallace B. A Sensitivity Analysis of (and Practitioners’ Guide to) Convolutional Neural Networks for Sentence Classification. 2015 Oct 13; arXiv preprint arXiv:1510.03820. [Google Scholar]
- 23.Pennington J, Socher R, Manning CD. Glove: Global Vectors for Word Representation. EMNLP. 2014 Oct 25;14:1532–43. [Google Scholar]
- 24.Bird S. NLTK: the natural language toolkit. Proceedings of the COLING/ACL on Interactive presentation sessions; 2006 Jul 17; Association for Computational Linguistics; pp. 69–72. [Google Scholar]
- 25.Filannino M, Nenadic G. Temporal expression extraction with extensive feature type selection and a posteriori label adjustment. Data & Knowledge Engineering. 2015;100:19–33. [Google Scholar]
- 26.Johnson AE, Pollard TJ, Shen L, Lehman LW, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG. MIMIC-III, a freely accessible critical care database. Scientific data. 2016:3. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J. Scikit-learn: Machine learning in Python. Journal of Machine Learning Research. 2011;12(Oct):2825–30. [Google Scholar]
- 28.Okazaki N. CRFsuite: a fast implementation of conditional random fields (CRFs) [Google Scholar]
- 29.Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association. 2011 Sep 1;18(5):552–6. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stubbs A, Filannino M, Uzuner Ö. De-identification of psychiatric intake records: Overview of 2016 CEGS N-GRID Shared Tasks Track 1. Journal of Biomedical Informatics. 2017 doi: 10.1016/j.jbi.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]