Abstract
The exciting commercial application potential of graphene materials may inevitably lead to their increasing release into the environment where they may pose ecological risks. This study focused on using carbon-14 labeled few-layer graphene (FLG) to determine whether the size of graphene plays a role in its uptake, depuration and biodistribution in adult zebrafish. After 48 h exposure to larger FLG (L-FLG) at 250 μg/L, the amount of graphene in the organism was close to 48 mg/kg fish dry mass, which was more than 170-fold greater than the body burden of those exposed to the same concentration of smaller FLG (S-FLG). The amount of uptake for both L-FLG and S-FLG increased by a factor of 2.5 and 16, respectively, when natural organic matter (NOM) was added in the exposure suspension. While the L-FLG mainly accumulated in the gut of adult zebrafish, the S-FLG was found in both the gut and liver after exposure with or without NOM. Strikingly, the S-FLG was able to pass through the intestinal wall and enter the intestinal epithelial cells and blood. The presence of NOM increased the quantity of S-FLG in these cells. Exposure to L-FLG or S-FLG also had a significantly different impact on the intestinal microbial community structure.
Keywords: graphene, zebrafish, natural organic matter, liver, intestinal epithelial cells, microbial community
In the last five years, the production industry of graphene family materials in China has been developing rapidly, and its total annual production capacity of graphene sheets and films exceeds 400,000 kg and 110,000 m2, respectively.1 This growth stems from the many potential applications of graphene in a range of industries.2–6 This will likely lead to graphene being released into the natural environment.7 Substantial research efforts have been recently conducted to understand the potential ecological and human health risks from graphene family materials.8–12 To date, the majority of studies have focused on the cellular and pulmonary toxicity of graphene,8–12 with only a limited number of studies on bioaccumulation by ecological receptors,13,14 a particularly important component of environmental risk assessment. In two recent bioaccumulation studies conducted using Daphnia magna and graphene, body burdens up to 1 % (by mass) were measured after exposure for 24 h to 250 μg/L,13 while graphene uptake was substantially reduced for graphene partly degraded by the Fenton reaction.15 Uptake of graphene has also been analyzed using oligochaete Limnodrilus hoffmeisteri and earthworm Eisenia foetida.16 In addition, the biodistribution of graphene oxide injected into zebrafish embryos and in mice after intratracheal instillation and oral gavage has been studied.14,17,18 However, no bioaccumulation and biodistribution studies, two key ecotoxicological parameters needed to fully characterize the potential impacts of graphene in aquatic ecosystems, have been conducted with graphene to our knowledge with any other ecologically relevant species. Furthermore, due to the antibacterial behavior of GO and graphene,9,19 the disturbance of biodiversity of intestinal flora may be essential key ecological effect induced when graphene is ingested by organisms such as fish.
Synthesized graphene often vary substantially in size, ranging between tens of nanometers to a few micrometers in diameter.13,20 Smaller graphene nanoparticles, which generally have enhanced stability, are popular for applications in surfactant and drug delivery.21,22 Studies have reported that environmental processes such as enzymatic reaction, photodegradation and Fenton reaction can cause graphene to fragment and decrease in size.15, 23–25 This is of key importance for understanding the potential toxicological effects of graphene because the bioavailability and toxicity of graphene are likely to be size dependent.26,27 For example, Ma et al. found the size-dependent M1 induction of macrophages and pro-inflammatory responses of graphene oxide in vitro and in vivo, and also a size-dependent interaction between graphene oxide and the plasma membrane.26 In a study with single-wall carbon nanotubes (SWCNTs), the smaller and more mobile fractions of the nanocarbon (< 18 nm in length) were determined to be much more toxic than the larger ones.27 However, the impact of graphene size on its bioaccumulation and biodistribution in aquatic organisms has not been assessed to date. That is largely because of challenges with quantifying graphene uptake in biological samples. In some studies, the surface of nanoparticles was modified by fluorescent dyes to quantitatively track its behavior in organs or cells.28–30 However, such kind of modification would change its physicochemical properties and behaviors, such as surface charge and stability in water. In addition, it is possible for the attached dye moiety to be released from the nanoparticle in biological fluids, which may hinder precisely quantifying the amount of nanoparticles.31 Therefore, we used 14C-labelled few layer graphene to avoid these potential biases and artifacts.
In this study, we explored the accumulation, distribution and depuration of few layer graphene (FLG) in a vertebrate after waterborne exposure. Adult zebrafish were exposed to radiolabeled FLG at three different concentrations and two different FLG sizes, and the uptake and distribution of radioactivity in the fish were then quantified. After being exposed to FLG, fish were transferred to water without FLG to examine elimination of FLG and the FLG distribution after depuration. The influence of the presence of natural organic matter (NOM), a compound ubiquitous in natural aquatic environments and known to impact the toxicity and uptake of nanoparticles,32 on graphene accumulation and distribution in zebrafish was also determined. Uptake of FLG into intestinal cells and the effects of FLG with different size on intestinal flora were also studied.
RESULTS AND DISCUSSION
Preparation of FLG with Distinct Lateral Sizes
Since the size range for graphene in applications is usually from ≈ 50 nm to ≈ 1,000 nm,33,34 our graphene samples were prepared in a comparable lateral size range. As shown in Figure 1A, the morphology of graphene samples was visualized by AFM. The size distribution analysis showed most (>95%) graphene sheets ranged from 20 nm to 70 nm with one major peak at ≈ 30 nm for S-FLG (smallest) and from 300 nm to 700 nm with one major peak at ≈ 500 nm for L- FLG (largest), respectively (Figure 1B). In addition, AFM showed that the average thickness was ≈ 1.05 nm and ≈ 1.4 nm for S- and L-FLG (Figure S1 in SI), indicating that they were mainly composed of 3 and 4 layers of graphene sheets respectively.13 These data indicate that we have successfully prepared graphene samples with two different average lateral sizes but similar thicknesses.
Figure 1.
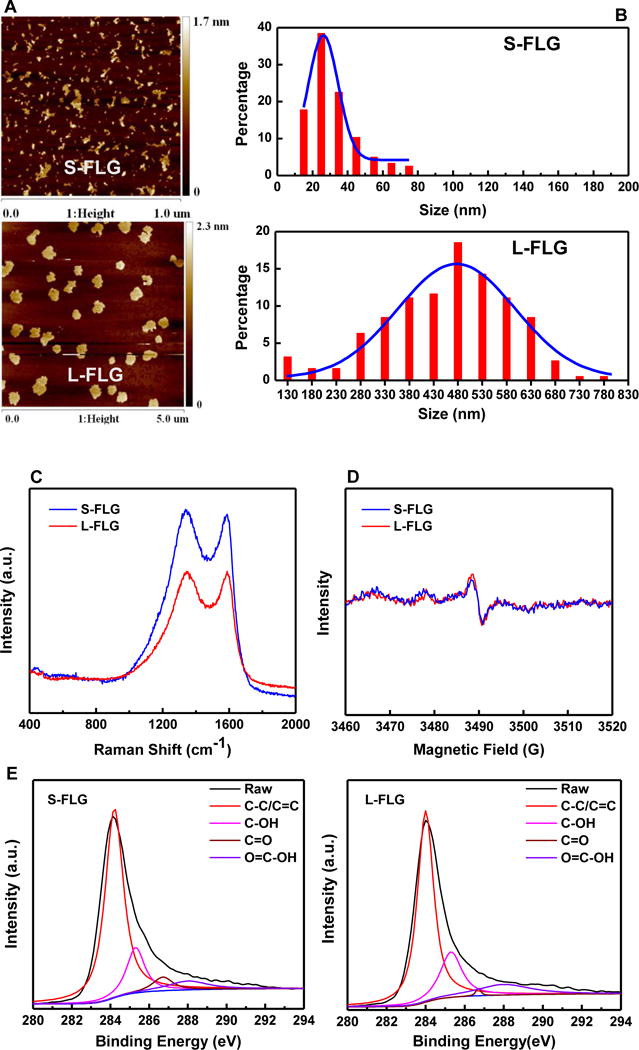
Morphological characterization of graphene samples. (A) Representative AFM topography of S-FLG and L-FLG. (B) Histogram of FLG size distribution. The histograms were developed by counting (280 to 300) sheets for each sample, with Gaussian fit curves shown in each histogram. (C) Raman spectrum and curve fitting of D band and G band. (D) The EPR spectra of S-FLG and L-FLG. (E) Characterization of surface composition of graphene samples by XPS. (F) Quantified data for the portions of functional groups (range: 0% to 100 %, n=3). Uncertainties for parts D and F are standard deviation values. (G) Electrophoretic mobility (EPM) of S-FLG and L-FLG (with and without the presence of NOM) (n=3).
Additional physicochemical characterization of the graphene samples was performed to assess if the sample preparation processes changed physicochemical properties in addition to the lateral size. Raman spectra showed similar basal structure profiles with characteristic D peak (1,331 cm−1) and G peak (1,596 cm−1) (Figure 1C) for both of the graphene samples, consistent with an earlier study.35 The D band means the presence of disorder in sp2 carbon rings, whereas the G band indicates stretching of C-C bonds in graphitic materials.36 The amount of structural defects in graphitic materials can be interpreted by the ratio of ID (intensity of D band)/IG (intensity of G band).37 A similar ID/IG ratio was found for both of the two graphene samples: 1.15 ± 0.13 and 1.08 ± 0.16 for S-FLG and L-FLG, respectively (uncertainties values here and throughout the paper indicate standard deviation values unless otherwise indicated); these values were not statistically different (p>0.05), indicating comparable structural defects in these graphene samples. Electron paramagnetic resonance (EPR) is a sensitive and specific technique for characterizing the formation of carbon radicals on the graphene surface.38 As shown in Figure 1D, S-FLG and L-FLG showed a similar single resonance peak of π-conjugated carbon radical with g = 2.0029, a result consistent with a previous report.38 As shown in Figure 1E, XPS for the graphene samples showed characteristic peaks at (284, 285.4, 286.8, 288.3) eV, representing C- C/C=C, C-OH, C=O and O=C-OH groups, respectively. The quantitative analysis of each group showed that the C-OH, C=O and O=C-OH content was increased in S-FLG relative to L-FLG (Figure 1F). The EPM value of L-FLG was lower than that of S-FLG (p=0.01), but the addition of NOM to the suspension made the difference between the EPM values of the S-FLG and L-FLG no longer statistically different (p>0.05) (Figure 1G). Taken together, these characterization data demonstrated that S-FLG and L-FLG samples showed comparable physicochemical properties except for the lateral size differences, EPM values in the absence of NOM, and XPS results.
