Abstract
Two-photon laser scanning microscopy of calcium dynamics using fluorescent indicators is a widely used imaging method for large scale recording of neural activity in vivo. Here we introduce volumetric Two-photon Imaging of Neurons using Stereoscopy (vTwINS), a volumetric calcium imaging method that employs an elongated, V-shaped point spread function to image a 3D brain volume. Single neurons project to spatially displaced “image pairs” in the resulting 2D image, and the separation distance between images is proportional to depth in the volume. To demix the fluorescence time series of individual neurons, we introduce a novel orthogonal matching pursuit algorithm that also infers source locations within the 3D volume. We illustrate vTwINS by imaging neural population activity in mouse primary visual cortex and hippocampus. Our results demonstrate that vTwINS provides an effective method for volumetric two-photon calcium imaging that increases the number of neurons recorded while maintaining a high frame-rate.
Introduction
Two-photon excitation laser scanning microscopy1 (TPM) enables high spatial resolution optical imaging in highly scattering tissue such as the mammalian brain. When combined with genetically-encoded calcium indicators2,3, or synthetic indicators that label neural populations4, intracellular calcium dynamics can be measured across a population of cells, providing a method for large scale recording of neural activity at cellular resolution4,5. In general, increasing the number of simultaneously recorded neurons is important because it increases the power of population analysis methods in studies of neural coding and dynamics. To increase the number of neurons recorded with two-photon calcium imaging, volumetric imaging methods, such as multi-plane imaging6, random access fluorescence microscopy7–9 and ultrasound lens scanning10, are under development.
In traditional TPM1, a diffraction-limited, high numerical aperture (NA) point spread function (PSF) is raster-scanned across a single plane. Volume imaging can be performed by sequentially moving the focal plane (or sample) up or down between each raster scan, repeating this pattern for each volume measurement. This method can be implemented with movable objectives, remote focusing11, or a liquid lens6. However, if the frame rate for single plane imaging is N frames/sec, and the number of planes imaged per volume is m, then the aggregate volume frame rate is reduced to N/m. Many calcium indicators12 have on-response kinetics below 0.1 s. To capture these dynamics, volume frame rates must remain close to 10 Hz. With current resonant scanner-based TPM (N ≈ 30 Hz), this implies that only a relatively low number of planes (m=3,4) can be used for multi-plane volumetric imaging.
Elongating the PSF of the focused excitation beam along the optical axis, using either a low-NA Gaussian beam focus or Bessel beam methods13, can be used with raster scanning to form a projection image of a volume14. This is useful in applications like functional imaging of dendritic spines in sample volumes with sparse neural expression of the indicator15. However, in samples with dense expression, such as those encountered in large-scale recording of a neural population in vivo, extending a single PSF axially causes neurons at different depths to be superimposed. Information about depth in the sample of individual neurons is lost, and demixing of fluorescence signals from individual neurons is compromised if their images significantly overlap.
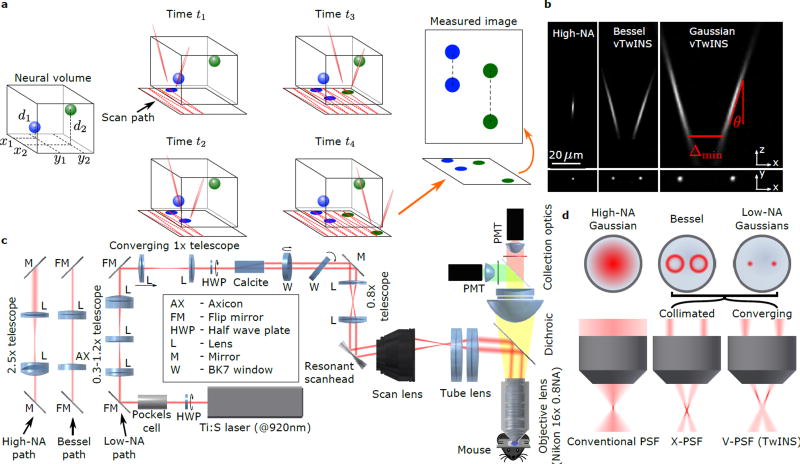
Our method addresses these limitations by splitting an elongated PSF into two excitation beams. These beams are spatially separated and angled inwards to create a stereoscopic “V”-shaped PSF configuration (Fig. 1a). Raster scanning with this PSF produces a 2D projection image that preserves information about neural activity at different depths. We refer to this method as volumetric Two-photon Imaging of Neurons using Stereoscopy (vTwINS). The intuition behind vTwINS is straightforward: the soma of any neuron in the 3D volume makes two contributions to the 2D projection image, one soma-shaped image for each arm of the V-shaped PSF. The spatial offset between these two images is equal to the distance between the two arms of the V at the neuron’s depth in the volume. This results in short distances between deep neurons, and longer distances for shallower neurons (Fig. 1a).
Figure 1.
vTwINS concept and design. (a) vTwINS uses a “V”-shaped PSF to image neural volumes. During scanning, the two PSF arms intersect neurons at different depths (e.g. the blue and green stylized neurons) at different time intervals. Deep neurons intersect the second arm shortly after the first. Shallow neurons take longer for the second arm to intersect. Each neuron thus appears twice, where the distance between images indicates depth. (b) Example PSFs for diffraction-limited (high-NA) TPM, and vTwINS microscopes using Bessel and low-NA Gaussian beams. (c) The vTwINS microscope consists of a beam-shaping module and a conventional two-photon microscope. The three optical paths generate the PSFs shown in (b). In the Bessel and Gaussian (low-NA) vTwINS paths, lenses adjust the PSF’s axial extent, and a birefringent block (calcite) splits the beam in two and sets the PSF angle. (d) The back aperture illumination profiles for the three paths in (c). In the high-NA (conventional TPM) path, the overfilled back aperture is focused to a point. In the Bessel and low-NA Gaussian paths, two beams are focused to form each arm of the PSF. The beam divergence is adjusted with the 1× telescope before the calcite block to separate the two arms of the X-PSF and form the V-PSF.
Although vTwINS ensures that all neurons have distinct “paired” spatial profiles in the projection image, identifying which pair of image regions reflect a single neuron’s activity is ill-posed from single images. Recent methods solve this problem by leveraging the temporal statistics of neural activity across frames (e.g. PCA/ICA16 and constrained non-negative matrix factorization; CNMF17,18). We describe a novel inference algorithm based on orthogonal matching pursuit that exploits both the expected shapes of neural spatial profiles (pairs of rings or disks displaced along the axis of the V-shaped PSF) and the sparseness of neural activity. The image pair separation Δ in a neuron’s identified spatial profile also determines its axial position via the relationship d = 0.5(Δ − Δmin)/tan(θ), where Δmin is the minimum inter-beam distance of the PSF and θ is the beam angle from the axial direction (Fig 1a,b). Thus, the demixing algorithm both provides the neuron’s fluorescence time course and location in the volume.
In the following, we describe the optics developed to produce the vTwINS PSF and demonstrate images and image time series produced using this method. We then present the algorithm that was developed for identifying active neurons in these time series and demixing fluorescence transients. Finally, using the combined imaging system and algorithm, we demonstrate large-scale recording of GCaMP-expressing neurons in visual cortex and hippocampus of awake mice
Results
vTwINS Optics
In a vTwINS microscope, the diffraction limited PSF (Fig. 1b) of traditional TPM is replaced with an elongated V-shaped PSF produced from two intersecting Gaussian beams (Fig. 1b), or Bessel beams (Fig. 1b). The strategy we used to create the V-shaped PSF was dual-beam excitation through a single objective lens (Fig. 1c,d). To produce a V-shaped PSF, the back-aperture is illuminated with a pair of small Gaussian beams or rings (Bessel beams19). Adjusting the separation distance, beam convergence, and beam parameters changes the angle, offset, and extension of the PSF, respectively (Online Methods, Supplementary Fig 1a., Supplementary Note 1).
