Abstract
Genome-Wide Association Studies (GWAS) and subsequent fine-mapping studies (> 50) have implicated single nucleotide polymorphisms (SNPs) located at the CCDC170/C6ORF97-ESR1 locus (6q25.1) as being associated with the risk of breast cancer. Surprisingly, our analysis using genome-wide differential allele-specific expression (DASE), an indicator for breast cancer susceptibility, suggested that the genetic alterations of CCDC170, but not ESR1, account for GWAS-associated breast cancer risk at this locus. Breast cancer-associated CCDC170 nonsense mutations and rearrangements have also been detected, with the latter being specifically implicated in driving breast cancer. Here we report that the wild type CCDC170 protein localizes to the region of the Golgi apparatus and binds Golgi-associated microtubules (MTs), and that breast cancer-linked truncations of CCDC170 result in loss of Golgi localization. Overexpression of wild type CCDC170 triggers Golgi reorganization, and enhances Golgi-associated MT stabilization and acetyltransferase ATAT1-dependent α-tubulin acetylation. Golgi-derived MTs regulate cellular polarity and motility, and we provide evidence that dysregulation of CCDC170 affects polarized cell migration. Taken together, our findings demonstrate that CCDC170 plays an essential role in Golgi-associated MT organization and stabilization, and implicate a mechanism for how perturbations in the CCDC170 gene may contribute to the hallmark changes in cell polarity and motility seen in breast cancer.
Keywords: CCDC170/C6ORF97, Golgi-associated microtubules, Tubulin acetylation, Polarized cell migration, Genome-wide association studies (GWAS), Differential allele specific expression (DASE)
Highlights
-
•
The CCDC170 protein localizes to the Golgi apparatus, and breast cancer-associated truncations result in loss of Golgi association.
-
•
The CCDC170 protein binds and stabilizes Golgi-associated perinuclear microtubules and contributes to Golgi organization.
-
•
Dysregulation of CCDC170 affects polarized cell migration by altering the Golgi-associated microtubule network.
A large body of evidence has suggested that mutations at the CCDC170 gene impact both breast cancer risk and progression. We determined that the protein encoded by the CCDC170 gene localizes to a cellular structure called the Golgi apparatus, and that mutations alter such localization. At the Golgi location, CCDC170 is able to attach to the cell framework (microtubules) that controls cell movement directions by specifying the front and back edges of the cells. Our findings support a mechanism whereby breast cancer-associated mutations in the CCDC170 gene may contribute to the hallmark changes in cell migration seen in breast cancer.
1. Introduction
Advances in single nucleotide polymorphism (SNP) array technologies and Next Generation Sequencing have made it practical to perform Genome-Wide Association Studies (GWAS), an unbiased genomic approach to identify genetic factors that account for cancer susceptibility. Following initial GWAS (Zheng et al., 2009, Turnbull et al., 2010), subsequent fine-mapping studies (> 50) have implicated genetic variants located at the CCDC170/C6ORF97-ESR1 locus (6q25.1) as being associated with the risk of breast cancer. Estrogen receptor α (ERα), the protein encoded by the ESR1 gene, binds to estrogen, and the estrogen-ERα axis promotes the growth of breast epithelial cells and thereby contributes to breast cancer risk (Ali and Coombes, 2000). It is therefore logical to hypothesize that the breast cancer-associated SNPs at the CCDC170/C6ORF97-ESR1 locus impact function of the ESR1 gene (Hein et al., 2012, Koller et al., 2013, Paternoster et al., 2013, Yang et al., 2013). However, thus far, few studies have identified any strong causal variants regulating ESR1 function or expression (Cai et al., 2011, Stacey et al., 2010). Interestingly, the CCDC170-ESR1 intergenic rs2146210 SNP was found to have stronger risk-association in ER- breast tumors than those in ER + breast tumors, which suggests that this risk variant is likely ESR1-independent (Hein et al., 2012, Mulligan et al., 2011, Stacey et al., 2010, Zheng et al., 2009).
Nonsense mutations (e.g. p.E48* and p.Q405*) in the CCDC170 gene have been reported in sporadic breast cancer and other cancers by both the Cancer Genome Atlas (TCGA) and Cancer Genome Project. Importantly, several tumor-specific gene rearrangements from the second noncoding exon of ESR1 to the sixth and/or seventh coding exon(s) of CCDC170 were also reported by several studies using high-throughput RNA-seq (Robinson et al., 2011, Sakarya et al., 2012, Veeraraghavan et al., 2014). This ESR1-CCDC170 gene arrangement presents in ~ 14% of ER + breast cancer and could be one of the most important recurrent gene fusions in breast cancer (Veeraraghavan et al., 2014). This recent study by Veeraghavan et al. demonstrated that N-terminally truncated CCDC170 proteins were produced as a result of this ESR1-CCDC170 rearrangement (Veeraraghavan et al., 2014). Ectopic expression of these truncated proteins increased breast cancer cell motility and enhanced the transformation of normal mammary epithelial cells (MECs) (Veeraraghavan et al., 2014), indicating the important role of CCDC170 gene abnormalities in breast cancer initiation and/or progression. Taken together, the findings from GWAS, TCGA, cell culture, and mouse xenograft studies strongly indicate that a variety of perturbations of the CCDC170 protein are capable of driving breast cancer. Despite the wealth of genetic information relating to the CCDC170 gene, nothing was known about the encoded protein.
Here, we initially show that the CCDC170 locus is associated with significant Differential Allele Specific Expression (DASE), which supports specifically a link to breast cancer risk. As nothing was known about the molecular function of the CCDC170 protein, the present work focused largely on identifying a potential molecular mechanism for CCDC170-associated breast cancer risk and progression. We demonstrate that the CCDC170 protein, a predicted coiled-coil domain containing (CCDC) protein, associates with the Golgi apparatus, stabilizes perinuclear microtubules (MTs), and plays an essential role in the known process of MT-dependent Golgi organization. Distinct Golgi-derived MTs that extend into the cytoplasm are now understood to contribute to cell polarity and directional migration. We hypothesized that breast cancer-related perturbations of the CCDC170 Golgi-MT network could lead to altered cell polarity and migration, and thereby drive breast cancer initiation and progression. We provide evidence that dysregulation of CCDC170 indeed affects polarized cell migration.
2. Materials and Methods
2.1. Cell Lines
Thirty human mammary epithelial cell (HMEC) lines were utilized as starting materials for DASE analysis at the CCDC170-ESR1 locus. Under an approved protocol by the Institutional of Review Board (IRB) at Fox Chase Cancer Center, we derived primary HMEC lines from adjacent or contralateral normal mammary tissue of breast cancer patients as described previously (Gao et al., 2012). Non-tumorigenic MEC lines, MCF-10A and -10F, and human breast cancer cell lines, MCF-7, T-47D, ZR-75-1, MDA-MB-231, HCC-1937, and SK-BR-3, were purchased from American Type Culture Collection (ATCC). Cell lines were maintained in medium recommended by ATCC at 37 °C in the presence of 5% CO2. MCF10ADCIS·COM cells were a gift from Dr. Fariba Behbod (University of Kansas Medical Center) and were maintained as previously described (Behbod et al., 2009). MCF-7 Tet-On® cells were purchased from Clontech and were maintained according to the manufacture's guidelines. U2OS cells were obtained from Dr. Sanjeevani Arora, Fox Chase Cancer Center. U2OS cell clones stably expressing WT GFP-CCDC170 were created by transfection, followed by single cell sorting. Single cell clones displayed heterogeneous CCDC170 levels and localization.
2.2. DASE Analysis at CCDC170-ESR1 Locus
Genomic DNA (gDNA), RNA and double-stranded cDNA (ds-cDNA) from primary HMEC lines were prepared as previously described (Gao et al., 2012). gDNA (quantified by PicoGreen assay) and ds-cDNA samples were subjected to whole genome application and fragmentation prior to Illumina HumanOmni5-quad BeadChip hybridization. To obtain the array probes that fall within the CCDC170 and ESR1 exons, SNP information from the HumanOmni5 BeadChip was retrieved and SNP coordinates were used. For each probe marker with the heterozygous genotype, scanned raw signal intensities were processed by GenomeStudio Software (Illumina) to generate X and Y intensity values for allelic expression at each marker position. Raw expression data for each SNP site were filtered (X + Y value > 1000). A total of 20 and 10 SNP probes mapping to CCDC170 or ESR1 exons, respectively, were obtained and used for final DASE analysis.
2.3. CCDC170 Expression Plasmids
The CCDC170 orf (RefSeq accession NM_025059) was obtained from OriGene Technologies in the pCMV6-AC-GFP C-terminal turboGFP fusion vector. The CCDC170 orf was transferred to the pCMV6-AN-mGFP N-terminal monomeric GFP fusion vector using the OriGene Precision Shuttle cloning sites Sgf I and Mlu I. The CCDC170 wild-type gene was subsequently subcloned into a pTRE-Tight (Clontech) at Kpn I and Not I sites. Fragments of CCDC170 1-48, 1-405, 1-591, 1-649, 1-689, 355-591, 355-689, 355-715, 593-715 were generated in the N-terminal GFP CCDC170 fusions by either gene synthesis (GenScript) or using the Agilent QuickChange II site-directed mutagenesis kit. Non-tag wild-type CCDC170 was also subcloned in the pRetroX-IRES-ZsGreen1 vector at the Not I site.
2.4. CRISPR/Cas9 Mediated CCDC170 Editing
CCDC170/C6orf97 knockout MCF-7 cells were generated using CRISPR/Cas9 technology. A guide RNA (gRNA-GTTCGGAAGTCCCGGTCACG) with predicted highest target specificity was selected using the CRISPR design tool from MIT (http://crispr.mit.edu). Guide RNA synthetic DNA fragments were ordered from Integrated DNA Technologies (IDT). The gRNA sequence was cloned into the px459 vector (Addgene plasmid #62988) (Ran et al., 2013). CCDC170−/− or CCDC170+/− clones were generated by single cell expansion. To detect the presence of out-of-frame insertions/deletions (indels) in all CCDC170 alleles, the genotypes of knock-out clones were screened and verified by Sanger DNA sequencing. CCDC170 protein levels were further examined by SDS PAGE/immunoblot analysis.
2.5. Immunofluorescence
Cells were plated on 4-well coverslips and transfected after 24 h with the indicated vectors using Lipofectamine® 3000 (Thermo Fisher Scientific) according to the manufacturer's recommendations. After 2 days of transfection, the cells were washed three times with PBS and fixed in 4% paraformaldehyde for 15 min at RT. Fixed cells were then washed with PBS three times for 5 min and permeabilized with 0.5% Triton X-100 for 15 min at RT. After three 5-minute washes, cells were incubated in 3% BSA in PBS containing 0.1% Tween 20 (PBS-T) for 30 min and washed twice in 1% BSA in PBS-T. Cells were incubated with primary antibodies at optimized dilutions for 1 h at RT if not specifically mentioned, washed four time for 5 min with 1% BSA in PBS-T and incubated with secondary Alexa 488/555/647 antibodies (1:100 Molecular Probes) for 1 h. The coverslips were washed four times for 5 min with PBS and Vectashield Mounting Medium with DAPI (Vector laboratories) was added. The cells were than visualized by confocal microscopy (Leica TCS SP8). The following primary antibodies were used for IF: AKAP9 rabbit antibody (1:100, #HPA008548, Sigma-Aldrich), CCDC170 rabbit antibody (1:100, HPA027114, Sigma-Aldrich). MAP4 rabbit antibody (1:139, #HPA038149, Sigma-Aldrich), TGN46 Rabbit antibody (1:100, #ab50595, Abcam), Mannosidase II rabbit antibody (1:100, #ab12277, Abcam), chicken GFP antibody (1:400, #A10262, Life Technologies), RCAS1 rabbit antibody (1:100, #12290, Cell Signaling), acetylated α-tubulin mouse antibody (1:2000, #sc-23950, Santa Cruz); α-tubulin mouse antibody (1:1000, #T5168, Sigma-Aldrich).
2.6. Electron Microscopy
Localization of CCDC170 was examined by EM in HeLa cells transiently expressing GFP-CCDC170. As described previously (Pimkina et al., 2009, Tokuyasu, 1980), cells were trypsinized, fixed in 6% formaldehyde, washed, cryoprotected with sucrose, and frozen in liquid nitrogen. Thin sections were cut at − 120 °C, collected on EM grids, and labeled with anti-GFP antibody ThermoFisher (#A-11122). Colloidal gold particles (10 nm) conjugated to goat anti-rabbit antibody was used as the secondary detection reagent. Samples were viewed in a FEI Tecnai 12 TEM operated at 80 kV and the images were recorded using an AMT digital camera.
2.7. Antibodies for Western Blots
The following primary antibodies were used for Western blots: GFP rabbit antibody (1:1000, #ab290, Abcam), CCDC170 rabbit antibody (1:500, #Ab97814, Abcam), RCAS1 rabbit antibody (1:100, #12290, Cell Signaling), acetylated α-tubulin mouse antibody (1:500, #sc-23950, Santa Cruz), α-tubulin mouse antibody (1:2000, #T5168, Sigma-Aldrich), and β-actin mouse monoclonal antibody (1:5000, Cat#A5316, Sigma-Aldrich). For secondary antibodies, ECL HRP-conjugated anti-mouse or anti-rabbit IgG (1:2000, GE Healthcare) were used.