Uptake Results
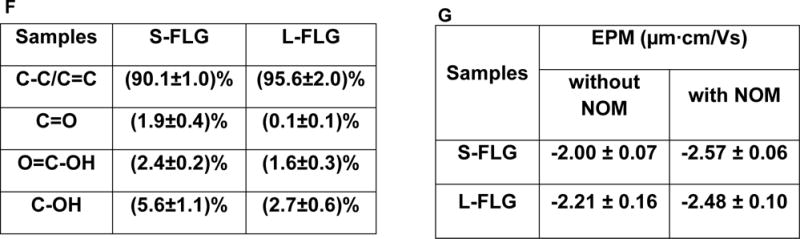
Our preliminary results indicated that (98.9 ± 2.3)% (n=3) of FLG was recovered after adding a volume of the FLG suspension to the dried organism, drying the mixture, combusting the sample using BO, and measuring the radioactivity using LSC. Immobilization of zebrafish being exposed for 72 h to a L-FLG/S-FLG concentration of 250 μg/L or in the control containers were not observed, thus indicating a lack of acute toxicity. As shown in Figure 2A and B, statistically significant uptake of L-FLG (or S-FLG) was observed during the exposure time under the tested concentration of 250 μg/L, 75 μg/L and 50 μg/L compared to the freshwater only control organisms. The uptake results at 24 h, 48 h and 72 h significantly differed among the exposure concentrations (p<0.05) (Figure 2A and B) for many of the time points, revealing that the uptake quantity of L-FLG (or S-FLG) was dependent on the exposure concentration. Statistical analysis of the body burden values (250 μg/L) for L-FLG and S-FLG indicated that the 4 h, 24 h, 48 h and 72 h time points generally differed from each other (p < 0.05). Uptake results for the tested concentration of 250 μg/L showed that a maximum accumulation was reached at 48 h for both L-FLG and S-FLG by showing a slight decrease from 48 h to 72 h. The change of the organism mass during the 72 h exposures was less than 5 % for the S-FLG and L-FLG exposures and thus changes in the organism mass cannot account for the changes in the body burden increase during the exposure period. Accumulation in the zebrafish after 48 h exposure to a 250 μg/L L-FLG suspension (≈ 48 μg/g dry mass) was over two orders of magnitude higher than that for the S-FLG (≈ 0.29 μg/g dry mass) for the same exposure duration and FLG concentration. This result indicates that L-FLG had a stronger tendency to be accumulated in zebrafish than that of S-FLG. The settling pattern of L-FLG and S-FLG was also assessed by measuring the radioactivity in the exposure solutions without zebrafish during the exposure period (see Figure S2). The settling rates of 50 μg/L and 75 μg/L L-FLG in the exposure solution with or without the presence of zebrafish were not significantly different while L-FLG settling at a concentration of 250 μg/L graphene was enhanced by the presence of zebrafish (Figure S2). For the S-FLG, minimal settling occurred (> 10 %) during the 72 h period in the absence or presence of zebrafish.
Figure 2.
Uptake of L-FLG and S-FLG with the absence (A, B) or presence (C, D) of NOM in zebrafish. Zebrafish were exposed to graphene in freshwater for 72 h with the absence or presence 10 mg (TOC)/L NOM and an initial suspended graphene concentration of (50, 75 or 250) μg/L. Data points with the same letter are not significantly different from one another; Tukey’s multiple comparisons test, p ≥ 0.05. Mean and standard deviation values were calculated from five samples.
To better understand the uptake of graphene under more environmentally relevant conditions, the uptake of L-FLG and S-FLG by zebrafish was further investigated in the presence of 10 mg (TOC)/L NOM. Uptake results across the range of tested concentrations in the presence of NOM still showed that a maximum accumulation was reached at 48 h with a slight decrease from 48 h to 72 h (Figure 2C and D). The body burden values at 12 h, 24 h, 48 h and 72 h significantly differed for FLG with and without the presence of NOM in the FLG exposure dispersions (250 μg/L, 75 μg/L and 50 μg/L) (p<0.05) (Figure 2 and Figure S3). The presence of NOM significantly enhanced the accumulation of both the L-FLG and S-FLG after 250 μg/L graphene exposure for 48 h by a factors of ≈ 2.5 and ≈ 16 times, respectively. It is possible that coating with NOM increased the consumption of FLG by the fish. This is a valuable topic that requires additional research. Similar to the results in the absence of NOM, there was a minimal change in the S-FLG concentration (> 10 %), and for the L-FLG, the presence of fish caused an increased in the degree of settling in the presence of fish compared to control experiments with the absence of fish (Figure S4). In addition, the presence of NOM caused a decrease in L-FLG settling in the absence of fish (compare Figures S2 and S4), a result in agreement with the findings of recent studies.39
Depuration Results
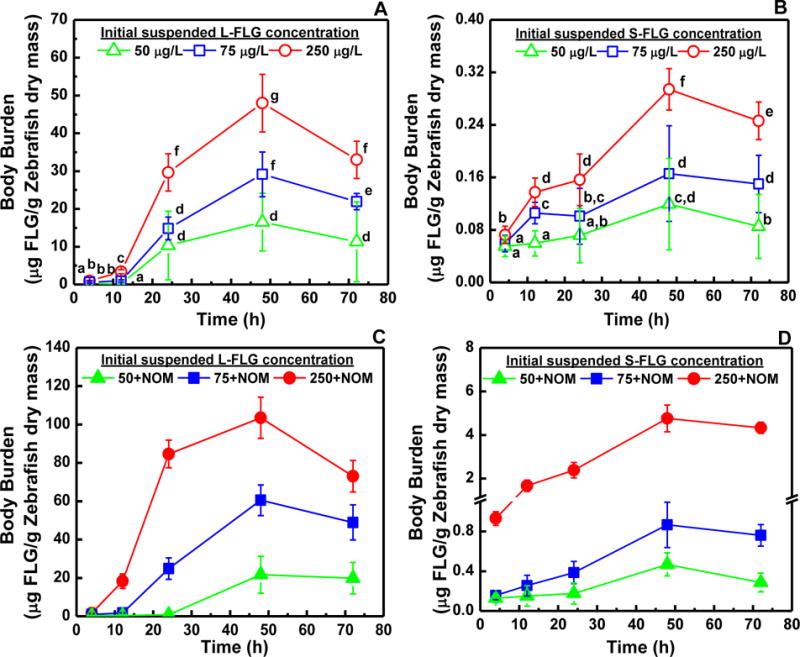
With the accumulation of FLG in zebrafish, the rate of FLG elimination by the organisms is essential for the understanding the potential toxicological effects of graphene. Therefore, we investigated the depuration of FLG in clean freshwater after waterborne exposure. As shown in Figure 3A, the accumulated graphene in the guts of zebrafish after being exposed for 48 h to the L-FLG at a concentration of 75 μg/L can be excreted for more than 95 % after 4 h depuration. The mass of graphene remaining in the zebrafish after depuration in clean water for 120 h was not statistically greater than 0, thus revealing that the zebrafish completely removed L-FLG from the gut tract. In contrast, zebrafish exposed to S-FLG (75 μg/L) were only able to discharge ≈ 30% of total graphene accumulated in their body during a depuration period of 4 h after which point the concentration in the fish did not change after depuration for an additional 68 h (Figure 3B).
Figure 3.
Depuration of L-FLG and S-FLG with the absence (A, B) or presence (C, D) of NOM in zebrafish. Zebrafish were exposed to graphene in freshwater for 48 h with the absence or presence 10 mg (TOC)/L NOM and an initial suspended graphene concentration of 75 μg/L. After acclimating fish for 5 d in aerated freshwater without graphene or NOM. Mean and standard deviation values were calculated from five samples; Tukey’s multiple comparisons test, p ≥ 0.05. The symbols (°) and (*) indicate not significantly or significantly different from zero, respectively.
Overall, the residual concentration of L-FLG and S-FLG in the zebrafish after depuration was increased when the zebrafish were exposed in the graphene solution with NOM. The depuration results for fish exposed to L-FLG in the presence of NOM showed that, after depuration for 72 in clean water, 8% of the accumulated L-FLG remained in the organism (Figure 3C). The amount of L-FLG remaining in the organism did not significantly change between 72 h and 120 h of depuration (p > 0.05). This contrasts with the L-FLG depuration results in the absence of NOM which showed complete elimination of the L-FLG. After 120 h depuration, the L-FLG and S-FLG concentration in zebrafish was about 4.84 μg and 0.18 μg graphene/g zebrafish dry mass, respectively (Figure 3D). Theses concentrations are statistically greater (p < 0.05) than the L-FLG and S-FLG concentrations in zebrafish after depuration for exposures in the absence of NOM (Figure 3B).