As an initial proof of principle that vTwINS could spatially localize objects in a 3D volume, we imaged fluorescent beads embedded in agarose (Supplementary Fig. 2, Supplementary Note 2). Using the beads, we localized the center position of the beads to 2.7±1.6 µm (N=31) against a reference image stack, more than accurate enough for imaging neuronal cell bodies. While beads were localized over a large axial range, we found that in vivo (Online Methods) the effective axial illumination length of each elongated beam is approximately 1/e full-width of the maximum intensity.
vTwINS Calcium Imaging
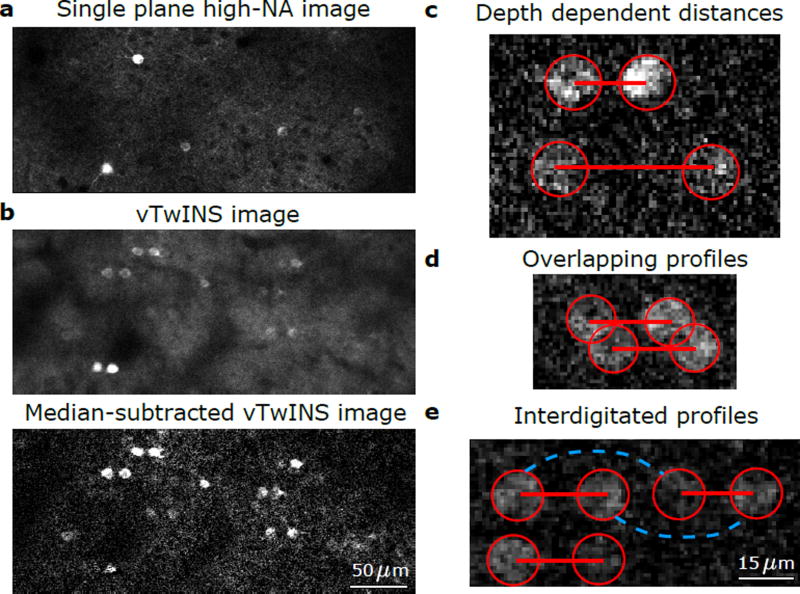
In diffraction-limited TPM (Fig. 2a), a single soma-shaped spatial profile of high fluorescence intensity is observed when calcium transients are produced in an active neuron and the cell soma of GCaMP-expressing quiescent cells can typically be resolved20. A vTwINS image is qualitatively different. Active neurons are represented as two bright soma shaped regions (disk or ring; Fig. 2b) and the images of quiescent neurons are typically not resolved because the projection produces an increased, and more uniform, background intensity. The geometry of the vTwINS PSF greatly reduces the signal of active neuropil and spreads it out over large regions, resulting in a broad, time-varying addition to the background intensity. When multiple cells are simultaneously active, many soma pairs become visible. Pairs from different cells have different spatial separations (Fig. 2c), representing different depths of the cell somas in the volume.
Figure 2.
Example vTwINS images. All images are averages of 5 consecutive frames taken at 30 Hz. (a) Diffraction-limited TPM single plane image of GCaMP6f in mouse visual cortex. (b) vTwINS scan of the same V1 area as (a) demonstrates paired somas of active neurons and reduced SNR as the background levels are much higher. Subtracting the temporal median at each pixel highlights neural activity. (c) Two fluorescing neurons imaged by vTwINS at different depths have different distances between the image pairs. Red circles indicate the different images and red lines connect corresponding image pairs. (d) vTwINS images typically have overlapping spatial profiles. (e) Neurons aligned in the direction parallel to the plane of the V-shaped PSF (which is the same as the fast scan direction in our implementation) can create ambiguity in the spatial profile image pair assignment. Both the solid red lines (the true pairing) and the dashed green lines indicate realizable distance pairings corresponding to different neuron positions, and temporal activity must be used to resolve this ambiguity.
The properties of vTwINS based calcium imaging data (Fig. 2) introduce a number of unique challenges in demixing spatial profiles of neural activity to extract the fluorescence time traces of individual cells. First, there is a lower SNR per cell due to the axially extended PSFs (Fig. 2b). Second, the spatial profiles of cells under vTwINS can partially overlap (Fig. 2d), and typically consist of disjoint regions. While the geometry of vTwINS reduces maximal overlap between profiles (Online Methods, Supplementary Note 3, Supplementary Fig. 3), the disjoint nature of the profiles violates the spatial locality assumption in current demixing methods16, 18 (Online Methods, Supplementary Fig. 4b). Third, neurons co-aligned in the fast-scan direction can create ambiguous, interdigitated spatial profile pairs (Fig. 2e). Finally, intensity differences between the two images in a pair may result from the non-uniform scattering between the two beam paths (e.g. due to varying tissue properties).
vTwINS Profile Identification and Demixing
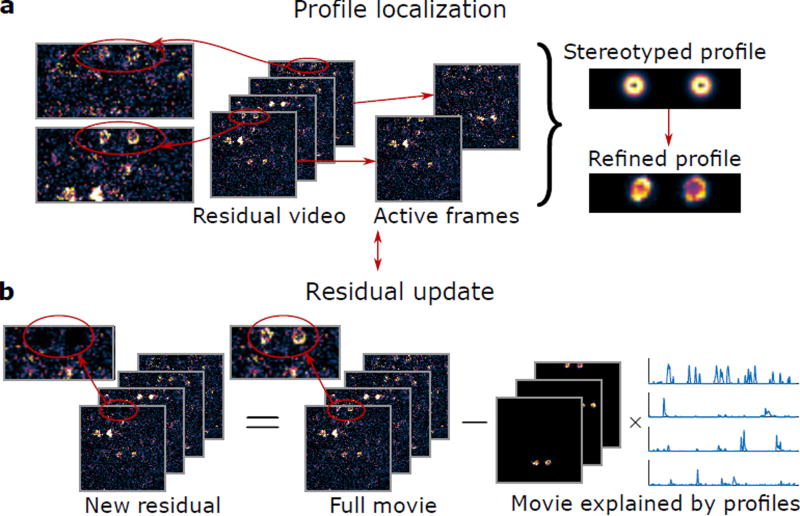
We addressed the challenges of analyzing vTwINS data with Sparse Convolutional Iterative Shape Matching (SCISM), a novel demixing method that explicitly seeks horizontally separated image pairs (Online Methods, Fig. 3, Supplementary Fig. 5). As a pre-processing step, we motion-corrected, temporally averaged, and spatially binned the raw image time series (Online Methods). At each iteration, candidate spatial profiles, consisting of stereotyped profiles (pairs of annuli separated in the fast-scan direction with different separation distances), are compared to the measured fluorescence frames across the field-of-view (FOV) (Fig. 3a, Supplementary Fig. 5a,b). Estimating pairs of images simultaneously increases the available signal to distinguish the neural profile from the noise fluctuations (Supplementary Fig. 6, Supplementary Note 4). The stereotyped profile most correlated with the data is then selected (Supplementary Fig. 5c), and the most highly correlated frames are used to refine the profile shape to better match the data (Supplementary Fig. 5d). This step allows SCISM to handle spatial profile pairs where one beam path has lower intensity. The new profile is added to the set of active spatial profiles, and the corresponding time-traces are estimated using non-negative LASSO21 (Supplementary Fig. 5e). Finally, the data residual is calculated by subtracting off the component of the data captured by the current set of active spatial profiles (Fig. 3b, Supplementary Fig. 5f); this residual is used in the step to determine the next spatial profile.
Figure 3.