2.8. Analysis of Nocodazole-Resistant MTs
HeLa or MCF-7 cells were plated on 4-well coverslips, grown to 50–75% confluence, and transfected with GFP or GFP-CCDC170 plasmids. After 20 h, transfected cells were treated with 10 μM nocodazole for the next 4 h to depolymerize MTs. At 24-hours, nocodazole was removed by washing with PBS three times, and fresh complete medium was added. Microtubules were allowed to re-polymerize for 0 or 30 min in a 5% CO2, 37 °C incubator. Cells were fixed and stained with anti-GFP, anti-α-tubulin, and anti-ac-α-tubulin antibodies as described in the IF section.
2.9. Quantitative Image Analysis
To analyze the fluorescence intensities in stained cells, we utilized ImageJ software (v1.48, NIH) to draw the target regions around cells, and additional regions were drawn in an area without fluorescent objects to be used for background subtraction. The area size and integrated density were obtained for corrected total cell fluorescence (CTCF) calculation: CTCF = Integrated Density − Area of selected cell × mean fluorescence of background readings. CTCF calculation is adapted from previous studies (Banerjee et al., 2013, Gavet and Pines, 2010, McCloy et al., 2014).
2.10. Live Cell Imaging for Motility Experiments
MCF-7 Tet-on cells (Clontech) were transfected with pTET-GFP-CCDC170 and GFP-control plasmids for 24 h, and then seeded into 6-well plates. After 24 h, cells were incubated in the presence of 500 ng/ml Doxycycline for another 24 h. After changing to regular media supplemented with 15 mM HEPES, cell images were taken in 5 minute time intervals using a Nikon Inverted TE300 camera, with stage and z-axis motor, for the next 12 h. Cell migration tracks were quantified using the ImageJ plugin “Manual Tracking” by tracing the center of the cells in the DIC (differential interference contrast) recordings of migrating single cells as reference points.
2.11. Wound Healing Assay
After cells were cultured in monolayer and reached 90–95% confluence, a scratch wound, cell-free region was created using a sterilized pipette tip. Cell debris was removed, and the edge of the scratch was smoothed by washing the cells twice with PBS. Media was re-applied and the dishes were placed in the incubator of 37 °C. The progress of cell migration into the scratch was recorded at every 24 h by image capture using an inverted microscope. The images are further analyzed quantitatively by using ImageJ software.
2.12. siRNA Transfection
Predesigned MAP4, AKAP9, and ATAT1 DsiRNAs and scrambled controls were purchased from IDT. DsiRNAs were transfected into cells using Oligofectamine™ or Lipofectamine 3000 reagents (Life Technologies).
2.13. BioID Method and Construction of Bait Plasmids
The BioID method (Kim et al., 2014) allows detection of candidate binding partners in live cells through proximity-dependent biotinylation. The bait protein plasmid encodes the gene of interest (e.g. CCDC170) fused to the promiscuous biotinylase BirA*. Cells are transfected with the bait protein plasmid, and the culture is incubated with biotin-containing media for 18–24 h. Cells can be lysed with SDS, as the bait-prey interactions do not have to be maintained. Biotinylated proteins are captured on streptavidin beads and subjected to identification by mass spectrometry.
For construction of BioID bait proteins, the CCDC170 orf (RefSeq accession NM_025059) from OriGene Technologies was transferred using the OriGene Precision Shuttle cloning sites Sgf I and Mlu I. The BirA* N- and C-terminal fusions vectors developed by the Roux Laboratory (Kim et al., 2014) were obtained from Addgene. The multi-cloning sites of these vectors, pcDNA3.1 MCS-BirA(R118G)-HA (Plasmid #36047) and pcDNA3.1 mycBioID (Plasmid #35700), were first reengineered to accept fragments from the OriGene Precision Shuttle system. In this way, an Sgf I-Mlu I CCDC170 orf derived from the pCMV6-AN-mGFP N-terminal monomeric GFP fusion vector was fused in frame to create a CCDC170 C-terminal BirA* fusion, and an Sgf I-Not I CCDC170 fragment was fused in frame to create a CCDC170 N-terminal BirA* fusion. We confirmed that the CCDC170 C-terminal BirA* fusion localized to the expected Golgi and perinuclear regions (Fig. S13). Furthermore, protein biotinylation at the Golgi and perinuclear regions was confirmed by probing fixed cells with streptavidin (Fig. S13).
2.14. BioID Mass Spectrometry and Data Analysis
CCDC170 BirA* fusion plasmids were used for transient transfection of HeLa cells, and BioID was carried out as described (Kim et al., 2014). For the initial set of experiments, BioID was carried out with duplicate transfections of plasmids expressing BirA* fused to the C-terminus of CCDC170, followed by a single transfection using plasmids expressing BirA* fused to the N-terminus of CCDC170. BioID tryptic digestion was performed using a method optimized from an earlier publication (Kim et al., 2014). Beads were resuspended in 50 μl of 8 M urea/50 mM ammonium bicarbonate, and proteins were reduced by adding 2 μl of 0.5 M Tris(2-carboxyethyl)phosphine (TCEP) to 50 μl of beads-proteins suspension mix. Proteins were reduced at 30 °C for 60 min, and the reaction was cooled to room temperature before alkylation by adding 4 μl of 0.5 M iodoacetamide at room temperature in the dark for 30 min. Sample volume was adjusted by adding 350 μl of 50 mM ammonium bicarbonate to dilute the 8 M urea to 1 M before trypsin digestion with mass spectrometry grade trypsin (Promega). Five micrograms of trypsin per sample was added, and incubation was overnight at 30 °C using Eppendorf Thermomixer at 700 rpm. Digested peptides were separated from beads by centrifugation and peptide digests were transferred to a new tube followed by an extra wash step with 50 μl of 50 mM ammonium bicarbonate. Formic acid was added to the peptide solution (to 2%), followed by desalting with C18 TopTip (Item# TT10C18.96, PolyLC), and finally drying in a SpeedVac. Tryptic peptides were re-suspended in 100 μl of 2% Acetonitrile in % 0.1 formic acid, and 10 μl of total tryptic peptides were utilized for the 1D LC-MSMS analysis in triplicate runs by on-line analysis of peptides by high-resolution, high-accuracy LC-MS/MS, consisting of an EASY-nLC 1000 HPLC Acclaim PepMap peptide trap, a 25-cm 2 μm Easy-Spray C18 column, Easy Spray Source, and a Q Exactive Plus mass spectrometer (all from Thermo Fisher Scientific). A 230-min acetonitrile gradient consisting of 5–16% in 140 min, 16–28% in 70 min, 28–38% in 10 min, and 38–85% in 10 min was used to separate the peptides. The total LC time was 250 min. The Q Exactive Plus is set to scan precursors at 70,000 resolution followed by data-dependent MS/MS at 17,500 resolution of the top 12 precursors.
For protein identification and data analysis, the LC-MSMS raw data of two technical replicates were combined and submitted to Sorcerer Enterprise v.3.5 release (Sage-N Research, Inc.) with SEQUEST algorithm as the search program for peptide/protein identification. SEQUEST was set up to search the target-decoy UniProt Human Reviewed (vs. March 2015) protein fasta database containing protein sequences using trypsin for enzyme with the allowance of up to 2 missed cleavages, Semi Tryptic search, fixed modification of 57 Da for cysteine to account for carboxyamidomethylation, and precursor mass tolerance of 50 ppm. Differential search includes 16 Da for methionine oxidation, and 226 Da on lysine for biotinylation. The search results were viewed, sorted, filtered, and statically analyzed by using comprehensive proteomics data analysis software, Peptide/Protein prophet v.4.02 (ISB). The minimum trans-proteomic pipeline (TPP) probability score for proteins was set to 0.9, to assure very low error (much less than FDR 2%) with reasonably good sensitivity. The differential spectral count analysis was done by QTools, an open source in-house developed tool for automated differential peptide/protein spectral count analysis (Brill et al., 2009). The protein prophet peptide report is utilized to report biotinylated peptides.
The LC-MSMS raw data were also submitted to Integrated Proteomics Pipelines (IP2) Version IP2 v.3 (Integrated Proteomics Applications, Inc.) with ProLucid algorithm as the search program (Xu et al., 2015, Xu et al., 2006) for peptide/protein identification. ProLucid search parameters were set up to search the UniProt Human Reviewed (vs. March 2015) protein fasta database including reversed protein sequences using trypsin for enzyme with the allowance of up to 2 missed cleavages, Semi Tryptic search fixed modification of 57 Da for cysteine to account for carboxyamidomethylation, and precursor mass tolerance of 50 ppm. Differential search includes 16 Da for methionine oxidation, and 226 Da on lysine for biotinylation. The search results were viewed, sorted, filtered, and statically analyzed by using DTASelect for proteins to have protein FDR rate of < 2.5% (Tabb et al., 2002). Differential label-free proteomics data analysis was done by IP2-Census, Protein Identification STAT COMPARE (Park et al., 2008) using two technical replicates. The result is a label-free quantification analysis, of duplicate technical data for each sample using spectral count analysis with t-test (Robinson et al., 2004).
2.15. Criteria for Identifying Candidate CCDC170 Binding Partners by BioID
The strategy for filtering candidate CCDC170 BioID protein hits is outlined in Fig. S13. The initial output of proteins detected by BioID were samples in which a protein was reported if one of two duplicates had a spectral count of 1 or greater. The minimal value reported was thereby 0.5. The first filters applied were that the protein was absent in the untransfected control, and that the average of spectral counts for the duplicates was 2 or greater in either of two independent bait transfections using BirA* fused to the C-terminus of CCDC170. The two independent transfection experiments (Sample 1, Sample 2) detected largely the same proteins. The signal strength in the Sample 2 data set was stronger, and therefore this sample was used to rank the hits. However, several proteins with low spectral counts were uniquely detected in Sample 1, and were regarded as hits as well. Using these initial criteria, 217 proteins, including the CCD170 bait protein itself, were detected. As in the original BioID method, we utilized untransfected cells as a control, as the non-fused BirA* has the potential to biotinylate proteins throughout the cell via transient collision. As such, a significant non-informative background of biotinylated proteins could be generated using non-fused BirA*. Therefore, we chose to filter the initial set of 217 protein hits against a second experimental set that included a nuclear BirA* bait protein, and a second independent untransfected HeLa cell control (data not included). When comparing the CCDC170 and our own nuclear bait protein data sets, we identified protein families that were common, including ribosomal proteins and RNA splicing factors (Fig. S13). Elimination of these common proteins reduced the hits to 62 proteins (Table S2). Although cytoskeletal proteins are common contaminants in affinity purification-mass spec experiments (and were also detected using the nuclear bait protein) they were not removed at this stage, as we had evidence that CCDC170 associated with microtubules. Instead, we further filtered the 62 proteins against published control and experimental data sets that used unrelated bait proteins that localize either to the nuclear lamina, tight junctions, or the nuclear pore (Fig. S13, Table S2). We also filtered the 62 proteins against the Contaminant Repository for Affinity Purification database (http://www.crapome.org/) (Fig. S13, Table S2), with a criteria that proteins found in > 25% of the 411 control experiments were removed. This stringent filtration reduced the number of protein hits to 32. Lastly, 14 of the 32 proteins were also detected in the independent transfection using BirA* fused to the N-terminus CCDC170 bait protein. We did not use such detection as a criterion, as the proximity-dependent biotinylation (10 nm) by BirA* positioned at either terminus of a predicted extended coiled-coil domain protein might be expected to detect differential partners. Notably, the bait protein with BirA* fused to the CCDC170 N-terminus produced a very strong signal among some proteins that failed other stringent criteria (Table S2). We do not exclude proteins as potential CCDC170 partners that were detected with the N-terminal fusion but failed other criteria, and they can be investigated in the future. We note that overall, our unbiased and high stringency filtration approach likely resulted in a high percentage of false negative hits overall. The final list of candidate CCDC170 protein (Fig. S13, Table S3) is highly enriched for Golgi, microtubule and centrosomal proteins.
2.16. Statistics
All data are represented as mean ± standard deviation (SD). The Wilcoxon signed rank test was used to compare differences between paired CCDC170 and ESR1 DASE data. IF and live cell imaging data were analyzed using one-way ANOVA for multiple post-hoc comparisons or the two-sample t-test. All tests were two-sided and used a Type I error of 5%. Unless mentioned specifically, all experiments were repeated in triplicate.
