Abstract
Type 1 diabetes (T1D) has been associated with both genetic and environmental factors. Increasing incidence of T1D worldwide is prompting researchers to adopt different approaches to explain the biology of T1D, beyond the presence and activity of autoreactive lymphocytes. In this review, we propose inflammatory pathways as triggers for T1D. Within the scope of those inflammatory pathways and in understanding the pathogenesis of disease, we suggest that viruses, in particular Coxsackieviruses, act by causing a type 1 interferonopathy within the pancreas and the microenvironment of the islet. As such, this connection and common thread represents an exciting platform for the development of new diagnostic, treatment and/or prevention options.
Keywords: Type 1 diabetes, Autoimmune disease, Interferon signature, Type 1 interferonopathies, Chronic virus infection, Coxsackieviruses
Highlights
-
•
Recent clinical evidence has identified that interferon signalling pathways are strongly associated with Type 1 diabetes.
-
•
Coxsackieviruses are strongly associated with the onset of Type 1 diabetes.
-
•
T1D likely results from a virus induced type 1 interferonopathy of the islets and this premise will guide novel treatments.
1. Introduction
Type 1 Diabetes (T1D) is a disease characterized by the discriminatory destruction of pancreatic beta cells (Gillespie, 2006). T1D is a process that requires both autoimmunity and autoinflammation where the pancreas is infiltrated by immune cells such as macrophages, dendritic cells and natural killer cells secreting pro-inflammatory cytokines, and autoreactive B and T cells specific for islet antigens such as insulin, glutamic acid decarboxylase (GAD)-65, and islet antigen(IA)-2 (Li et al., 2014, Arvan et al., 2012). Although no single etiology is known for T1D, epidemiological and genome-wide association studies have linked T1D with both genetic factors i.e. polymorphisms in human leukocyte antigen (HLA) haplotypes, and environmental factors such as viral infections (Christoffersson et al., 2016, Richardson and Horwitz, 2014). While T1D is canonically considered a T-cell mediated disease (Berry and Waldner, 2013, Kelly et al., 2003), research has demonstrated that the mere presence of autoreactive T cells is not the initiating factor but rather a determinant of disease progression (Laitinen et al., 2014, Serreze et al., 2000). Additionally, the presence of autoantibodies can be detected years before clinical disease, and not all islet autoantibody-positive individuals develop T1D (Reynier et al., 2010, Kulmala et al., 1998). Thus, we propose the inflammatory pathway as a focus for understanding early triggering events in T1D, and viral infections in acceleration of an established autoimmune/inflammatory process. Here, we review T1D inflammation as it relates to the interferon (IFN) signature, and establish a link with Type 1 interferonopathies (type 1 IFN-opathies) and viruses, specifically Coxsackieviruses. Type 1 IFN-opathies constitute a group of human diseases associated with overproduction of the pro-inflammatory type I IFNs, and are widely thought to be controlled by genetics (Crow and Manel, 2015, Lee-Kirsch et al., 2015). These diseases, often systemic and clinically symptomatic, are associated with destructive inflammation as well as development of autoimmune phenomena. We propose that persistent or chronic Coxsackievirus infections in pancreatic islet cells simulate a local type 1 IFN-opathy in the islet microenvironment that is clinically silent before diabetes onset. As several of the candidate susceptibility genes are involved in IFN responses and because virus infections would amplify genetic tendencies for a heightened IFN response, we propose that the underlying molecular mechanisms associated with type 1 IFN-opathies can serve as a foundation or reference point for the evaluation of inflammatory processes of T1D.
With the increasing incidence of T1D worldwide (Tuomilehto, 2013, Patterson et al., 2009), deciphering the inflammatory pathways represents a direct and relevant approach for the development of new targeted therapeutics to prevent, interrupt, or overturn progression of disease.
2. The Interferon Signature
IFNs are a group of pro-inflammatory cytokines. To date, three types of IFNs have been identified: type I (which includes IFNα, IFNβ, IFNε, IFNκ and IFNώ), type II (IFNγ), and the recently discovered type III (IFNλ) (Donnelly and Kotenko, 2010, Sheppard et al., 2003).
The “IFN signature” refers to expression of genes that are known to be regulated by IFNs. Overall, the repertoire of responses induced by IFNs covers a wide range of activities. These activities can go from: priming an antiviral state in local cells, to secretion of cytokines and chemokines that recruit and arm effector cells of the adaptive immune system to regulation of neuronal connectivity, but also T cell dysfunction and establishment of chronic inflammation (Filiano et al., 2016, Odendall and Kagan, 2015, Teijaro et al., 2013, Hultcrantz et al., 2007, Curtsinger et al., 2005). Some recognized type I IFN-stimulated gene products include: IFIT1 (interferon-induced protein with tetratripeptide repeats 1), OAS1/2 (2′-5′-oligoadenylate synthetase 1/2), MX1 (myxovirus resistance protein 1/MxA), ISG15 (interferon stimulated protein 15), and CCL5 (chemokine C-C motif ligand 5). Other gene products stimulated by both type I and II IFNs include: STAT1/2 (signal transducer and activator of transcription 1/2), and CXCL9 (chemokine C-X-C motif ligand 9) (Carrero et al., 2014, Zhang et al., 2014, Carrero et al., 2013, Li et al., 2008). Expression of the aforementioned genes is suggestive of either an ongoing viral infection that elicited production of IFNs or an inherent defect in controlling IFN production.
In the literature, “IFN signature” and “type I IFN signature” have been used interchangeably, suggesting that the signature depends mainly on type I IFNs. It is noteworthy that the contribution of type II and III IFNs should not be discounted. Specifically, type III IFN-inducible genes have not been investigated as extensively as for type I/II IFNs, but appear to induce a signature almost identical to type I IFN (Domsgen et al., 2016, Egli et al., 2014, Donnelly and Kotenko, 2010). Type I, II and III IFNs are reported to use the JAK/STAT signalling pathway, and a recent study revealed that type I and III IFNs use redundant pathways for the induction of an antiviral response against influenza A virus (Crotta et al., 2013, Liu et al., 2012a, Ank et al., 2008). Still, plasmacytoid dendritic cells and epithelial cells appear to be the primary responders to type III IFNs, thus the affects of type III IFNs may be confined whereas those of type I IFNs would apply to almost all tissues (Odendall and Kagan, 2015, Ank et al., 2008, Sommereyns et al., 2008). Nonetheless, recent investigations with Coxsackievirus B3-infected human pancreatic islets in vitro have revealed expression of type I and III IFNs with an associated IFN signature (Domsgen et al., 2016). Therefore, this area warrants further investigations to decipher the involvement of type III IFNs in T1D.
Systemic Lupus Erythematosus (SLE) can be considered the prototypic platform for the study of inflammatory profiles associated with an IFN signature. SLE is an autoimmune disease hallmarked by overexpression of type I IFNs, specifically IFNα. Studies using a variety of methods such as microarrays, quantitative polymerase-chain reaction, and laser-capture isolation of kidney cells were instrumental in solidifying the idea of an IFN signature in SLE. These studies demonstrated the IFN signature both in the peripheral blood and the kidney of SLE patients but not control patients. (Peterson et al., 2004, Han et al., 2003, Baechler et al., 2003). Taken together, these clinical studies demonstrate an underlying inflammatory process not only systemically but also at the level of end-organ autoimmunity. Importantly, these studies in SLE also support the concept of inflammation as a crucial component for understanding the pathogenesis of additional autoimmune disorders such as T1D, beyond the presence of autoreactive lymphocytes.
