Abstract
Maintaining functional telomeres is important for long-term proliferation of cells. About 15% of cancer cells are telomerase-negative and activate the alternative-lengthening of telomeres (ALT) pathway to maintain their telomeres. Recent studies have shown that the human CTC1/STN1/TEN1 complex (CST) plays a multi-faceted role in telomere maintenance in telomerase-expressing cancer cells. However, the role of CST in telomere maintenance in ALT cells is unclear. Here, we report that human CST forms a functional complex localizing in the ALT-associated PML bodies (APBs) in ALT cells throughout the cell cycle. Suppression of CST induces telomere instabilities including telomere fragility and elevates telomeric DNA recombination, leading to telomere dysfunction. In addition, CST deficiency significantly diminishes the abundance of extrachromosomal circular telomere DNA known as C-circles and t-circles. Suppression of CST also results in multinucleation in ALT cells and impairs cell proliferation. Our findings imply that the CST complex plays an important role in regulating telomere maintenance in ALT cells.
Keywords: Telomere, CTC1/STN1/TEN1, ALT
1. Introduction
Telomeres, the physical termini at linear chromosomes, preserve genome stability by distinguishing natural chromosomal termini from broken DNA ends and protecting chromosomes from degradation and inappropriate repair activities [1–3]. Maintenance of functional telomeres is essential for long-term cell proliferation. In the majority of immortalized cell lines, including germline cells and ~85% of cancer cells, telomeres are maintained by telomerase-catalyzed de novo addition of telomeric repeats, allowing for indefinite cellular proliferation [4,5]. The remaining ~15% of human tumor cells lack telomerase activity, and maintain their telomeres using the alternative lengthening of telomeres (ALT) pathway [6–8].
Telomeric DNA, consisting of repetitive double-stranded (TTAGGG/AATCCC)n repeats and single-stranded G-rich 3′ overhangs, is bound by a group of proteins that play an important role in maintaining telomere stability. The shelterin complex, consisting of TRF1, TRF2, POT1, TPP1, TIN2, and RAP1, shields chromosome ends from being recognized as damaged DNA, as evidenced by numerous studies showing that shelterin components prevent the activation of ATM/ATR damage response pathways at telomeres (reviewed in [9]). Another important telomere maintenance complex is the CTC1-STN1-TEN1 (CST) complex, a trimeric protein complex that binds to single-stranded DNA with high affinity [10–16]. Several recent studies have shown that the CST complex is important for telomere maintenance in a multifaceted manner. First and foremost, it facilitates efficient replication of telomeric DNA, thereby preventing catastrophic telomere loss [12–14]. As a result, suppression of individual components of CST increases the frequencies of fragile telomeres and leads to telomere loss in human somatic cells and mammalian cells [12–14]. CST is also involved in the late S/G2-specific synthesis of telomeric C-strands referred to as C-strand fill-in, and depletion of CST results in excessively long G-overhangs [12–15]. Additionally, CST may compete with shelterin POT1-TPP1 for binding to telomeric DNA and restrict telomerase extension of telomeres [17]. The importance of CST in maintaining telomere stability is underscored by genetic studies showing that CTC1 and STN1 mutations cause the Coats Plus syndrome and dyskeratosis congenita [18–22], two diseases that are associated with telomere maintenance defects.
To date, the role of CST in telomere maintenance has mainly been investigated in non-ALT cells, and its role in ALT-mediated telomere maintenance is largely unknown. Although it appears that ALT cells employ homology-directed synthesis for telomere maintenance [8,23–25], the mechanism for the ALT pathway remains largely elusive. In addition, ALT cells show several characteristics distinct from non-ALT cells. First, ALT cells contain ALT-associated promyelocytic leukemia (PML) bodies (APBs), which are special PML nuclear bodies containing telomeric DNA, shelterin proteins and repair factors [25–27]. Mounting evidence suggests a model that APBs may provide sites to accumulate telomeric repeats and relevant proteins to facilitate telomere synthesis events [26,28–30]. Second, ALT cells show high frequency of telomere sister chromatid exchange (T-SCE), which is presumably caused by elevated levels of homology-directed repair (HDR) events at telomere repeats [31,32]. Third, telomere lengths in ALT cells are extremely heterogeneous [6,7]. Moreover, abundant extrachromosomal telomere repeats (ECTRs) are detectable in ALT cells. These ECTR molecules are predominantly composed of double-stranded telomeric circles named t-circles (TCs) and partially single-stranded circles referred to as C-circles (CCs) or G-circles that possess intact continuous C- or G-rich strands [24,33]. CCs are much more abundant than G-circles, and are found to be more specific and quantifiable to ALT activity than G-circles and TCs [24]. Lastly, while only 3′ G-rich overhangs are detectable in non-ALT cells, abundant 5′ C-rich overhangs are present in ALT cells [34]. These 5′ C-overhangs have been implicated in the telomere recombination pathway [35].
In this study we set out to examine the effect of CST suppression on telomere maintenance in ALT cells. We report that CTC1 and STN1 display punctate nuclear staining that colocalizes with APBs in ALT cells. Suppression of CST significantly decreases CC and TC abundance, elevates telomere abnormalities including T-SCE and fragile telomeres, and induces telomere DNA damage. Moreover, CST suppression limits ALT cell proliferation and dramatically increases the formation of multinucleated polyploid cells. Our results demonstrate that CST plays an important role in telomere maintenance in ALT cells, and suggest that targeting CST may be a potential therapeutic approach for inhibiting the growth of ALT-positive cancer cells.
2. Materials and methods
2.1. Cell culture
U2OS cells stably expressing Flag-CTC1 were constructed by retroviral transduction of pBabe- Flag-CTC1 [36], followed by hygromycin selection. All cells were cultured at 37 °C under 5% CO2 in DMEM supplemented with 10% fetal bovine serum or cosmic calf serum (HyClone). Double thymidine block was used to synchronize U2OS cells. Briefly, exponentially growing cells were treated with thymidine (2 mM) for 14 h, followed by a cell wash with prewarmed DMEM (three times) and then released into fresh media for 10 h. The second thymidine (2 mM) was then added to medium for 12–16 h, followed by a cell wash with pre-warmed DMEM (three times) before cells were released into fresh medium containing serum. Cells were collected at different time points for analyzing DNA contents using a Beckman Coulter EPICS® XL™ flow cytometer.
2.2. Antibodies
The following primary antibodies were used: rabbit α-OBFC1/STN1 (Santa Cruz), mouse α-OBFC1/STN1 (Sigma), rabbit α-CTC1 (ThermoFisher), rabbit α-TRF2 (Santa Cruz), rabbit α-PML (Santa Cruz), mouse α-PML (Santa Cruz), rabbit α-FLAG (Cell Signaling), mouse α-FLAG (Sigma), mouse α-actin (Millipore). Secondary antibodies were horseradish peroxidase conjugated anti-mouse IgG and anti-rabbit IgG (BD Biosciences) for western blotting, Dylight 488-anti-mouse IgG (ThermoFisher) and Dylight 549-anti-rabbit IgG (ThermoFisher) for immunofluorescence.