3. Results
3.1. The CCDC170/C6orf97 Locus is Associated With Significant Differential Allele Specific Expression (DASE)
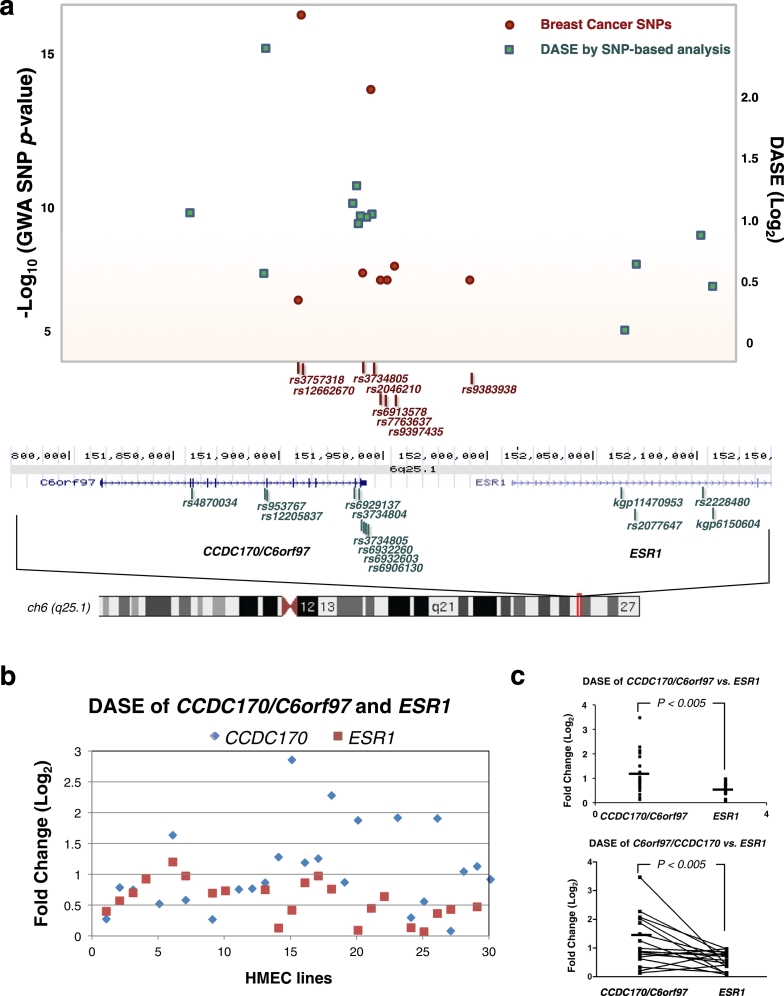
DASE is detected using allele-specific, non-breast cancer-associated polymorphic SNP markers. Using this approach, we previously demonstrated that germline, breast cancer-associated mutations in BRCA1/2 caused allele-specific nonsense-mediated mRNA decay (NMD) leading to DASE (Chen et al., 2008). In addition to NMD mutations, multiple genetic and epigenetic alterations, such as mutations in promoter regions, alteration in microRNA binding sites on 3′UTRs, and changes in DNA methylation status can contribute to DASE. Therefore we have hypothesized that DASE is a functional index for cis-acting germline variants and pathogenic mutations that affect mRNA levels, and that global profiling of DASE in breast cancer precursor tissues can be used to uncover causative alleles for breast cancer susceptibility (Gao et al., 2012). To investigate the proposed role of the CCDC170 gene in breast cancer risk, we first performed DASE analysis at the CCDC170-ESR1 locus using a set of 30 primary human mammary epithelial cell (HMEC) lines derived from adjacent normal breast tissue from breast cancer patients. DASE analysis detected significant allele-specific differences in CCDC170 mRNA levels, but not in ESR1 mRNA levels (Fig. 1a). Furthermore, the higher total number of, and most statistically significant, GWAS breast cancer risk-associated SNPs were linked to the CCDC170 versus ESR1 locus (Fig. 1a). Lastly, individual analysis of each of the 30 HMEC lines derived from breast cancer patients revealed levels of DASE for the CCDC170 gene that were significantly higher than those for the ESR1 gene (Log2 mean comparison: 1.15 vs. 0.56, P < 0.002 and matched pair comparison: 1.22 vs. 0.54, P < 0.009) (Fig. 1b and c, Table S1). Taken together, our global DASE analysis of the CCDC170-ESR1 locus showed that CCDC170, but not ESR1, is a gene with DASE in breast cancer precursor tissues. These results suggest that one mechanism by which the GWAS causal SNPs may contribute to the breast cancer is by affecting CCDC170 mRNA expression levels. To gain insight into how altered levels of CCDC170 gene expression, or breast cancer-associated somatic mutations of CCDC170, may contribute to breast cancer, we next investigated the function of the unstudied protein encoded by the CCDC170 gene.
Fig. 1.
The CCDC170/C6orf97 locus is associated with significant Differential Allele Specific Expression (DASE) and Genome Wide Association (GWA) signals.
a. SNPs at the CCDC170/C6orf97-ESR1 locus that are associated with breast cancer and DASE. Upper plot shows regional GWA and DASE plots of the chromosome 6q25.1 loci CCDC170/C6orf97 and ESR1. Results of Log10 (P-value) are shown for SNPs associated with breast cancer (red dots) for the region of 151.8–152.2 Mb. P-values plotted for SNPs reported by multiple studies are shown as average P-values. Results of DASE (green squares) at Log2 (fold-mRNA expression differences from one allele to the other) are calculated and averaged for primary HMEC lines (n = 30) by SNP-based analysis (Gao et al., 2012). Gene locations and SNPs used for DASE analyses (lower map) are from the UCSC Genome Browser assembly. b. Results of DASE analysis of CCDC170 (in blue) or ESR1 (in red) at Log2 (fold-changes) are calculated by gene-based analysis in individual primary HMEC lines (n = 30). DASE could not be calculated for all samples because the heterozygous DASE-detection SNPs were not available for some samples. c. DASE comparison between CCDC170 and ESR1 genes (upper panel: t-test; lower panel: Matched Pairs test).
3.2. The CCDC170 Protein Associates With the Golgi Apparatus
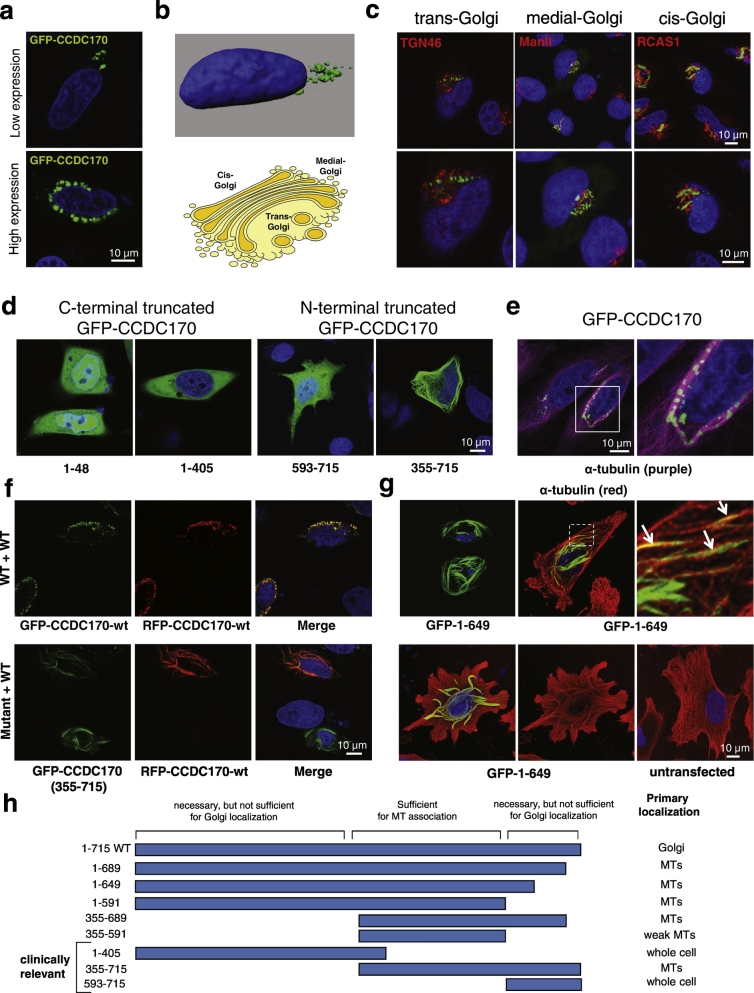
To determine the subcellular localization of CCDC170, we initially employed N- and C-terminal GFP-tagged versions of the protein. In HeLa cells, these tagged proteins localized to a cytoplasmic membranous structure that appeared to be the Golgi apparatus (Fig. 2a and b, only the N-terminal GFP fusion is shown). At high CCDC170 expression levels, the Golgi-like structures spread, forming a perinuclear ring (discussed below) (Fig. 2a). The CCDC170 gene is named based on the prediction that it is a coiled-coil domain-containing (CCDC) protein, and numerous resident Golgi structural proteins are characterized by long coiled-coil regions (Munro, 2011). Co-staining GFP-tagged CCDC170 with multiple Golgi markers (trans, medial and cis) confirmed that CCDC170 localizes primarily to the region of the Golgi apparatus, although not within the Golgi cisternae (Fig. 2c). GFP-CCDC170 was not associated with other intracellular organellar compartments (Fig. S1), and untagged CCDC170 also localized to the Golgi region (Fig. S2). Furthermore, transmission electron microscopy showed that GFP-tagged CCDC170 localizes with Golgi cisternae-like structures (Fig. S3). We found that a subset of ER + breast cancer cell lines express high levels of CCDC170 mRNA relative to normal MEC lines (Fig. S4a), consistent with a previous report (Dunbier et al., 2011). In these ER + breast cancer lines, the CCDC170 protein levels were also found to be elevated relative to MEC lines, with the protein localizing at the Golgi in a similar manner to GFP-tagged CCDC170 (Fig. S4c and Fig. 2a). In normal breast cells, endogenous CCDC170 was found to be associated with the Golgi as well (Fig. S4b). Regarding the specificity of the CCDC170 antibodies used here, we observed similar CCDC170 localization with two independent antibodies (Figs. S2 and S4b). Notably, endogenous CCDC170 mRNA and protein were undetectable in HeLa cells (data not shown). Therefore, HeLa cells were appropriate for further localization studies, as the potential for coiled-coil associations between GFP-tagged and endogenous CCDC170 is eliminated.
Fig. 2.
CCDC170 localizes to the Golgi apparatus and has microtubule (MT)-binding activities.
a. Subcellular localization of the GFP-CCDC170 fusion protein in HeLa cells. b. Top panel: Z-stack images were obtained using Leica SP8 confocal microscope. A 3D-rendering was created using IMARIS 7.5.0 software. Nucleus is shown in blue, with GFP-CCDC170 shown in green. The GFP-tagged protein localized to a membranous structure suspected to be the Golgi apparatus. Lower panel: Diagram of Golgi apparatus. c. GFP-CCDC170 Golgi localization was confirmed in HeLa cells by co-staining with an antibody to each of three resident Golgi proteins (in red): TGN46 (trans-Golgi), ManII (medial-Golgi), and RCAS1 (cis-Golgi). The nuclei are shown in blue (DAPI) as a reference. d. Subcellular localization of truncation mutants of N-terminally GFP-tagged CCDC170 in HeLa cells. All of these breast cancer-associated truncations of CCDC170 resulted in the loss of Golgi localization. Clinically relevant truncations are referenced from Cancer Genome Project and a recent report (Veeraraghavan et al., 2014). e. CCDC170 associates with perinuclear MTs in HeLa cells. GFP-CCDC170 (green) and α-tubulin (purple) are shown. f. Upper panels: The WT GFP-CCDC170 was coexpressed with WT RFP-CCDC170 in HeLa cells. Bottom Panels: The GFP-CCDC170 (355–715) fragment was coexpressed with WT RFP-CCDC170 in HeLa cells. A dramatic relocalization of the wild type protein from the Golgi to MTs was observed. g. Upper panels: GFP CCDC170 1–649 fragment (green) expressed in HeLa cells localizes to MTs (red). Bottom panels: A comparison of transfected and untransfected cells in the same culture indicates that overexpression of the GFP CCDC170 1–649 fragment (green) can induce MT bundling (red). h. Diagram summarizing mapping of Golgi- and MT-association domains, based on truncated constructs. Additional data supporting these mapping studies are shown in Fig. S6.