3. Experimental Evidence for an Interferon Signature in T1D
IFNα is known to stimulate expression of class I major histocompatibility complex (MHC-I) molecules at the surface of exposed cells. Hyper-expression of MHC-I molecules on islets along with detection of IFNα in pancreases of T1D patients compared to non-diabetic patients was an early suggestion that IFNs may be pathogenic (Huang et al., 1995, Foulis et al., 1987b). This hypothesis was also supported by earlier experiments in mice and rats indicating the potential nefarious role of type I IFNs in mammals (Gresser et al., 1980). Since then, many teams have led investigations into the role of IFNs in the pathogenesis of T1D using both human samples and mouse models such as the non-obese diabetic (NOD) mouse. Consequently, T1D has been related to the IFN signature discussed above, further supporting a role for inflammation as an initial triggering event during T1D. However, it is noteworthy that human data for an interferon signature is limited and this is likely due to the limited expression in the microenvironment of the islet.
Comparisons of gene expression profiles in pancreatic lymph node CD4+ T cells of NOD mice (which spontaneously develop diabetes ~ 12 weeks of age) and NOD/BDC2.5 T cell receptor transgenic mice (where more than 90% of T cell receptors are islet-antigen reactive and the mice develop diabetes ~ 3 weeks of age) identified the up-regulation of IFN-stimulated genes in the mice. mRNA expression for IFN-stimulated genes included IFIT1, IFIT3, ISG15, and OAS1. Moreover, the up-regulated IFN-stimulated genes positively correlated with age of the mice where levels were higher in 6 week-old mice compared to 2 week-old mice (Li et al., 2008). Similarly, Planas et al. reported a data set of whole genome transcription profiles of human T1D pancreases and purified islets, which revealed an overall overexpression of both innate immunity and IFN-responsive genes (Planas et al., 2010). Even though the study by Planas and colleagues only included a small population (4 T1D patient pancreases and 7 non-T1D pancreases), the findings represented a valuable platform for organ-specific transcriptomic analysis in T1D and established a significant ground for additional large-scale investigations of locally relevant inflammation.
A study, by Diana et al., analyzing initiation of diabetes in the NOD mice and the non-autoimmune prone C57Bl/6 and BALB/c mice observed IFNα and IFNα-stimulated gene products in NOD mice only (Diana et al., 2013). Additionally, the study established plasmacytoid dendritic cells (pDCs) as crucial players in the IFN signature in NOD mice, since depletion of pDCs in 2 week-old NOD mice delayed development of diabetes up to 30 weeks. While Diana et al.'s work supports the case for IFN-α as a pivotal component for the IFN signature, we should underline that pDCs are also potent secretors of type III IFNs (Ank et al., 2008). Therefore, the positive effect of pDC depletion may not be explained solely through IFNα. Similarly, another study encompassing microarray analysis and qPCR examined chronological changes in gene expression in pancreatic islets of NOD mice from 2 weeks of age until development of diabetes ~ 20 weeks and up. To strengthen their examination, the researchers also used non-diabetic NOD.Rag1 −/− mice which lack a functional repertoire of adaptive T and B cells. The results demonstrated a unique age-correlated IFN signature in the NOD mice that was not present in the NOD.Rag1 −/−, implying that the absence of B and T cells negatively affects the induction of an IFN signature. Notably, at two weeks of age, there were no discernable differences in immune responses between NOD and NOD.Rag1 −/− mice. Overall, the study suggests a synergistic interaction between innate and adaptive arms of the immune response for the establishment of inflammation and an IFN signature overtime, attesting to the multifactorial complexity of T1D development (Carrero et al., 2013).
Importantly, Marro et al. used a viral mouse model to demonstrate that progression to T1D can be directly modulated through IFN-α signaling. In their model, Rip-LCMV, C57Bl/6 mice expressed a transgene of the lymphocytic choriomeningitis virus (LCMV) glycoprotein under the control of the rat insulin promoter only within the islets. Subsequent infection with LCMV led to the development of diabetes in these mice (Marro et al., 2017). Marro and colleagues showed that administration of antibodies against IFN- α-β receptor (IFNAR) and IFN-α species, but not IFN-β, prevented the virally-induced development of diabetes in the Rip-LCMV mice (Marro et al., 2017). These findings provided further support for the involvement of IFN driven inflammation in diabetes development.
Concurrently, two independent longitudinal studies have been following a cohort of German and Finnish children with genetically determined predisposition to T1D. These cohorts are part of the BABYDIET and DIPP studies, respectively. In the DIPP study, subjects (n = 28) are matched with autoantibody-negative controls based on date/place of birth, sex, and HLA genotype. The BABYDIET study includes 109 genetically susceptible children, along with 49 individuals recently diagnosed with T1D (disease duration of at least 3 years), 15 adult individuals with longstanding T1D, 93 healthy adult volunteers, and 25 patients with SLE. Within the course of these studies, peripheral blood is collected from participants and microarray analysis is performed on RNA from PBMCs or whole blood to determine transcriptional profiles. Both studies reported presence of an IFN signature before the detection of autoantibodies in the children. The number of subjects who manifested the IFN signature was small, likely owing to the localized nature of the process or affects of chronic IFN response as well as age-related changes. However, despite these differences and the fact magnitude of the IFN signature varied among the susceptible children, recent onset diabetic cases and individuals with longstanding diabetes, their overall IFN signature overlapped with what was already reported for SLE patients and the signature was not observed in healthy controls (Ferreira et al., 2014, Kallionpää et al., 2014).
The BABYDIET and DIPP cohorts constitute two epidemiologically relevant landmark studies that support the early involvement of an IFN response during T1D. Moreover, these reports of IFN response are directly in line with other studies demonstrating hyper-expression/dysregulation of MHC-I molecules in human pancreases during T1D (Foulis et al., 1987a). Specifically, Richardson et al. (2016) provided strong evidence for involvement of MHC-I molecules (both at the protein and the RNA level) and inflammatory pathway during T1D. Using human samples from nPOD (network for pancreatic organ donors with diabetes), the Diabetes Virus Detection study, and from a collection of recent-onset T1D samples from the United Kingdom, Richardson et al. demonstrated that classical and non-classical MHC-I hyper-expression is pertinent to patients with T1D but not the controls (Richardson et al., 2016). Marroqui and colleagues also reported comparable effects of IFN-α on MHC-I overexpression in human beta cells (Marroqui et al., 2017). Meanwhile, it is important to mention that Richardson et al.'s and Marroqui et al.'s studies are contradictory to an earlier report by Skog et al. (2015). Skog et al.'s investigations did not conclusively define any considerable differences in MHC-I expression between T1D patients and non-T1D patients (Skog et al., 2015). These discrepancies further attest to the multifaceted complexity of T1D development and may indeed be a testament to the heterogeneity of T1D presentation.
Finally, the concept that IFNs can play a crucial role in T1D is also supported by numerous reports of T1D development following IFN-α therapy for viral hepatitis, often associated with other autoimmune disorders as well. Some of these reports have recently been reviewed (Zornitzki et al., 2015). Altogether these aforementioned reports establish a strongly relevant basis for more extensive incorporation of inflammatory pathways in attempts to explain the pathogenesis of T1D and proposition of alternative treatment options.