Sparse convolutional iterative shape matching (SCISM) for demixing vTwINS data. (a) Example stereotyped neuron image pairs with different distances are matched across frames to determine the most likely pair. The new profile is refined by locally masking and averaging frames closely aligned with the stereotyped spatial profile. (b) The new profile is added to the set of spatial profiles, and the time-traces for all spatial profiles are calculated via non-negative LASSO. The residual movie is re-computed by subtracting the contribution of the current set of spatial profiles (the sum of outer products of the spatial profiles and their time traces). The algorithm then finds the next spatial profile by restarting and operating on the new residual.
This procedure iteratively selects spatial profiles greedily in order of correlation strength with the data, using both spatial and temporal statistics to determine the most likely spatial profile at each iteration. Specifically, SCISM leverages sparsity in neural activity as well as the spatial constraint that each spatial profile consists of two areas separated in the fast-scanning direction. Sparse neural activity is particularly important as it permits minimal cross-contamination due to spatially overlapping neurons. Once spatial profiles are determined with SCISM, full resolution time trace estimates are obtained using non-temporally averaged data via non-negative LASSO.
Large Scale Recording in Mouse Visual Cortex
Head-restrained GCaMP6f-expressing transgenic mice, running on a spherical treadmill, were presented with a visual stimulus sequence consisting of randomly placed Gabor patches (Online Methods). vTwINS imaging was performed in layer 2/3 of primary visual cortex (V1). Images were acquired in a 550 µm × 550 µm area with a 45 µm-long inverted-V PSF (FWHM, 60 µm 1/e full-width) at 30 Hz frame rate over a 14 minute imaging session.
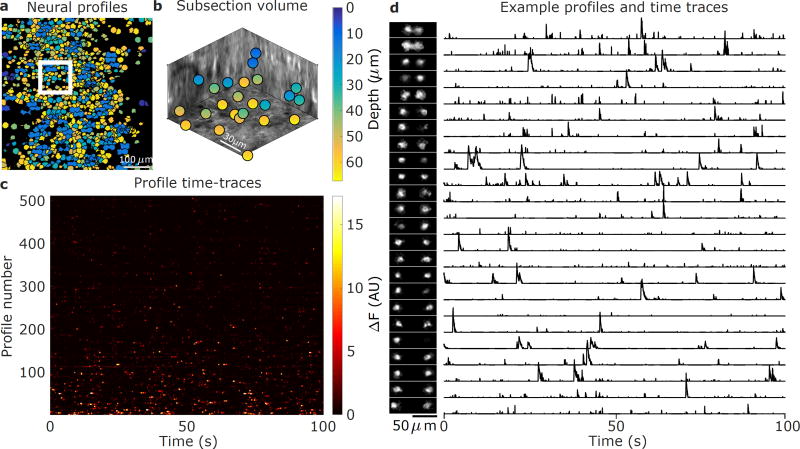
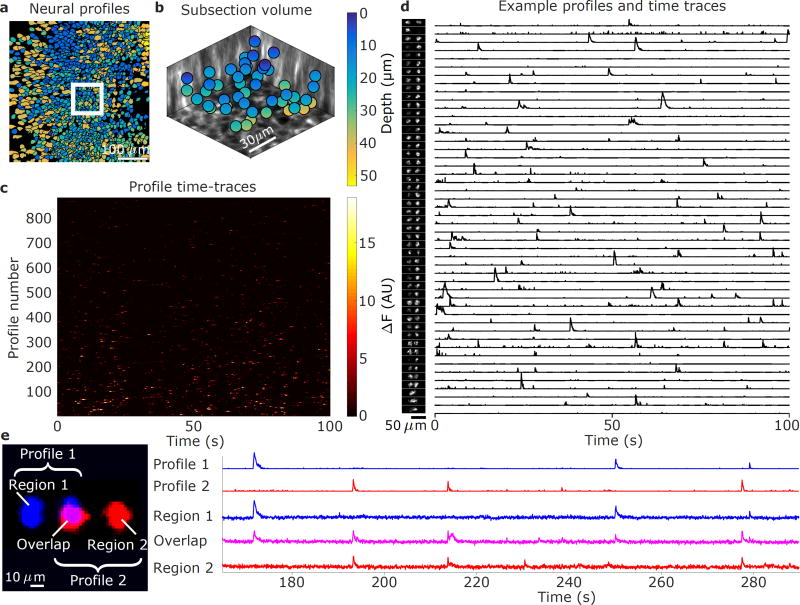
The time series fluorescence data was preprocessed with rigid motion-correction and spatio-temporal averaging (Online Methods, Supplementary Fig. 7a, Supplementary Video 1,2). The 511 spatial profiles obtained via SCISM (Fig. 4) show significant overlap, as expected from the relatively high density of GCaMP-expressing cells and the vTwINS PSF. Given the spatial profiles, we used the vTwINS PSF to extract the 3D cell positions (Online Methods, Fig 4a,b). The demixed spatial profile activity traces (Fig. 4c, Supplementary Fig. 8) have the expected temporal statistics of sparsely firing neurons. Because SCISM is an iterative method that extracts highly active spatial profiles first, the time traces are ordered by how correlated the profiles are with the data.
Figure 4.
Demixed spatial profiles and calcium activity in mouse visual cortex. (a) Full set of spatial profiles, color-coded by depth, show significant overlap. (b) 3D locations of the spatial profiles located in the white box in (a) show that spatial profiles are found at different depths. (c) Time-traces of spatial profiles show sporadic activity in the 0–100s time interval. (d) Example subset of spatial profiles (chosen from the white inset box in (a) and sorted by depth) and corresponding normalized time traces show rich activity patterns. The increasing separation distance as a function of depth reflects the inverted V shape of the PSF used in this recording.
The spatial profile volumetric locations (Fig. 4b) indicates that vTwINS records activity across the entire axial extent. The range of axial depths captured by vTwINS is further illustrated by plotting the spatial profiles in a 107 µm × 107 µm subsection of the FOV (Fig. 4d), sorted by inferred depth, (Fig. 4d) and their corresponding position in a 3D anatomical volume (Online Methods, Supplementary Fig. 9). The corresponding spatial profile activity traces (Fig. 4d) also show that cell transients are well isolated, despite the highly overlapping spatial profiles.
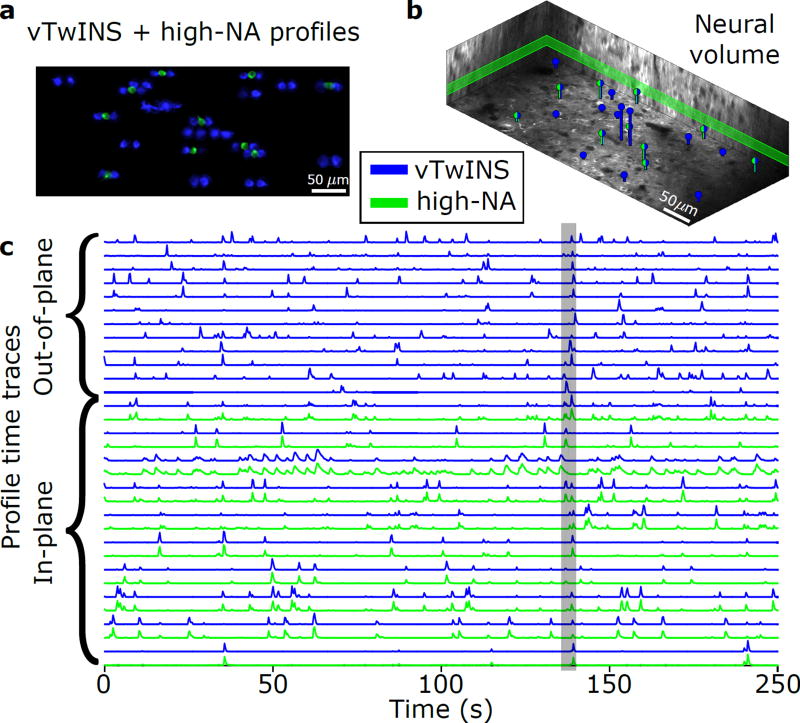
To validate that neural activity recorded with vTwINS is comparable to standard methods, we simultaneously imaged an entire neural volume with vTwINS, and a single slice of the volume with diffraction-limited TPM. Both datasets were collected at 30 Hz over a 470 µm × 200 µm overlapping area (Supplementary Fig. 1b, 6c, Supplementary Video 3,4). vTwINS data was demixed using SCISM while we extracted spatial profiles and activity traces from the single-plane data using a modified CNMF algorithm22 as an independent comparison (Online Methods).