3.3. Clinically Relevant Truncations Disrupt CCDC170 Golgi Association and Reveal a Microtubule (MT)-Binding activity
The discovery that the breast cancer-related CCDC170 protein associated with the Golgi apparatus suggested that such localization is linked to its normal function. We therefore next examined the effects of breast cancer truncations of CCDC170 on Golgi association. C-terminal truncations (e.g. E48* and Q405*) were reported by both the Cancer Genome Atlas (TCGA) and Cancer Genome Project. As noted above, tumor-specific CCDC170 gene rearrangements were reported in a subset of luminal B breast tumors, and the expression of the predicted N-terminally truncated CCDC170 proteins (e.g. 355–715 and 593–715) was shown to increase cell motility and drive tumorigenicity of normal MEC cells (Robinson et al., 2011, Sakarya et al., 2012, Veeraraghavan et al., 2014). As shown in Fig. 2d, we found that the GFP-tagged 1–48 or 1–405 C-terminal truncations resulted in loss of Golgi association without re-localization to any other identifiable structures. Golgi association of CCDC170 was also disrupted by the N-terminal truncations (Fig. 2d). The N-terminally truncated 593–715 fragment localized throughout the cell. However the 355–715 fragment localized to tubular structures, primarily in the perinuclear region. Another member of the coiled-coil domain containing (CCDC) protein family, CCDC165/MTCL1 (Sato et al., 2014), had been found to stabilize perinuclear microtubules (MTs) on the Golgi membrane and thereby help organize the Golgi structure at the nuclear periphery. We suspected that CCDC170 had a similar organizational role, and that this role was impacted by the 355–715 truncation, resulting in loss of CCDC170 Golgi association and retention of MT association. These results predicted that the WT CCDC170 protein also associates with MTs. As shown in Fig. 2e and Fig. S5, ectopically expressed and endogenous WT CCDC170 could be detected at perinuclear MTs in a pattern similar to that seen with the CCDC165 Golgi-MT tethering protein (Sato et al., 2014).
Based on structure-function and localization of the wild type CCDC170, and breast cancer-associated CCDC170 fragments, we hypothesized that a potential dominant cancer mechanism could involve the functional impact of truncated CCDC170 forms on WT CCDC170. As mentioned, both the 355–715 and 593–715 CCDC170 fragments have been implicated as cancer drivers (Veeraraghavan et al., 2014) and as shown, the 355–715 fragment localizes to MTs while the 593–715 fragment localizes throughout the cell (Fig. 2d). When the CCDC170 355–715 fragment was coexpressed with WT CCDC170 in HeLa cells (lacking endogenous CCDC170), a dramatic relocalization of the wild type protein from the Golgi to MTs was observed (Fig. 2f). In the case of the 593–715 fragment, coexpression resulted in loss of whole cell distribution, with the fragment colocalizing with WT CCDC170 at the Golgi (data not shown). CCDC170 is strongly predicted to form parallel coiled-coil dimer domains over 70% of its length, and truncated forms are likely capable of forming mixed dimers with the full length protein. We speculate that the 355–715 fragment may interfere with WT CCDC170 function by triggering its mislocalization, while the mechanism through which the 593–715 fragment may affect the WT protein is not obvious from these localization studies.
To further map and confirm CCDC170 domains responsible for MT and Golgi association, multiple fragments were generated by site-directed mutagenesis. A GFP tagged 1–649 fragment lost Golgi localization, and instead showed dramatic association with perinuclear MTs (Fig. 2g). Furthermore, by comparing transfected and untransfected cells in the same culture, it appeared that the 1–649 fragment was inducing MT bundling (Fig. 2g and h). As the 1–649, 355–715 and 1–591 fragments localize to MTs (Fig. 2d, g and Fig. S6) and the 1–405 and 593–715 fragments localize throughout the cell, the sequences between 355 and 591 are predicted to contribute to MT binding (Fig. 2h). Regarding Golgi association, the CCDC170 C-terminus is necessary, but is not sufficient (e.g. compare WT, 1–591 and 593–715; summarized Fig. 2h).These results fit well with the known roles of resident Golgi coiled-coil proteins in forming complex bridging networks between the Golgi, MTs and the Golgi anchor, the centrosome (Rios, 2014).
3.4. Evidence for a Role of CCDC170 in Golgi Organization
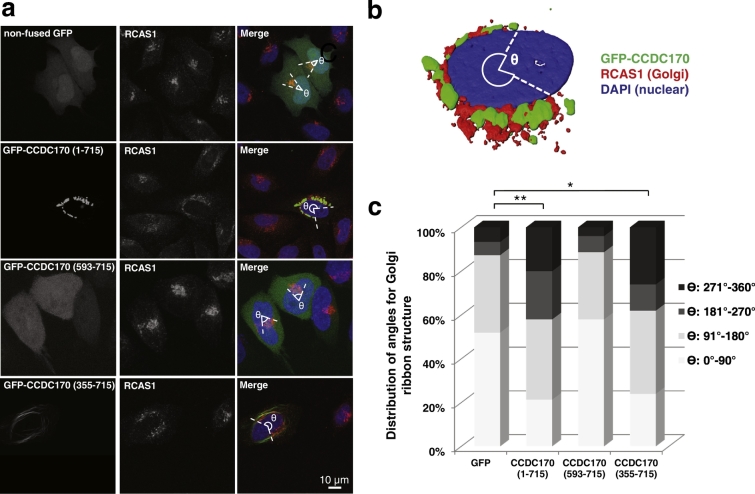
As CCDC170 is aberrantly expressed in breast tumors (i.e. high in ER + and low in ER − breast cancer), we next addressed how over- or under-expression of full length WT CCDC170 might impact Golgi and MT organization, and cellular functions. The Golgi apparatus consists of stacks of cisternae that are joined by lateral tubular connections to form the “Golgi ribbon”. We tested the hypothesis that CCDC170 perinuclear localization, MT-binding, and Golgi association (Fig. 2) provide a bridging function for Golgi ribbon organization. We had initially observed that when expressed at high levels, CCDC170 promoted an expansion, or fragmentation of the Golgi, resulting in a perinuclear ring (Fig. 2a). To investigate this phenomenon further, we quantitated the effect of WT CCDC170 overexpression on reorganization of the Golgi. For these experiments we overexpressed GFP-CCDC170 and determined the fraction of cells showing perinuclear Golgi spreading, as well as the extent of spreading, as measured by the angle of spreading in individual cells (Sato et al., 2014). We defined spreading as an angle of > 180°. HeLa cells (naturally lacking CCDC170) were transfected with GFP-tagged WT and truncated forms of CCDC170, in parallel with a vector encoding a non-fused GFP as a control. As shown in Fig. 3a and b, overexpression of WT GFP-CCDC170 resulted in a significant increase in the percentage of cells with spreading of the Golgi ribbon as measured by the Golgi marker RCAS1. In GFP-CCDC170-expressing cells, 42.0% showed Golgi spreading versus 12.7% in cells expressing non-fused GFP (P < 0.001). The diffusely localizing GFP- 593-715 fragment failed to induce Golgi spreading (Fig. 3c). We also validated GFP-CCDC170-induced Golgi spreading using an inducible MCF-7 Tet-On cell line (Fig. S7). Although MCF-7 cells normally express significant levels of endogenous CCDC170, transfection of GFP-CCDC170 triggered Golgi spreading as seen in HeLa cells. At present, we have no evidence that high levels of CCDC170 can be a cancer driver, rather, overexpression has been informative for identifying the function of CCDC170 through its ability to promote Golgi spreading. Interestingly, the CCDC170 355–715 N-terminal truncated form was still capable of promoting Golgi spreading even though it does not localize to the Golgi (Fig. 2d). This 355–715 fragment is capable of binding to, and stabilizing, MTs (Fig. S6, also see Fig. 4) and a subpopulation of MTs (i.e. Golgi-associated MTs) is known to regulate the positioning of the Golgi (Miller et al., 2009, Zhu and Kaverina, 2013). We therefore hypothesize that CCDC170-induced Golgi spreading is mediated through perinuclear MTs.
Fig. 3.
CCDC170 overexpression promotes spreading or fragmentation of the Golgi ribbon structure.
a. To examine whether overexpression of GFP-CCDC170 promoted reorganization of the Golgi, HeLa cells were transfected with non-fused GFP as a negative control, GFP-CCDC170, or clinical relevant truncation constructs (355–715 and 593–715). Quantitative results showed that CCDC170 overexpression can trigger spreading of the cis-Golgi ribbon structure as measured by both the fraction of CCDC170 positive cells showing perinuclear Golgi spreading (RCAS1 marker), as well as the angle of spreading of the cis-Golgi in individual cells (Ɵ). The diffusely expressed 593–715 fragment had no effect on Golgi spreading, while the 355–715 fragment showed an effect similar to the full length protein. b. 3D-rendering showing that CCDC170 overexpression triggers a dramatic spreading or fragmentation of the Golgi ribbon structure. c. Distribution of cells with different ranges of Golgi ribbon angle (Ɵ: 0°–360°), in cells expressing non-fused GFP, GFP-CCDC170, GFP-593-715 and GFP-355-715 [cell number (n) = 100 for each construct; vs. non-fused GFP (180°–360°); *P < 0.01, **P < 0.001].
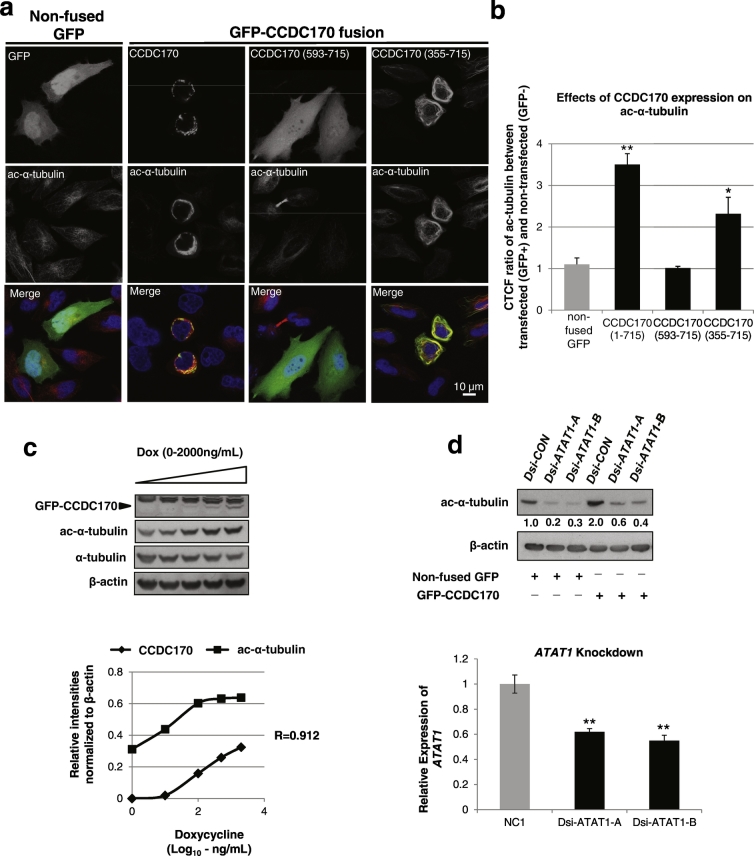
Fig. 4.
CCDC170 enhances alpha-tubulin acetylation.
a. HeLa cells were transiently transfected with GFP-CCDC170 full length, the clinically relevant N-terminally truncated forms (GFP-593-715, GFP-355-715) or a non-fused GFP negative control, and were subjected to immunofluorescence for acetylated alpha-tubulin (ac-α-tubulin) (red). b. Corrected Total Cell Fluorescence (CTCF) counts from panel a. Data represent CTCF ratio of red signal (ac-α-tubulin) between GFP-positive and GFP-negative cells (ratio of red signal in GFP Positive: GFP Negative) as compared to the negative control of non-fused GFP (n = 100, *P < 0.01, **P < 0.001). c. MCF-7-CCDC170-GFP Tet-On cells (Clontech) were treated with doxycycline (Dox) at 0, 10, 100, 500, or 2000 ng/ml and cell lysates for Western analysis were collected at the 24 h after Dox treatment. Upper panel: Western blot with indicated antibodies; lower panel: Quantification of Western blot. d. Western blot of HeLa cells with indicated antibodies. After reaching 50% confluence, HeLa cells were first treated with DsiRNA-scramble control (Dsi-CON) and Dsi-ATAT1. After 24 h, cells were transfected with non-fused GFP control and GFP-CCDC170 vectors for another 24 h and cell lysates were collected for Western blotting. ATAT1 mRNA knockdown effects were measured by RT-qPCR, as shown in graph below (vs. NC1; *P < 0.01, **P < 0.001).
3.5. CCDC170 Promotes Acetylation of α-Tubulin and Enhances MT Stabilization
The Golgi apparatus primarily functions in the processing and sorting of proteins (Wilson et al., 2011). More recently the Golgi has been identified as a MT-organizing center (MTOC) and mediates the nucleation of a subset of non-centrosomal MTs (Zhu and Kaverina, 2013). Such MTs function to position the Golgi (Miller et al., 2009) and also control cell polarity and motility (Miller et al., 2009, Zhu and Kaverina, 2013). As we found that CCDC170 localizes to both the Golgi and MTs, we next investigated a role for CCDC170 in MT function. Alpha tubulin acetylation (ac-α-tubulin) is a marker for stable MTs (Wloga and Gaertig, 2010, Janke and Bulinski, 2011). Strikingly, GFP-WT CCDC170 and GFP-355-715 overexpression caused a significant increase in acetylated MTs in the perinuclear region (Fig. 4a). No such effect was seen with negative controls: the GFP-593-715 fragment that localizes throughout the cell, and non-fused GFP. Quantitative image analysis using Corrected Total Cell Fluorescence (CTCF) showed that in GFP-CCDC170 overexpressing cells, the level of ac-α-tubulin increased about 2.5-fold as compared the negative controls (Fig. 4b) (P < 0.001). An inducible CCDC170 MCF-7-tet-on cell line was used to demonstrate that over-accumulation of CCDC170 could trigger parallel increases in ac-α-tubulin levels (Fig. 4c). To further assess this role of CCDC170 in MT function, we used CRISPR/Cas9 to disable the CCDC170 gene in the MCF-7 breast cancer cell line (Fig. S8a). Western blotting confirmed that the endogenous CCDC170 protein was depleted (Fig. S8c). In these CCDC170 knockout cells, the expected decrease in ac-α-tubulin levels was detected by IF and Western blotting, as compared to the parental cells (Figs. S8b and S8c). Thus, the experiments in Fig. 2, Fig. 4, and Fig. S8 using both overexpression and knockout of CCDC170, provide reciprocal evidence that CCDC170 promotes MT bundling and concomitantly increases ac-α-tubulin levels. We next assessed a role for the major tubulin acetyltransferase in mammals, ATAT1, in CCDC170-mediated MT acetylation. DsiRNA knockdown of ATAT1 significantly decreased both baseline and CCDC170-induced ac-α-tubulin in HeLa cells (Fig. 4d). This result suggests that CCDC170-induced α-tubulin acetylation is mediated by ATAT1 activities.