4. Type 1 Interferonopathies and Link to T1D
4.1. Type 1 Interferonopathies
Type 1 interferonopathies (type 1 IFN-opathies) classically refer to a group of monogenic Mendelian disorders defined by a dysfunction in the innate immune system. In type 1 IFN-opathies, there is an overproduction of type I IFNs associated with signs of both autoinflammation and autoimmunity. Currently recognized type 1 IFN-opathies encompass mutations in 12 genes: TREX1, RNASEH2 A/B/C, SAMHD1, ADAR, TMEM173, ACP5, ISG15, DDX58, PSMB8, and IFIH1 (Crow and Manel, 2015, Lee-Kirsch et al., 2015). These mutations, summarized in Table 1, all include an inflammatory response, with a characteristic IFN signature that extends to autoimmune traits relevant to established autoimmune disorders such as T1D. Of particular relevance to T1D are mutations in IFIH1 which encodes the intracellular pattern recognition receptor (PRR), melanoma differentiation-associated protein 5 (MDA5). MDA5 has been reported to be expressed at low levels in human pancreatic islets (Hultcrantz et al., 2007) and polymorphisms in IFIH1 have previously been correlated with risk of T1D development (Cen et al., 2013, Winkler et al., 2011, Smyth et al., 2006). For example, the IFIH1 nonsynonymous single nucleotide polymorphism (SNP) rs1990760 genotype TT is considered a T1D risk genotype while the TC genotype is associated with protection from disease (Smyth et al., 2006). Recently, Domsgen et al. demonstrated modulation of the IFN signature in primary human pancreatic islets by rs1990760, as well as an involvement of type III IFNs following infection with Coxsackievirus B3 in vitro. Infection led to significantly increased expression of both type I IFNs (IFNβ) and type III IFNs (IFNλ1 and 2), and IFN-inducible genes such as CXCL-10, MxA, and MDA5 itself, independent of the rs1990760 genotype of donors when comparing infected islets to uninfected islets. However, pancreatic islets from donors heterozygous for the IFIH1 SNP genotype (TC) displayed significantly increased expression of type III IFN mRNA, as well as some IFN-inducible genes when compared to islets from donors homozygous for the IFIH1 SNP genotype (TT) (Domsgen et al., 2016). Also, our own research has shown that MDA5−/− mice on the NOD background are protected from spontaneous autoimmune diabetes, while MDA5+/− mice (that produced nearly 48% less MDA5 protein than wild-type MDA5+/+) have a reduced incidence of spontaneous disease when compared to MDA5+/+ mice. Additionally, Coxsackievirus B4-infected MDA5+/− mice were fully protected from virus-mediated diabetes while age-matched infected MDA5+/+ mice succumbed to disease with an incidence of 50% (Lincez et al., 2015).
Table 1.
Current identified single-gene mutations characterizing Type 1 interferonopathies.
| Gene | Description and effect of mutations |
|---|---|
| TREX1 | Encodes the 3′ repair exonuclease which recognizes cytosolic single-stranded-(ss)DNA molecules for degradation. Mutations in TREX1 are thought to lead to a build-up of ssDNA molecules in the cytosol which are perceived as a danger signal and hence stimulate the type 1 IFN response. |
| RNASEH2 A, RNASEH2 B, RNASEH2C | Encode the A, B, and C subunits of the ribonuclease H2 complex. Hypomorphic mutations in these genes negatively impact the ability of the RNASEH2 complex to degrade RNA that has been inappropriately inserted in DNA hybrids. The low-level activity of the RNASEH2 complex thus leads to a continuous loss of genomic integrity, and simultaneously continued DNA repair system whose metabolites are hypothesized to activate the type 1 IFN response. |
| SAMHD1 | Encode the SAM domain and HD domain containing protein 1 with reported dNTP triphosphohydrolase, triphosphatase, and nuclease activity. Mutations in SAMHD1 are thought to result in an imbalance in the cytosolic dNTP pool leading to genomic instability and a chronic DNA damage response similar to mutations in RNASEH2 genes. |
| ADAR | Encodes the adenosine deaminase, RNA-specific protein, whose role involves editing double-stranded(ds)-RNA species, thus rendering them immunosuppressive. Mutations in ADAR interfere with the ability of the protein to edit dsRNA species. Non-edited, immunoreactive dsRNA can subsequently activate the type 1 IFN response. |
| IFIH1 | Encodes the melanoma differentiation-associated protein 5 (MDA5). MDA5 is a cytosolic dsRNA sensor and gain of function mutations in IFIH1 lead to an increased affinity of MDA5 for dsRNA, hence amplifying the induction of the type 1 IFN response. |
| TMEM173 | Encodes the stimulator of interferon genes (STING) adaptor molecule. Gain of function mutations in TMEM173 result in the constitutive activation of STING even in the absence of its ligand cGAMP. That constitutive activation of STING ensures the continuous activation of the IFNβ promoter. |
| ACP5 | Encodes the tartrate-resistant acid phosphatase type 5 (TRAP5). Mutations in ACP5 are associated with the induction of the type 1 IFN response. |
| ISG15 | Encodes the interferon-stimulated protein 15. Mutations in ISG15 results in an enhancement of the type I IFN signalling. ISG15 has been reported to stabilize the Ubl carboxy-terminal hydrolase 18 (USP18), a known negative regulator of type I IFN-induced responses. |
| DDX58 | Encodes the retinoic-acid inducible gene 1 (RIG-1) protein that senses cytoplasmic dsRNA similar to MDA5. Also, resembling IFIH1 mutations, gain of function mutations in DDX58 cause constitutive activation of RIG-1 and continuous induction of the type I IFN response. |
| PSMB8 | Encodes the proteasome subunit β type 8 or subunit β5i. Although the specifics of PSMB8 activity in the type 1 IFN response remain elusive, mutations in PSMB8 associate with an interferon signature. |
Other examples of T1D risk genes or candidate genes that are also linked to the interferon pathway and responses to viral infections are TLR7, TLR8, and PSMB8 (Qiu et al., 2014, Cooper et al., 2009). Specifically, although PSMB8's exact role in the type 1 IFN response remains to be elucidated, mutations in PSMB8 have been associated with an IFN response, characteristic of a T 1 IFN-opathy (Liu et al., 2012b). Altogether, the results from these studies support the argument for using type 1 IFN-opathies as a reference point for the evaluation of inflammatory processes in T1D as well as the viral involvement in T1D.
Type 1 IFN-opathies have previously been proposed to overlap with SLE (Crow, 2003), and seeing that the IFN signature in SLE already overlaps with T1D, we propose to also extend type 1 IFN-opathies to T1D. Not all gene mutations in type 1 IFN-opathies show 100% penetrance, and the genotypes are for the most part recessive (Tüngler et al., 2014, Vogt et al., 2013). Such penetrance varieties may help explain the heterogeneity seen in T1D disease, why not all autoantibody positive individuals progress to T1D, or why there is a difference in magnitude of the IFN signature across T1D development. Studies examining IFN levels and/or IFN responsiveness may uncover individuals that will progress to T1D. Also, the overlay of virus infection may exacerbate inflammation already present in islets and help explain a rapid progression towards T1D before onset of clinical disease. There is also the option that combinations of genotypes that affect production and/or response to IFN may promote differential responses. Alternatively, the diversity in T1D may relate to the fact that among other factors yet to be determined, a certain threshold of IFN dysfunction must be reached. When assessing triggers of T1D initiation and the relationship to type 1 IFN-opathies, it would also be useful to investigate mutations in genes that control DNA activation and repression. For example, a recent paper showed that the methyl-CpG-binding protein Mbd2 is involved in priming DCs for a Th2 response (Cook et al., 2015). It is likely that Mbd2 and other epigenetic factors influence DC activation and their capacity to secrete IFNs. Hence, mutations in such crucial epigenetic markers would influence IFN activation and response pathways. Therefore, research into those mutations is likely to discover new players involved in establishment of an IFN signature, hence expanding the area of type 1 IFN-opathies and providing additional framework for the inflammatory pathways of T1D.