Distribution Results
To explore the impact of the mechanism of NOM on the uptake and the depuration of FLG, the biodistribution of FLG was also examined after exposing fish for 48 h to a FLG concentration of 75 μg/L. The recovery of graphene in each organism tissue was determined to be in the range of 95.1 % to 98.3 %; the reported data for each tissue was corrected by the recovery. As shown in Figure 4A, (98.3 ± 1.3) % of the accumulated L-FLG was in the digestive tract of zebrafish while > 2 % of the accumulated graphene was detected in the gills. No L-FLG was detected in the brain, liver, roe or carcass tissues. The lack of detectable transfer out of the gut tract for the L-FLG is similar to what has been previously observed in carbon nanotube bioaccumulation experiments.40–42 The distribution results of S-FLG in zebrafish after being depurated for 48 h suggest that ≈ 52 % of the accumulated S-FLG (in zebrafish after being exposed for 48 h to a concentration of 75 μg/L) was still retained in the gut. The S-FLG concentration in the liver was ≈ 2.18 μg/g liver dry mass (liver’s dry mass was ~0.5 % of that of the whole fish), a concentration significantly different from the control (Figure 4B). The S-FLG concentration in the liver did not significantly change during the 48 h depuration period. In contrast, Maes et al. investigated the uptake and distribution of 14C labeled multiwall carbon nanotubes (MWCNTs) in zebrafish and found that MWCNTs also mainly accumulated in the gut of fish and were not found in the liver, the gonads, and the brain.43 Bisesi tracked and quantified SWCNTs in fathead minnow using near infrared fluorescence and found that it was not detectable in the liver, spleen, gall bladder, and reproductive organs.44 The lengths of the tested MWCNTs and SWCNTs were ≈ 500 μm and ≈ 150 nm, respectively, which are larger than that of the S-FLG (lateral diameter of ≈ 30 nm) tested in this study but similar to the size of the L-FLG (lateral diameter of ≈ 500 nm). Therefore, our hypothesis is that the size of the carbon nanomaterial plays a key role in its potential for absorption across the gut tract in fish.
Figure 4.
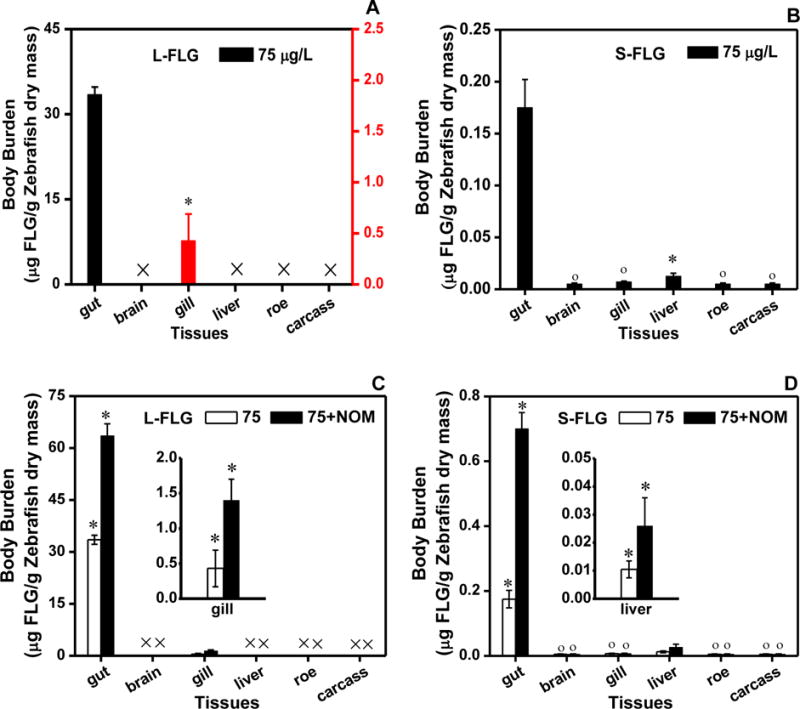
Distribution of L-FLG and S-FLG with the absence (A, B) or presence (C, D) of NOM in zebrafish. The left ordinate in Figure 4A is for gut while the right one is for the gill. Zebrafish were exposed to graphene in freshwater for 48 h with the absence or presence 10 mg (TOC)/L NOM and an initial suspended graphene concentration of 75 μg/L. Mean and standard deviation values were calculated from five samples; Tukey’s multiple comparisons test, p ≥ 0.05. The symbols (°) and (*) indicate not significantly or significantly different from zero, respectively.
The distribution results for exposures in the presence of NOM suggested that the increased body burdens mainly resulted from the significantly increased accumulation of graphene in the gut (Figure 4C and D). Accumulation of L-FLG in the gills and S-FLG in the liver was also found to have increased. However, the L-FLG was still not transferred into other tissues such as liver. This result is similar to a study of SWCNT uptake by Daphnia magna, which showed a lack of absorption through the gut tract even when the SWCNTs had different surface coatings including NOM.40 Figure S4 reveals that the S-FLG suspension was stable with NOM presence during the exposure time regardless of the presence or absence of zebrafish. The presence of zebrafish in the L-FLG and NOM exposure solution enhanced the settling rates of L-FLG causing approximately 30% and 70 % of the L-FLG to settle from the exposure solution after 72 h for the 75 μg/L and 250 μg/L conditions, respectively.
Cellular Association and Toxic Response of S-FLG in the Liver
To further explore the distribution of S-FLG in the liver, hepatocytes in the liver of zebrafish exposed to 250 μg/L S-FLG suspension for 48 h were isolated and analyzed. Images of these hepatocytes were taken using the light microscope and shown in Figure 5A. Figure 5B indicates that 76.3 % ± 6.0 % of the accumulated S-FLG in liver was found in the hepatocytes while the rest was detected in the residual tissues of liver. The obtained hepatocytes were sectioned and analyzed using TEM. As it shown in Figure 5C, black particles were observed in cells by the low magnification TEM. These particles were confirmed to be FLG by the results of selected area electron diffraction patterns (SADP) that the well-defined diffraction spots and the six-fold symmetry typical of graphene particles were observed for the particles (SADP image of pristine FLG is presented in Figure S5 of SI). Figure 5D and 5E show SADPs image taken from the blank control (p1 position) and black spots (p2 positions), which further confirmed the presence of FLG in the hepatocytes. High resolution TEM revealed visible ordered graphite lattices with interlayer distances of ≈ 0.344 nm (Figure 5F and 5G), which is the interlayer distance of graphite.13 The sectioned hepatocytes isolated from the liver of the zebrafish of the control untreated group were also analyzed using TEM. The six-fold symmetry typical of graphene particles were not observed (see Figure S6A and S6B). To further confirm the location of the S-FLG in hepatocytes, the sectioned hepatocytes were analyzed with Raman spectroscopy. As shown in Figure 5H, typical D and G bands of FLG were found in the S-FLG treated group, while D and G bands were not observed in the control group (fish not exposed to FLG). These results indicate that S-FLG was phagocytized by the hepatocyte cells after uptake into the liver.
Figure 5.
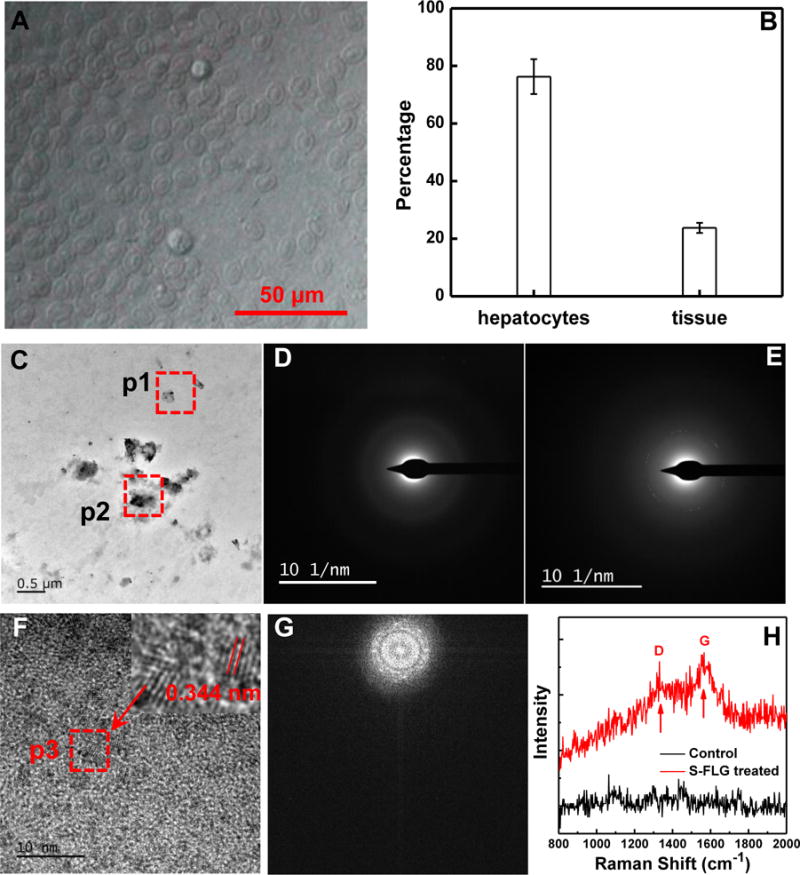
(A) Microscope image of the obtained hepatocytes isolated from the liver of the zebrafish of the S-FLG treated group; (B) The relative distribution of the S-FLG in hepatocytes from residual tissues of liver collected from the zebrafish, which was exposed for 48 h to a S-FLG suspension of 250 μg/L. Background radioactivity was subtracted. Data are presented as mean ± standard deviation (n=5).; (C) TEM image of the sectioned hepatocytes; (D) and (E) SADPs taken from p1 (control) and p2 positions, respectively. Note: six fold symmetry was observed in (E); (F) High resolution TEM image taken from p2 position in (C) (the red arrow pointed to an enlarged view of p3 position); (F) Fourier transfer spectra of Figure F; (H) Raman spectra of the sectioned hepatocytes of zebrafish treated with S-FLG.
To explore the potential toxic response of S-FLG in the liver, histopathological and biochemical (malondialdehyde (MDA), glutathione content (GSH), superoxide dismutase (SOD) and catalase (CAT)) changes to the liver tissues were analyzed. Results in Figure S7 suggest that the values of SOD, CAT, GSH and MDA in the liver (after 24, 48 and 72 h exposure) had no significant difference between the control and treatment group (250 μg/L), revealing that no obvious oxidative stress was induced by the S-FLG accumulated in the hepatocytes. The results of hematoxylin and eosin (HE) staining in Figure S8 showed that no noticeable toxic effect on liver was observed at our tested FLG condition. This results contrasts with a previous study which showed that graphene oxide (GO) induced oxidative stress and tissue damage when zebrafish exposed to GO (10 mg/L) for 14 d.45 It is likely attributed to the fact that toxic effects induced by graphene are related to its exposure time and dosage, namely that the exposure concentration used in this study was almost two orders of magnitude lower than the previous study.