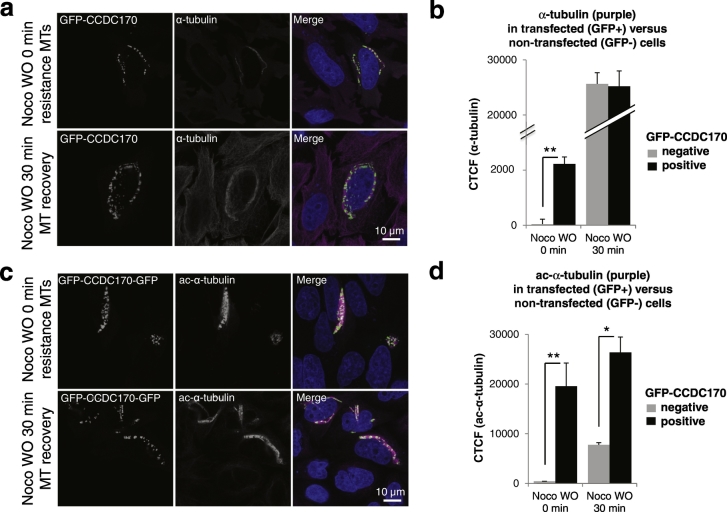
To confirm and extend the concept that CCDC170 stabilizes MTs, we implemented a standard approach using ac-α-tubulin as a marker to determine whether GFP-CCDC170 overexpression could promote stabilization of MTs after challenge with the MT-depolymerization drug, nocodazole (De Brabander et al., 1976). Non-transfected HeLa cells (GFP-negative) in the same culture served as negative controls. As expected, in non-transfected cells, MTs were depolymerized by 4 h of nocodazole treatment and were able to re-polymerize by 30 min after wash-out (Fig. 5a and b). However, in transfected cells, a subset of perinuclear MTs that colocalize with GFP-CCDC170 were found to be resistant to 4 h of nocodazole treatment (Fig. 5a and b). As expected, this perinuclear subset of nocodazole-resistant MTs were highly acetylated (Fig. 5c and d). Co-staining showed that both ac-α-tubulin and α-tubulin display punctations in regions corresponding to GFP-CCDC170 (Fig. S9). From these experiments we conclude that CCDC170 stabilizes MTs and promotes resistance of MTs to depolymerization by nocodazole.
Fig. 5.
CCDC170 enhances MT stabilization.
a and b. HeLa cells were transfected with GFP-CCDC170, and after 20 h were treated with nocodazole for 4 h to depolymerize MTs and observe potential CCDC170-induced resistance to nocodazole-induced MT depolymerization. Cells were then fixed and stained with anti-GFP and anti-α-tubulin (purple) antibodies 0 min after Nocodazole treatment (Noco WO 0 min) or 30 min after nocodazole washout to observe MT re-polymerization (Noco WO 30 min). As shown in panel d, after 4 h of nocodazole treatment, cells expressing GFP-CCDC170 (GFP-positive) showed nocodazole-resistant perinuclear MTs, as compared to neighboring nontransfected cells (GFP-negative). The GFP-CCDC170-induced nocodazole-resistance of MTs was quantitated by Corrected Total Cell Fluorescence (CTCF) counts (Panel b). Data represents absolute CTCF signal in GFP-positive versus GFP-negative cells (n = 100; *P < 0.01, **P < 0.001). Such quantitation was able to detect a significant difference in the α-tubulin signal between transfected (GFP-positive) and non-transfected (GFP-negative) cells at the WO 0 min time point, while 30 min after washout (Noco WO 30 min), the difference was negligible. c and d. The experiment was performed as in panels d and e except that anti-ac-α-tubulin was used. In this case, a more robust difference in the anti-ac-α-tubulin signal in GFP-positive and GFP-negative cells was seen at both time points.
3.6. CCDC170 Mediates Golgi-Associated Polarized Cell Migration
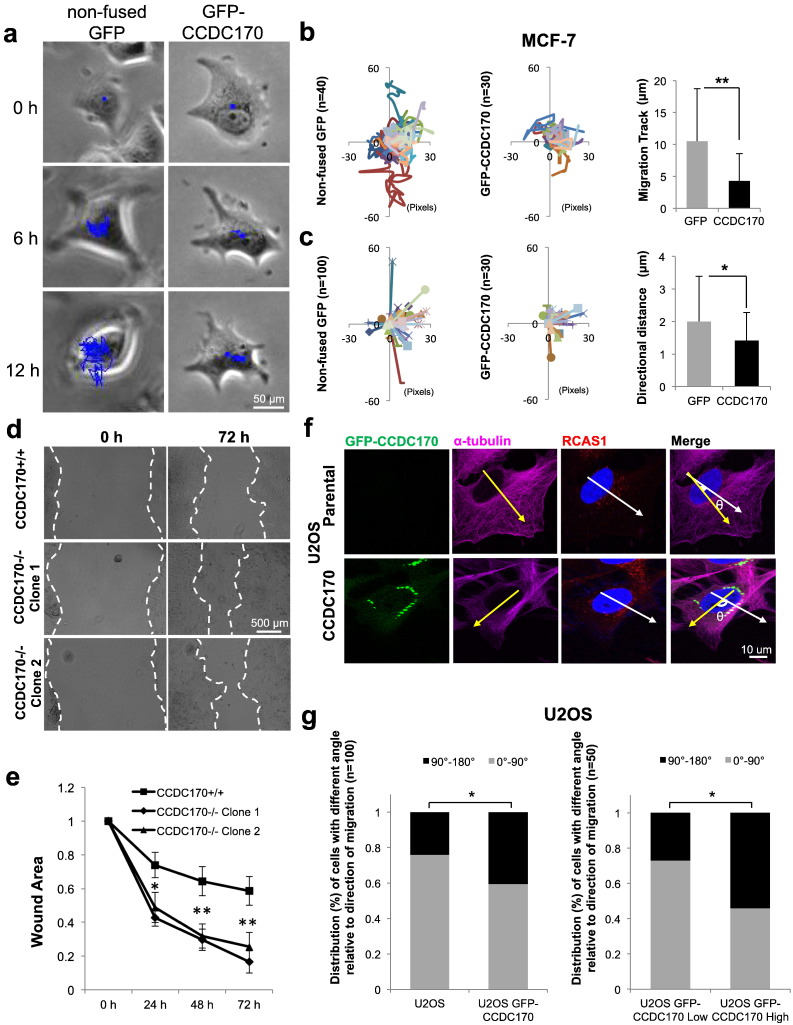
Golgi-associated MTs have been found to be essential for regulating directional cellular migration (Sato et al., 2014, Rios, 2014). We tested the hypothesis that CCDC170 plays a role in polarized cell migration by affecting the Golgi-associated MT network. As shown in Fig. 6a and b, live cell imaging analysis demonstrated that induced GFP-CCDC170-positive MCF-7 Tet-On cells migrated less than half of the total distance of induced control cells expressing non-fused GFP (P = 0.0003). Directional migration of each cell was also quantified as the direct distance from start to end point (Fig. 6c). Again, GFP-CCDC170-positive cells showed a shorter directional migration distance than those in the cells transfected with non-fused GFP (P = 0.04). We next examined cell migration using a standard wound healing assay. As shown in Fig. 6d and e, CCDC170-knockout MCF-7 cells needed significantly less time for wound closure than parental cells. Two independent CRISPR/Cas9 CCDC170−/− or cell clones were tested and the results were similar (Fig. 6d and e). We further compared CRISPR/Cas9 knockout CCDC170−/− and parental +/+ cells to +/− monoallelic knockout cells and detected statistically significant differences in directional migration in each case (Fig. S10). In contrast to the effects of the CCDC170 knockout, CCDC170-overexpressing U2OS cells required longer time for wound closure than parental U2OS cells (P < 0.01) (Fig. S11). Taken together, these results suggested that CCDC170 overexpression suppresses cell migration while depletion increases migration.
Fig. 6.
CCDC170 protein levels affect the rate of 2D cell migration.
a. Live cell imaging was carried out to monitor migratory activity of MCF-7 cells expressing either GFP-CCDC170 or non-fused GFP as a negative control. Cell migration tracks (blue) were quantified with ImageJ software by tracing the center of the cells. b. Average migration distance between GFP vs. GFP-CCDC170 overexpressing cells were compared (t-test, *P < 0.01, **P < 0.001). c. Directional migration of each cell was quantified as the direct distance from start to end point and the average distances were compared (t-test). The cells that directionally moved > 10 pixels (1.82 um) were defined as “fast”, less than that were defined as “slow”. The percentage of fast and slow cells was compared between each group. d. The relative wound closure of parental, and CRISPR/Cas9 MCF-7-CCDC170−/− cells was quantified from 6 different fields for 72 h. Images from experiments utilizing two different cells clones are shown. The images were representative of wound healing of each group, and white dotted lines labeled the wound edges. e. The wound healing process was evaluated at 0, 24, 48 and 72 h and the relative wound areas (vs. 0 h) are shown as mean ± SD (n = 3, *P < 0.01, **P < 0.001). f. The effects of CCDC170 on the positioning of the Golgi with respect to the migratory leading edge of the nucleus were examined using U2OS cells stably overexpressing GFP-CCDC170. Directional migration of U2OS cells was initiated by wounding the monolayer and directional migration is defined as facing the wound. After cells began migrating into the wound, the cells were fixed and stained with anti-tubulin (purple), anti-RCAS1 (red, Golgi), and DAPI (nucleus). The yellow arrows point toward the wound (directional cell migration), while the white arrows describe the center of the nuclear-Golgi axis. In parental migrating U2OS cells, the directional cell migration and nuclear-Golgi axis largely overlapped, and an angle of 0 to 90° between the two was defined as the normal range. An angle of > 90° was defined as an uncoupling of Golgi positioning at the leading edge of nucleus. One example of uncoupling is shown. The angles were measured using ImageJ software. g. Quantitative results showing the distribution (%) of cells with different angle relative to direction of migration (*P < 0.01). Parental cells were compared to GFP-overexpressing cells (left). High and low GFP-CCDC170-expressing cells were also compared (right).
The Golgi is usually positioned at the side of the nucleus corresponding to the leading edge of migrating cells (Yadav and Linstedt, 2011), and Golgi-derived MTs are required for polarized migration (Miller et al., 2009). We therefore evaluated the effect of CCDC170 overexpression on Golgi-associated directional migration through measurement of the angles between the direction of cell migration and the nuclear-Golgi axis (Fig. 6f) (Yadav et al., 2009). An angle > 90° indicates that Golgi positioning does not correlate with the direction of cell migration. As shown in Fig. 6g, U2OS cells expressing high levels of GFP-CCDC170 displayed larger angles than those with low level of GFP-CCDC170 or cells expressing non-fused GFP (P < 0.01). The findings of Hurtado et al. indicate that the positioning of the Golgi at the centrosome is the major determinant of cell polarity and directional migration (Hurtado et al., 2011). Furthermore, mislocalization of the Golgi was found to result in failure of the centrosome to re-orientate, suggesting that the Golgi contributes to centrosome organization. We therefore expected that centrosome polarization might be defective in CCDC170-overexpressing cells due to the spreading of the Golgi, and this is indeed what we observed (Fig. S12). These findings support the interpretation that GFP-CCDC170-induced Golgi-spreading de-polarizes Golgi positioning and thereby disrupts directional cell migration.