4.2. Viruses and Type 1 Interferonopathy in T1D
As mentioned earlier, incidence of T1D is rising worldwide and among environmental factors that may explain the positive trend, epidemiological studies have implicated viruses such as enteroviruses, mumps virus, and congenital rubella virus (Filippi and Von Herrath, 2008, Viskari et al., 2002). Enteroviruses, specifically Coxsackieviruses, are increasingly and more frequently being associated with T1D than any other virus. As early as 1969, researchers observed that the sera of recent onset T1D patients tended to present with higher antibody titers to Coxsackieviruses than patients with established T1D (more than 2 years) or healthy controls (Gamble et al., 1969). Although there lacks a definitive causal link between enteroviral infections and development of T1D in humans, a significant body of research is showing positive correlations between these viruses and disease. These correlations are supported by the detection of enteroviral RNA in both blood and pancreas of prediabetic, recent-onset, and established T1D patients and the presence of anti-enteroviral antibodies, as well as in vitro studies showing specific enteroviral tropism for pancreatic islet cells (Krogvold et al., 2015, Anagandula et al., 2014, Salvatoni et al., 2013, Oikarinen et al., 2011). Additionally, in vivo work with mouse models implicates Coxsackieviruses as a direct agent for the development and/or acceleration of diabetes onset (Serreze et al., 2005, Horwitz et al., 2004). Importantly, type I IFNs have been implicated in establishment of viral latency during Epstein-Barr virus infection, which is epidemiologically associated with the autoimmune disease, Multiple Sclerosis (Salamon et al., 2012, Xu et al., 2006). This therefore suggests a similar role for type I IFNs in maintenance of viral persistence and chronicity of infection in the pancreas, contributing to T1D. Alternatively, considering the natural history of Coxsackievirus infections, it is possible that multiple Coxsackievirus infections are harbored in the pancreas, leading to differential viral clearance and hence affecting the IFN signature itself.
Although there is some discussion as to how Coxsackieviruses might contribute to T1D, it has been proposed that one such mechanism may revolve around persistent infection of pancreatic beta cells (Jaïdane and Hober, 2008). More recently, researchers showed that naturally occurring 5′ terminal deletions in the genomes of Coxsackieviruses play key roles in the maintenance of persistence in heart and pancreas of humans and mice (Tracy et al., 2015, Chapman et al., 2008). Considering these findings, we are elaborating on the hypothesis of persistent viral infection as a mechanism for T1D pathogenesis. We propose that persistently infected beta cells constitute a local microenvironmental type 1 IFN-opathy during T1D, where persistent infection constitutes a continuous source of pathogen-associated molecular patterns (PAMPs) that promote host pattern recognition receptor (PRRs) signalling for the induction of interferon secretion (Zuniga et al., 2015). This is supported by previous work associating Coxsackievirus infection with:
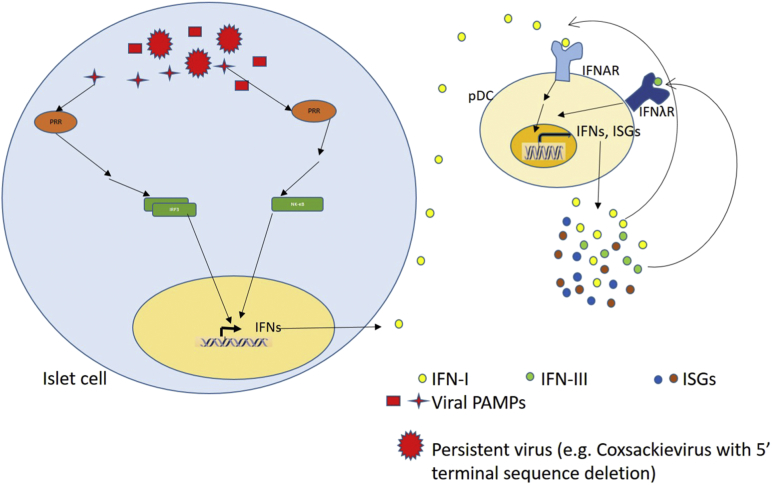
1) A continuous local production of type I IFNs (Hultcrantz et al., 2007, Huang et al., 1995) that can steadily activate resident monocytes, specifically pDCs which are themselves potent secretors of type I and III interferons. Local IFN signature in turn maintains a positive feedback loop of local inflammation with secretion of chemokines that recruit both anti-viral and self-reactive effector lymphocytes (Fig. 1).
Fig. 1.
Persistence of virus within islet cells maintains a continuous source of viral PAMPs which lead to the induction of type 1 and III IFN production. The secreted IFNs potentiate the activation of resident pDCs, hence creating a positive feedback for the secretion of more type I and III IFNs from the pDCs, as well as ISGs. Some of the ISGs serve as cytokines and chemokines to recruit T cells. The established type 1 IFN environment, in turn, helps T cells overcome the tolerance threshold to become pathogenic.
2) Persistent presence of virus within the beta cells leads to intrinsic changes that alter the resolution of IFN-induced responses over time. The alteration is likely to involve viral proteins interacting with cellular proteins that are known components of the IFN response. Therefore, when considering the pancreas as a local type 1 IFN-opathy during T1D, the currently known type 1 IFN-opathies provide researchers with a substantial list of candidate proteins that are likely modified by persistent infection. Hence, type 1 IFN-opathies represent a platform for the investigation and evaluation of already hypothesized mechanisms of viral involvement in T1D pathogenesis.
While presence of enteroviral proteins correlates with onset of T1D in some patients (Willcox et al., 2011), it is also established that type 1 IFN-opathies are reminiscent of congenital infections (Jepps et al., 2008, Sanchis et al., 2005). As such, an argument can be made for an existing axis between type 1 IFN-opathies, T1D, and viruses. Hence, at least in a subgroup of T1D patients, viral infections in the pancreas alter the profiles of gene products that actively contribute to establishment of an inflammatory response for the initiation and/or progression of clinical disease.
5. Type I IFNs and Type 1 IFN-Opathies: Implications for T cell Responses?
As previously mentioned, T1D is historically characterized as a T-cell mediated disease. Here, we extend our discussions to suggest that the established IFN response that precedes T1D onset, together with viral-induced type 1 IFN-opathy in the pancreas and microenvironment of the islet, serves to fine-tune subsequent T cell responses during T1D. Several lines of research have demonstrated correlations of IFNs and T cells, supporting our proposal. Specifically, concurrent publications by Crouse et al. and Xu et al. have shown that, within the context of viral infections in mice, signalling through the type I IFN receptor (IFNAR) promoted survival of CD8 T cells and prevented their killing by NK cells (Crouse et al., 2014, Xu et al., 2014). Importantly, a paper by Curtsinger et al. found that, in addition to influencing CD8 T cells through CD4 T helper cells and cytokines such as interleukin-5, type I IFNs could directly interact with CD8 T cells. Type I IFNs could provide the third signal (in addition to antigen stimulation, and costimulatory molecules) needed for complete CD8 T cell expansion and activation of cytolytic functions (Curtsinger et al., 2005). For example, when CD8T cells from OT-1 mice were exposed to antigen, costimulatory signals, and IFNα, there was increased clonal expansion, increased lytic activity, and increased interferon-gamma secretion by the CD8 T cells compared to antigen and costimulatory signals alone. Conversely, cells from OT-1/IFNAR-deficient mice had reduced functionality (Curtsinger et al., 2005). Together with the previously proven capacity of type I IFNs to promote survival of activated T cells (Marrack et al., 1999), Curtsinger et al.'s findings imply that persistent/chronic infections in the pancreas could be providing the continued IFN signal needed for autoreactive T cells to overcome the tolerance threshold and become pathogenic during T1D.