2.3. RNAi
STN1 siRNA sequences targeting GAUCCUGUGUUUCUAGCCU (siSTN1-1) and GCUUAACCUCACAACUUAA (siSTN1-2) were described previously [13]. STN1 shRNA sequences targeting GAUCCUGUGUUUCUAGCCU (shSTN1-1) GCUUAACCUCACAACUUAA (shSTN1-2) [13], GGACUGCCAGAAACCAAAT (shSTN1-4), CTC1 shRNA sequences targeting GTGTTTCCTTTGACCATCA (shCTC1-1), GAAAGTCTTGTCCGGTATT (shCTC1-2), and control siRNA targeting luciferase (shLUC) were described previously [36].
2.4. Immunofluorescence (IF), Immunofluorescence-FISH (IF-FISH)
For IF or IF-FISH, cells grown on chamber slides were fixed with 4% paraformaldehyde and permeabilized in 0.15% Triton X-100. In experiments with pre-permeabilization, cells were treated with 0.1% Triton X-100 for 5 min on ice prior to paraformaldehyde fixation [10]. Cells were then blocked for 1 h in 3% BSA at 37 °C, incubated with primary antibodies for overnight at 4 °C, washed 3 times with PBS and incubated with fluorescence-conjugated secondary antibodies for 1 h at 37 °C. Cells were washed again and counterstained with DAPI. Images were taken under Zeiss AxioImager M2 epifluorescence microscope. For IF-FISH, after staining for protein antigens, slides were re-fixed with 4% paraformaldehyde for 10 min at r.t. Slides were then denatured in hybridization buffer [10 mM Tris pH 7.5, 70% formamide, 0.5% blocking solution (Roche)] at 90 °C for 5 min and hybridized with peptide nucleic acid (PNA) probes Alexa488-OO-(TTAGGG)3 (TelG) or Cy5-OO-(CCCTAA)3 (TelC) at r.t. for 2 h as described previously [13]. Slides were then washed with 10 mM Tris pH7.5/70% formamide for 15 min, followed by washing two times in 0.1 M Tris pH7.5/0.15 M NaCl/0.08% Tween-20 for 5 min each. Slides were dried and DAPI containing mounting medium (Vector Labs) was applied for microscopy visualization.
2.5. Chromosome-oriented FISH (CO-FISH)
CO-FISH was performed as described previously [37]. Briefly, cells were cultured in the presence of 10 μM BrdU and BrdC (3:1) for one population doubling prior to colcemid treatment. Slides containing metaphase spreads were incubated with Hoechst 33258 for 15 min, exposed to UV light for 30 min, treated with Exonuclease III (Promega) for 10 min at room temperature, and then sequentially hybridized to PNA Alexa488-OO-(TTAGGG)3 and Cy3-OO-(CCCTAA)3 probes at the room temperature for 2 h. Slides were then washed and dehydrated in ethanol series. DNA was counterstained with DAPI, and images were taken under Zeiss AxioImager M2 epifluorescence microscope.
2.6. C-Circle assay (CC assay)
CC assay was performed as described previously [24]. Briefly, 40 ng of genomic DNA was digested with AluI and HaeIII, and then subjected to rolling circle amplification in the presence of 1 mM of each dATP, dGTP, dTTP, with or without 10 U Φ29 DNA polymerase (NEB). Reaction was incubated at 30 °C for 8 h, then at 65 °C for 20 min. Equal amount of reaction solution from each sample was subjected to agarose gel electrophoresis under non-denaturing condition to detect amplified long G-strand products that migrated minimally from the wells. For quantitative CC assay, an aliquot of reaction products was diluted with 2×SSC and slot-blotted onto Hybond-N+ membrane (GE Healthcare). DNA was UV-crosslinked and hybridized with the 32P-(CCCTAA)3 probe. Meanwhile, an equal amount of reaction products was denatured in 0.1 M NaOH at 65 °C for 10 min and then hybridized with 32P-Alu probe on slot blot. Membranes were exposed to Phosphor screens to obtain signals in the linear range, and were scanned on a STORM 860 (GE Healthcare) with the ImageQuant software. Signals from telomeric probe hybridization (native membrane) were normalized to denatured Alu signals to obtain CC abundance. All signals in each experiment were then normalized to the mean value of CC abundance in the control sample derived from three independent experiments to obtain relative CC abundance.
2.7. TRF analysis and pulsed-field gel electrophoresis (PFGE)
TRF was performed with standard PFGE using a Bio-Rad CHEF MAPPER™ apparatus. Genomic DNA was digested with a mixture of four enzymes (AluI, HaeIII, HinfI, MspI), separated with PFGE on a 0.8% agarose gel in 0.5×TBE for 20 h at 20 °C with the following setting: 1 s initial switch time, 12 s final switch time, 120° angle, 6 V cm−1. Following electrophoresis, the gel was denatured in denaturing solution (0.5 M NaOH, 1.5 M NaCl) for 45 min at r.t., dried, neutralized in neutralization solution (1.5 M NaCl, 0.5 M Tris pH 8.0), and then hybridized with the telomeric 32P-(TTAGGG)3 probe. Hybridization was performed as described previously [38].
2.8. 2D gel analysis for detecting t-circles
For 2D gel analysis, 10 μg of digested genomic DNA was resolved in the first dimension in a 0.4% agarose gel at 1 V cm−1 and stained with ethidium bromide. The gel slice was then cut out, casted into a second 1% agarose gel containing EtBr and resolved at 6 V cm−1. The gel was denatured in denaturing solution (0.5 M NaOH, 1.5 M NaCl) for 45 min at r.t., dried, neutralized in neutralization solution (1.5 M NaCl, 0.5 M Tris pH 8.0), and then hybridized to the telomeric 32P-(TTAGGG)3 probe. Hybridization was performed as described previously [38]. The gel was then exposed to a Phosphor screen to obtain signals in the linear range, and was scanned on a STORM 860 (GE Healthcare) with the ImageQuant software. To quantitate relative TC abundance, signals from t-circle were first normalized to total telomere signals, followed by normalizing to the mean value of shLUC control from three independent experiments.
2.9. Cell proliferation assay (MTS assay)
Cells were seeded onto 96-well microplates and treated with STN1 siRNA or control siRNA for 72 h. The determination of cell growth was performed with the CellTiter 96® Aqueous One Solution Cell Proliferation Assay kit (Promega), following the manufacturer’s instruction.