Cellular Uptake of L-FLG and S-FLG in Gut
To further explore the cellular uptake of L-FLG and S-FLG in the gut tract, the gut of zebrafish (after being exposed for 48 h to a graphene concentration of 250 μg/L) was obtained and its intestinal content was washed using water and ethanol six times. The residual radioactivity in the gut tissues and intestinal content was then quantified. The L-FLG concentration in zebrafish guts was (69.8 ± 3.4) μg graphene/g dry zebrafish mass, but were contained entirely in the gut contents with the concentration in the gut tissue being below the detection limit (see Figure S9 in SI). This result revealed that the residual L-FLG after depuration was removable by washing and likely trapped by the intestinal villus.46
After being exposed in 250 μg/L of the S-FLG graphene suspension, (367 ± 11) ng (n=3) was detected in zebrafish gut. After washing the gut tract, (40 ± 2.5) ng S-FLG was still retained in the gut, which was 11% of the body burden (see Figure S8). To explore the cellular uptake of the S-FLG, intestinal epithelial cells from graphene treated zebrafish were collected, washed, centrifuged and the cellular precipitate was resuspended for LSC analysis. Approximately (16.8 ± 3.4) ng graphene was detected in the intestinal epithelial cells of the S-FLG treated fish (Figure 6A). The control experiment showed a lack of detectable artifacts from the separation process;18 the radioactivity in cells from control fish that were briefly mixed with suspended S-FLG for 5 min before conducting the separation process was below the detection limit. Notably, the quantity of S-FLG entering into the intestinal cells was magnified by a factor of 2.8 when the exposure suspension contained NOM. Interestingly, there was an increase in the bioaccumulation factor for earthworms exposed to FLG when the FLG had been wrapped with proteins.16 One hypothesis for this finding was that the protein coating caused the FLG to interact differently with the microvilli in the gut tract, which would be similar to results observed in this study. Figure 6B shows an image of intestinal epithelial cells (obtained from the S-FLG exposure sample), which were subsequently sectioned for TEM analysis. A low magnification TEM image of the cell shows the presence of black particles (Figure 6C). These particles were confirmed to be FLG using SADP and high resolution TEM (Figure 6E, 6F and 6G).18 The sectioned intestinal epithelial cells isolated from the gut of the zebrafish of the control untreated group were also analyzed using TEM. The six-fold symmetry typical of graphene particles were also not observed (see Figure S10A and S10B). Similar to the confirmation of S-FLG in hepatocytes, the sectioned intestinal epithelial cells were also analyzed with Raman spectroscopy and D and G bands were measured in the S-FLG treated sample. S-FLG was found in the intestinal epithelial cells, which indicated S-FLG was phagocytized by the cells after uptake into the intestines. The blood from fish exposed to 250 μg/L of S-FLG for 48 h with or without NOM was collected; analyzed and considerable radioactivity was detected in the blood of the S-FLG treated zebrafish (see Figure S11). The gut has the function to adsorb nutrients enabling them to enter into blood circulation. The considerable S-FLG amounts measured in blood, liver and intestinal epithelial cells indicate that S-FLG may pass through the intestinal tissues into blood and then be delivered to liver (see Scheme S1 in SI).
Figure 6.
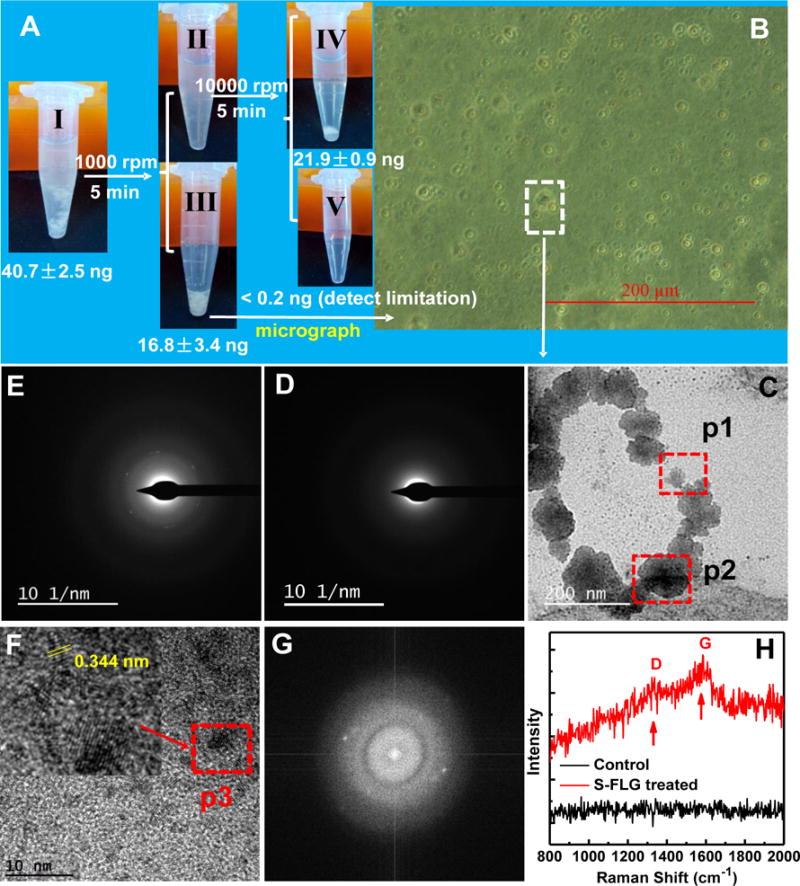
(A) S-FLG content in intestinal epithelial cells collected from the zebrafish of the S-FLG treated group, background radioactivity was subtracted. Tube I, gut tissues (after removal of the intestinal contents) were incubated with enzyme solution (collagenase and dispase). Tube II, mixture after the removing of cellular precipitate. Tube III, cellular precipitate. Tube IV, residual tissues. Tube V, mixture solution. Data are presented as mean ± standard deviation (n=5).; (B) microscope of the obtained cells; (C) TEM image of the intestinal epithelial cells; (D) and (E) SADPs taken from p1 (control) and p2 positions, respectively. Note: six fold symmetry was observed in (E); (F) High resolution TEM image taken from p2 position in (C) (the red arrow pointed to an enlarged view of p3 position); (G) Fourier transfer spectra of Figure F; (H) Raman spectra of the sectioned intestinal epithelial cells of zebrafish treated by S-FLG.
The intestinal contents from fish exposed to L-FLG or S-FLG were also analyzed using Raman spectroscopy. The presence of graphene in the intestinal content was confirmed through the observed D and G bands (distinctive of graphitic materials) (see Figure S12). Chemical peaks indicative of the formation of degradation products in the intestines were not found in the extraction solution using either HPLC or GC-MS. In addition, the radioactivity of the extraction solutions did not statistically differ from the background. Therefore, L-FLG (or S-FLG) degradation was not detectable in gut. Together with the confirmation of graphene in intestinal cells, we thus conclude that the radioactivity detected in the blood and liver was from graphene not its metabolites. These results do not exclude the potential of modifications to the L-FLG or S-FLG particles in the gut, but a robust extraction and characterization procedure is not yet available to conclusively make this measurement. Our data suggests the uptake of graphene in epithelial cells of fish, although uptake of FLG into mammalian cells has been observed.17,18,26 It was observed that CNTs, which possess a tubular shape, were able to interact with lipid bilayers and cross into the cytoplasm.47 S-FLG with a smaller size and more planar shape has a greater possibility of penetration through membranes which may explain its detection in the intestinal epithelial cells and blood.48
Toxic Response of Gut after FLG Uptake
According to the distribution results we described above, the FLG was mainly accumulated in the gut, regardless of whether the exposure media contained NOM or the FLG size. In particular, S-FLG was found in the intestinal epithelial cells. To explore the potential toxic response of FLG in the gut, histopathological changes to the gut tissues were analyzed. The results of HE staining in Figure S13 showed that the gut had no toxic histopathological changes as the result of the L-FLG or S-FLG exposure at our tested conditions.
Ingestion of FLG may also impact the gut microbiota which play critical roles in immunity, health maintenance and especially energy metabolism.49,50 Therefore, we also analyzed the impact of exposure to S-FLG or L-FLG on the biodiversity of intestinal flora. Alpha diversity, including Abundance Coverage-based Estimator (ACE) and Simpson’s diversity index were calculated to analyze the differences in bacterial communities among the treatment groups.51,52 No significant difference in ACE was found among the control, L-FLG and S-FLG treated groups, which indicated that L-FLG and S-FLG exposure induced no significant changes in community richness compared to the control group (Figure 7A). Significant difference of Simpson’s diversity index was found between control and S-FLG treated group (p < 0.05), which suggested S-FLG exposure increased the community diversity, while no significant differences were observed between control and L-FLG treated group (Figure 7B). To quantify the impact of FLG exposure on gut microbial community structure of the zebrafish, non-metric multidimensional scaling (NMDS) plots of the bacterial communities based on Bray-Curtis dissimilarities was performed.53,54 A difference in community structures were observed between the control and S-FLG treated groups (p < 0.05) but not between the control and L-FLG treated groups (Figure 7C). The result was consistent with the NMDS plot that the between-group distances were significantly higher than the within-group distances for control and S-FLG treated group,55 but not for the control and L-FLG treated group (p = 0.43) (Figure 7D). These data indicated that the microbial community structures between control and S-FLG treated group were significantly different.
Figure 7.