Comparison of spatial profiles from the simultaneous recordings (Fig. 5) indicates that vTwINS captures both neural activity in the single slice TPM and activity at other depths. Overall, in a ten minute imaging session, 454 spatial profiles were found in the volume using vTwINS, as compared to 169 spatial profiles found in the single plane diffraction-limited data. Activity traces corresponding to the found spatial profiles of cells identified in both the single plane and the volume, restricted to cells active in a 5 s window (Fig. 5a,b), show very high correlation between the two imaging modalities. Of the single-slice spatial profiles, 116 spatial profiles had >1 transient per minute. Of these, 98 (84%) had a matching spatial profile in the vTwINS data (Supplementary Fig. 10). Of the remaining single-slice spatial profiles, many had very low SNR, suggesting that that activity fell below the vTwINS’ lower SNR level. These correlations indicate that vTwINS still captures most of the activity at any given depth while also capturing additional activity elsewhere in the volume.
Figure 5.
Simultaneous imaging of visual cortex using conventional two-photon (green) and vTwINS (blue). (a) Max-projection of 5 s of activity for vTwINS (blue) and single-plane imaging (green) demonstrates that vTwINS records neural activity at multiple depths. (b) Volumetric depiction of vTwINS extracted spatial profile locations and depth. Green/blue profiles indicate location of cells that were matched in single-plane activity. The single-plane slice is outlined in green. (c) Time traces corresponding to cells in (a) show that for vTwINS with diffraction-limited TPM counterparts, the temporal activity traces match. The gray bar indicates the 5 s period of activity used to isolate cells.
Large Scale Recording in Mouse Hippocampus
As a more challenging application of vTwINS, we recorded and demixed activity from the CA1 region of mouse hippocampus. In this region, neuronal cell soma are densely packed in a well-defined layer, producing high spatial overlap in vTwINS data. To induce activity in CA1, water-restricted mice were trained to run down a linear track in a virtual reality system23 for water rewards (Online Methods). Images were collected over a 14 minute session in a 470 µm × 470 µm area with a 35 µm long vTwINS PSF (FWHM, 45 µm 1/e full-width, non-inverted V) at 30 Hz (Supplementary Fig. 7b, Supplementary Video 5,6). CA1 recordings were processed and analyzed using the same pre-processing and SCISM demixing as described for the V1 data.
The 3D positions for each of the 882 spatial profiles found using SCISM were calculated (Fig. 6a,b) and their activity was demixed (Fig. 6c, Supplementary Fig. 11). Interestingly, the tendency for shallower neurons towards the center of the FOV and deeper neurons towards the edges of the FOV indicates that the vTwINS spatial profiles are capturing the curvature of CA1 (Fig. 6a). A subset of spatial profiles in a 92 µm × 92 µm section (Fig. 6b,d) of the FOV demonstrate that the inferred 3D locations match well to the anatomical z-stack (Supplementary Fig. 12).
Figure 6.
Demixed spatial profiles and calcium activity in mouse hippocampus. (a) Full set of spatial profiles, color-coded by depth, show more overlap in CA1 than in cortical recordings. (b) 3D locations of the spatial profiles from the white box in (a) show found spatial profiles at various depths. (c) Time-traces of spatial profiles in (a) show sporadic activity in the 0–100s time interval. (d) Example subset of spatial profiles (chosen from the white inset box in (a) and sorted by depth) and corresponding normalized time traces show rich activity patterns. Note that some profiles have very sparse activity and do not contain transients in the displayed 100 s range. (e) Example demixed spatially overlapping profiles. Profile 1 (blue) and Profile 2 (red) spatially overlap yet have demixed time traces (right). Averaged raw fluorescence traces from pixels in the overlapping region (Overlap) are a linear combination of the traces from Profile 1 (Region 1) and Profile 2 (Region 2). The Region 2 time-trace also contains a transient from yet another profile at 230 s.
Despite the highly overlapping spatial profiles due to the vTwINS PSF and high neural density, SCISM successfully demixed spatial profiles in CA1. Fluorescence time courses in different regions of two overlapping spatial profiles illustrate the demixed time traces (Fig. 6e). The trace from the overlapped region of the two cells contains transients from both non-overlap regions, while the demixed traces contain only the single-profile region activity. Interestingly, one transient at 230 s in Region 2’s trace is missing from the overlap trace, indicating that this transient originated from a third profile and was successfully demixed in Profile 2’s time trace.
We also explored in mouse CA1 a variation of vTwINS where we alternate between each half of the vTwINS PSF on consecutive frames (Online Methods, Supplementary Fig. 1c). Illumination with a single beam (40 µm-long PSF; FWHM, 56 µm 1/e full-width) increases SNR per frame, however each beam can only be recorded at half the framerate (15 Hz). By merging the 30Hz interpolated recording, we used SCISM to locate spatial profiles separated by the width of the image (Supplementary Fig. 13a–c, 14). The temporal separation of the two channels was observed to reduce the total background neuropil, improving the SNR of the recorded transients (Supplementary Fig. 13d–f).
Discussion
Early strategies for large scale recording using calcium imaging generally used the spatial resolution of the optical instrumentation to ensure that the fluorescence from individual neurons formed spatially independent, disjoint sets. Spatial separation was the basis for hand selection of neural regions of interest, which has been widely used as a mask for extracting the time traces of individual cells. In practice, however, perfect separation of adjacent cell signals has been difficult to achieve for densely labeled cells. As a result, demixing algorithms16,17,22 have been developed to identify spatial profiles by allowing individual pixels to have contributions from multiple neurons. vTwINS (and also a recent multi-plane technique24) take this mixing assumption as a starting point for the development of the optical instrumentation. The vTwINS V-shaped PSF increases signal mixing in individual pixels, but also ensures that each neuron will have a unique spatial profile that can be efficiently used in the co-designed demixing algorithm to extract the time traces and 3D location of individual cells. We anticipate that this strategy in which optical instrumentation and demixing algorithm are co-designed for large scale recording may generalize to other excitation geometries (e.g. 3 beams, multiple objectives).
vTwINS required the ability to seek specific spatial profile shapes while maintaining the flexibility to adapt to variations in neuronal shape. SCISM permitted the specification of these shapes as guides to locate relevant activity while still balancing the general expected temporal statistics of neural activity. Current automated methods do not use such detailed spatial information, focusing instead on temporal demixing25–29 with no spatial constraints16,17 or utilizing generic locality assumptions (i.e. spatial profiles must be fully contained in a constrained region)22,30. The ability of SCISM to adapt profiles to the data also differentiates it from standard matching pursuit-style algorithms31–33, which assume a fixed dictionary of features. Although we designed SCISM to seek features specific to vTwINS imaging, it can easily accommodate other spatial profile shapes in future imaging methods.