3. Results
3.1. CTC1 and STN1 formed punctate nuclear staining complex colocalizing with APBs
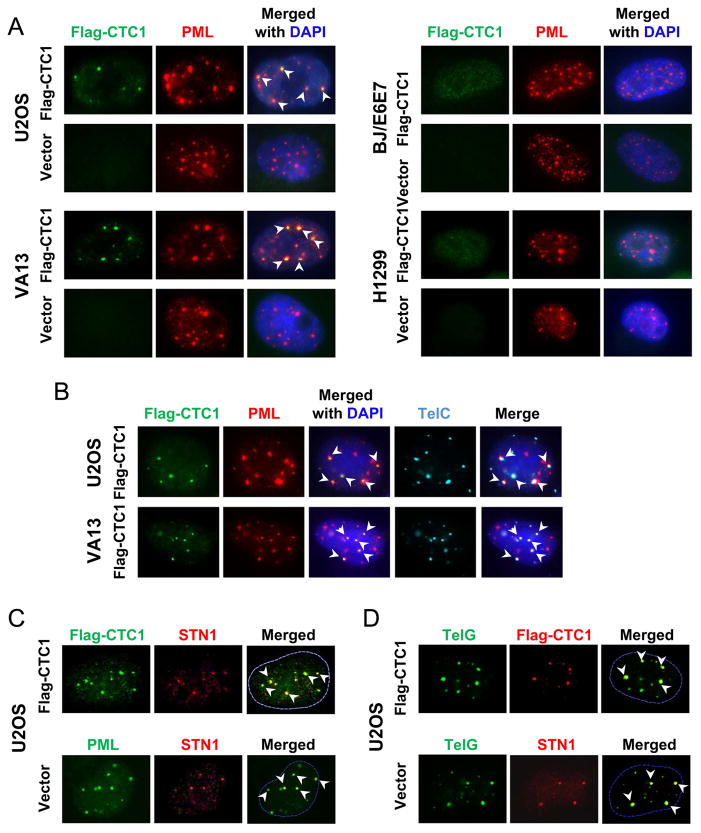
To investigate whether CST plays a functional role in ALT cells, we stably expressed Flag-tagged CTC1 in the U2OS ALT cell line by retroviral transduction, and then detected the cellular localization of CST components. Immunofluorescence with anti-Flag showed that Flag-CTC1 localized in nuclei in U2OS and VA13 cells (Fig. 1A). Notably, a subset of CTC1 formed distinct punctate nuclear foci in both U2OS and VA13, and such punctate foci colocalized with PML (Fig. 1A). To determine whether the punctate foci formation and PML colocalization are specific to ALT cells, we expressed Flag-CTC1 in two non-ALT cell lines [H1299 (telomerase positive) and BJ/E6E7 (BJ primary fibroblasts expressing HPV16 E6 and E7 proteins)]. In both cell lines, Flag-CTC1 showed diffused and weak nuclear staining with complete absence of punctate foci (Fig. 1A). No CTC1 colocalization with PML was observed in either H1299 or BJ/E6E7 (Fig. 1A). Since previously it has been reported that pre-permeabilization with Triton X-100 prior to fixation helps reveal foci of STN1 and CTC1 [10], we then tested this condition. We found similar foci formation of CTC1 as well as CTC1 colocalization with PML in ALT cells (Suppl Fig. S1). No punctate CTC1 foci formation was observed in either H1299 or telomerase-immortalized BJ cells (BJ/hTERT) under pre-permeabilization conditions, and CTC1 showed diffused nuclear staining in both cells (Suppl Fig. S1). Collectively, our results suggest that at least a subset of CTC1 forms punctate foci that colocalize with PML specifically in ALT cells.
Fig. 1.
CTC1 and STN1 formed punctate nuclear staining complex colocalizing in APBs. (A) IF for Flag-CTC1 (green) and PML (red) in two ALT cell lines U2OS and VA13 and two non-ALT cell lines H1299 and BJ/E6/E7. Arrowheads indicate colocalizations (yellow). (B) Flag-CTC1 located in APBs in U2OS and VA13. Cells were stained with Flag antibody (green) and PML antibody (red), followed by hybridization with telomeric PNA probe (TelC, teal). Arrowheads indicate colocalizations. (C) Flag-CTC1 and STN1 formed a complex that colocalized with PML. U2OS cells expressing Flag-CTC1 or vector alone were co-stained with anti-Flag and STN1, or PML and STN1 as indicated. (D) Flag-CTC1 and STN1 colocalized with telomeric DNA. U2OS cells expressing Flag-CTC1 or vector alone were co-stained with anti-Flag and telomeric probe (TelG), or STN1 and TelG as indicated.
Since ALT cells contain APBs that are comprised of special PML nuclear bodies, telomeric DNA, and telomere binding proteins [26,27], the observed specific colocalization of CTC1 with PML in ALT cells but not in non-ALT cells suggests that the colocalized foci may be in APBs. To test this, we performed combined immunofluorescence and FISH (IF-FISH) analysis, and found that the punctate foci of Flag-CTC1 colocalized with PML as well as telomeric DNA in both U2OS and VA13 cells (Fig. 1B), suggesting that CTC1 is localized in APBs. By immunostaining with anti-STN1, we also found that the punctate CTC1 foci contained STN1 (Fig. 1C, top panel). Similar to CTC1, STN1 foci colocalized with PML (Fig. 1C) as well as with telomeric DNA (Fig. 1D). In addition, U2OS without Flag-CTC1 expression displayed similar STN1 punctate nuclear staining that colocalized with PML (Fig. 1C, the “vector” sample) and telomeric DNA (Fig. 1D, the “vector” sample), arguing against the possibility that the localization of Flag-CTC1 and STN1 in APBs is due to CTC1 overexpression.