3.7. Candidate Functional Interaction Partners of CCDC170
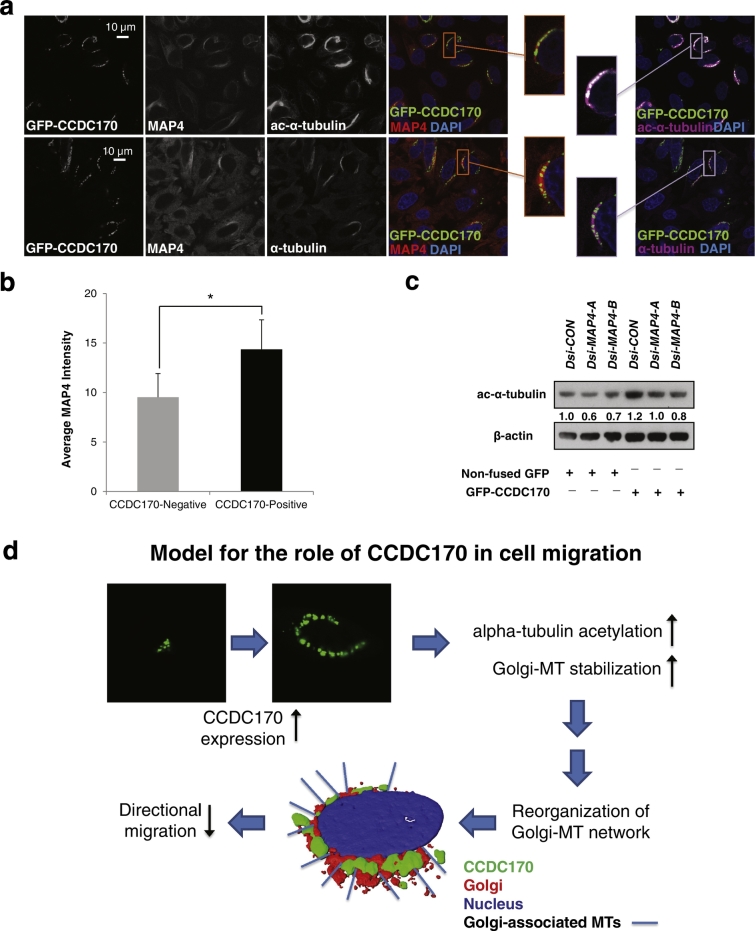
To gain further insights into the mechanisms by which CCDC170 contributes to the Golgi-MT network, and how its dysfunction may contribute to breast cancer, we identified candidate binding partners of CCDC170 using the BioID method (Fig. S13) (Roux et al., 2013, Kim et al., 2014). Of the final 32 candidates (Table S3), 18 are known to localize to the Golgi (e.g. AKAP9), MTs (e.g. MAP4) or the centrosome (e.g. PCM1). AKAP9 and MAP4 were of particular interest as AKAP9 was previously found to serve as a Golgi anchor (Tonucci et al., 2015, Sanders and Kaverina, 2015), in particular for CCDC165/MTCL1 (Sato et al., 2014), while MAP4 functions to stabilize MTs (Zahnleiter et al., 2015, Andersen, 2000). As shown in Fig. S14, native AKAP9 colocalized with GFP-CCDC170 in the expanded Golgi structure driven by CCDC170 overexpression. However, knockdown of AKAP9 had no detectable effects on native CCDC170 localization in U2OS cells, ectopic GFP-CCDC170 localization in HeLa cells, or GFP-CCDC170-induced α-tubulin acetylation (data not shown). Notably, in HeLa cells overexpressing GFP-CCDC170, MAP4 is often enriched and co-localized with CCDC170 in the perinuclear regions (Fig. 7a). Quantitative image analysis showed that MAP4 localization to GFP-CCDC170-enriched perinuclear areas was increased about 70% as compared to CCDC170-negative areas (Fig. 7b) (P < 0.01). Furthermore, knockdown of MAP4 decreased the levels of ac-α-tubulin in GFP-CCDC170-expressing HeLa cells (Fig. 7c). The CCDC170-driven increase in tubulin acetylation is likely the result of MT stabilization by CCDC170. A role for CCDC170 recruitment of MAP4 as part of this process remains to be investigated.
Fig. 7.
MAP4, a candidate functional interaction partner of CCDC170 identified by BioID.
a. MAP4 was detected as a candidate CCDC170 functional binding partner using BioID (Fig. S13, Tables S2 and S3). MAP4 is a microtubule-associated protein and plays a major role in the regulation of MT stabilization. To determine if MAP4 plays a role in CCDC170-mediated α-tubulin acetylation, HeLa cells were transfected with GFP-CCDC170 and after 20 h were treated with nocodazole for 4 h to promote MT disassembly. Cells were then immediately fixed and stained: GFP signal (green), MAP4 (red), and ac-α-tubulin (purple, upper panel) or total α-tubulin (purple, lower panel). Representative images show that in cells overexpressing GFP-CCDC170, perinuclear MTs become resistant to nocodazole-driven disassembly, and MAP4 becomes enriched and co-localized with CCDC170 at these perinuclear regions. b. Quantitative image analysis of MAP4 in GFP-CCDC170-expressing areas vs CCDC170-negative areas at perinuclear regions. Such quantitation was able to detect a significant difference in the MAP4 signal between CCDC170-positive and -negative areas at perinuclear regions (n = 50 cells, *P < 0.01). c. After reaching 50% confluency, HeLa cells were first treated with DsiRNA-scramble control (Dsi-CON) and Dsi-MAP4. After 24 h, cells were transfected with non-fused GFP control and GFP-CCDC170 vectors for another 24 h. Cell lysates were collected at 24 h after transfection for Western analysis. Experiment was repeated in triplicate. d. Diagram of proposed roles for CCDC170 in MT dynamics and Golgi-mediated polarized migration.
4. Discussion
The lifetime risk of breast cancer is greatly increased by the presence of breast cancer-driver or -modifier gene mutations that are carried in the germline. These mutations, in these breast cancer susceptibility genes, are also expected to contribute to the initiation and progression of sporadic breast cancer, which accounts for the majority of breast cancer cases. The availability of large DNA sequence data sets from normal and breast cancer tissues has allowed a more detailed and comprehensive assessment of these fundamental principles. Such analysis also poses a problem in that critical somatic driver mutations are difficult to identify among numerous passenger mutations, other than by recurrence (Martincorena and Campbell, 2015). GWAS studies had implicated the CCDC170-ESR1 locus in breast cancer risk, and identified germline SNPs located primarily between the two genes (Zheng et al., 2009, Turnbull et al., 2010). There was also evidence for a role of the CCDC170-ESR1 locus in sporadic breast cancer, driven by recurrent DNA rearrangements that are predicted to result in N-terminal truncations of the CCDC170 protein (Veeraraghavan et al., 2014). Taken together, these findings suggested that this locus may be involved in inherited risk for breast cancer, as well as in sporadic breast cancer, and that the CCDC170 gene plays a role. If so, multiple mechanisms impacting CCDC170 function may contribute: increased or decreased CCDC170 mRNA expression through the effects of noncoding SNPs, abnormal protein function through nonsynonymous coding SNPs, and somatic loss- or gain-of-function through gene rearrangements, missense, or nonsense mutations. Based on the unbiased detection of the CCDC170 locus in both GWAS and somatic gene rearrangement surveys, there was a strong indication that defects in CCDC170 gene could drive of breast cancer.
Here, we first analyzed the ESR1-CCDC170 locus-associated germline SNPs that had been detected in numerous GWAS analyses as being involved in breast cancer risk. In general, GWAS SNPs map mainly to noncoding regions, and their mechanisms of action cannot always be linked to a specific gene (Tycko, 2010). Although the ESR1 gene attracted the most attention, few studies had identified any strong causal SNPs impacting ESR1 function, and several were found to have stronger risk-association in ER(−) versus ER(+) cases (Fejerman et al., 2014, Han et al., 2011, Mulligan et al., 2011). Our DASE analysis detected allele-specific differences in CCDC170 mRNA levels in adjacent normal tissues from breast cancer patients, and suggested that GWAS breast cancer-associated SNPs could impact CCDC170 versus ESR1 mRNA expression at the ESR1-CCDC170 locus (Fig. 1). These findings suggested that germline SNPs might promote imbalanced mRNA expression levels of CCDC170 and thereby contribute to breast cancer susceptibility.
Despite the vast genetic data pointing to a role for the CCDC170 gene in breast cancer, nothing was known about the protein encoded by this gene. Here we report that the CCDC170 protein associates with the Golgi apparatus. The Golgi is an organelle previously linked to cancer through effects on Golgi enzymes that control glycosylation of the Golgi cargo, e.g., secretory and membrane proteins (Petrosyan, 2015). Here we show that CCDC170 is an intrinsic, Golgi-associated structural component, rather than an enzyme or cargo. The first clue in determining the function of CDCC170 came from the analysis of cells overexpressing GFP-tagged CCDC170. The Golgi is typically found in a focal area at one side of the nucleus in association with the centrosome, with some spreading around the nuclear periphery. We observed that when overexpressed, GFP-CCDC170 localized around the nuclear periphery (Fig. 2). We found that an endogenous Golgi marker became similarly distributed along with GFP-CCDC170 in CCDC170-overexpressing cells (Fig. 3). We conclude that overexpression of CCDC170 promotes formation or reorganization of Golgi structures around the nucleus, and that CCDC170 is thereby a critical Golgi organizing protein. Indeed, CCDC170 is predicted to be structurally similar to many other Golgi proteins that are characterized by long coiled-coil domains (golgins) (Munro, 2011). Regarding the effect of CCDC170 overexpression on Golgi organization, we cannot distinguish between stretching of the Golgi ribbon, with the stacks still joined by lateral tubular connections, versus de novo formation of independent Golgi structures.
We also found that CCDC170 can associate with, stabilize, and bundle, perinuclear MTs (Fig. 2, Fig. 4), possibly creating a perinuclear “track” for Golgi organization (Fig. 3 and Fig. S7). We also showed both by imaging and Western blotting that overexpression of CCDC170 leads to increases in the level of ac-α-tubulin, a marker for stable MTs. Expression of CCDC170 stabilized MTs even in the presence of the MT depolymerizing agent nocodazole (Fig. 4, Fig. 5). In addition, we observed MT-bundling/stabilization after expression of CCDC170 fragments 1-649 and 1-591, as well as the clinically relevant 355–715 fragment (Fig. 2 and Fig. S6). Lastly, we identified the MAP4 MT-stabilizing protein (Holmfeldt et al., 2009) as a potential functional binding partner of CCDC170 (Fig. 7). Although our results clearly show that CCDC170 can stabilize MTs, the experimental approaches described here do not distinguish between roles in dynamic growth of MTs versus stabilization of existing MTs.
As we found that CCDC170 can associate with both the Golgi and MTs, we propose that its normal function is to link the Golgi to, and stabilize, perinuclear MTs. As such, the function of CCDC170 seems to be similar to another CCDC protein that associates with Golgi, CCDC165/MTCL1 (Sato et al., 2014). Overexpression analysis of CCDC170 was informative in terms of its function, as it revealed a role for CCDC170 in organizing the Golgi and MTs. Also, these results suggest that changes in the level, or other perturbations, of CCDC170 could affect Golgi organization. Our findings suggest that the regulation of the Golgi-MT network by CCDC proteins might be impacted in disease. Regarding a breast cancer mechanism, the Golgi is now understood to control cell polarity and directional migration through Golgi-derived MTs that extend throughout the cytoplasm (discussed below).
As CCDC170 functions as a structural and organizational protein, we assessed how breast cancer-associated CCDC170 truncations might impact Golgi and MT associations, and we also performed functional mapping of CCDC170 Golgi and MT binding domains (Fig. 2 and Fig. S6). We found that clinically relevant truncations of CCDC170 (1–48, 1–405, 355–715, 593–715) lost Golgi localization. Combined with our mapping analyses, it was evident that the extreme C-terminal region of CCDC170 was necessary, but not sufficient, for Golgi localization. For example the 1–649 and 1–591 truncations resulted in loss of Golgi localization, with retention of MT association. However, the 593–715 fragment was not sufficient for Golgi association and showed whole cell localization, as did the 1–405 fragment. These findings indicate that positioning of CCDC170 at the Golgi requires multiple protein anchors, while MT-association was more modular and mapped between positions 405 and 591.
Taken together, our findings implicate a clear model for how CCDC170 alterations might play a role in the breast cancer cell phenotype. As CCDC170 contributes to Golgi organization through MT-binding and stabilization, we hypothesized that breast cancer-associated changes in CCDC170 could affect Golgi organization. Disorganization of the Golgi could, in turn, affect the positioning of Golgi-derived MTs that control cell polarity and migration, as follows. The Golgi has recently been identified as a MT-organizing center (MTOC) and mediates the nucleation of a subset of non-centrosomal MTs (Zhu and Kaverina, 2013). Unlike dynamic centrosomal MTs, Golgi-derived microtubules are stable and acetylated (Skoufias et al., 1990, Thyberg and Moskalewski, 1993). As mentioned, these stable microtubules function in the positioning of the Golgi, but more importantly, Golgi-derived MTs have been found to extend into the cytoplasm and be essential for maintaining cell polarity. Furthermore, in migrating cells, the Golgi is normally positioned at the side of the nucleus facing the leading edge, and motile cells lacking Golgi-derived MTs are defective for polarized migration (Miller et al., 2009, Rios, 2014). Overall, very little is known about how dysfunction of Golgi-derived MTs can lead to cellular defects or disease. We tested whether CCDC170 levels could affect polarized cell migration and found that overexpression could slow directional migration, while knockout increased wound healing (Fig. 6). Furthermore, in cells overexpressing CCDC170, there was an uncoupling between Golgi positioning and directional migration (Fig. 6f). Thus, regarding the mechanisms by which CCDC170 may contribute to breast cancer, we hypothesize, and provide evidence that, CCDC170 regulates cell polarity and motility and that CCDC170 MT-stabilizing activity may be critical for these processes (Fig. 7d). In addition to a role of CCDC170 in Golgi organization (with coordinated impact on Golgi-associated MT organization), it is possible that CDCC170 perturbations may affect cell mobility or invasion by influencing the exocytic pathway (Eaton and Martin-Belmonte, 2014). We are currently investigating this possibility.