When compared to the most similar current approaches, vTwINS offers a number of advantages. Unlike fast point scanning strategies10, vTwINS has a substantially lower peak nonlinear excitation (Supplementary Note 5), allowing for scanning in larger volumes. Additionally, the vTwINS excitation configuration is comparatively power efficient34, and has high total two-photon excitation for a given average power. Compared to other multiplexed two-photon approaches24, vTwINS requires no additional recordings to initialize components, and guarantees uniqueness of spatial profiles. Finally, when compared to volumetric TPM using single, elongated PSFs, the use of two beams in vTwINS decreases ambiguity between spatial profiles (Supplementary Fig. 3, 4a).
vTwINS, like other TPM techniques, is subject to practical concerns of imaging in vivo. For our recordings, motion correction was not compromised due to the elongated PSF and sufficient high spatial frequency features in the imaged regions remained for accurate motion correction. Alternatively, in brain regions with low spatial structure, nuclearly localized RFP may be used for accurate motion correction. Nonlinear phototoxicity35,36 in vTwINS is reduced (Supplementary Note 5) as compared to high-NA TPM, while heating37 is limited to 100mW per excitation beam.
Additional work can further optimize vTwINS for other applications. vTwINS has additional background fluorescence from neuropil contamination, but might work particularly well with a nuclear localized GCaMP38, which would significantly reduce background fluorescence from neuropil and improve SNR. Furthermore, more flexible spatial information can be implemented in SCISM to improve demixing of neural signals from neuropil contamination. For brain regions with limited optical access (e.g. hippocampus23 or MEC39), smaller angles between arms or separation distance may be necessary. The choice between Bessel beams and Gaussian beams requires additional study. Bessel beams offer flexibility in controlling the axial profile and lateral resolution40. Gaussian beams, while less flexible, are simpler to implement and have higher two-photon excitation (Online Methods, Supplementary Fig. 1). Finally, as a complementary method, vTwINS can be paired with existing improvements, such as sequential plane imaging (e.g. remote focusing11, or liquid lens6) to image larger volumes or take advantage of the improvement to SNR that a lower repetition rate laser offers34.
Online Methods
Microscope Design
The vTwINS microscope was modeled in ZEMAX (Zemax LLC) and custom MATLAB (Mathworks) scripts. The microscope (Fig. 1c) was constructed as a modification of a resonant scanning two-photon microscope. A beam shaping module to produce the V-shaped PSF for vTwINS was designed to be inserted between the laser and microscope. This strategy was used so that the module could, in principle, straightforwardly be adapted for any existing standard two-photon microscope. The beam-shaping module consisted of three optical paths that could be switched via flip-mount mirrors between: 1) a standard high-NA path for standard two-photon imaging, 2) a vTwINS path using low-NA Gaussian beams, or 3) a vTwINS path using Bessel beams.
The collimated Gaussian laser beam entering the beam-shaping module had a measured knife-edge width (10/90 percent) of 1.3mm which corresponds to a 1/e2 diameter of 2 mm. The high-NA path consisted of a 2.5× beam expander (AC254-40-B and AC254-100-B, Thorlabs). The Gaussian vTwINS path consisted of a 0.3–1.2× variable telescope (G06-203-525 AC 140/31,5 Linos, LC1120 and AC254-125-B, Thorlabs). When aligning the Gaussian vTwINS path, care was taken to avoid focusing the laser beam directly onto the scanners. The Bessel vTwINS path consisted of an axicon and achromat lens pair (179.2° BK7 Axicon, Altechna and AC254-200-B, Thorlabs) to generate the ring-shaped excitation for the Bessel beam. The specific choice of axicon and achromat lens pair was based on tradeoff between lateral resolution and two-photon excitation efficiency. For the Bessel beams to be correctly formed within the sample, the rear pupil of the objective needs to be illuminated with well focused annuli of light. For this reason, the back aperture of the objective is conjugate to the achromatic lens front focal plane of the Axicon-Achromat pair. If collimated, parallel beams are used, the two branches of the PSF form a X-shape. The PSF V-shape was obtained by introducing a slight beam convergence at the objective back-aperture created and tuned by a 1× telescope (2× AC254-100-B, Thorlabs). When the vTwINS modalities were used, the beam was split in two parallel beams with a half-wave plate and a Calcite beam displacer (AHWP05M-980 and BD27, Thorlabs). The half-wave plate was oriented such that the fluorescence intensities of the two images are equal. The birefringent beam displacer was mounted in a rotation mount and oriented such that the two beams lie in a plane perpendicular to the resonant (fast) scanning mirror axis of rotation. This is to guarantee that the two images formed of a fluorescent object lie on the same scanned line. A pair of BK7 windows mounted on orthogonal rotation axes was used to adjust and center the lateral position of the beams on the scanners. The beams separation (2.7 mm out of the Calcite beam displacer) was further reduced using a 0.8× telescope (AC254-100-B and AC254-80-B, Thorlabs). This specific choice, in combination with the magnification of the microscope (X3.75) and the 12.5 mm focal length of the water immersion Nikon objective resulted in an angle of 43° between the two branches of the PSF. This choice of angle resulted in an accurate axial localization of the cell bodies (Supplementary Fig. 2). When the high-NA path was used for conventional two-photon imaging, the half-wave plate was rotated to zero the power of one of the emerging beams, and the two BK7 windows were oriented to center the remaining beam on the optical axis of the microscope.
A Ti:Sapphire laser (Chameleon Vision II, Coherent) at 920 nm was used for two-photon excitation, and dispersion compensation in the laser was adjusted to maximize the two-photon signal. A Pockels cell (Model 350–80 with 302RM driver, Conoptics) was used to modulate laser intensity and a half-wave plate plus polarizing beamsplitter cube (Thorlabs) was used to adjust the maximum laser intensity. The two-photon microscope body consisted of a resonant scanning head (6215/CRS 8 kHz, Cambridge Technologies), a 100 mm f − θ scan lens (4401-464-000, Linos) and a 375 mm achromat pair tube lens (2× PAC097, Newport), and an objective lens (N16XLWD-PF, Nikon41). The excitation and emission were separated by a shortpass dichroic (T680-DCSPXR-UF3 52 mm × 75 mm × 3 mm, Chroma), and the collection optics (ACL7560-A, LC1611-A, ACL25416U-A, Thorlabs) focused the emitted light onto two PMTs (H10770PA-40, Hamamatsu), separated into red and green channels (FF555-Di03-40×54, FF01-720/SP-50, FF02-525/40-32, FF01-593/40-32, Semrock). The PMT signal was amplified with an 80MHz preamplifier (DHPCA-100, Femto) and digitized with a FPGA (NI PXIe-7961R and NI 5732 DAQ, National Instruments). Scanning and data acquisition were controlled with Scanimage 2015 (Vidrio). Average power during vTwINS data acquisition varied between 150 mW and 200 mW at 920 nm, and average power during high-NA acquisition was between 50 mW and 70 mW at 920 nm. Images here were typically acquired at 30Hz with an image size 512×512 pixels at with a 90% spatial cutoff, corresponding to an image size of 470 µm × 470 µm (2.8 zoom) or 550 µm × 550 µm (2.4 zoom). Nearly simultaneous calcium imaging using rapid switching between vTwINS excitation and the traditional focused high-NA Gaussian PSF was performed using an alternate optical setup (Supplementary Fig. 1b). A galvanometer (6210H, Cambridge Technologies) was used to select between high-NA and vTwINS (38 µm-long FWHM PSF) paths, which were recombined downstream with a (50 µm, 0.88° optical) offset. A modified Scanimage analog control was used to switch between the two paths at every frame (≈ 17 ms). For each modality, images were acquired at 30 Hz with a 512×256 pixel image size. Rapid alternation between two low-NA Gaussian beams was performing using a similar alternate optical setup (Supplementary Fig. 1c). Each of the two paths were identical variable low-NA Gaussian beams separated by a fixed distance imaged onto the scanners. The fixed separation distance set the relative angle of the two axially extended beams.