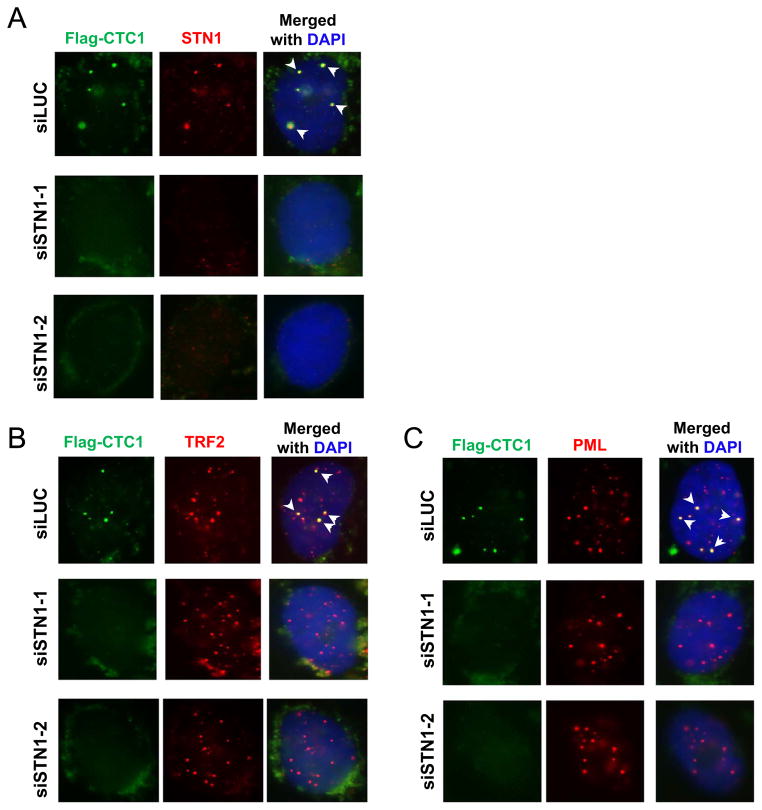
Since the three components of CST form a trimeric complex [10], we next tested whether the complex formation was required for CTC1 and STN1 foci formation in ALT cells. We depleted STN1 in U2OS cells with two distinct sequences of siRNA, and then carried out co-immunostaining. As shown in Fig. 2, both siRNA sequences efficiently reduced STN1 expression, and STN1 depletion did not impact TRF2 or PML staining. Noticeably, STN1 depletion reduced punctate foci formation of Flag-CTC1 (Fig. 2), suggesting that STN1 or perhaps the complex formation of CST is needed for CTC1 foci formation. Consequently, CTC1 colocalization with PML or TRF2 was completely abolished (Fig. 2B, C). Taken together, our results suggest that CTC1 and STN1 are components of APBs in ALT cells, implying a role of human CST in ALT cells.
Fig. 2.
Knock down of STN1 in U2OS abolished CTC1 punctate nuclear staining pattern. U2OS cells expressing Flag-CTC1 were treated with siLUC or siSTN1 for 72 h before IF staining for (A) Flag-CTC1 and STN1, (B) TRF2 and Flag-CTC1, or (C) PML and Flag-CTC1 as indicated. Arrow heads indicate colocalizations.
3.2. CST localized at telomeres in ALT cells throughout the cell cycle
In non-ALT telomerase-positive cells, CST association with telomeres fluctuates in the cell cycle [17]. We therefore determined whether this was the case in ALT cells. U2OS cells stably expressing Flag-CTC1 were synchronized at G1/S border with double-thymidine block, and then released into S and G2 phases (Suppl Fig. S2A). Cells were fixed at 0 h (G1), 3 h (early S), 6 h (mid S), and 9 h (G2) after release and IF-FISH was performed. In G1 phase, the majority of telomeric DNA was in APBs that were co-stained for PML and telomeres, and CTC1 predominantly colocalized with telomeres in APBs (Suppl Fig. S2B, 0 h). During the S phase, nearly all CTC1 colocalized with telomeric DNA, with a substantial portion of CTC1 colocalizing with PML-free telomeres, while the remaining CTC1 colocalizing with APBs (Suppl Fig. S2B, 3 h and 6 h). We also noticed that more telomeric DNA became dissociated from PML in early and mid S phase. Such increase in PML-free telomeres is consistent with the dynamics of PML nuclear bodies during the cell cycle, which is unstable in early S phase due to PML fission and then is reconstructed in late S/G2 [39]. In G2 phase, the majority of CTC1 regained colocalization with APBs (Suppl Fig. S2B, 9 h). Thus, CTC1 colocalizes with telomeres in all stages of the cell cycle.
3.3. Depletion of STN1 or CTC1 reduced the abundance of CCs and TCs
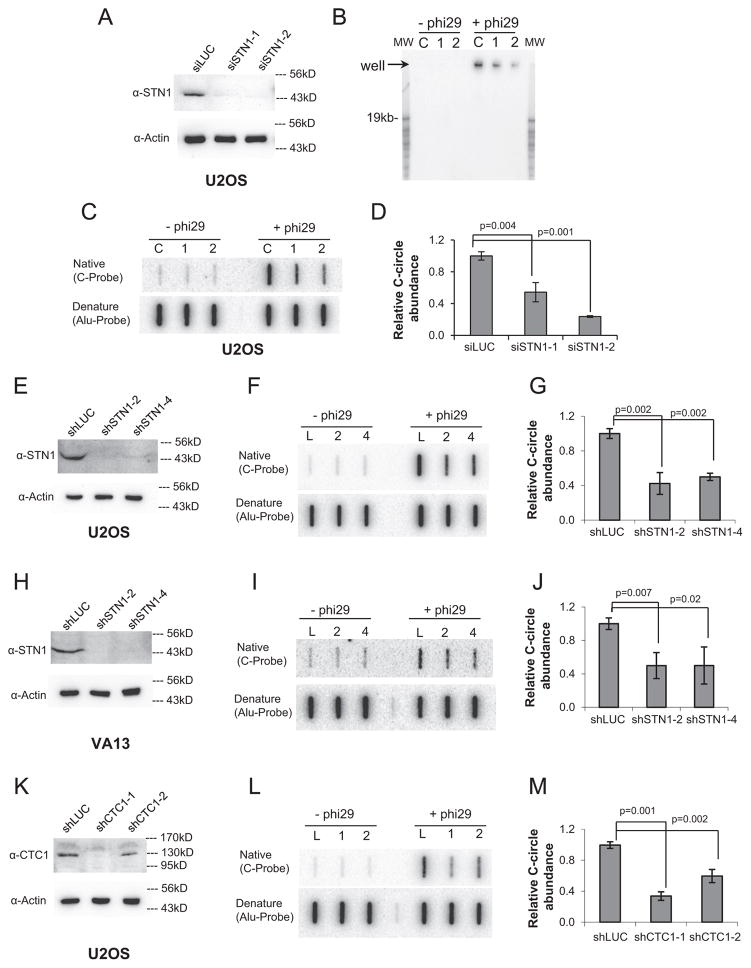
Our observation that CTC1 and STN1 localized in APBs prompted us to study the potential role of CST in the ALT pathway. Because CC is a specific and quantifiable marker of ALT activity [24], we set out to determine whether CST deficiency affected CC abundance using the previously described quantitative C-circle assay (CC assay) [24]. In CC assay, the partially single-stranded CCs act as self-priming templates for Φ29 polymerase to amplify via rolling circle amplification, producing single-stranded G-rich products of > 70 kb that can be hybridized with a radioactively labeled telomeric C-rich probe under native conditions. The long amplified products can be detected either by agarose gel electrophoresis or by slot blot, allowing for rapid and quantitative detection of CCs [24]. We found that both transient and stable depletion of STN1 in U2OS cells significantly reduced CC abundance (Fig. 3A–G). Depletion of STN1 in another ALT cell line VA13 showed similar CC reduction (Fig. 3H–J), indicating that this effect was not cell line specific. Likewise, stable depletion of CTC1 significantly diminished the amount of CCs (Fig. 3K–M).