The clear genetic link between breast cancer and the CCDC170 gene, and the earlier identification of another CCDC protein (MTCL1/CCDC165) that mediates the formation of a stable Golgi-derived microtubule network (Sato et al., 2014), provided a solid foundation for further exploration of the role of CCDC170 in breast cancer. We provide some insight into one possible mechanism, whereby the clinically relevant, and potential driver CCDC170 355–715 truncation (Veeraraghavan et al., 2014) localizes to MT rather than the Golgi. We found that when co-expressed, the WT CCDC170 protein relocalizes to MTs with the 355–715 truncation. Thus, the 355–715 fragment may either dysregulate MT function or disable WT CCDC170. Clearly, disease-associated overexpression or underexpression of the WT CCDC170 protein could also impact cell polarity or migration. Taken together, the work presented here along with previous publications suggest that CCDC170 could function as either an oncogene or a tumor suppressor gene, or both. An exciting possibility is that perturbations of CCDC170 may underlie germline breast cancer predisposition, as well as drive somatic, sporadic breast cancer.
In summary, a variety of GWAS studies, as well as our own DASE analyses, have implicated the CCDC170 gene in breast cancer risk. Our identification a function for the CCDC170 protein in Golgi-associated MT dynamics suggests a potential cancer mechanism related to a dysfunction in polarized cell migration. Our findings thereby support the idea that CCDC170 plays an essential role in Golgi-MT network and explain how perturbations in CCDC170 could trigger altered cell polarity and thereby drive breast cancer initiation and progression.
Funding Sources
This work was kindly partially supported by the Susan G. Komen for the Cure (KG100274 to XC) in data analysis, the Eileen Stein Jacoby Fund (to XC) in data collection, FCCC (P30-CA006927) in data collection, as well as by an appropriation from the State of Pennsylvania.
Conflicts of Interest
The authors declare no competing financial interests.
Author Contributions
XC and RAK designed the experiments and wrote the manuscript. MD contributed to the experimental design. PJ and YL performed tubulin acetylation analysis. YL and ML carried out quantitative imaging of Golgi reorganization. PJ carried out cell migration studies, MAP4 experiments, and created knockout cells. NB and AP performed initial subcellular localization experiments. CMS helped with cell culture, IF and Western blot experiments. VM carried out the BioID experiments. YZ (Yongchao) and NB performed initial cloning of deletion mutants. YZ (Yan) helped with the BioID data analysis. TP performed IF analyses of deletion mutants, created the stable cell line, and assisted in site-directed mutagenesis. JW designed, and assisted with, site-directed mutagenesis experiments. AE performed the electron microscopy. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank Kelly Dunlevy and Jessica Goldshteyn for assistance in vector construction and site-directed mutagenesis, Alison Biester for assistance in generation of stable cell lines, and Sullena Staton for assistance with transfection experiments. The authors also thank the following Core Facilities at FCCC for their help: Bioimaging, BioSample Repository, Cell Sorting, Genomics, High Throughput, and Biostatistics and Bioinformatics. Bio ID mass spec was carried out by the Proteomics Facility at Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA, with funding from the NCI Cancer Center Support Grant P30 CA030199. We thank the Facility Director, Dr. Khatereh Motamedchaboki, for her advice during the course of this work. We also thank Dr. Michal Jarnik for assistance and guidance for the electron microscopy experiments.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2017.06.024.
Contributor Information
Pengtao Jiang, Email: Pengtao.Jiang@fccc.edu.
Yueran Li, Email: liyueran1986@126.com.
Andrey Poleshko, Email: poleshko@mail.med.upenn.edu.
Valentina Medvedeva, Email: Valentina.Medvedeva@fccc.edu.
Natalia Baulina, Email: tati.90@mail.ru.
Yongchao Zhang, Email: zhangycqh@yahoo.com.
Yan Zhou, Email: Yan.Zhou@fccc.edu.
Carolyn M. Slater, Email: Carolyn.Slater@fccc.edu.
Trinity Pellegrin, Email: Trinity.Pellegrin@fccc.edu.
Jason Wasserman, Email: tue48797@temple.edu.
Michael Lindy, Email: michaelnlindy@gmail.com.
Andrey Efimov, Email: Andrey.Efimov@fccc.edu.
Mary Daly, Email: Mary.Daly@fccc.edu.
Richard A. Katz, Email: Richard.Katz@fccc.edu.
Xiaowei Chen, Email: Xiaowei.Chen@fccc.edu.
Appendix A. Supplementary data
Supplementary Figures and Tables.
References
- Ali S., Coombes R.C. Estrogen receptor alpha in human breast cancer: occurrence and significance. J. Mammary Gland Biol. Neoplasia. 2000;5:271–281. doi: 10.1023/a:1009594727358. [DOI] [PubMed] [Google Scholar]
- Andersen S.S. Spindle assembly and the art of regulating microtubule dynamics by MAPs and Stathmin/Op18. Trends Cell Biol. 2000;10(7):261. doi: 10.1016/s0962-8924(00)01786-4. [DOI] [PubMed] [Google Scholar]
- Banerjee A., Sahana A., Lohar S., Hauli I., Mukhopadhyay S.K., Safin D.A., Babashkina M.G., Bolte M., Garcia Y., Das D. A rhodamine derivative as a "lock" and SCN- as a "key": visible light excitable SCN- sensing in living cells. Chem. Commun. (Camb.) 2013;49:2527–2529. doi: 10.1039/c3cc40582f. [DOI] [PubMed] [Google Scholar]
- Behbod F., Kittrell F.S., Lamarca H., Edwards D., Kerbawy S., Heestand J.C., Young E., Mukhopadhyay P., Yeh H.W., Allred D.C., Hu M., Polyak K., Rosen J.M., Medina D. An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast Cancer Res. 2009;11:R66. doi: 10.1186/bcr2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill L.M., Motamedchaboki K., Wu S., Wolf D.A. Comprehensive proteomic analysis of Schizosaccharomyces pombe by two-dimensional HPLC-tandem mass spectrometry. Methods. 2009;48:311–319. doi: 10.1016/j.ymeth.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Wen W., Qu S., Li G., Egan K.M., Chen K., Deming S.L., Shen H., Shen C.Y., Gammon M.D., Blot W.J., Matsuo K., Haiman C.A., Khoo U.S., Iwasaki M., Santella R.M., Zhang L., Fair A.M., Hu Z., Wu P.E., Signorello L.B., Titus-Ernstoff L., Tajima K., Henderson B.E., Chan K.Y., Kasuga Y., Newcomb P.A., Zheng H., Cui Y., Wang F., Shieh Y.L., Iwata H., Le Marchand L., Chan S.Y., Shrubsole M.J., Trentham-Dietz A., Tsugane S., Garcia-Closas M., Long J., Li C., Shi J., Huang B., Xiang Y.B., Gao Y.T., Lu W., Shu X.O., Zheng W. Replication and functional genomic analyses of the breast cancer susceptibility locus at 6q25.1 generalize its importance in women of Chinese, Japanese, and European ancestry. Cancer Res. 2011;71:1344–1355. doi: 10.1158/0008-5472.CAN-10-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Weaver J., Bove B.A., Vanderveer L.A., Weil S.C., Miron A., Daly M.B., Godwin A.K. Allelic imbalance in BRCA1 and BRCA2 gene expression is associated with an increased breast cancer risk. Hum. Mol. Genet. 2008;17:1336–1348. doi: 10.1093/hmg/ddn022. [DOI] [PubMed] [Google Scholar]
- De Brabander M.J., Van De Veire R.M., Aerts F.E., Borgers M., Janssen P.A. The effects of methyl (5-(2-thienylcarbonyl)-1H-benzimidazol-2-yl) carbamate, (R 17934; NSC 238159), a new synthetic antitumoral drug interfering with microtubules, on mammalian cells cultured in vitro. Cancer Res. 1976;36:905–916. [PubMed] [Google Scholar]
- Dunbier A.K., Anderson H., Ghazoui Z., Lopez-Knowles E., Pancholi S., Ribas R., Drury S., Sidhu K., Leary A., Martin L.A., Dowsett M. ESR1 is co-expressed with closely adjacent uncharacterised genes spanning a breast cancer susceptibility locus at 6q25.1. PLoS Genet. 2011;7:e1001382. doi: 10.1371/journal.pgen.1001382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S., Martin-Belmonte F. Cargo sorting in the endocytic pathway: a key regulator of cell polarity and tissue dynamics. Cold Spring Harb. Perspect. Biol. 2014;6:a016899. doi: 10.1101/cshperspect.a016899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejerman L., Ahmadiyeh N., Hu D., Huntsman S., Beckman K.B., Caswell J.L., Tsung K., John E.M., Torres-Mejia G., Carvajal-Carmona L., Echeverry M.M., Tuazon A.M., Ramirez C., Consortium C., Gignoux C.R., Eng C., Gonzalez-Burchard E., Henderson B., Le Marchand L., Kooperberg C., Hou L., Agalliu I., Kraft P., Lindstrom S., Perez-Stable E.J., Haiman C.A., Ziv E. Genome-wide association study of breast cancer in Latinas identifies novel protective variants on 6q25. Nat. Commun. 2014;5:5260. doi: 10.1038/ncomms6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Devarajan K., Zhou Y., Slater C.M., Daly M.B., Chen X. Identifying breast cancer risk loci by global differential allele-specific expression (DASE) analysis in mammary epithelial transcriptome. BMC Genomics. 2012;13:570. doi: 10.1186/1471-2164-13-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O., Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev. Cell. 2010;18:533–543. doi: 10.1016/j.devcel.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W., Woo J.H., Yu J.H., Lee M.J., Moon H.G., Kang D., Noh D.Y. Common genetic variants associated with breast cancer in Korean women and differential susceptibility according to intrinsic subtype. Cancer Epidemiol. Biomark. Prev. 2011;20:793–798. doi: 10.1158/1055-9965.EPI-10-1282. [DOI] [PubMed] [Google Scholar]
- Hein R., Maranian M., Hopper J.L., Kapuscinski M.K., Southey M.C., Park D.J., Schmidt M.K., Broeks A., Hogervorst F.B., Bueno-De-Mesquita H.B., Muir K.R., Lophatananon A., Rattanamongkongul S., Puttawibul P., Fasching P.A., Hein A., Ekici A.B., Beckmann M.W., Fletcher O., Johnson N., Dos Santos Silva I., Peto J., Sawyer E., Tomlinson I., Kerin M., Miller N., Marmee F., Schneeweiss A., Sohn C., Burwinkel B., Guenel P., Cordina-Duverger E., Menegaux F., Truong T., Bojesen S.E., Nordestgaard B.G., Flyger H., Milne R.L., Perez J.I., Zamora M.P., Benitez J., Anton-Culver H., Ziogas A., Bernstein L., Clarke C.A., Brenner H., Muller H., Arndt V., Stegmaier C., Rahman N., Seal S., Turnbull C., Renwick A., Meindl A., Schott S., Bartram C.R., Schmutzler R.K., Brauch H., Hamann U., Ko Y.D., Network G., Wang-Gohrke S., Dork T., Schurmann P., Karstens J.H., Hillemanns P., Nevanlinna H., Heikkinen T., Aittomaki K., Blomqvist C., Bogdanova N.V., Zalutsky I.V., Antonenkova N.N., Bermisheva M., Prokovieva D., Farahtdinova A., Khusnutdinova E., Lindblom A., Margolin S., Mannermaa A., Kataja V., Kosma V.M., Hartikainen J., Chen X., Beesley J., Kconfab I., Group A., Lambrechts D., Zhao H., Neven P., Wildiers H., Nickels S., Flesch-Janys D., Radice P., Peterlongo P., Manoukian S., Barile M., Couch F.J., Olson J.E., Wang X. Comparison of 6q25 breast cancer hits from Asian and European genome wide association studies in the Breast Cancer Association Consortium (BCAC) PLoS One. 2012;7:e42380. doi: 10.1371/journal.pone.0042380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt P., Sellin M.E., Gullberg M. Predominant regulators of tubulin monomer-polymer partitioning and their implication for cell polarization. Cell. Mol. Life Sci. 2009;66:3263–3276. doi: 10.1007/s00018-009-0084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado L., Caballero C., Gavilan M.P., Cardenas J., Bornens M., Rios R.M. Disconnecting the Golgi ribbon from the centrosome prevents directional cell migration and ciliogenesis. J. Cell Biol. 2011;193:917–933. doi: 10.1083/jcb.201011014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Bulinski J.C. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2011;12:773–786. doi: 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- Kim D.I., Birendra K.C., Zhu W., Motamedchaboki K., Doye V., Roux K.J. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E2453–E2461. doi: 10.1073/pnas.1406459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller D.L., Zheng H.F., Karasik D., Yerges-Armstrong L., Liu C.T., Mcguigan F., Kemp J.P., Giroux S., Lai D., Edenberg H.J., Peacock M., Czerwinski S.A., Choh A.C., Mcmahon G., St Pourcain B., Timpson N.J., Lawlor D.A., Evans D.M., Towne B., Blangero J., Carless M.A., Kammerer C., Goltzman D., Kovacs C.S., Prior J.C., Spector T.D., Rousseau F., Tobias J.H., Akesson K., Econs M.J., Mitchell B.D., Richards J.B., Kiel D.P., Foroud T. Meta-analysis of genome-wide studies identifies WNT16 and ESR1 SNPs associated with bone mineral density in premenopausal women. J. Bone Miner. Res. 2013;28:547–558. doi: 10.1002/jbmr.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I., Campbell P.J. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–1489. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- McCloy R.A., Rogers S., Caldon C.E., Lorca T., Castro A., Burgess A. Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle. 2014;13:1400–1412. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P.M., Folkmann A.W., Maia A.R., Efimova N., Efimov A., Kaverina I. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat. Cell Biol. 2009;11:1069–1080. doi: 10.1038/ncb1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan A.M., Couch F.J., Barrowdale D., Domchek S.M., Eccles D., Nevanlinna H., Ramus S.J., Robson M., Sherman M., Spurdle A.B., Wappenschmidt B., Lee A., Mcguffog L., Healey S., Sinilnikova O.M., Janavicius R., Hansen T., Nielsen F.C., Ejlertsen B., Osorio A., Munoz-Repeto I., Duran M., Godino J., Pertesi M., Benitez J., Peterlongo P., Manoukian S., Peissel B., Zaffaroni D., Cattaneo E., Bonanni B., Viel A., Pasini B., Papi L., Ottini L., Savarese A., Bernard L., Radice P., Hamann U., Verheus M., Meijers-Heijboer H.E., Wijnen J., Gomez Garcia E.B., Nelen M.R., Kets C.M., Seynaeve C., Tilanus-Linthorst M.M., Van Der Luijt R.B., Van Os T., Rookus M., Frost D., Jones J.L., Evans D.G., Lalloo F., Eeles R., Izatt L., Adlard J., Davidson R., Cook J., Donaldson A., Dorkins H., Gregory H., Eason J., Houghton C., Barwell J., Side L.E., Mccann E., Murray A., Peock S., Godwin A.K., Schmutzler R.K., Rhiem K., Engel C., Meindl A., Ruehl I., Arnold N., Niederacher D., Sutter C., Deissler H., Gadzicki D., Kast K., Preisler-Adams S., Varon-Mateeva R., Schoenbuchner I., Fiebig B., Heinritz W., Schafer D., Gevensleben H., Caux-Moncoutier V., Fassy-Colcombet M., Cornelis F., Mazoyer S., Leone M., Boutry-Kryza N., Hardouin A., Berthet P., Muller D., Fricker J.P., Mortemousque I., Pujol P. Common breast cancer susceptibility alleles are associated with tumour subtypes in BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2. Breast Cancer Res. 2011;13:R110. doi: 10.1186/bcr3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a005256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S. K., Venable, J. D., Xu, T. & Yates, J. R., 3RD 2008. A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Methods, 5, 319–22. [DOI] [PMC free article] [PubMed]
- Paternoster L., Lorentzon M., Lehtimaki T., Eriksson J., Kahonen M., Raitakari O., Laaksonen M., Sievanen H., Viikari J., Lyytikainen L.P., Mellstrom D., Karlsson M., Ljunggren O., Grundberg E., Kemp J.P., Sayers A., Nethander M., Evans D.M., Vandenput L., Tobias J.H., Ohlsson C. Genetic determinants of trabecular and cortical volumetric bone mineral densities and bone microstructure. PLoS Genet. 2013;9:e1003247. doi: 10.1371/journal.pgen.1003247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosyan A. Onco-Golgi: is fragmentation a gate to cancer progression? Biochem. Mol. Biol. J. 2015;1 doi: 10.21767/2471-8084.100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimkina J., Humbey O., Zilfou J.T., Jarnik M., Murphy M.E. ARF induces autophagy by virtue of interaction with Bcl-xl. J. Biol. Chem. 2009;284:2803–2810. doi: 10.1074/jbc.M804705200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios R.M. The centrosome-Golgi apparatus nexus. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.R., Kalyana-Sundaram S., Wu Y.M., Shankar S., Cao X., Ateeq B., Asangani I.A., Iyer M., Maher C.A., Grasso C.S., Lonigro R.J., Quist M., Siddiqui J., Mehra R., Jing X., Giordano T.J., Sabel M.S., Kleer C.G., Palanisamy N., Natrajan R., Lambros M.B., Reis-Filho J.S., Kumar-Sinha C., Chinnaiyan A.M. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat. Med. 2011;17:1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson P.N., Wollstein A., Bohme U., Beattie B. Ontologizing gene-expression microarray data: characterizing clusters with Gene Ontology. Bioinformatics. 2004;20:979–981. doi: 10.1093/bioinformatics/bth040. [DOI] [PubMed] [Google Scholar]
- Roux K.J., Kim D.I., Burke B. BioID: a screen for protein-protein interactions. Curr. Protoc. Protein Sci. 2013;74 doi: 10.1002/0471140864.ps1923s74. (Unit 19 23) [DOI] [PubMed] [Google Scholar]
- Sakarya O., Breu H., Radovich M., Chen Y., Wang Y.N., Barbacioru C., Utiramerur S., Whitley P.P., Brockman J.P., Vatta P., Zhang Z., Popescu L., Muller M.W., Kudlingar V., Garg N., Li C.Y., Kong B.S., Bodeau J.P., Nutter R.C., Gu J., Bramlett K.S., Ichikawa J.K., Hyland F.C., Siddiqui A.S. RNA-Seq mapping and detection of gene fusions with a suffix array algorithm. PLoS Comput. Biol. 2012;8:e1002464. doi: 10.1371/journal.pcbi.1002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders A.A., Kaverina I. Nucleation and dynamics of Golgi-derived microtubules. Front. Neurosci. 2015;9:431. doi: 10.3389/fnins.2015.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Hayashi K., Amano Y., Takahashi M., Yonemura S., Hayashi I., Hirose H., Ohno S., Suzuki A. MTCL1 crosslinks and stabilizes non-centrosomal microtubules on the Golgi membrane. Nat. Commun. 2014;5:5266. doi: 10.1038/ncomms6266. [DOI] [PubMed] [Google Scholar]
- Skoufias D.A., Burgess T.L., Wilson L. Spatial and temporal colocalization of the Golgi apparatus and microtubules rich in detyrosinated tubulin. J. Cell Biol. 1990;111:1929–1937. doi: 10.1083/jcb.111.5.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey S.N., Sulem P., Zanon C., Gudjonsson S.A., Thorleifsson G., Helgason A., Jonasdottir A., Besenbacher S., Kostic J.P., Fackenthal J.D., Huo D., Adebamowo C., Ogundiran T., Olson J.E., Fredericksen Z.S., Wang X., Look M.P., Sieuwerts A.M., Martens J.W., Pajares I., Garcia-Prats M.D., Ramon-Cajal J.M., De Juan A., Panadero A., Ortega E., Aben K.K., Vermeulen S.H., Asadzadeh F., Van Engelenburg K.C., Margolin S., Shen C.Y., Wu P.E., Forsti A., Lenner P., Henriksson R., Johansson R., Enquist K., Hallmans G., Jonsson T., Sigurdsson H., Alexiusdottir K., Gudmundsson J., Sigurdsson A., Frigge M.L., Gudmundsson L., Kristjansson K., Halldorsson B.V., Styrkarsdottir U., Gulcher J.R., Hemminki K., Lindblom A., Kiemeney L.A., Mayordomo J.I., Foekens J.A., Couch F.J., Olopade O.I., Gudbjartsson D.F., Thorsteinsdottir U., Rafnar T., Johannsson O.T., Stefansson K. Ancestry-shift refinement mapping of the C6orf97-ESR1 breast cancer susceptibility locus. PLoS Genet. 2010;6:e1001029. doi: 10.1371/journal.pgen.1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb, D. L., Mcdonald, W. H. & Yates, J. R., 3RD 2002. DTASelect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res., 1, 21–6. [DOI] [PMC free article] [PubMed]
- Thyberg J., Moskalewski S. Relationship between the Golgi complex and microtubules enriched in detyrosinated or acetylated alpha-tubulin: studies on cells recovering from nocodazole and cells in the terminal phase of cytokinesis. Cell Tissue Res. 1993;273:457–466. doi: 10.1007/BF00333700. [DOI] [PubMed] [Google Scholar]
- Tokuyasu K.T. Immunochemistry on ultrathin frozen sections. Histochem. J. 1980;12:381–403. doi: 10.1007/BF01011956. [DOI] [PubMed] [Google Scholar]
- Tonucci F.M., Hidalgo F., Ferretti A., Almada E., Favre C., Goldenring J.R., Kaverina I., Kierbel A., Larocca M.C. Centrosomal AKAP350 and CIP4 act in concert to define the polarized localization of the centrosome and Golgi in migratory cells. J. Cell Sci. 2015;128:3277–3289. doi: 10.1242/jcs.170878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C., Ahmed S., Morrison J., Pernet D., Renwick A., Maranian M., Seal S., Ghoussaini M., Hines S., Healey C.S., Hughes D., Warren-Perry M., Tapper W., Eccles D., Evans D.G., Breast Cancer Susceptibility C., Hooning M., Schutte M., Van Den Ouweland A., Houlston R., Ross G., Langford C., Pharoah P.D., Stratton M.R., Dunning A.M., Rahman N., Easton D.F. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko B. Mapping allele-specific DNA methylation: a new tool for maximizing information from GWAS. Am. J. Hum. Genet. 2010;86:109–112. doi: 10.1016/j.ajhg.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeraraghavan J., Tan Y., Cao X.X., Kim J.A., Wang X., Chamness G.C., Maiti S.N., Cooper L.J., Edwards D.P., Contreras A., Hilsenbeck S.G., Chang E.C., Schiff R., Wang X.S. Recurrent ESR1-CCDC170 rearrangements in an aggressive subset of oestrogen receptor-positive breast cancers. Nat. Commun. 2014;5:4577. doi: 10.1038/ncomms5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C., Venditti R., Rega L.R., Colanzi A., D'angelo G., De Matteis M.A. The Golgi apparatus: an organelle with multiple complex functions. Biochem. J. 2011;433:1–9. doi: 10.1042/BJ20101058. [DOI] [PubMed] [Google Scholar]
- Wloga D., Gaertig J. Post-translational modifications of microtubules. J. Cell Sci. 2010;123:3447–3455. doi: 10.1242/jcs.063727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Park S.K., Venable J.D., Wohlschlegel J.A., Diedrich J.K., Cociorva D., Lu B., Liao L., Hewel J., Han X., Wong C.C., Fonslow B., Delahunty C., Gao Y., Shah H., Yates J.R., 3rd ProLuCID: an improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J. Proteome. 2015;129:16–24. doi: 10.1016/j.jprot.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Venable J., Park S.K., Cociorva D., Lu B., Liao L., Wohlschlegel J., Hewel J., Yates J.R., 3rd ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Cell Proteom. 2006;5:S174. [Google Scholar]
- Yadav S., Linstedt A.D. Golgi positioning. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a005322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S., Puri S., Linstedt A.D. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol. Biol. Cell. 2009;20:1728–1736. doi: 10.1091/mbc.E08-10-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Shen J., Cao Z., Wang B. Association between a novel polymorphism (rs2046210) of the 6q25.1 locus and breast cancer risk. Breast Cancer Res. Treat. 2013;139:267–275. doi: 10.1007/s10549-013-2494-1. [DOI] [PubMed] [Google Scholar]
- Zahnleiter D., Hauer N.N., Kessler K., Uebe S., Sugano Y., Neuhauss S.C., Giessl A., Ekici A.B., Blessing H., Sticht H., Dorr H.G., Reis A., Thiel C.T. MAP4-dependent regulation of microtubule formation affects centrosome, cilia, and Golgi architecture as a central mechanism in growth regulation. Hum. Mutat. 2015;36:87–97. doi: 10.1002/humu.22711. [DOI] [PubMed] [Google Scholar]
- Zheng W., Long J., Gao Y.T., Li C., Zheng Y., Xiang Y.B., Wen W., Levy S., Deming S.L., Haines J.L., Gu K., Fair A.M., Cai Q., Lu W., Shu X.O. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Kaverina I. Golgi as an MTOC: making microtubules for its own good. Histochem. Cell Biol. 2013;140:361–367. doi: 10.1007/s00418-013-1119-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Tables.