Transgenic Mice
All experimental procedures were approved by the Princeton University Institutional Animal Care and Use Committee. Transgenic GCaMP6f-expressing mice were produced by crossing Emx1-Cre (B6.129S2 − Emx1tm1(cre)Krj/J, Jax #005628), CaMK2-tTA (B6.Cg-Tg(Camk2a-tTA)1Mmay/DboJ, Jax #007004) and TITL-GCaMP6f (Ai93; B6. Cg − Igs7tm93.1(tetOGCaMP6f)Hze/J, Jax #024103) strains42. Male or female transgenics heterozygous for all three genes were used for all experiments.
Imaging Mouse Visual Cortex
For imaging in mouse visual cortex, mice underwent surgery under isoflurane anesthesia for implantation of imaging windows and head-plates. A 5 mm diameter craniotomy was made over one hemisphere of parietal cortex (centered 2 mm caudal, 1.7 mm lateral to bregma). A custom titanium head-plate and optical window (#1 thickness, 5 mm diameter glass coverslip, Warner Instruments) bonded to a steel ring (0.5 mm thickness, 5 mm diameter, SS316 ring, Ziggy’s Tubes and Wires, Inc.) were attached to the mouse’s skull with dental cement (Metabond, Parkell). The location of V1 was estimated using a separate widefield imaging microscope to record retinotopic responses in fluorescence activity as the mouse viewed horizontally and vertically drifting bars on a 32” monitor43. Boundaries between the primary and secondary visual areas were defined using an automated algorithm to locate reversals in the retinotopic gradients44. Five days after surgery, mice were trained to run on a spherical treadmill (8 inch diameter Styrofoam ball) surrounded by a 270° toroidal screen45. Visual stimuli were generated using the Psychophysics Toolbox46–48 and displayed on the toroidal screen using a DLP projection system (Mitsubishi HC3000), consisting of ≈100 randomly placed/oriented Gabor patches, with visual field size 5 – 10°, updated at 4 Hz. To prevent light from the projected display from entering the fluorescence collection system, the region between the base of the objective lens and the head-plate was light-proofed using a black rubber tube prior to imaging. The rubber tube was glued to a silicone ring and the ring itself attached to the titanium headplate with silicone elastomer (Body Double, Smooth On Inc.). Examples of images from cortical imaging are depicted in Supplementary Figure 7a,c and Supplementary Video (1–4).
Imaging Mouse Hippocampus
For imaging in mouse hippocampus, mice underwent surgery under isoflurane anesthesia for implantation of an imaging window and a head-plate for head-restraint in virtual reality49. Optical access to the hippocampus was obtained as described previously23. Briefly, a ≈ 3 mm diameter circular craniotomy over the left hemisphere was performed, centered 1.8 mm lateral to the midline, and 2.0 mm posterior to bregma. The cortical tissue overlying the hippocampus was aspirated, and a circular metal cannula with a #1 coverslip bonded to the bottom was implanted, with a thin layer of Kwik-sil (WPI) between the hippocampus and coverslip. During the surgery, a titanium headplate was attached to the skull with Metabond. After recovery, mice were water restricted for five days and then trained to run on a 4 m virtual linear track using a virtual reality setup50. Visually distinct towers were placed every 1m and 4 µ L water rewards given at 1.6 m and 3.6 m down the track. Mice ran on a 6 inch diameter Styrofoam cylinder (The Baker’s Kitchen) whose position was detected by an angular encoder. Mice were trained for a 60 minute session per day and were given 1–1.5 mL of water a day total (including behavioral training and supplemental water). The virtual reality projection system was as described previously44,48 and controlled with ViRMEn51. Lightproofing around the objective was performed as described for experiments in visual cortex. Examples of images from hippocampal imaging are depicted in Supplementary Figure 7b and Supplementary Video (5,6).
Motion Correction and Pre-processing
All video sequences were first subject to a normalized cross-correlation-based motion correction algorithm. This algorithm, implemented via the template matching function of OpenCV52, found the best horizontal and vertical shifts for each frame to match a fixed template. The template used was set to the median across frames. Shifts were set to have a maximum allowable value (set to 10 pixels for the V1 data and 15 pixels for the CA1 data). Videos were cropped to remove edge rows and columns with missing data due to shifting. To improve SNR and run-time, five-fold temporal running averages and two-fold spatial binning were applied post-motion correction.
For the alternating-beam variation of vTwINS, we have two interleaved videos Yright and Yleft To use SCISM, we pre-process the data by linearly interpolating each video temporally up to twice the frame rate. The frames at each time-step were concatenated side-by-side creating a vTwINS video of twice the width where the minimum distance between pairs, dmin, was the actual distance between beams plus the entire width of the FOV. In the second interleaved video (Supplementary Fig. 4, 18d), dmin = 15pixels, approximating a typical vTwINS movie.
vTwINS Orthogonal Matching Pursuit
In this section, we describe the mathematical details of the vTwINS Sparse Convolutional Iterative Shape Matching (SCISM) demixing algorithm (Fig. 3, Supplementary Fig. 5, Supplementary Video 7). Let Y ∈ RN×T denote the calcium video sequence, X ∈ RN×K denote the neural spatial components (spatial profiles), and S ∈ RT×K denote the neural temporal activity traces, where N is the number of pixels in each image, T is the number of images (or time points), and K is the number of neurons. Thus, the columns of Y represent single frames of the video, the columns of X represent individual spatial profiles, and the columns of S represent temporal activity traces of single neurons. We model background activity with a set of B background components Xbg ∈ RN×B and denote the (inferred) background temporal activity Sbg ∈ RT×B.
Our algorithm is designed to exploit a priori knowledge of both the spatial profile shapes as well as neural firing statistics. Specifically, the algorithm seeks to factor the full movie matrix Y into the set of spatial profiles X and time-traces S such that
The sum of outer products of spatial profiles and time traces explains the observed data (Y ≈ XST).
The time-traces S are sparse in time.
The spatial profiles are shaped like pairs of neuronal soma (disks or annuli), offset horizontally by a small separation distance. The dark center in each soma is due to the lack of GCaMP6f in the nucleus.
Few latent sources (active neurons) relative to the size of the data generate activity in the observed data, making the fluorescence movie low-rank. This constraint captures the physical density constraints on neuron tissue.
The optimization program that includes all these terms is
| (1) |
where sk is the kth column of S, representing the activity trace of neuron k, is the squared-Frobenius norm, D is a matrix whose columns represent all possible expected neural spatial profile shapes, λd is the trade-off parameter for penalizing the deviation of spatial profile shapes X from the idealized shapes in D, λgs is the group-sparse penalization parameter for ensuring that not all spatial profiles are active and λsp is the penalization parameter that encourages sparsity of the time traces.. We set the spatial profiles dk to be annuli separated by a depth-dependent distance (Fig. 3a, Supplementary Note 6).
Direct optimization of Equation (1) can be inefficient due to the problem size and the large search space (number of possible spatial profiles). We thus approximated a solution to Equation (1) with a greedy, iterative approach wherein spatial profiles are selected sequentially. Our method alternates between finding the best element of D that approximates Y given the sparsity constraints (Fig. 3b,c, Supplementary Note 6), updating that profile to the data (Fig. 3d, Supplementary Note 6), and inferring the temporal activity of each spatial profile (Fig. 3e, Supplementary Note 6). The first step sets X = D and solves for the best single trace to approximate Y (solving the first and third terms). The shape refinement step then uses the first two terms with the newly found time-trace to allow the spatial profile xk to deviate from its mean dk. SCISM is in essence a modification of the orthogonal matching pursuit (OMP) method for greedy sparse signal estimation31,53. Our method extends OMP by including an additional temporal sparsity penalty and a shape refinement step that allows for deviations from the stereotyped neuronal shapes (whereas traditional OMP assumes a fixed dictionary of features).