Fig. 3.
Suppression of CST reduced C-circle abundance. (A) Immunoblot showing knockdown of STN1 in U2OS by transient depletion of STN1 with siRNA for 72 h. (B) Non-denaturing agarose gel electrophoresis to detect of CC assay products. Genomic DNA was isolated from U2OS cells treated with siSTN1 and 40 ng genomic DNA was then subjected to CC amplification with Φ29 polymerase. The amplification products were separated on agarose gel electrophoresis, and G-strand products were detected with the 32P-(CCCTAA)3 probe under native conditions. Amplification of C-circles produced large DNA molecules that migrated minimally from the wells. Signals at the wells represent the presence of C-circles. L: siLUC, 1: siSTN1-1, 2: siSTN1-2. (C) Representative slot blot of CC assay products after transient knockdown of STN1 with siRNA for 72 h. (D) Quantification of CC abundance in U2OS expressing STN1 siRNA. (E) Immunoblot showing stable knockdown of STN1 with shRNA in U2OS cells. (F) Representative slot blot of CC assay in U2OS. L: shLUC, 2: shSTN1-2, 4: shSTN1-4. (G) Quantification of CC abundance after stable STN1 depletion. (H) Immunoblot showing knockdown of STN1 in VA13 cells. (I) Representative slot blot of CC assay of VA13 depleted of STN1. (J) Quantification of CC abundance in VA13 expressing STN1 shRNA. (K) Immunoblot showing knockdown of CTC1 in U2OS cells. (L) Representative slot blot of CC assay with genomic DNA isolated from U2OS with stable expression of shLUC or shCTC1. L: shLUC, 1: shCTC1-1, 2: shCTC1-2. (M) Quantification of CC abundance in U2OS expressing CTC1 shRNA. Results in each quantification were from at least three independent experiments. Two-tailed t-tests were used to calculate p values. Error bars: s.d.
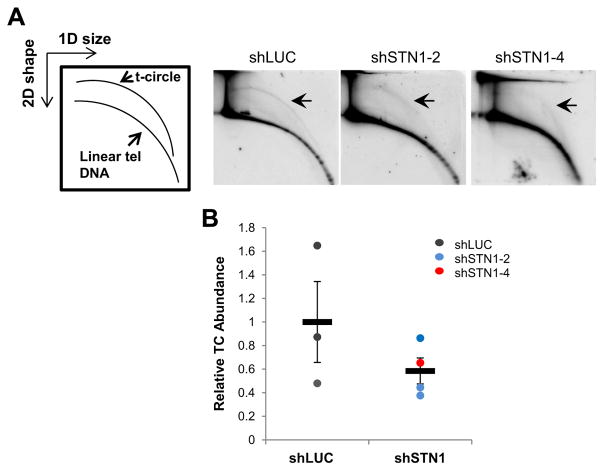
Like CCs, TCs are also abundant in ALT cells. Although the origin of CCs is unknown, it has been proposed that CCs may result from nucleolytic degradation of the G-rich strand of TCs [40]. Thus, we then determined whether CST deficiency altered TC abundance. Using 2D gel analysis, we found that the amount of total TCs was decreased upon STN1 depletion (Fig. 4). Collectively, these results suggest that the CST complex likely regulates CC and TC homeostasis in ALT cells.
Fig. 4.
Suppression of STN1 reduced the abundance of t-circles. (A) Genomic DNA isolated from U2OS stably expressing shLUC or shSTN1 was separated with 2D gel electrophoresis analysis (first dimension: molecular weight; second dimension: DNA conformation). Arrows indicate extrachromosomal t-circles. (B) Relative abundance of t-circles in control and shSTN1 cells. Error bars: SEM.
3.4. Deficiency in STN1 increased telomere damage, telomere fragility, and frequency of T-SCE, but had no effect on APB formation or global telomere length
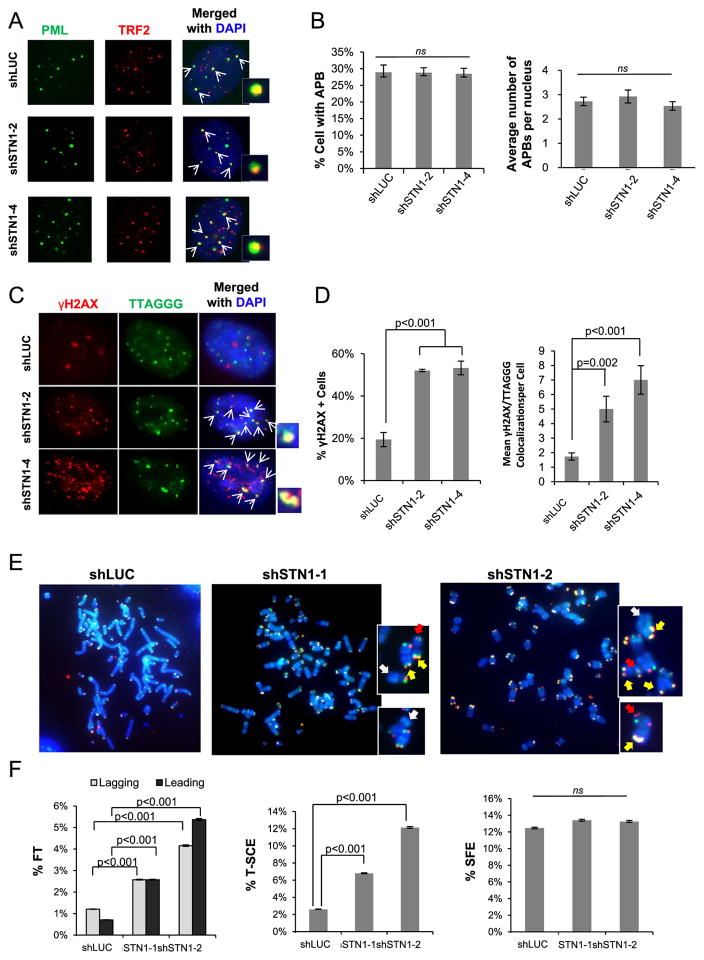
Next, we tested whether CST deficiency affected other aspects of telomere maintenance, including APB formation, frequency of T-SCE, and telomere length. Lines of evidence suggest that APBs has a functional role in promoting replication- and/or HDR-mediated extension of ALT telomeres, probably via enriching repair/replication proteins, enhancing the activities of these proteins, and providing sites for telomere clustering [26,28]. Because CTC1 and STN1 form punctate nuclear staining complex colocalizing with APBs (Fig. 1), we then determined whether CST played a role in APB formation. We stably depleted STN1 in U2OS with shRNA and cells were stained with antibodies against PML and TRF2 to reveal APBs. Neither the percentage of APB-positive cells (with > 3 APBs per nucleus) nor the average number of APBs per nucleus was perturbed by STN1 knockdown (Fig. 5A and B), suggesting that CST is not needed for APB formation.