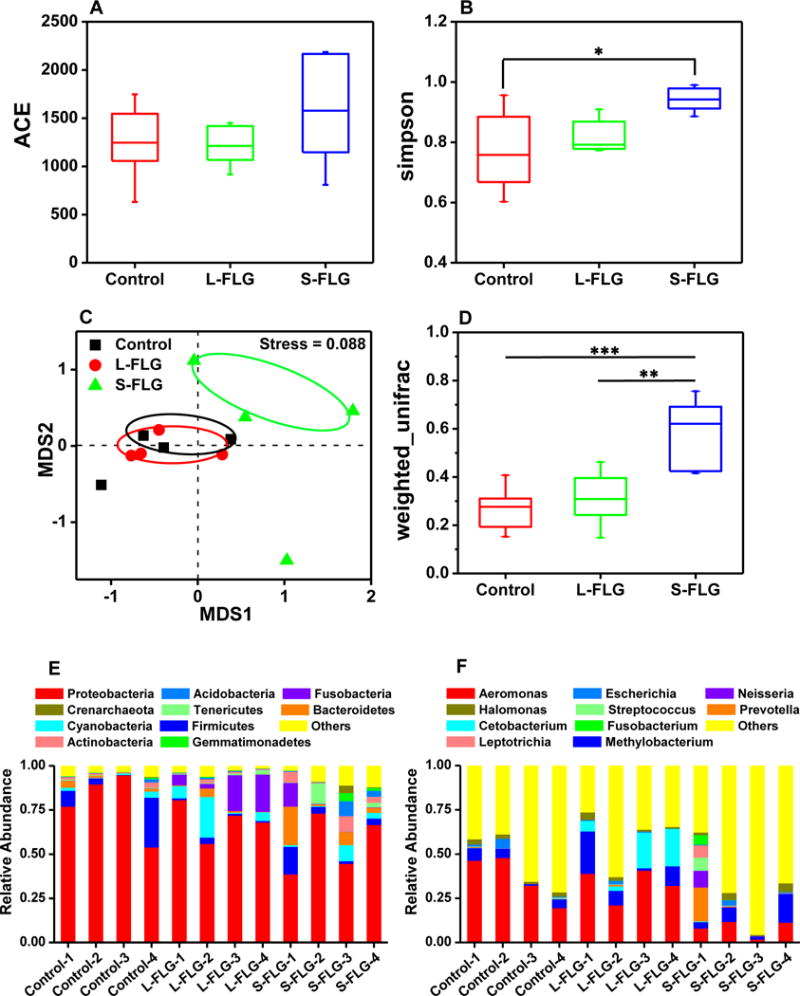
Differences in bacterial community richness, diversity and structures among three groups (control, L-FLG and S-FLG). (A) Abundance coverage-based estimator (ACE). (B) Simpson’s diversity index. (C) Non-metric multi-dimensional scaling (NMDS) plots of the bacterial communities based on Bray-Curtis dissimilarities. (D) Comparison of the weighted Unifrac distances (** p < 0.01, *** p < 0.001). Microbial composition of three groups in phylum (E) and genus (F) level. Date are presented as mean ± standard deviation (n=4).
Data in Figure 7E further suggest that the main microbial community in gut of the control group are Proteobacteria, which account for (78 ± 18)% of the total gut flora and was consistent with previous results from zebrafish studies.56 While the dominant community of Proteobacteria was the same between the control and S-FLG-treated zebrafish, its relative abundance in the gut microbial community statistically differed. The relative abundance of Proteobacteria in zebrafish exposed to S-FLG was significantly decreased to (55 ± 16)% compared to the control group while the relative abundance in zebrafish exposed to L-FLG was not significantly changed (70 ± 10) %. At the genus level, the quantity of Aeromonas was similar between the control and L-FLG-treated zebrafish, but the relative abundance of Aeromonas in S-FLG-treated zebrafish statistically differed and decreased from (36 ± 13)% to (8 ± 5)% (Figure 7F).
It has been reported that the production of carbon radicals on the graphene surfaces is associated with bactericidal effects.19 However, as shown in Figure 1D, the carbon radical densities on the surfaces of S-FLG and L-FLG were not significantly different. To further investigate the cause of the above changes, 4 bacterial strains of the Aeromonas (E. coli, A. caviae, M. extorquens and S. mutans) were selected as model bacteria of the zebrafish intestinal flora and incubated with S-FLG or L-FLG. E. coli, A. caviae and M. extorquens were used as models for Gram-negative bacteria and S. mutans was used as model for Gram-positive bacteria. The MTT assay was conducted to measure the viability loss of bacteria induced by FLG. As shown in Figure 8 (A), after 1 d incubation with various concentrations of the S-FLG, the viability of A. caviae decreased to > 80 %, while no viability loss was observed after incubation with L-FLG. For S. mutans and M. extorquens, the viability loss induced by S-FLG was statistically more severe than that from exposure to L-FLG (p<0.05). However, E. coli was killed by both of S-FLG and L-FLG (not statistically significant different) after 1 d incubation, a result consistent with the negligible Escherichia proportion change at the genus level of microbial community (Figure 7F). The viability loss of 4 bacteria strains after incubation with FLG may explain the microbial composition disturbance induced by FLG.
Figure 8.
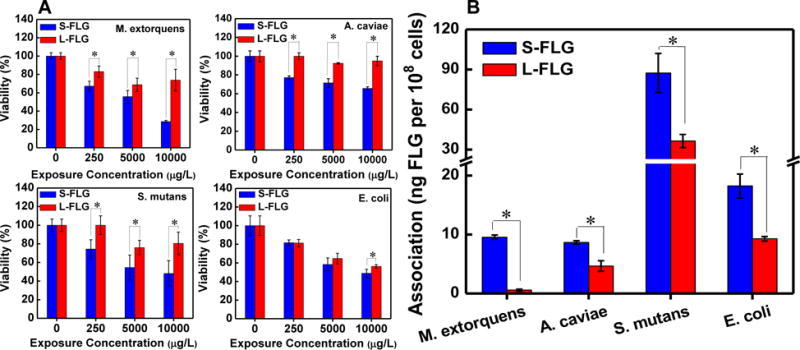
The effect of S-FLG and L-FLG on cell viability (A) and the association (B) of S-FLG and L-FLG by bacterial strains selected from various genus of zebrafish intestinal flora. Data are presented as mean ± standard deviation (n=4). Statistically significant differences between the S-FLG and L-FLG were testing using t-tests and asterisks (*) indicate statistically significant differences.
Experiments were also conducted to evaluate the association of bacteria with FLG. Control experiments were first conducted to assess the efficiency of the extraction process to separate FLG and bacteria. Bacteria were spiked with FLG and then the separation process was immediately performed. These results showed that > 94.5 % ± 0.2 % of the FLG remained suspended in the suspension. To calculate the amount of FLG that was associated with the bacteria after a 2 h exposure, the amount from the 0 h exposure described above was subtracted from the results for the 2 h exposure. As shown in Figure 8 (B), association of S-FLG and L-FLG was found in E. coli, A. caviae, S. mutans, while the quantity of S-FLG associated with the three bacterial was about magnified by a factor of 2 compared with that of L-FLG. Notably, 9.6 ng ± 0.4 ng S-FLG but no considerable amount of L-FLG (not statistically significant difference with 0) was detected in M. extorquens (108 cells) after being incubated with 250 μg/L of the FLG for 2 h. The lack of detectable uptake of L-FLG is similar to the results shown above where exposure to L-FLG did not statistically change the gut microbial community. Taken together, the decreasing of the species in Figure 7 is likely attributed to the highest accumulation of S-FLG in the gut microbiota.
CONCLUSIONS
If they are released into the environment, graphene family materials will come into contact with a range of ecological receptors. This study answers fundamental questions about the bioaccumulation and biodistribution of graphene family material after uptake by fish through conducting experiments with two FLG sizes and three concentrations using carbon-14 labeled FLG to enable accurate quantification of FLG in the fish tissues. The increased uptake and decreased depuration rates from the presence of NOM highlight the key importance of conducting uptake studies under more environmentally relevant conditions such as with NOM. Studies conducted in water without NOM may underestimate the bioaccumulation of graphene and provide different information about the biodistribution and depuration rates. This suggests that bioaccumulation studies with other carbon nanomaterials should strongly consider testing the effects of NOM especially given the range of different types of NOM in different environmental systems. While the results for L-FLG were largely similar to those observed in previous studies with fish and carbon nanotubes, the results for the S-FLG differed with S-FLG being detected in the blood and liver. Even if graphene materials are released into the environment with larger sizes similar to the L-FLG, the potential for degradation of FLG if the material stays suspended in the water column for a sufficiently long period could lead to bioaccumulation behaviors more similar to the S-FLG. Therefore, environmental modeling of the environmental impacts of FLG needs to consider that the bioaccumulation behaviors may change as a result of degradation processes.
EXPERIMENTAL SECTION
Graphene Preparation and Physiochemical Characterization
In our previous study, we described the synthesis, purification, and characterization of 14C-labeled few layer graphene (FLG).13 A synthesis process based upon graphitization and exfoliation of sandwich-like FePO4/dodecylamine hybrid nanosheets was used to synthesize the FLG. Iron catalysts were then removed from these materials through purification using hydrochloric acid. The specific radioactivity of the purified graphene was (16.12 ± 0.59) mCi/g (n=3; uncertainties always indicate standard deviation values). Vacuum filtration (SHZIII, Nanjing Keer Instrument Equipment Co., Ltd., China) with a 0.22 μm pore size membrane (cellulose acetate, SCBB-207, 50 mm, Anpel Scientific Instrument Co., Ltd., China) was used to further treat the as-prepared FLG samples. This sample was named larger-sized graphene samples (L-FLG), and was used as the substrate for the preparation of smaller-sized graphene (namely S-FLG). S-FLG was prepared through probe sonication (Ultrasonic cell disrupter. JY88-II, Nanjing Immanuel Instrument Equipment Co, China) with cycles of 3 s work and 2 s rest with ultrasonic power 100 W in an ice-water bath for 60 h. After sonication, the S-FLG was washed sequentially using methanol and n-hexane, and then obtained by centrifugation at 49 000 g (4 °C; Biofuge Stratos, Kendro Laboratory Products Co., US). Mass spectrometry and liquid scintillation counting (LSC) analyses were formed as described in our previous studies13 to test for the formation of carbon- 14 byproducts during all steps of the processing of the L-FLG and S_FLG (synthesis, purification, or dispersion). No such byproducts were detected.