SCISM was implemented in MATLAB and made use of the TFOCS library54 to solve the weighted, non-negative LASSO optimization step. Typical analysis ran at a rate of approximately 20 s per profile found, applied to 10–15 minutes of 256×256 raw imaging data.
vTwINS and High-NA Spatial Profile Registration
High resolution anatomical z-stacks (median of 200–300 frames per slice at 2.5–4 µm slice separation taken with the high-NA beam path) were taken for each vTwINS imaging volume to align the vTwINS spatial profiles to anatomical positions. Alignment between the anatomical z-stack and the vTwINS imaging volume was performed in two steps: 1. The 3D position of cells was estimated to their position within the vTwINS volume. 2. The estimated 3D positions were offset to the anatomical volume. First, the centroids of each half of the spatial profile were used to calculate the 3D cell position via d = 0.5(Δ − Δmin)/tan(θ), where Δmin is the minimum inter-beam distance of the PSF and θ is the beam angle from the axial direction. A correction to the xy position was made for any differences in θ between the two halves of the vTwINS PSF. Second, a 3D offset between the estimated positions and anatomical z-stack positions was either automatically or manually calculated. For automatic alignment, the anatomical stack was first deconvolved (Lucy-Richardson) with the high-NA PSF and then convolved with the vTwINS PSF. A 3D cross-correlation was then calculated between the convolution stack and the median vTwINS image and the peak of the cross-correlation was used as the offset between the vTwINS images and the anatomical z-stack. For manual alignment, highly active cells with similar cell shapes between the vTwINS spatial profiles and high-NA anatomical z-stacks were located manually and used to estimate the offset between the vTwINS images and anatomical z-stack.
For simultaneous vTwINS and conventional TPM imaging, neural activity was independently extracted from raw images with separate analyses. Neural activity underlying calcium dynamics for conventional TPM was estimated using the Constrained Non-negative Matrix Factorization and deconvolution algorithm (CNMF) to demix contributions from possibly overlapping cells22,55. Spatial profiles extracted using CNMF were manually selected for regions that approximated a cell shape (roughly circular, 10–15 µm in diameter). To compare number of spatial profiles between imaging modalities, spatial profiles from either method were only included if their center position was within 20 pixels (18 µm) of the × (fast-scanning) edge of the acquisition region. This is to prevent bias from clipping half of a single vTwINS profile near the edges of the image.
Spatial profiles and time traces extracted using vTwINS SCISM and CNMF were paired off by their normalized time trace Pearson correlation (Supplementary Fig. 10), subject to the constraint that the extracted spatial profile center positions were within 5 pixels (4.5 µm) in the y (slow-scanning) direction and 40 pixels (37 µm) in the × (fast-scanning) direction. This distance is roughly equal to half the maximum separation distance between vTwINS spatial profile image pairs, which does not restrict pairing of CNMF spatial profiles to vTwINS spatial profiles with a single blocked beam. Spatial profiles and time traces were paired off until the correlation dropped below a 5σ excess of the average correlation between any two time traces. Only high activity cells with >1 statistically significant transient/min were included for this analysis56. A transient was considered statistically significant if its peak was >3σ above the average noise levels.
Code and Data Availability
The SCISM source code (written in MATLAB) and its documentation on its usage is available on Bitbucket (https://bitbucket.org/adamshch/scism). Sample data and the datasets used are available on Open Science Framework (osf.io/z6bd3).
Supplementary Material
Acknowledgments
We thank C. Domnisoru, R. Low and B. Scott for their insightful thoughts and comments. We also thank J. Homann for his assistance using Psychophysics Toolbox. This work was supported by NIH grants R01MH083868 and U01NS09054, and the Simons Collaboration on the Global Brain. A.C. was supported by an NIH NRSA Training Grant in Quantitative Neuroscience (T32MH065214). J.W.P was supported by grants from the McKnight Foundation, Simons Collaboration on the Global Brain (SCGB AWD1004351) and NSF CAREER Award (IIS-1150186).
Footnotes
Author Contributions
D.W.T. conceived the project. A.S. and S.Y.T. designed and constructed the vTwINS microscope. S.A.K. and J.L.G. performed the surgery on the mice. A.S. trained the mice and performed the imaging experiments. A.S.C. and J.W.P. designed the SCISM algorithm. A.S.C. implemented SCISM and applied the method to the vTwINS data. A.S. and A.S.C. performed the analysis on the results. A.S., A.S.C., J.W.P., and D.W.T wrote the manuscript, with comments and contributions from all authors. J.W.P and D.W.T supervised the project.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–6. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 2.Mank M, Griesbeck O. Genetically encoded calcium indicators. Chem. Rev. 2008;108:1550–64. doi: 10.1021/cr078213v. [DOI] [PubMed] [Google Scholar]
- 3.Tian L, Akerboom J, Schreiter ER, Looger LL. Neural activity imaging with genetically encoded calcium indicators. Prog. Brain. Res. 2012;196:79–94. doi: 10.1016/B978-0-444-59426-6.00005-7. [DOI] [PubMed] [Google Scholar]
- 4.Stosiek C, Garaschuk O, Holthoff K, Konnerth A. In vivo two-photon calcium imaging of neuronal networks. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7319–7324. doi: 10.1073/pnas.1232232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dombeck DA, Khabbaz AN, Collman F, Adelman TL, Tank DW. Imaging large-scale neural activity with cellular resolution in awake, mobile mice. Neuron. 2007;56:43–57. doi: 10.1016/j.neuron.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grewe BF, Voigt FF, van ’t Hoff M, Helmchen F. Fast two-layer two-photon imaging of neuronal cell populations using an electrically tunable lens. Biomed. Opt. Express. 2011;2:2035–46. doi: 10.1364/BOE.2.002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gobel W, Kampa BM, Helmchen F. Imaging cellular network dynamics in three dimensions using fast 3D laser scanning. Nat. Methods. 2007;4:73–9. doi: 10.1038/nmeth989. [DOI] [PubMed] [Google Scholar]
- 8.Duemani Reddy G, Kelleher K, Fink R, Saggau P. Three-dimensional random access multiphoton microscopy for functional imaging of neuronal activity. Nat. Neurosci. 2008;11:713–20. doi: 10.1038/nn.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirkby PA, Srinivas Nadella KM, Silver RA. A compact Acousto-Optic Lens for 2D and 3D femtosecond based 2-photon microscopy. Opt. Express. 2010;18:13721–45. doi: 10.1364/OE.18.013720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong L, et al. Continuous volumetric imaging via an optical phase-locked ultrasound lens. Nat. Methods. 2015;12:759–762. doi: 10.1038/nmeth.3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Botcherby EJ, Juskaitis R, Booth MJ, Wilson T. An optical technique for remote focusing in microscopy. Opt. Commun. 2008;281:880–887. [Google Scholar]
- 12.Chen T-W, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Botcherby EJ, Juskaitis R, Wilson T. Scanning two photon fluorescence microscopy with extended depth of field. Opt. Commun. 2006;268:253–260. [Google Scholar]
- 14.Theriault G, Cottet M, Castonguay A, McCarthy N, De Koninck Y. Extended two-photon microscopy in live samples with Bessel beams: steadier focus, faster volume scans, and simpler stereoscopic imaging. Front. Cell. Neurosci. 2014;8:139. doi: 10.3389/fncel.2014.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu R, et al. Video-rate volumetric functional imaging of the brain at synaptic resolution. 2016 doi: 10.1038/nn.4516. Preprint at http://biorxiv.org/content/early/2016/06/12/058495. [DOI] [PMC free article] [PubMed]
- 16.Mukamel EA, Nimmerjahn A, Schnitzer MJ. Automated analysis of cellular signals from large-scale calcium imaging data. Neuron. 2009;63:747–760. doi: 10.1016/j.neuron.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maruyama R, et al. Detecting cells using non-negative matrix factorization on calcium imaging data. Neural Networks. 2014;55:11–19. doi: 10.1016/j.neunet.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Pnevmatikakis E, Paninski L. Sparse nonnegative deconvolution for compressive calcium imaging: algorithms and phase transitions. Adv. Neural Inf. Process. Syst. 2013:1250–1258. [Google Scholar]
- 19.McGloin D, Dholakia K. Bessel beams: diffraction in a new light. Contemp. Phys. 2005;46:15–28. [Google Scholar]
- 20.Apthorpe NJ, et al. Automatic Neuron Detection in Calcium Imaging Data Using Convolutional Networks. 2016 Preprint at https://arxiv.org/abs/1606.07372.