6. Treatment Avenues
Altogether, our review suggests a two-phase affect of virus and IFN in T1D. The first phase is the probable initiation or propagation of cellular immune responses including cytotoxic T lymphocyte and macrophage activation in the islet microenvironment, either initiated or expanded by virus and IFN response. In the second phase of T1D, after onset, there is a loss of cellular immunity but a persistence of beta cells with insulin as well as virus. When considering treatment options for T1D, use of anti-IFN receptor antibodies to remove IFN negative influences upon beta cell function could prove effective in targeting the second phase of disease development. Indeed, biological research and development companies have been conducting trials with a potential monoclonal antibody against the type I IFN receptor (IFNAR) for the treatment of lupus, with potential crossover to other autoimmune diseases such as T1D (Hultquist, 2015). While the prospect of using anti-IFNAR monoclonal antibodies is promising, considering persistent/chronic viral infections as contributors to establishment and/or pathogenesis of T1D provides exciting alternatives for more targeted treatment options, such as antivirals. One avenue of scrutiny involves the removal of virus/PAMPs to subvert signalling through host PRRs, hence interrupting continual production of IFNs and subsequent inflammatory processes. As research continues to demonstrate an association between viruses and T1D, investigating the use of antiviral treatments is increasingly proving necessary. Also, continued research into viral infections and the inflammatory pathways during T1D may provide a crucial platform for the consideration of off-target effects of vaccines and how vaccines can be used to prevent the development of T1D. Within the scope of vaccine development, a recent preclinical study by Larsson et al. demonstrated that a non-adjuvanted, formalin-inactivated prototype vaccine against Coxsackievirus B1 was indeed efficacious and safe in NOD mice. Vaccination led to significantly higher titers of anti-Coxsackievirus B1 neutralising antibodies, accompanied by significantly reduced viral titers in the blood and pancreas, when compared to mock-treatment. Importantly, while infection accelerated diabetes onset in non-vaccinated prediabetic NOD mice, there was no acceleration of disease in vaccinated mice (Larsson et al., 2015). Larsson et al.'s paper reinforces the association of enteroviruses like Coxsackieviruses with T1D but also supports the proposition for the removal of virus/PAMPS in investigating treatment options for T1D.
Additionally, establishing an archive of type 1 IFN-opathy related genes would be useful in studying the molecular mechanisms that drive inflammation in T1D. The data will be specifically relevant at the level of the pancreas in terms of determining events that sustain the loss of beta cells. We also envisage that insights into the single gene mutations will dictate the development of more specific monoclonal antibodies, targeting proteins upstream of IFN production as well as upstream proteins in IFN-signalling. Such new therapeutics will allow a more directed treatment approach.
7. Search Strategy and Selection Criteria
Data for this review were identified through PubMed and references from relevant articles using the search terms “interferon signature”, “type 1 interferon signature”, “type 1 diabetes”, “Coxsackieviruses”, “type 1 interferonopathy”, “autoimmune diseases”. Articles between 1969 and 2016 were included, with emphasis on those published in the last 5 years as appropriate.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Acknowledgements
We would like to thank Michelle Krakowski for critical reading and discussion of the manuscript. We would also like to think other colleagues whose work facilitated the discussion and structure of this review, but were nonetheless not cited here due to restrictions on the number of references.
References
- Anagandula M., Richardson S.J., Oberste M.S., Sioofy-Khojine A.-B., Hyöty H., Morgan N.G., Korsgren O., Frisk G. Infection of human islets of Langerhans with two strains of Coxsackie B virus serotype 1: assessment of virus replication, degree of cell death and induction of genes involved in the innate immunity pathway. J. Med. Virol. 2014;86:1402–1411. doi: 10.1002/jmv.23835. [DOI] [PubMed] [Google Scholar]
- Ank N., Iversen M.B., Bartholdy C., Staeheli P., Hartmann R., Jensen U.B., Dagnaes-Hansen F., Thomsen A.R., Chen Z., Haugen H., Klucher K., Paludan S.R. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J. Immunol. 2008;180:2474–2485. doi: 10.4049/jimmunol.180.4.2474. [DOI] [PubMed] [Google Scholar]
- Arvan P., Pietropaolo M., Ostrov D., Rhodes C.J. Islet autoantigens: structure, function, localization, and regulation. Cold Spring Harb. Perspect. Med. 2012;2 doi: 10.1101/cshperspect.a007658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baechler E.C., Batliwalla F.M., Karypis G., Gaffney P.M., Ortmann W.A., Espe K.J., Shark K.B., Grande W.J., Hughes K.M., Kapur V., Gregersen P.K., Behrens T.W. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc. Natl. Acad. Sci. U. S. A. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry G., Waldner H. Accelerated type 1 diabetes induction in mice by adoptive transfer of diabetogenic CD4 + T cells. J. Vis. Exp. 2013:e50389. doi: 10.3791/50389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero J.A., Calderon B., Towfic F., Artyomov M.N., Unanue E.R. Defining the transcriptional and cellular landscape of type 1 diabetes in the NOD mouse. PLoS One. 2013;8:e59701. doi: 10.1371/journal.pone.0059701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero J.A., Calderon B., Towfic F., Artyomov M.N., Unanue E.R. Correction: defining the transcriptional and cellular landscape of Type 1 Diabetes in the NOD mouse. PLoS One. 2014;9 doi: 10.1371/journal.pone.0059701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen H., Wang W., Leng R.X., Wang T.Y., Pan H.F., Fan Y.G., Wang B., Ye, D. Q. Association of IFIH1 rs1990760 polymorphism with susceptibility to autoimmune diseases: a meta-analysis. Autoimmunity. 2013;46:455–462. doi: 10.3109/08916934.2013.796937. [DOI] [PubMed] [Google Scholar]