L-FLG and S-FLG were analyzed by X-ray photoelectron spectroscopy (XPS, PHI 5000 VersaProbe with a monochromatic Al Ka X-ray source), Raman spectroscopy (XploRA PLUS system, Horiba Scientific, 532 nm incident radiation), and atomic force microscope (AFM, Bruker, German) as described in Supporting Information (SI).25 The electrophoretic mobility (EPM) of FLG samples were measured in the absence and presence of NOM via the Malvern Nano ZS instrument; preparation of the NOM solution is described in the “Uptake Experiments” section. For each FLG suspension condition, ten measurements of the EPM were conducted for each of three samples. Electron paramagnetic resonance (EPR) measurements of L-FLG and S-FLG (0.25 mg/mL) were carried out at room temperature by an X-band Bruker EMX-10/12 spectrometer with the frequency around 9.7709 GHz, a magnetic field at 3480 G, and a g value at 2.0029.
Test Organisms
Wild-type adult zebrafish (> 3 months old, Danio rerio, AB strain, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China) with body lengths of (34.2 ± 1.6) mm and weights of (42.8 ± 16.4) mg (dry weight) (n=50) were acclimated to the experimental conditions ((28 ± 0.5) °C; 14 h: 10 h, light:dark; aerated freshwater (aeration time >3 d); daily water change) for at least 5 d before the uptake and depuration experiments.57 No abnormal behavior or death was observed during the uptake and depuration experiments.
Uptake Experiments
In brief, one zebrafish was added to each container containing 250 mL of graphene suspension (50 μg/L, 75 μg/L and 250 μg/L); full details are presented in the SI. There was no feeding during these experiments. Triplicate control containers without zebrafish but with 250 mL of the exposure media with varying concentrations of graphene were used to monitor graphene settling during the exposure period. At each predetermined interval ((4, 12, 24, 48, 72) h), five containers were sampled; organism mortality was not observed for any exposure condition. After removal from the container, each zebrafish was washed, freeze-dried (using a lyophilizer for 5 d), weighed (Mettler Toledo) and combusted in the biological oxidizer (BO) (OX-500; Zinsser Analytic, Germany). The samples were oxidized for 4 min at 900 °C under an oxygen gas stream running at 360 mL/min. The combustion process caused the release of 14CO2 which was captured in 10 mL alkaline scintillation cocktail (Zinsser Analytic, Germany). These samples were then analyzed using LSC. The radioactivity from control samples (i.e. zebrafish unexposed to graphene) was subtracted from all of the uptake results. After fish removal, aqueous-phase radioactivity was also measured as described above to determine the concentration of graphene remaining in the suspension. The minimum detection limit of BO is detected as 0.2 ng 14C graphene; this value was determined by measuring multiple blank samples and then adding three times the standard deviation of these samples to the mean of the blank samples.
Suwannee River NOM (RO isolation, International Humic Substances Society) was used as a model NOM and a stock solution of SRNOM was prepared with a total organic carbon (TOC) concentration of (860 ± 1) mg/L (n=3) (details for the TOC analysis are available in the SI). Graphene suspensions (50 μg/L, 75 μg/L and 250 μg/L) with a NOM concentration of 10 mg (TOC)/L were utilized to examine the extent to which the presence of NOM influences graphene uptake by zebrafish using the same exposure setup and procedure described above.
Depuration Experiments
Elimination experiments were conducted similarly to the uptake experiments. After acclimating fish for 5 d in aerated freshwater without graphene, 30 containers (each with 1 organism) were prepared and then exposed for 48 h to 75 μg/L graphene suspension with or without the presence of NOM (10 mg TOC/L). Five fish were sampled, treated, dried, weighed and their radioactivities determined as described above. Each of the remaining fish (25 organisms in total) was transferred to container with 250 mL clean freshwater. At each predetermined interval ((4, 12, 24, 72, 120) h), five fish were sampled from the depuration freshwater and sacrificed to measure graphene concentration in the body, respectively.
Distribution of Graphene in Adult Zebrafish
Ten replicates (each with 10 fish) were exposed for 48 h to 250 mL of 75 μg/L S-FLG (or L-FLG) suspensions with or without 10 mg (TOC)/L NOM for each condition (40 total replicates) and then washed by pipetting clean water onto them. Five sets of organisms (10 zebrafish in each set) were sacrificed and their tissues (including brain, liver, gill, gut, roe and carcass tissues) were harvested. Each tissue in one set (10 zebrafish) was combined, freeze-dried, weighed, combusted using BO and then the radioactivity was determined using LSC. The remaining 5 sets of organisms (10 zebrafish in each set) were added to clean freshwater for 48 h depuration and then were sacrificed. Their tissues, including brain, liver, gill, gut, roe and carcass were harvested. Then, each tissue was freeze-dried, weighed, combusted using BO and the radioactivity was determined as described above.
Cellular Uptake of S-FLG in Liver and Toxicity Response
After 48 h exposure in a 250 mL 250 μg/L S-FLG suspension, the hepatocytes were isolated by modifying a previous method.58,59 Generally, zebrafish were quickly euthanized in melting ice and the liver was carefully removed, transferred onto a glass dish, and rinsed three times with phosphate buffered saline (PBS: 8.0 g/L NaCl, 0.4 g/L KCl, 0.06 g/L KH2PO4, 0.06 g/L Na2HPO4, 0.35 g/L NaHCO3, pH = 7.4) to remove the blood. Then, the liver was cut into small pieces, and the tissue was digested for 20 min at 28 °C with PBS containing 0.1 % collagenase I (Sigma) on a shaker. The softened liver tissue was mixed and filtered through a 70 μm nylon mesh (Sangon Biotech, China). The resulting hepatocyte cell suspension was collected and centrifuged three times at 100 g for 5 min at 4 °C. The cellular precipitate was combusted using BO and LSC was used to quantify the radioactivity in the cell suspension. A fraction of the obtained cells (from the samples without NOM) were prepared for TEM analysis by washing them with physiological saline, prefixing at 4 °C overnight in 3 % (mass fraction) glutaraldehyde, and then post-fixing in 1 % osmium tetroxide. Cells were sectioned to 50 nm to 60 nm thick pieces after dehydration and resin embedding. Then, the pieces were stained with uranyl acetate and lead citrate as previously described.18,60 High resolution TEM imaging and selective area electron diffraction patterns (SADP) was performed on a 200kV FEI Tecnai TF20 FEG-TEM18,40 to identify FLG in the cells. The presence of graphene in these cells was also evaluating using Raman spectroscopy (see SI). The cells obtained from the untreated zebrafish (not exposed to FLG) were subjected to the identical analysis.
Zebrafish were exposed to 250 μg/L S-FLG suspension (250 mL) and then tested for biochemical and histopathological changes. At predetermined exposure time (24 h, 48 h and 72 h), MDA, GSH, SOD and CAT was determined to assess the toxic response of the liver (a full description of the methods used is provided in the SI). Liver after 0 h or 72 h exposure was sampled, sectioned and observed using an optical microscope equipped with a digital camera (a full description of the method is provided in the SI).
Cellular Uptake of L-FLG and S-FLG in Gut
After 48 h exposure in 250 mL of a 250 μg/L S-FLG (or L-FLG) suspension with or without NOM, 3 replicates (each with 5 fish) per condition were sampled and their blood and gut tract were harvested. Blood was combined from all of the fish in each replicate, freeze-dried, combusted using BO, and then the radioactivity was determined using LSC. The intestinal contents in the harvested guts was removed and collected by washing the gut tract with water and methanol 6 times each and then the gut contents were combined and freeze-dried for subsequent Raman spectroscopy analysis or for the analysis of potential metabolic products. The obtained intestinal contents were freeze-dried and extracted in sequence using dichloromethane (5 mL) and n-hexane (5 mL) to determine if metabolic products were formed from the FLG. The two solutions were recombined, and water was removed using anhydrous sodium sulfate. For HPLC and GC-MS analysis, the sample was dried using a gentle nitrogen stream and reconstituted in a 4:1 ratio by volume of methanol and dichloromethane (additional details are provided in the SI). Both the extraction liquids and residues were also analyzed using LSC although the residues of the gut contents after extraction were first combusted using BO.
The obtained gut tissues after removal of the intestinal contents were further washed three times using Hanks’ buffered salt solution (HBSS, low calcium formulation).61 Parts of the gut tract tissues were weighed and then analyzed using BO and LSC to quantify the radioactivity in the tissues after the contents were removed. Then, the remaining pieces of the cleaned gut tissues were cut into 1 mm to 2 mm length pieces, transferred into a 1.5 mL centrifuge tube with 1 mL enzyme solution (collagenase and dispase), and shaken vigorously for 20 min at 28 °C.61 The solution was pipetted vigorously ≈ 50 times and centrifuged for 5 min at 2100 g to obtain the cellular precipitate. The cellular precipitate was combusted using BO and LSC was used to quantify the radioactivity in the cell suspension. A fraction of the obtained cells (from the samples without NOM) were prepared for TEM analysis. The samples were analyzed using high resolution TEM imaging and SADP, and then further analyzed with Raman spectroscopy (shown in SI); the sample preparation and TEM analysis methods are the same as used for the liver hepatocyte cell analysis. Control experiments were performed to assess if a quantifiable amount of FLG adsorbed on the cell membranes during the separation process or if the FLG was incompletely removed. Cells obtained from control zebrafish (not exposed to FLG) were mixed with 200 μL of a 14C labeled FLG suspension (250 μg/L). Within 5 min after mixing, low speed centrifugation was used to separate the cells from the FLG suspension. Then, the cellular precipitate was washed using water and methanol 6 times and then washed using HBSS, and lastly, the radioactivity was quantified as described above. Gut tissues after fish were exposed to 250 μg/FLG for 72 h were also sampled, sectioned and observed using an optical microscope equipped with a digital camera (details were shown in SI).
Toxic Response of Gut after FLG Uptake
Forty zebrafish were exposed in a single container with 12 L of aerated water as a control group, while groups of 40 zebrafish were exposed with 12 L of either L-FLG or S-FLG suspension (5 mg/L). The exposure suspension was sonicated four times per day using a probe tip sonicator for 5 min to keep the settling < 30 %, and the zebrafish were transferred to a separate container during this period. After exposure for 2 weeks, forty zebrafish of each group were randomly divided to be four groups of ten zebrafish. The germ free feces of each group were collected from their rectums, combined, and stored in −80 °C. Total genomic DNA was extracted from each of the three groups and analyzed as described in SI for details.