- 21.Tibshirani R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Series B Stat. Methodol. 1996:267–288. [Google Scholar]
- 22.Pnevmatikakis E, et al. A structured matrix factorization framework for large scale calcium imaging data analysis. 2014 Preprint at https://arxiv.org/abs/1409.2903.
- 23.Dombeck DA, Harvey CD, Tian L, Looger LL, Tank DW. Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat. Neurosci. 2010;13:1433–1440. doi: 10.1038/nn.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang W, et al. Simultaneous Multi-plane Imaging of Neural Circuits. Neuron. 2016;89:269–84. doi: 10.1016/j.neuron.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yaksi E, Friedrich RW. Reconstruction of firing rate changes across neuronal populations by temporally deconvolved Ca2+ imaging. Nat. Methods. 2006;3:377–383. doi: 10.1038/nmeth874. [DOI] [PubMed] [Google Scholar]
- 26.Vogelstein JT, et al. Spike inference from calcium imaging using sequential Monte Carlo methods. Biophys. J. 2009;97:636–655. doi: 10.1016/j.bpj.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deneux T, et al. Accurate spike estimation from noisy calcium signals for ultrafast three-dimensional imaging of large neuronal populations in vivo. Nat. Commun. 2016;7 doi: 10.1038/ncomms12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jon O, Schultz SR, Dragotti PL, et al. A finite rate of innovation algorithm for fast and accurate spike detection from two-photon calcium imaging. J. Neural Eng. 2013;10:046017. doi: 10.1088/1741-2560/10/4/046017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grewe BF, Langer D, Kasper H, Kampa BM, Helmchen F. High-speed in vivo calcium imaging reveals neuronal network activity with near-millisecond precision. Nat. Methods. 2010;7:399–405. doi: 10.1038/nmeth.1453. [DOI] [PubMed] [Google Scholar]
- 30.Pachitariu M, et al. Extracting regions of interest from biological images with convolutional sparse block coding. Adv. Neural Inf. Process. Syst. 2013:1745–1753. [Google Scholar]
- 31.Pati Y, Rezaiifar R, Krishnaprasad P. Orthogonal matching pursuit: Recursive function approximation with applications to wavelet decomposition. Asilomar Conf. Signals Syst. Comput. 1993:40–44. [Google Scholar]
- 32.Needell D, Tropp JA. CoSaMP: Iterative signal recovery from incomplete and inaccurate samples. Appl. Comput. Harmon. Anal. 2009;26:301–321. [Google Scholar]
- 33.Donoho DL, Tsaig Y, Drori I, Starck J-L. Sparse solution of underdetermined systems of linear equations by stagewise orthogonal matching pursuit. IEEE T. Inform. Theory. 2012;58:1094–1121. [Google Scholar]
- 34.Prevedel R, et al. Fast volumetric calcium imaging across multiple cortical layers using sculpted light. Nat. Methods. 2016;13:1021–1028. doi: 10.1038/nmeth.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hopt A, Neher E. Highly nonlinear photodamage in two-photon fluorescence microscopy. Biophys. J. 2001;80:2029–2036. doi: 10.1016/S0006-3495(01)76173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji N, Magee JC, Betzig E. High-speed, low-photodamage nonlinear imaging using passive pulse splitters. Nat. Methods. 2008;5:197–202. doi: 10.1038/nmeth.1175. [DOI] [PubMed] [Google Scholar]
- 37.Podgorski K, Ranganathan GN. Brain heating induced by near infrared lasers during multi-photon microscopy. J. Neurophysiol. 2016 doi: 10.1152/jn.00275.2016. jn.00275.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim CK, et al. Prolonged, brain-wide expression of nuclear-localized GCaMP3 for functional circuit mapping. Front. Neural Circuits. 2014;8:138. doi: 10.3389/fncir.2014.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Low RJ, Gu Y, Tank DW. Cellular resolution optical access to brain regions in fissures: imaging medial prefrontal cortex and grid cells in entorhinal cortex. Proc. Natl. Acad. Sci. U.S.A. 2014;111:18739–18744. doi: 10.1073/pnas.1421753111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cizmár T, Dholakia K. Axial intensity shaping of a Bessel beam. Proc. SPIE. 2009:74001Q–74001Q. [Google Scholar]
- 41.Watanabe KNC. Microscope Objective Lens. 2005-189732. Japanese Patent No. 2005
- 42.Madisen L, et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron. 2015;85:942–58. doi: 10.1016/j.neuron.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marshel J, Garrett M, Nauhaus I, Callaway E. Functional Specialization of Seven Mouse Visual Cortical Areas. Neuron. 2011;72:1040–1054. doi: 10.1016/j.neuron.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garrett ME, Nauhaus I, Marshel JH, Callaway EM. Topography and Areal Organization of Mouse Visual Cortex. J. Neurosci. 2014;34:12587–12600. doi: 10.1523/JNEUROSCI.1124-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rickgauer JP, Deisseroth K, Tank DW. Simultaneous cellular-resolution optical perturbation and imaging of place cell firing fields. Nat. Neurosci. 2014;17:1816–24. doi: 10.1038/nn.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brainard DH. The Psychophysics Toolbox. Spat. Vis. 1997;10:433–436. [PubMed] [Google Scholar]
- 47.Pelli DG. The VideoToolbox software for visual psychophysics: Transforming numbers into movies. Spat. Vis. 1997;10:437–442. [PubMed] [Google Scholar]
- 48.Kleiner M, Brainard D, Pelli DG. What’s new in Psychtoolbox-3? Perception. 2007;36:1–16. [Google Scholar]
- 49.Harvey CD, Collman F, Dombeck DA, Tank DW. Intracellular dynamics of hippocampal place cells during virtual navigation. Nature. 2009;461:941–6. doi: 10.1038/nature08499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Domnisoru C, Kinkhabwala AA, Tank DW. Membrane potential dynamics of grid cells. Nature. 2013;495:199–204. doi: 10.1038/nature11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aronov D, Tank DW. Engagement of neural circuits underlying 2D spatial navigation in a rodent virtual reality system. Neuron. 2014;84:442–456. doi: 10.1016/j.neuron.2014.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bradski G. Dr. Dobb’s Journal of Software Tools. 2000. [Google Scholar]
- 53.Swirszcz G, Abe N, Lozano A. Grouped orthogonal matching pursuit for variable selection and prediction. Adv. in Neural Inf. Process. Syst. 2009:1150–1158. [Google Scholar]
- 54.Becker S, Candes E, Grant M. TFOCS: flexible first-order methods for rank minimization. SIAM Conf. Optim. 2011 [Google Scholar]
- 55.Machado TA, Pnevmatikakis E, Paninski L, Jessell TM, Miri A. Primacy of flexor locomotor pattern revealed by ancestral reversion of motor neuron identity. Cell. 2015;162:338–350. doi: 10.1016/j.cell.2015.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harvey CD, Coen P, Tank DW. Choice-specific sequences in parietal cortex during a virtual-navigation decision task. Nature. 2012;484:62–68. doi: 10.1038/nature10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.