- Chapman N.M., Kim K.-S., Drescher K.M., Oka K., Tracy S. 5′ terminal deletions in the genome of a coxsackievirus B2 strain occurred naturally in human heart. Virology. 2008;375:480–491. doi: 10.1016/j.virol.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffersson G., Rodriguez-Calvo T., von Herrath M. Recent advances in understanding Type 1 Diabetes [version 1; referees: 2 approved] F1000Res. 2016 doi: 10.12688/f1000research.7356.1. 5 (F1000 Faculty Rev):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P.C., Owen H., Deaton A.M., Borger J.G., Brown S.L., Clouaire T., Jones G.-R., Jones L.H., Lundie R.J., Marley A.K., Morrison V.L., Phythian-Adams A.T., Wachter E., Webb L.M., Sutherland T.E., Thomas G.D., Grainger J.R., Selfridge J., Mckenzie A.N.J., Allen J.E., Fagerholm S.C., Maizels R.M., Ivens A.C., Bird A., Macdonald A.S. A dominant role for the methyl-CpG-binding protein Mbd2 in controlling Th2 induction by dendritic cells. Nat. Commun. 2015;6:6920. doi: 10.1038/ncomms7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J.D., Walker N.M., Smyth D.J., Downes K., Healy B.C., Todd J.A., Type I.D.G.C. Follow-up of 1715 SNPs from the Wellcome Trust Case Control Consortium genome-wide association study in type I diabetes families. Genes Immun. 2009;10(Suppl. 1):S85–S94. doi: 10.1038/gene.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotta S., Davidson S., Mahlakoiv T., Desmet C.J., Buckwalter M.R., Albert M.L., Staeheli P., Wack A. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 2013;9:e1003773. doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse J., Bedenikovic G., Wiesel M., Ibberson M., Xenarios I., Von Laer D., Kalinke U., Vivier E., Jonjic S., Oxenius A. Type I interferons protect T cells against NK cell attack mediated by the activating receptor NCR1. Immunity. 2014;40:961–973. doi: 10.1016/j.immuni.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Crow Y.J. Cree encephalitis is allelic with Aicardi-Goutieres syndrome: implications for the pathogenesis of disorders of interferon alpha metabolism. J. Med. Genet. 2003;40:183–187. doi: 10.1136/jmg.40.3.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow Y.J., Manel N. Aicardi–Goutières syndrome and the type I interferonopathies. Nat. Rev. Immunol. 2015;15:429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- Curtsinger J.M., Valenzuela J.O., Agarwal P., Lins D., Mescher M.F. Type I IFNs provide a third signal to CD8 T cells to stimulate clonal expansion and differentiation. J. Immunol. 2005;174:4465–4469. doi: 10.4049/jimmunol.174.8.4465. [DOI] [PubMed] [Google Scholar]
- Diana J., Simoni Y., Furio L., Beaudoin L., Agerberth B., Barrat F., Lehuen A. Crosstalk between neutrophils, B-1a cells and plasmacytoid dendritic cells initiates autoimmune diabetes. Nat. Med. 2013;19:65–73. doi: 10.1038/nm.3042. [DOI] [PubMed] [Google Scholar]
- Domsgen E., Lind K., Kong L., Huhn M.H., Rasool O., Van Kuppeveld F., Korsgren O., Lahesmaa R., Flodstrom-Tullberg M. An IFIH1 gene polymorphism associated with risk for autoimmunity regulates canonical antiviral defence pathways in Coxsackievirus infected human pancreatic islets. Sci Rep. 2016;6:39378. doi: 10.1038/srep39378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly R.P., Kotenko S.V. Interferon-Lambda: a new addition to an old family. J. Interf. Cytokine Res. 2010;30:555–564. doi: 10.1089/jir.2010.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli A., Santer D.M., O'shea D., Tyrrell D.L., Houghton M. The impact of the interferon-lambda family on the innate and adaptive immune response to viral infections. Emerg. Microbes & Infect. 2014;3:e51. doi: 10.1038/emi.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R.C., Guo H., Coulson R.M.R., Smyth D.J., Pekalski M.L., Burren O.S., Cutler A.J., Doecke J.D., Flint S., Mckinney E.F., Lyons P.A., Smith K.G.C., Achenbach P., Beyerlein A., Dunger D.B., Clayton D.G., Wicker L.S., Todd J.A., Bonifacio E., Wallace C., Ziegler A.G. A type I interferon transcriptional signature precedes autoimmunity in children genetically at risk for type 1 diabetes. Diabetes. 2014;63:2538–2550. doi: 10.2337/db13-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiano A.J., Xu Y., Tustison N.J., Marsh R.L., Baker W., Smirnov I., Overall C.C., Gadani S.P., Turner S.D., Weng Z., Peerzade S.N., Chen H., Lee K.S., Scott M.M., Beenhakker M.P., Litvak V., Kipnis J. Unexpected role of interferon-gamma in regulating neuronal connectivity and social behaviour. Nature. 2016;535:425–429. doi: 10.1038/nature18626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi C.M., Von Herrath M.G. Viral trigger for type 1 diabetes: pros and cons. Diabetes. 2008;57:2863–2871. doi: 10.2337/db07-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulis A.K., Farquharson M.A., Hardman R. Aberrant expression of class II major histocompatibility complex molecules by B cells and hyperexpression of class I major histocompatibility complex molecules by insulin containing islets in type 1 (insulin-dependent) diabetes mellitus. Diabetologia. 1987;30:333–343. doi: 10.1007/BF00299027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulis A.K., Farquharson M.A., Meager A. Immunoreactive alpha-interferon in insulin-secreting beta cells in type 1 diabetes mellitus. Lancet (London, England) 1987;2:1423–1427. doi: 10.1016/s0140-6736(87)91128-7. [DOI] [PubMed] [Google Scholar]
- Gamble D.R., Kinsley M.L., Fitzgerald M.G., Bolton R., Taylor K.W. Viral antibodies in diabetes mellitus. Br. Med. J. 1969;3:627–630. doi: 10.1136/bmj.3.5671.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie K.M. Type 1 diabetes: pathogenesis and prevention. CMAJ. 2006;175:165–170. doi: 10.1503/cmaj.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresser I., Morel-Maroger L., Rivière Y., Guillon J.C., Tovey M.G., Woodrow D., Sloper J.C., Moss J. Interferon-induced disease in mice and rats. Ann. N. Y. Acad. Sci. 1980;350:12–20. doi: 10.1111/j.1749-6632.1980.tb20602.x. [DOI] [PubMed] [Google Scholar]
- Han G.M., Chen S.L., Shen N., Ye, S., Bao, C. D. & Gu, Y. Y. Analysis of gene expression profiles in human systemic lupus erythematosus using oligonucleotide microarray. Genes Immun. 2003;4:177–186. doi: 10.1038/sj.gene.6363966. [DOI] [PubMed] [Google Scholar]
- Horwitz M.S., Ilic A., Fine C., Balasa B., Sarvetnick N. Coxsackieviral-mediated diabetes: induction requires antigen-presenting cells and is accompanied by phagocytosis of beta cells. Clin. Immunol. 2004;110:134–144. doi: 10.1016/j.clim.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Huang X., Yuang J., Goddard A., Foulis A., James R.F., Lernmark A., Pujol-Borrell R., Rabinovitch A., Somoza N., Stewart T.A. Interferon expression in the pancreases of patients with type I diabetes. Diabetes. 1995;44:658–664. doi: 10.2337/diab.44.6.658. [DOI] [PubMed] [Google Scholar]
- Hultcrantz M., Hühn M.H., Wolf M., Olsson A., Jacobson S., Williams B.R., Korsgren O., Flodström-Tullberg M. Interferons induce an antiviral state in human pancreatic islet cells. Virology. 2007;367:92–101. doi: 10.1016/j.virol.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Hultquist M. Lupus treatment: new hope for lupus patients. 2015. https://www.medimmune.com/about-us/media/new-hope-for-lupus-patients-20150811.html [Online]. Available: (Accessed March 10 2017)
- Jaïdane H., Hober D. Role of coxsackievirus B4 in the pathogenesis of type 1 diabetes. Diabete Metab. 2008;34:537–548. doi: 10.1016/j.diabet.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Jepps H., Seal S., Hattingh L., Crow Y.J. The neonatal form of Aicardi-Goutières syndrome masquerading as congenital infection. Early Hum. Dev. 2008;84:783–785. doi: 10.1016/j.earlhumdev.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Kallionpää H., Elo L.L., Laajala E., Mykkänen J., Ricaño-Ponce I., Vaarma M., Laajala T.D., Hyöty H., Ilonen J., Veijola R., Simell T., Wijmenga C., Knip M., Lähdesmäki H., Simell O., Lahesmaa R. Innate immune activity is detected prior to seroconversion in children with HLA-conferred type 1 diabetes susceptibility. Diabetes. 2014;63:2402–2414. doi: 10.2337/db13-1775. [DOI] [PubMed] [Google Scholar]