4 bacterial strains, including Escherichia coli (E. coli), Aeromonas caviae (A. caviae), Streptococcus mutans (S. mutans), Methylobacterium extorquens (M. extorquens), were obtained from BeNa Culture Collection. Each of the bacteria was cultured and grown in a defined culture medium (the culture media preparation and components are described in the SI). The cultures were suspended in 10 mM PBS buffer (pH 7.4) and the suspension was diluted to 5×108 cell mL−1. The effect of S-FLG or L-FLG on cell viability was measured by MTT assay.62 A detailed experimental description is provided in the SI. S-FLG or L-FLG was added to the bacterial culture yielding a final nominal graphene concentration of 250 μg/L. Bacteria were then incubated at 30 °C and 250 rpm (26 rad/s). After 0 or 2 h incubation, 5 mL culture solution was transferred to sterile centrifuge tubes (polypropylene 10 mL) which contained 0.1% Tween 80 (final concentration) and centrifuged at 8600 g for 5 min. The purpose of the 0.1 % Tween 80 was to facilitate separation between the FLG and the bacteria through keeping the FLG suspended while the bacteria settled. Supernatant was removed and the remained bacterial was further washed by vortexing (5 s) in 5 mL of PBS containing 0.1% Tween 80 for 5 times to remove the possible free graphene and loosely absorbed graphene. The optical density of obtained bacteria was measured and the radioactivity associated with the cells was quantified by LSC.
Statistical Analyses
SPSS 18.0 (PASW Statistics, IBM Company) was applied for statistical analyses; differences were considered statistically significant at p < 0.05. One-way analysis of variance (ANOVA) and Tukey’s post hoc test was used for data analysis.
Supplementary Material
Acknowledgments
We acknowledge the financial support from the National Natural Science Foundation of China (21377049 and 21677074) and a Foundation for the Author of National Excellent Doctoral Dissertation of PR China (201355).
Footnotes
Supporting Information
Additional description of certain experimental procedures; Figure S1-S11. These materials are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ren W, Cheng HM. The Global Growth of Graphene. Nat Nanotechnol. 2014;9:726–730. doi: 10.1038/nnano.2014.229. [DOI] [PubMed] [Google Scholar]
- 2.Allen MJ, Tung VC, Kaner RB. Honeycomb Carbon: A Review of Graphene. Chem Rev. 2010;110:132–145. doi: 10.1021/cr900070d. [DOI] [PubMed] [Google Scholar]
- 3.Geim AK, Novoselov KS. The Rise of Graphene. Nat Mater. 2007;6:183–191. doi: 10.1038/nmat1849. [DOI] [PubMed] [Google Scholar]
- 4.Stankovich S, Dikin DA, Dommett GHB, Kohlhaas KM, Zimney EJ. Graphene- Based Composite Materials. Nature. 2006;442:282–286. doi: 10.1038/nature04969. [DOI] [PubMed] [Google Scholar]
- 5.Singh V, Joung D, Zhai L, Das S, Khondaker SI. Graphene Based Materials: Past, Present and Future. Prog Mater Sci. 2011;56:1178–1271. [Google Scholar]
- 6.Stoller MD, Park S, Zhu Y, An J, Ruoff RS. Graphene-Based Ultracapacitors. Nano Lett. 2008;8:3498–3502. doi: 10.1021/nl802558y. [DOI] [PubMed] [Google Scholar]
- 7.Selck H, Handy RD, Fernandes TF, Klaine SJ, Petersen EJ. Nanomaterials in the Aquatic Environment: A European Union-United States Perspective on the Status of Ecotoxicity Testing, Research Priorities, and Challenges Ahead. Environ Toxicol Chem. 2016;35:1055–1067. doi: 10.1002/etc.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu M, Zhu J, Wang F, Xiong Y, Wu Y, Wang Q, Weng J, Zhang Z, Chen W, Liu S. J Improved in vitro and in vivo Biocompatibility of Graphene Oxide Through Surface Modification: Poly(Acrylic Acid)-Functionalization Is Superior to PEGylation. ACS Nano. 2016;10:3267–3281. doi: 10.1021/acsnano.6b00539. [DOI] [PubMed] [Google Scholar]
- 9.Akhavan O, Ghaderi E. Toxicity of Graphene and Graphene Oxide Nanowalls against Bacteria. ACS Nano. 2010;4:5731–5736. doi: 10.1021/nn101390x. [DOI] [PubMed] [Google Scholar]
- 10.Yang K, Li YJ, Tan XF, Peng R, Liu Z. Behavior and Toxicity of Graphene and Its Functionalized Derivatives in Biological Systems. Small. 2013;9:1492–1503. doi: 10.1002/smll.201201417. [DOI] [PubMed] [Google Scholar]
- 11.Bianco A. Graphene: Safe or Toxic? The Two Faces of the Medal. Angew Chem Int edit. 2013;52:4986–4997. doi: 10.1002/anie.201209099. [DOI] [PubMed] [Google Scholar]
- 12.Akhavan O, Ghaderi E, Hashemi E, Akbari E. Dose-Dependent Effects of Nanoscale Graphene Oxide on Reproduction Capability of Mammals. Carbon. 2015;95:309–317. [Google Scholar]
- 13.Guo XK, Dong SP, Petersen EJ, Gao SX, Huang QG, Mao L. Biological Uptake and Depuration of Radio-Labeled Graphene by Daphnia Magna. Environ Sci Technol. 2013;47:12524–12531. doi: 10.1021/es403230u. [DOI] [PubMed] [Google Scholar]
- 14.Jeong JO, Cho HJ, Choi M, Lee WS, Chung BH, Lee JS. In vivo Toxicity Assessment of Angiogenesis and the Live Distribution of Nano-Graphene Oxide and Its PEGylated Derivatives Using the Developing Zebrafish Embryo. Carbon. 2015;93:431–440. [Google Scholar]
- 15.Feng Y, Lu K, Mao L, Guo X, Gao S, Petersen EJ. Degradation of 14C-Labeled Few Layer Graphene via Fenton Reaction: Reaction Rates, Characterization of Reaction Products, and Potential Ecological Effects. Water Res. 2015;84:49–57. doi: 10.1016/j.watres.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 16.Mao L, Liu C, Lu K, Su Y, Gu C, Huang Q, Petersen EJ. Exposure of Few Layer Graphene to Limnodrilus Hoffmeisteri Modifies the Graphene and Changes Its Bioaccumulation by Other Organisms. Carbon. 2016;109:566–574. doi: 10.1016/j.carbon.2016.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Yang JZ, Huang Q, Zhang Y, Peng C, Zhang YJ, He Y, Shi J, Li W, Hu J, Fan C. Biodistribution and Pulmonary Toxicity of Intratracheally Instilled Graphene Oxide in Mice. NPG Asia Mater. 2013;5:e44. [Google Scholar]
- 18.Mao L, Hu M, Pan BC, Xie Y, Petersen EJ. Biodistribution and Toxicity of Radio-Labeled Few Layer Graphene in Mice after Intratracheal Instillation. Part Fibre Toxicol. 2016;13:7. doi: 10.1186/s12989-016-0120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li R, Mansukhani ND, Guiney LM, Ji Z, Zhao Y, Chang CH, French CT, Miller JF, Hersam MC, Nel AE, Xia T. Identification and Optimization of Carbon Radicals on Hydrated Graphene Oxide for Ubiquitous Antibacterial Coatings. ACS Nano. 2016;10:10966–10980. doi: 10.1021/acsnano.6b05692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gurunathan S, Han JW, Kim J. Green Chemistry Approach for the Synthesis of Biocompatible Graphene. Int J Nanomed. 2013;8:2719–2732. doi: 10.2147/IJN.S45174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo ZJ, Geng HZ, Zhang X, Du B, Ding EX, Wang J, Lu Z, Sun B, Wang J, Liu J. A Timesaving, Low-Cost, High-Yield Method for the Synthesis of Ultrasmall Uniform Graphene Oxide Nanosheets and Their Application in Surfactants. Nanotechnology. 2016;27:055601. doi: 10.1088/0957-4484/27/5/055601. [DOI] [PubMed] [Google Scholar]
- 22.Caruthers SD, Wickline SA, Lanza GM. Nanotechnological Applications in Medicine. Curr Opin Biotechnol. 2007;18:26–30. doi: 10.1016/j.copbio.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Hou WC, Chowdhury I, Goodwin DG, Henderson M, Fairbrother DH, Bouchard D, Zepp RG. Photochemical Transformation of Graphene Oxide in Sunlight. Environ Sci Technol. 2015;49:3435–3443. doi: 10.1021/es5047155. [DOI] [PubMed] [Google Scholar]
- 24.Hu X, Zhou M, Zhou Q. Ambient Water and Visible-Light Irradiation Drive Changes in Graphene Morphology, Structure, Surface Chemistry, Aggregation and Toxicity. Environ Sci Technol. 2015;49:3410–3418. doi: 10.1021/es503003y. [DOI] [PubMed] [Google Scholar]
- 25.Lu K, Huang Q, Wang P, Mao L. Physicochemical Changes of Few-Layer Graphene in Peroxidase-Catalyzed Reactions: Characterization and Potential Ecological Effects. Environ Sci Technol. 2015;49:8558–8565. doi: 10.1021/acs.est.5b02261. [DOI] [PubMed] [Google Scholar]
- 26.Ma J, Liu R, Wang X, Liu Q, Chen Y, Valle RP, Zuo YY, Xia T, Liu S. A Crucial Role of Lateral Size for Graphene Oxide in Activating Macrophages and Stimulating Pro- Inflammatory Responses in Cells and Animals. ACS Nano. 2015;9:2155–2175. doi: 10.1021/acsnano.5b04751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Templeton RC, Ferguson PL, Washburn KM, Scrivens WA, Chandler GT. Life-Cycle Effects of Single-Walled Carbon Nanotubes on an Estuarine Meiobenthic Copepod. Environ Sci Technol. 2006;40:7387–7393. doi: 10.1021/es060407p. [DOI] [PubMed] [Google Scholar]