Fig. 5.
Effect of CST deficiency on APB formation and telomere stability. (A) Representative images for APB staining in U2OS after STN1 depletion. Cells were stained with PML antibody (green) and TRF2 antibody (red). APBs (yellow) are labeled with arrows. (B) Percentage of cells with APBs and the average numbers of APBs per nucleus after stable expression of STN1 shRNA in U2OS. (C) IF-FISH of γ-H2AX (red) colocalization with telomeres (green) in U2OS after STN1 suppression. Colocalizations (yellow) are indicated by white arrows and shown in amplified images. (D) Quantification of dysfunctional telomeres. (E) Representative images of CO-FISH in U2OS. Chromosomes are shown in amplified images. Arrows indicate T-SCE (yellow), fragile telomeres (red), and signal free ends (white). (F) Quantification of T-SCE, signal-free ends (SFEs), and fragile telomeres (FTs) in U2OS after STN1 suppression. Frequency of each event per chromosome end was plotted. Three independent experiments were performed. Approximately 2000 chromosomes were scored in each sample in each independent experiment. All means were evaluated using a one-way ANOVA with a post hoc Tukey test. Proportions were evaluated with binomial z-statistic pairwise comparison with a Holm-Bonferroni correction to control for familywise error over multiple comparison. Error bars: SEM.
Using IF-FISH, we also found that STN1 knockdown induced telomere damage, as evidenced by increased colocalization of γ-H2AX with telomere signals (Fig. 5C, D). This suggests that CST is important for maintaining functional telomeres in ALT cells. We then performed the chromosome-oriented FISH (CO-FISH) analysis to examine telomere dysfunction induced by CST deficiency. STN1 suppression increased telomere fragility (Fig. 5E, F), consistent with previous report that CST is important for efficient replication of telomeric DNA [14]. In addition, STN1 deficiency increased the frequency of T-SCE (Fig. 5F), suggesting that CST might be important for preventing excessive telomere recombination. No change in signal free ends was observed in STN1 knockdown cells (Fig. 5F).
We then performed TRF analysis with pulse-field electrophoresis to detect telomere length change in STN1 depleted U2OS cells. After stable expression of STN1 shRNAs, U2OS cells were cultured for about 7 weeks. STN1 suppression had no noticeable impact on global telomere length as revealed by TRF analysis (Suppl Fig. S3A). Western blotting showed that STN1 silencing was effectively maintained during the long-term cell culture (Suppl Fig. S3B), suggesting that the absence of telomere length change was not due to failure of knockdown. Although CST deficiency-induced telomere fragility was expected to lead to telomere breakage and contribute to sudden loss of telomeres [13,41], it is possible that the elevated telomere recombination events induced by CST deficiency (Fig. 5F) might counteract telomere loss in ALT cells.
3.5. Acute STN1 suppression limited cell proliferation and induced multinucleation in U2OS cells
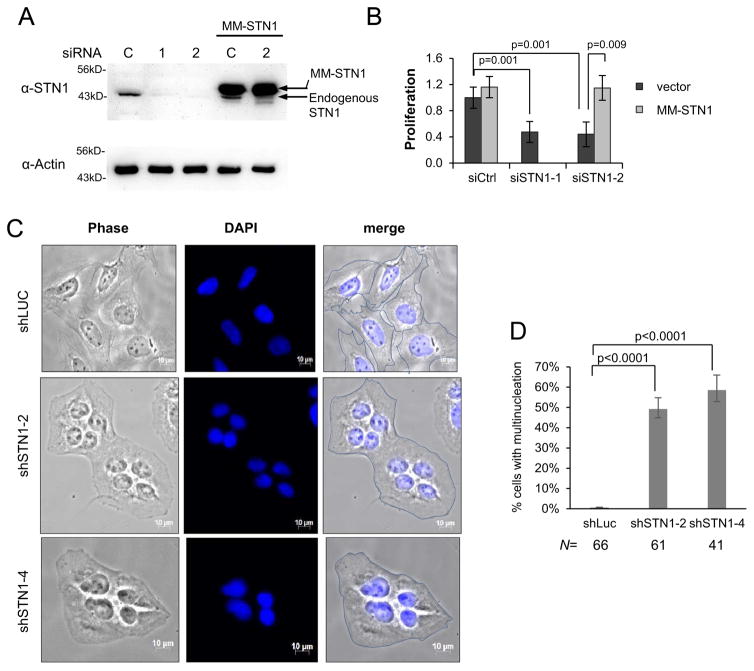
Next, we determined the effect of CST deficiency on cell proliferation. STN1 was depleted in U2OS with siRNA (Fig. 6A), and cells at 72 h post siRNA treatment were subjected to the MTS assay to measure cell viability. As shown in Fig. 6B, acute STN1 knockdown in U2OS cells limited cell proliferation compared to control knockdown. SiRNA treatment of U2OS in presence of RNAi-resistant STN1 (MM-STN1) has no obvious effect on cell proliferation (Fig. 6B), suggesting that the knock-down was specific. This is consistent with the previous report that U2OS shSTN1 clones showed slower proliferation rate [14]. Simultaneously, we observed a sharp increase of enlarged multinucleated cells, with about 50% of cells displaying multinucleation two days after STN1 depletion (Fig. 6C, D, and Suppl Fig. S4). Although the cause of multinucleation in STN1 deficient U2OS is unknown, lines of evidence support that telomere dysfunction promotes endoreplication. Davoli et al. show that persistent telomere damage bypasses mitosis and induces endoreplication, leading to polyploidy [42]. Endoreplication can also be contributed by telomere replication defects, which induces abortive cytokinesis due to failed chromosome segregation or the persistence of anaphase bridges [43,44]. Given that CST deficiency induces telomere replication defect, we consider that multinucleation might be induced by perturbation of telomere replication.
Fig. 6.
STN1 deficiency limited proliferation and resulted in multinucleation in U2OS cells. (A) Immunoblot showing STN1 knockdown with siRNA. (B) MTS assay of cell proliferation. U2OS expressing RNAi-resistant STN1 (MM-STN1) or vector alone was treated with siRNA for 72hr before MTS staining. Results were from 6 independent experiments. Error bars: s.d. All values were normalized to control siRNA treated cells. The final value for U2OS with vector only and treated with control siRNA was set to 1. (C) Representative phase images of multinucleated cells in U2OS after STN1 knockdown. Cells were fixed on slides two days after puromycin selection, counterstained with DAPI, and images were taken with phase contrast. The edge of each cell was outlined in merged images. Enlarged multinucleated images are shown in Supplemental Fig. S4. (D) Quantification of % multinucleated cells. U2OS cells were counted as multinucleated if they contained > 1 nucleus per cytoplasm. Proportion of multinucleated cells was evaluated using a binomial z-statistic pairwise comparison, with a Holm-Bonferroni correction to control for familywise error over multiple comparisons. N indicates the number of cells analyzed in each sample.