- Kelly M.A., Rayner M.L., Mijovic C.H., Barnett A.H. Molecular aspects of type 1 diabetes. Mol. Pathol. 2003;56:1–10. doi: 10.1136/mp.56.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogvold L., Edwin B., Buanes T., Frisk G., Skog O., Anagandula M., Korsgren O., Undlien D., Eike M.C., Richardson S.J., Leete P., Morgan N.G., Oikarinen S., Oikarinen M., Laiho J.E., Hyöty H., Ludvigsson J., Hanssen K.F., Dahl-Jørgensen K. Detection of a low-grade enteroviral infection in the islets of langerhans of living patients newly diagnosed with type 1 diabetes. Diabetes. 2015;64:1682–1687. doi: 10.2337/db14-1370. [DOI] [PubMed] [Google Scholar]
- Kulmala P., Savola K., Petersen J.S., Vähäsalo P., Karjalainen J., Löppönen T., Dyrberg T., Akerblom H.K., Knip M. Prediction of insulin-dependent diabetes mellitus in siblings of children with diabetes. A population-based study. The Childhood Diabetes in Finland Study Group. J. Clin. Invest. 1998;101:327–336. doi: 10.1172/JCI119879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen O.H., Honkanen H., Pakkanen O., Oikarinen S., Hankaniemi M.M., Huhtala H., Ruokoranta T., Lecouturier V., André P., Harju R., Virtanen S.M., Lehtonen J., Almond J.W., Simell T., Simell O., Ilonen J., Veijola R., Knip M., Hyöty H. Coxsackievirus B1 is associated with induction of β-cell autoimmunity that portends type 1 diabetes. Diabetes. 2014;63:446–455. doi: 10.2337/db13-0619. [DOI] [PubMed] [Google Scholar]
- Larsson P.G., Lakshmikanth T., Laitinen O.H., Utorova R., Jacobson S., Oikarinen M., Domsgen E., Koivunen M.R., Chaux P., Devard N., Lecouturier V., Almond J., Knip M., Hyoty H., Flodstrom-Tullberg M. A preclinical study on the efficacy and safety of a new vaccine against Coxsackievirus B1 reveals no risk for accelerated diabetes development in mouse models. Diabetologia. 2015;58:346–354. doi: 10.1007/s00125-014-3436-0. [DOI] [PubMed] [Google Scholar]
- Lee-Kirsch M.A., Wolf C., Kretschmer S., Roers A. Type I interferonopathies—an expanding disease spectrum of immunodysregulation. Semin. Immunopathol. 2015;37:349–357. doi: 10.1007/s00281-015-0500-x. [DOI] [PubMed] [Google Scholar]
- Li Q., Xu B., Michie S.A., Rubins K.H., Schreriber R.D., Mcdevitt H.O. Interferon- initiates type 1 diabetes in nonobese diabetic mice. Proc. Natl. Acad. Sci. 2008;105:12439–12444. doi: 10.1073/pnas.0806439105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Song L.-J., Qin X.-Y. Advances in the cellular immunological pathogenesis of type 1 diabetes. J. Cell. Mol. Med. 2014;18:749–758. doi: 10.1111/jcmm.12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincez P.J., Shanina I., Horwitz M.S. Reduced expression of the MDA5 Gene IFIH1 prevents autoimmune diabetes. Diabetes. 2015;64:2184–2193. doi: 10.2337/db14-1223. [DOI] [PubMed] [Google Scholar]
- Liu M.-Q., Zhou D.-J., Wang X., Zhou W., Ye L., Li J.-L., Wang Y.-Z., Ho W.-Z. IFN-λ3 inhibits HIV infection of macrophages through the JAK-STAT pathway. PLoS One. 2012;7:e35902. doi: 10.1371/journal.pone.0035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ramot Y., Torrelo A., Paller A.S., Si N., Babay S., Kim P.W., Sheikh A., Lee C.-C.R., Chen Y., Vera A., Zhang X., Goldbach-Mansky R., Zlotogorski A. Mutations in proteasome subunit β type 8 cause chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature with evidence of genetic and phenotypic heterogeneity. Arthritis Rheum. 2012;64:895–907. doi: 10.1002/art.33368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrack P., Kappler J., Mitchell T. Type I interferons keep activated T cells alive. J. Exp. Med. 1999;189:521–530. doi: 10.1084/jem.189.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marro B.S., Ware B.C., Zak J., De La Torre J.C., Rosen H., Oldstone M.B. Progression of type 1 diabetes from the prediabetic stage is controlled by interferon-alpha signaling. Proc. Natl. Acad. Sci. U. S. A. 2017;114:3708–3713. doi: 10.1073/pnas.1700878114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marroqui L., Dos Santos R.S., Op De Beeck, A., Coomans De Brachene, A., Marselli, L., Marchetti, P. & Eizirik, D. L. Interferon-alpha mediates human beta cell HLA class I overexpression, endoplasmic reticulum stress and apoptosis, three hallmarks of early human type 1 diabetes. Diabetologia. 2017;60:656–667. doi: 10.1007/s00125-016-4201-3. [DOI] [PubMed] [Google Scholar]
- Odendall C., Kagan J.C. The unique regulation and functions of type III interferons in antiviral immunity. Curr. Opin. Virol. 2015;12:47–52. doi: 10.1016/j.coviro.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikarinen S., Martiskainen M., Tauriainen S., Huhtala H., Ilonen J., Veijola R., Simell O., Knip M., Hyöty H. Enterovirus RNA in blood is linked to the development of type 1 diabetes. Diabetes. 2011;60:276–279. doi: 10.2337/db10-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson C.C., Dahlquist G.G., Gyürüs E., Green A., Soltész G. Incidence trends for childhood type 1 diabetes in Europe during 1989–2003 and predicted new cases 2005–20: a multicentre prospective registration study. Lancet. 2009;373:2027–2033. doi: 10.1016/S0140-6736(09)60568-7. [DOI] [PubMed] [Google Scholar]
- Peterson K.S., Huang J.-F., Zhu J., D'agati V., Liu X., Miller N., Erlander M.G., Jackson M.R., Winchester R.J. Characterization of heterogeneity in the molecular pathogenesis of lupus nephritis from transcriptional profiles of laser-captured glomeruli. J. Clin. Invest. 2004;113:1722–1733. doi: 10.1172/JCI19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas R., Carrillo J., Sanchez A., De Villa M.C.R., Nuñez F., Verdaguer J., James R.F.L., Pujol-Borrell R., Vives-Pi, M. Gene expression profiles for the human pancreas and purified islets in type 1 diabetes: new findings at clinical onset and in long-standing diabetes. Clin. Exp. Immunol. 2010;159:23–44. doi: 10.1111/j.1365-2249.2009.04053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y.H., Deng F.Y., Li M.J., Lei S.F. Identification of novel risk genes associated with type 1 diabetes mellitus using a genome-wide gene-based association analysis. J. Diabetes Investig. 2014;5:649–656. doi: 10.1111/jdi.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynier F., Pachot A., Paye M., Xu Q., Turrel-Davin F., Petit F., Hot A., Auffray C., Bendelac N., Nicolino M., Mougin B., Thivolet C. Specific gene expression signature associated with development of autoimmune type-I diabetes using whole-blood microarray analysis. Genes Immun. 2010;11:269–278. doi: 10.1038/gene.2009.112. [DOI] [PubMed] [Google Scholar]
- Richardson S.J., Horwitz M.S. Is type 1 diabetes “going viral”? Diabetes. 2014;63:2203–2205. doi: 10.2337/db14-0510. [DOI] [PubMed] [Google Scholar]