- 28.Scharf A, Piechulek A, von Mikecz A. Effect of Nanoparticles on the Biochemical and Behavioral Aging Phenotype of the Nematode Caenorhabditis Elegans. ACS Nano. 2013;7:10695–10703. doi: 10.1021/nn403443r. [DOI] [PubMed] [Google Scholar]
- 29.Kim KM, Kim MK, Paek HJ, Choi SJ, Oh JM. Stable Fluorescence Conjugation of ZnO Nanoparticles and Their Size Dependent Cellular Uptake. Colloids Surfa B. 2016;145:870–877. doi: 10.1016/j.colsurfb.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 30.Bertoli F, Garry D, Monopoli MP, Salvati A, Dawson KA. The Intracellular Destiny of the Protein Corona: A Study on Its Cellular Internalization and Evolution. ACS Nano. 2016;10:10471–10479. doi: 10.1021/acsnano.6b06411. [DOI] [PubMed] [Google Scholar]
- 31.Müller RH, Rühl D, Lück M, Paulke BR. Influence of Fluorescent Labelling of Polystyrene Particles on Phagocytic Uptake, Surface Hydrophobicity, and Plasma Protein Adsorption. Pharm Res. 1997;14:18–24. doi: 10.1023/a:1012043131081. [DOI] [PubMed] [Google Scholar]
- 32.Pakarinen K, Petersen EJ, Alvila L, Waissi-Leinonen GC, Akkanen J, Leppänen MT, Kukkonen JV. AA Screening Study on the Fate of Fullerenes (nC60) and Their Toxic Implications in Natural Freshwaters. Environ Toxicol Chem. 2013;32:1224–1232. doi: 10.1002/etc.2175. [DOI] [PubMed] [Google Scholar]
- 33.Maity AR, Chakraborty A, Mondal A, Jana NR. Carbohydrate Coated, Folate Functionalized Colloidal Graphene as a Nanocarrier for Both Hydrophobic and Hydrophilic Drugs. Nanoscale. 2014;6:2752–2758. doi: 10.1039/c3nr05431d. [DOI] [PubMed] [Google Scholar]
- 34.Kim H, Lee D, Kim J, Kim T, Kim WJ. Photothermally Triggered Cytosolic Drug Delivery via Endosome Disruption Using a Functionalized Reduced Graphene Oxide. ACS Nano. 2013;7:6735–6746. doi: 10.1021/nn403096s. [DOI] [PubMed] [Google Scholar]
- 35.Jorio A, Cancado LG. Perspectives on Raman Spectroscopy of Graphene-Based Systems: from the Perfect Two-Dimensional Surface to Charcoal. Phys Chem Chem Phys. 2012;14:15246–15256. doi: 10.1039/c2cp42621h. [DOI] [PubMed] [Google Scholar]
- 36.Pimenta MA, Dresselhaus G, Dresselhaus MS, Cancado LG, Jorioa A, Saitoe R. Studying Disorder in Graphite-Based Systems by Raman Spectroscopy. Phys Chem Chem Phys. 2007;9:1276–1291. doi: 10.1039/b613962k. [DOI] [PubMed] [Google Scholar]
- 37.Ferrari AC. Raman Spectroscopy of Graphene and Graphite: Disorder, Electron-Phonon Coupling, Doping and Nonadiabatic Effects. Solid State Commun. 2007;143:47–57. [Google Scholar]
- 38.Yang L, Zhang R, Liu B, Wang J, Wang S, Han MY, Zhang Z. Pi-Conjugated Carbon Radicals at Graphene Oxide to Initiate Ultrastrong Chemiluminescence. Angew Chem Int Ed. 2014;53:10109–10113. doi: 10.1002/anie.201405295. [DOI] [PubMed] [Google Scholar]
- 39.Su Y, Yang G, Lu K, Petersen EJ, Mao L. Colloidal Properties and Stability of Aqueous Suspensions of Few-Layer Graphene: Importance of Graphene Concentration. Environ Pollut. 2016;220:469–477. doi: 10.1016/j.envpol.2016.09.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgington AJ, Petersen EJ, Herzing AA, Podila R, Rao A, Klaine SJ. Microscopic Investigation of Single-Wall Carbon Nanotube Uptake by Daphnia Magna. Nanotoxicology. 2014;8:2–10. doi: 10.3109/17435390.2013.847504. [DOI] [PubMed] [Google Scholar]
- 41.Edgington AJ, Roberts AP, Taylor LM, Alloy MM, Reppert J, Rao AM, Mao J, Klaine SJ. The Influence of Natural Organic Matter on the Toxicity of Multiwalled Carbon Nanotubes. Environ Toxicol Chem. 2010;29:2511–2518. doi: 10.1002/etc.309. [DOI] [PubMed] [Google Scholar]
- 42.Petersen EJ, Huang Q, Weber WJ., Jr Ecological Uptake and Depuration of Carbon Nanotubes by Lumbriculus variegatus. Environ Health Persp. 2008;116:496. doi: 10.1289/ehp.10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maes HM, Stibany F, Giefers S, Daniels B, Deutschmann B, Baumgartner W, Schaffer A. Accumulation and Distribution of Multiwalled Carbon Nanotubes in Zebrafish (Danio rerio) Environ Sci Technol. 2014;48:12256–12264. doi: 10.1021/es503006v. [DOI] [PubMed] [Google Scholar]
- 44.Bisesi JH, Merten J, Liu K, Parks AN, Afrooz ARMN, Glenn JB, Klaine SJ, Kane AS, Saleh NB, Ferguson PL, Sabo-Attwood T. Tracking and Quantification of Single-Walled Carbon Nanotubes in Fish Using Near Infrared Fluorescence. Environ Sci Technol. 2014;48:1973–1983. doi: 10.1021/es4046023. [DOI] [PubMed] [Google Scholar]
- 45.Chen M, Yin J, Liang Y, Yuan S, Wang F, Song M, Wang H. Oxidative Stress and Immunotoxicity Induced by Graphene Oxide in Zebrafish. Aquat Toxicol. 2016;174:54–60. doi: 10.1016/j.aquatox.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 46.Ragnarsson EGE, Schoultz I, Gullberg E, Carlsson AH, Tafazoli F, Lerm M, Magnusson K, Söderholm JD, Artursson1 P. Yersinia Pseudotuberculosis Induces Transcytosis of Nanoparticles across Human Intestinal Villus Epithelium via Invasin-Dependent Macropinocytosis. Lab Invest. 2008;88:1215–1226. doi: 10.1038/labinvest.2008.86. [DOI] [PubMed] [Google Scholar]
- 47.Lacerda L, Raffa S, Prato M, Bianco A, Kostarelos K. Cell-Penetrating CNTs for Delivery of Therapeutics. Nano Today. 2007;2:38–43. [Google Scholar]
- 48.Zhang YB, Ali SF, Dervishi E, Xu Y, Li ZR, Casciano D, Biris AS. Cytotoxicity Effects of Graphene and Single-Wall Carbon Nanotubes in Neural Phaeochromocytoma-Derived PC12 cells. ACS Nano. 2010;4:3181–3186. doi: 10.1021/nn1007176. [DOI] [PubMed] [Google Scholar]
- 49.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A Core Gut Microbiome in Obese and Lean Twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tremaroli V, Backhed F. Functional Interactions between the Gut Microbiota and Host Metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 51.Chao A, Hwang WH, Chen Y, Kuo C. Estimating the Number of Shared Species in Two Communities. Stat Sin. 2000;10:227–246. [Google Scholar]
- 52.Hunter PR, Gaston MA. Numerical Index of the Discriminatory Ability of Typing Systems: An Application of Simpson’s Index of Diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bray JR, Curtis JT. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol Monogr. 1957;27:325–349. [Google Scholar]
- 54.Clarke KR. Nonparametric Multivariate Analyses of Changes in Community Structure. Aust J Ecol. 1993;18:117–143. [Google Scholar]
- 55.Lozupone C, Knight R. UniFrac: a New Phylogenetic Method for Comparing Microbial Communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stephens WZ, Burns AR, Stagaman K, Wong S, Rawls JF, Guillemin K, Bohannan BJ. The Composition of the Zebrafish Intestinal Microbial Community Varies across Development. ISME J. 2016;10:644–654. doi: 10.1038/ismej.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diekmann M, Hultsch V, Nagel R. On the Relevance of Genotoxicity for Fish Populations I: Effects of a Model Genotoxicant on Zebrafish (Danio rerio) in a Complete Life-Cycle Test. Aquat Toxicol. 2004;68:13–26. doi: 10.1016/j.aquatox.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 58.Eide M, Rusten M, Male R, Jensen KHM, Goksoyr A. A Characterization of the ZFL Cell Line and Primary Hepatocytes as in vitro Liver Cell Models for the Zebrafish (Danio rerio) Aquat Toxicol. 2014;147:7–17. doi: 10.1016/j.aquatox.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 59.Reschly EJ, Bainy ACD, Mattos JJ, Hagey LR, Bahary N, Mada SR, Ou J, Venkataramanan R, Krasowski MD. Functional Evolution of the Vitamin D and Pregnane X Receptors. BMC Evol Biol. 2007;7:222. doi: 10.1186/1471-2148-7-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kolosnjaj-Tabi J, Hartman KB, Boudjemaa S, Ananta JS, Morgant G, Szwarc H, Wilson LJ, Moussa F. In vivo behavior of Large Doses of Ultrashort and Full-Length Single-Walled Carbon Nanotubes after Oral and Intraperitoneal Administration to Swiss Mice. ACS Nano. 2010;4:1481–92. doi: 10.1021/nn901573w. [DOI] [PubMed] [Google Scholar]
- 61.Evans GS, Flint N, Somers AS, Eyden B, Potten CS. The Development of a Method for the Preparation of Rat Intestinal Epithelial Cell Primary Cultures. J Cell Biol. 1992;101:219–231. doi: 10.1242/jcs.101.1.219. [DOI] [PubMed] [Google Scholar]
- 62.Almutary A, Sanderson BJ. The MTT and Crystal Violet Assays: Potential Confounders in Nanoparticle Toxicity Testing. Int J Toxicol. 2016;35:454–462. doi: 10.1177/1091581816648906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