4. Discussion
Recent studies on CST mainly focus on its role in telomere maintenance in non-ALT cells. In telomerase-positive cancer cells and normal fibroblasts, the primary function of CST is to promote efficient replication of telomeric DNA [13,14] and facilitate replication restart at GC-rich repetitive sequences [36]. CST deficiency results in defective telomere replication, leading to telomere breakage and contributing to sudden loss of telomeres [13,14]. This study and previous report [14] both show that CST deficiency affects telomere replication in ALT cells. Thus, it is reasonable to conclude that CST is important for efficient and faithful replication of telomeric DNA regardless of cell types. Moreover, this study reveals additional telomeric functions of CST that are specific to ALT cells. First, our results demonstrate that CTC1 and STN1 form a punctate nuclear staining complex that colocalizes in APBs, while such punctate staining is completely absent in non-ALT cells. We also demonstrate that CST localizes at ALT telomeres throughout the cell cycle. Furthermore, CST deficiency elevates T-SCE and induces telomere dysfunction. Additionally, functional CST is needed for the production of CCs and TCs. Acute knocking down of STN1 limits cell proliferation and induces multinucleation in ALT cells. We propose that CST is critical for ALT-mediated telomere maintenance and viability of ALT cells, and targeting CST may provide a novel approach to inhibit the growth of ALT cancer cells.
Elevated T-SCE and ECTRs are generally considered as two major hallmarks of ALT. Interestingly, CST suppression reduces ECTRs but increases T-SCE (Figs. 3–5F). How can these seemingly conflictive observations be reconciled? To date, little is known about the mechanism for ECTRs formation. While it has been proposed that ECTRs are intermediates resulting from t-loop resolution [45] and CCs may result from nucleolytic degradation of the G-rich strands of TCs [40], recent evidence highlights the importance of replication in ECTR formation. Blocking replication fork progression with aphidicolin or hydroxyurea significantly attenuates TC and CC production in RTEL1- or ASF1-deficient cells [35,46], suggesting an essential role of replication fork passing through telomeric regions in ECTR formation. Thus, it is possible that CST deficiency reduces ECTRs through perturbation of telomeric DNA replication. Meanwhile, replication stress has been shown to induce sister-chromatid bridging at fragile sites in mitosis [47]. Given that telomeres are fragile sites, increased telomere replication stress by CST deficiency may promote sister telomere bridging that can facilitate T-SCE. Alternatively, telomere replication defect may lead to sudden loss of telomeres, generating short telomeres that tend to be more recombinogenic [48]. We suggest that the underlying reason for both T-SCE increase and ECTR reduction in CST-deficient cells could all be due to defective telomere replication.
It should be noted that those commonly described ALT “markers”, including T-SCE, ECTRs, APBs, and heterogeneous telomere length, are not always linked to each other. For example, depletion of MUS81 reduces T-SCE but has no impact on ECTR signals or telomere length [49]. Another study shows that while XRCC3 and NBS1 are required for ECTR production, knocking down these two genes does not change telomere length in ALT cells nor affect cell proliferation [50]. We suggest that until the relationship between ALT-associated phenotypic characteristics is fully understood, caution should be exercised when using these markers to interpret ALT activities. Meanwhile, understanding how various genes regulate specific types of ALT characteristics, as well as further dissecting their functions will be critical to identifying targets and devising potential drugs for ALT-positive tumors.
Supplementary Material
Acknowledgments
We thank Dr. S. Chang for providing the 2D gel protocol. This work was in part supported by NIH R01GM112864 and R56AG046292 to WC.
Abbreviations
- ALT
alternative lengthening of telomeres
- CST
the CTC1/STN1/TEN1 trimeric complex
- APBs
ALT-associated promyelocytic leukemia bodies
- PML
promyelocytic leukemia
- ECTR
extrachromosomal circular telomere repeats
- TC
t-circle
- CC
C-circle
- HDR
homology-directed recombination
- T-SCE
telomere sister chromatid exchange
- SFE
signal free ends
- FT
fragile telomere
- CO-FISH
chromosome-oriented FISH
- FISH
fluorescent in situ hybridization
- PFGE
pulse-field gel electrophoresis
- RCA
rolling circle amplification
- TRF
telomere restriction fragment analysis
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.yexcr.2017.03.058.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Author Contributions
CH, PJ, OS, MC, WC performed experiments and analyzed data. WC directed the study. CH and WC wrote the manuscript.
References
- 1.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 2.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu Rev Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 3.O’Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 5.Holt SE, Wright WE, Shay JW. Regulation of telomerase activity in immortal cell lines. Mol Cell Biol. 1996;16:2932–2939. doi: 10.1128/mcb.16.6.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- 8.Dilley RL, Verma P, Cho NW, Winters HD, Wondisford AR, Greenberg RA. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature. 2016;539:54–58. doi: 10.1038/nature20099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lange T. How Shelterin Solves the Telomere End-Protection Problem. Cold Spring Harb Symp Quant Biol. 2011 doi: 10.1101/sqb.2010.75.017. [DOI] [PubMed] [Google Scholar]
- 10.Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell. 2009;36:193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell. 2009;36:207–218. doi: 10.1016/j.molcel.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu P, Min JN, Wang Y, Huang C, Peng T, Chai W, Chang S. CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J. 2012;31:2309–2321. doi: 10.1038/emboj.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang C, Dai X, Chai W. Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res. 2012;22:1681–1695. doi: 10.1038/cr.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart JA, Wang F, Chaiken MF, Kasbek C, Chastain PD, 2nd, Wright WE, Price CM. Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J. 2012;31:3537–3549. doi: 10.1038/emboj.2012.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F, Stewart JA, Kasbek C, Zhao Y, Wright WE, Price CM. Human CST has independent functions during telomere duplex replication and C-strand fill-in. Cell Rep. 2012;2:1096–1103. doi: 10.1016/j.celrep.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan M, Qin J, Songyang Z, Liu D. OB-fold containing protein 1 (OBFC1), a human homologue of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J Biol Chem. 2009;284:26725–26731. doi: 10.1074/jbc.M109.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen LY, Redon S, Lingner J. The human CST complex is a terminator of telomerase activity. Nature. 2012;488:540–544. doi: 10.1038/nature11269. [DOI] [PubMed] [Google Scholar]