- Richardson S.J., Rodriguez-Calvo T., Gerling I.C., Mathews C.E., Kaddis J.S., Russell M.A., Zeissler M., Leete P., Krogvold L., Dahl-Jorgensen K., Von Herrath M., Pugliese A., Atkinson M.A., Morgan N.G. Islet cell hyperexpression of HLA class I antigens: a defining feature in type 1 diabetes. Diabetologia. 2016;59:2448–2458. doi: 10.1007/s00125-016-4067-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamon D., Adori M., Ujvari D., Wu L., Kis L.L., Madapura H.S., Nagy N., Klein G., Klein E. Latency type-dependent modulation of Epstein-Barr virus-encoded latent membrane protein 1 expression by type I interferons in B cells. J. Virol. 2012;86:4701–4707. doi: 10.1128/JVI.06829-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatoni A., Baj A., Bianchi G., Federico G., Colombo M., Toniolo A. Intrafamilial spread of enterovirus infections at the clinical onset of type 1 diabetes. Pediatr. Diabetes. 2013;14:407–416. doi: 10.1111/pedi.12056. [DOI] [PubMed] [Google Scholar]
- Sanchis A., Cerveró L., Bataller A., Tortajada J.L., Huguet J., Crow Y.J., Ali M., Higuet L.J., Martínez-Frías M.L. Genetic syndromes mimic congenital infections. J. Pediatr. 2005;146:701–705. doi: 10.1016/j.jpeds.2005.01.033. [DOI] [PubMed] [Google Scholar]
- Serreze D.V., Ottendorfer E.W., Ellis T.M., Gauntt C.J., Atkinson M.A. Acceleration of type 1 diabetes by a coxsackievirus infection requires a preexisting critical mass of autoreactive T-cells in pancreatic islets. Diabetes. 2000;49:708–711. doi: 10.2337/diabetes.49.5.708. [DOI] [PubMed] [Google Scholar]
- Serreze D.V., Wasserfall C., Ottendorfer E.W., Stalvey M., Pierce M.A., Gauntt C., O'donnell B., Flanagan J.B., Campbell-Thompson M., Ellis T.M., Atkinson M.A. Diabetes acceleration or prevention by a coxsackievirus B4 infection: critical requirements for both interleukin-4 and gamma interferon. J. Virol. 2005;79:1045–1052. doi: 10.1128/JVI.79.2.1045-1052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T.E., Kuestner R., Garrigues U., Birks C., Roraback J., Ostrander C., Dong D., Shin J., Presnell S., Fox B., Haldeman B., Cooper E., Taft D., Gilbert T., Grant F.J., Tackett M., Krivan W., Mcknight G., Clegg C., Foster D., Klucher K.M. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- Skog O., Korsgren S., Wiberg A., Danielsson A., Edwin B., Buanes T., Krogvold L., Korsgren O., Dahl-Jorgensen K. Expression of human leukocyte antigen class I in endocrine and exocrine pancreatic tissue at onset of type 1 diabetes. Am. J. Pathol. 2015;185:129–138. doi: 10.1016/j.ajpath.2014.09.004. [DOI] [PubMed] [Google Scholar]
- Smyth D.J., Cooper J.D., Bailey R., Field S., Burren O., Smink L.J., Guja C., Ionescu-Tirgoviste C., Widmer B., Dunger D.B., Savage D.A., Walker N.M., Clayton D.G., Todd J.A. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat. Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- Sommereyns C., Paul S., Staeheli P., Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro J.R., Ng C., Lee A.M., Sullivan B.M., Sheehan K.C., Welch M., Schreiber R.D., De La Torre J.C., Oldstone M.B. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 2013;340:207–211. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy S., Smithee S., Alhazmi A., Chapman N. Coxsackievirus can persist in murine pancreas by deletion of 5′ terminal genomic sequences. J. Med. Virol. 2015;87:240–247. doi: 10.1002/jmv.24039. [DOI] [PubMed] [Google Scholar]
- Tüngler V., Schmidt F., Hieronimus S., Reyes-Velasco C., Lee-Kirsch M.A. Phenotypic variability in a family with aicardi-gouti& #232; res syndrome due to the common A177T & lt; i& gt; RNASEH2B& lt; /i& gt; mutation. Case Rep. Clin. Med. 2014;03:153–156. [Google Scholar]
- Tuomilehto J. The emerging global epidemic of type 1 diabetes. Curr. Diabetes Rep. 2013;13:795–804. doi: 10.1007/s11892-013-0433-5. [DOI] [PubMed] [Google Scholar]
- Viskari H.R., Roivainen M., Reunanen A., Pitkäniemi J., Sadeharju K., Koskela P., Hovi T., Leinikki P., Vilja P., Tuomilehto J., Hyöty H. Maternal first-trimester enterovirus infection and future risk of type 1 diabetes in the exposed fetus. Diabetes. 2002;51:2568–2571. doi: 10.2337/diabetes.51.8.2568. [DOI] [PubMed] [Google Scholar]
- Vogt J., Agrawal S., Ibrahim Z., Southwood T.R., Philip S., Macpherson L., Bhole M.V., Crow Y.J., Oley C. Striking intrafamilial phenotypic variability in Aicardi-Goutières syndrome associated with the recurrent Asian founder mutation in RNASEH2C. Am. J. Med. Genet. Part A. 2013;161A:338–342. doi: 10.1002/ajmg.a.35712. [DOI] [PubMed] [Google Scholar]
- Willcox A., Richardson S.J., Bone A.J., Foulis A.K., Morgan N.G. Immunohistochemical analysis of the relationship between islet cell proliferation and the production of the enteroviral capsid protein, VP1, in the islets of patients with recent-onset type 1 diabetes. Diabetologia. 2011;54:2417–2420. doi: 10.1007/s00125-011-2192-7. [DOI] [PubMed] [Google Scholar]
- Winkler C., Lauber C., Adler K., Grallert H., Illig T., Ziegler A.G., Bonifacio E. An interferon-induced helicase (IFIH1) gene polymorphism associates with different rates of progression from autoimmunity to type 1 diabetes. Diabetes. 2011;60:685–690. doi: 10.2337/db10-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Brumm K., Zhang L. The latent membrane protein 1 of Epstein-Barr virus (EBV) primes EBV latency cells for type I interferon production. J. Biol. Chem. 2006;281:9163–9169. doi: 10.1074/jbc.M511884200. [DOI] [PubMed] [Google Scholar]
- Xu H.C., Grusdat M., Pandyra A.A., Polz R., Huang J., Sharma P., Deenen R., Kohrer K., Rahbar R., Diefenbach A., Gibbert K., Lohning M., Hocker L., Waibler Z., Haussinger D., Mak T.W., Ohashi P.S., Lang K.S., Lang P.A. Type I interferon protects antiviral CD8 + T cells from NK cell cytotoxicity. Immunity. 2014;40:949–960. doi: 10.1016/j.immuni.2014.05.004. [DOI] [PubMed] [Google Scholar]
- Zhang X., Bogunovic D., Payelle-Brogard B., Francois-Newton V., Speer S.D., Yuan C., Volpi S., Li Z., Sanal O., Mansouri D., Tezcan I., Rice G.I., Chen C., Mansouri N., Mahdaviani S.A., Itan Y., Boisson B., Okada S., Zeng L., Wang X., Jiang H., Liu W., Han T., Liu D., Ma T., Wang B., Liu M., Liu J.-Y., Wang Q.K., Yalnizoglu D., Radoshevich L., Uzé G., Gros P., Rozenberg F., Zhang S.-Y., Jouanguy E., Bustamante J., García-Sastre A., Abel L., Lebon P., Notarangelo L.D., Crow Y.J., Boisson-Dupuis S., Casanova J.-L., Pellegrini S. Human intracellular ISG15 prevents interferon-α/β over-amplification and auto-inflammation. Nature. 2014;517:89–93. doi: 10.1038/nature13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zornitzki T., Malnick S., Lysyy L., Knobler H. Interferon therapy in hepatitis C leading to chronic type 1 diabetes. World J. Gastroenterol. 2015;21:233–239. doi: 10.3748/wjg.v21.i1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga E.I., Macal M., Lewis G.M., Harker J.A. Innate and adaptive immune regulation during chronic viral infections. Annu Rev Virol. 2015;2:573–597. doi: 10.1146/annurev-virology-100114-055226. [DOI] [PMC free article] [PubMed] [Google Scholar]