- 18.Polvi A, Linnankivi T, Kivela T, Herva R, Keating JP, Makitie O, Pareyson D, Vainionpaa L, Lahtinen J, Hovatta I, et al. Mutations in CTC1, encoding the CTS telomere maintenance complex component 1, cause cerebroretinal microangiopathy with calcifications and cysts. Am J Hum Genet. 2012;90:540–549. doi: 10.1016/j.ajhg.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson BH, Kasher PR, Mayer J, Szynkiewicz M, Jenkinson EM, Bhaskar SS, Urquhart JE, Daly SB, Dickerson JE, O’Sullivan J, et al. Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat Genet. 2012;44:338–342. doi: 10.1038/ng.1084. [DOI] [PubMed] [Google Scholar]
- 20.Keller RB, Gagne KE, Usmani GN, Asdourian GK, Williams DA, Hofmann I, Agarwal S. CTC1 Mutations in a patient with dyskeratosis congenita. Pediatr Blood Cancer. 2012;59:311–4. doi: 10.1002/pbc.24193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walne AJ, Bhagat T, Kirwan M, Gitiaux C, Desguerre I, Leonard N, Nogales E, Vulliamy T, Dokal IS. Mutations in the telomere capping complex in bone marrow failure and related syndromes. Haematologica. 2013;98(8):334. doi: 10.3324/haematol.2012.071068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simon AJ, Lev A, Zhang Y, Weiss B, Rylova A, Eyal E, Kol N, Barel O, Cesarkas K, Soudack M, et al. Mutations in STN1 cause Coats plus syndrome and are associated with genomic and telomere defects. J Exp Med. 2016;213:1429–1440. doi: 10.1084/jem.20151618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 24.Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, Pickett HA, Reddel RR. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol. 2009;27:1181–1185. doi: 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- 25.Nabetani A, Ishikawa F. Alternative lengthening of telomeres pathway: recombination-mediated telomere maintenance mechanism in human cells. J Biochem. 2011;149:5–14. doi: 10.1093/jb/mvq119. [DOI] [PubMed] [Google Scholar]
- 26.Chung I, Osterwald S, Deeg KI, Rippe K. PML body meets telomere: the beginning of an ALTernate ending? Nucleus. 2012;3:263–275. doi: 10.4161/nucl.20326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- 28.Draskovic I, Arnoult N, Steiner V, Bacchetti S, Lomonte P, Londono-Vallejo A. Probing PML body function in ALT cells reveals spatiotemporal requirements for telomere recombination. Proc Natl Acad Sci USA. 2009;106:15726–15731. doi: 10.1073/pnas.0907689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nabetani A, Yokoyama O, Ishikawa F. Localization of hRad9, hHus1, hRad1, and hRad17 and caffeine-sensitive DNA replication at the alternative lengthening of telomeres-associated promyelocytic leukemia body. J Biol Chem. 2004;279:25849–25857. doi: 10.1074/jbc.M312652200. [DOI] [PubMed] [Google Scholar]
- 30.Wu G, Jiang X, Lee WH, Chen PL. Assembly of functional ALT-associated promyelocytic leukemia bodies requires Nijmegen Breakage Syndrome 1. Cancer Res. 2003;63:2589–2595. [PubMed] [Google Scholar]
- 31.Bailey SM, Brenneman MA, Goodwin EH. Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res. 2004;32:3743–3751. doi: 10.1093/nar/gkh691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Londono-Vallejo JA, Der-Sarkissian H, Cazes L, Bacchetti S, Reddel RR. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 2004;64:2324–2327. doi: 10.1158/0008-5472.can-03-4035. [DOI] [PubMed] [Google Scholar]
- 33.Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by freetelomeric circles and heterogeneous t-loops. Mol Cell Biol. 2004;24:9948–9957. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oganesian L, Karlseder J. Mammalian 5′ C-rich telomeric overhangs are a mark of recombination-dependent telomere maintenance. Mol Cell. 2011;42:224–236. doi: 10.1016/j.molcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Sullivan RJ, Arnoult N, Lackner DH, Oganesian L, Haggblom C, Corpet A, Almouzni G, Karlseder J. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat Struct Mol Biol. 2014;21:167–174. doi: 10.1038/nsmb.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chastain M, Zhou Q, Shiva O, Whitmore L, Jia P, Dai X, Huang C, Fadri-Moskwik M, Ye P, Chai W. Human CST facilitates genome-wide RAD51 recruitment to GC-Rich repetitive sequences in response to replication stress. Cell Rep. 2016;16:1300–1314. doi: 10.1016/j.celrep.2016.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH. Strand-specific postreplicative processing of mammalian telomeres. Science. 2001;293:2462–2465. doi: 10.1126/science.1062560. [DOI] [PubMed] [Google Scholar]
- 38.Sampathi S, Bhusari A, Shen B, Chai W. Human flap endonuclease I is in complex with telomerase and is required for telomerase-mediated telomere maintenance. J Biol Chem. 2009;284:3682–3690. doi: 10.1074/jbc.M805362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dellaire G, Ching RW, Dehghani H, Ren Y, Bazett-Jones DP. The number of PML nuclear bodies increases in early S phase by a fission mechanism. J Cell Sci. 2006;119:1026–1033. doi: 10.1242/jcs.02816. [DOI] [PubMed] [Google Scholar]
- 40.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 41.Boccardi V, Razdan N, Kaplunov J, Mundra JJ, Kimura M, Aviv A, Herbig U. Stn1 is critical for telomere maintenance and long-term viability of somatic human cells. Aging Cell. 2015;14:372–381. doi: 10.1111/acel.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141:81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Remeseiro S, Cuadrado A, Carretero M, Martinez P, Drosopoulos WC, Canamero M, Schildkraut CL, Blasco MA, Losada A. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 2012;31:2076–2089. doi: 10.1038/emboj.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pampalona J, Frias C, Genesca A, Tusell L. Progressive telomere dysfunction causes cytokinesis failure and leads to the accumulation of polyploid cells. PLoS Genet. 2012;8:e1002679. doi: 10.1371/journal.pgen.1002679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 46.Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 47.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Erdmann N, Giannone RJ, Wu J, Gomez M, Liu Y. An increase in telomere sister chromatid exchange in murine embryonic stem cells possessing critically shortened telomeres. Proc Natl Acad Sci USA. 2005;102:10256–10260. doi: 10.1073/pnas.0504635102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng S, Xiang T, Pandita TK, Gonzalez-Suarez I, Gonzalo S, Harris CC, Yang Q. Telomere recombination requires the MUS81 endonuclease. Nat Cell Biol. 2009;11:616–623. doi: 10.1038/ncb1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Compton SA, Choi JH, Cesare AJ, Ozgur S, Griffith JD. Xrcc3 and Nbs1 are required for the production of extrachromosomal telomeric circles in human alternative lengthening of telomere cells. Cancer Res. 2007;67:1513–1519. doi: 10.1158/0008-5472.CAN-06-3672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.