Abstract
The thiCOGE genes of Rhizobium etli code for enzymes involved in thiamin biosynthesis. These genes are transcribed with a 211-base untranslated leader that contains the thi box, a 38-base sequence highly conserved in the 5′ regions of thiamin biosynthetic and transport genes of Gram-positive and Gram-negative organisms. A deletion analysis of thiC-lacZ fusions revealed an unexpected relationship between the degree of repression shown by the deleted derivatives and the length of the thiC sequences present in the transcript. Three regions were found to be important for regulation: (i) the thi box sequence, which is absolutely necessary for high-level expression of thiC; (ii) the region immediately upstream to the translation start codon of thiC, which can be folded into a stem-loop structure that would mask the Shine-Dalgarno sequence; and (iii) the proximal part of the coding region of thiC, which was shown to contain a putative Rho-independent terminator. A comparative phylogenetic analysis revealed a possible folding of the thi box sequence into a hairpin structure composed of a hairpin loop, two helixes, and an interior loop. Our results show that thiamin regulation of gene expression involves a complex posttranscriptional mechanism and that the thi box RNA structure is indispensable for thiCOGE expression.
Thiamin pyrophosphate (TPP), also known as cocarboxylase, is the cofactor of key enzymes of carbon metabolism such as pyruvate dehydrogenase, α-ketoglutarate dehydrogenase, transketolase, pyruvate decarboxylase, and others. TPP is made by the enzymatic coupling of two independently synthesized precursors, 4-amino-5-hydroxymethyl-2-methyl pyrimidine pyrophosphate and 5-(2-hydroxyethyl)-4-methyl thiazole monophosphate (1).
In Escherichia coli and Salmonella enterica serovar Typhimurium, several genes are known to participate in TPP biosynthesis (2). thiC and thiD are required for 4-amino-5-hydroxymethyl-2-methyl pyrimidine pyrophosphate synthesis (3, 4), whereas dxs (5, 6), thiF, thiS, thiG, thiH, and thiI are involved in 5-(2-hydroxyethyl)-4-methyl thiazole monophosphate synthesis (3, 7, 8). In addition, the product of the thiE gene couples 4-amino-5-hydroxymethyl-2-methyl pyrimidine pyrophosphate and 5-(2-hydroxyethyl)-4-methyl thiazole monophosphate to give thiamin monophosphate, which undergoes another phosphorylation catalyzed by the product of the thiL gene to form TPP, the biologically active form of vitamin B1 (9, 10). A salvage enzyme (thiamin kinase) encoded by thiK, which incorporates exogenous thiamin into TPP (11), also exists. In both organisms the thi genes are located throughout the chromosome and are arranged in three operons and four single gene loci (2). thiCEFSGH, thiMD (ThiM and ThiD are involved in salvage of 5-(2-hydroxyethyl)-4-methyl thiazole and 4-amino-5-hydroxymethyl-2-methyl pyrimidine from the culture medium, respectively; ThiD also catalyzes the phosphorylation of 4-amino- 5-hydroxymethyl-2-methyl pyrimidine monophosphate and thiBPQ (which code for an ABC type transport system for thiamin and TPP) are the operons transcriptionally regulated by TPP, whereas the single loci are not regulated (4, 12, 13). Although some point mutations have been isolated that affect the transcriptional regulation of thi genes, they map at min 10 on the S. enterica chromosome and at least one of them is an allele of thiL gene encoding thiamin monophosphate kinase, suggesting that TPP is the effector molecule involved in regulation of thi gene expression (10, 12). No regulatory genes have been identified so far in any bacterial species.
In Rhizobium etli, which nodulates and fixes nitrogen in a symbiosis with bean plants, we have characterized a cluster of four genes named thiCOGE whose products are involved in thiamin biosynthesis and that are presumably organized as an operon (14). thiC, thiG, and thiE are homologous to E. coli genes. thiO is a gene that codes for a predicted flavoprotein with homology to d-amino acid oxidases probably involved in 5-(2-hydroxyethyl)-4-methyl thiazole monophosphate production (14). The transcription start site (tss) of the thiC gene lies 211 bases upstream of thiC start codon. The RNA untranslated leader contains a 38-base sequence, named thi box, that is highly conserved in the upstream region of thi genes of organisms as diverse as E. coli, Synechocystis, Bacillus subtilis, and Mycobacterium tuberculosis (14).
In this work, we found by functional analysis of the untranslated RNA leader that repression by thiamin is a posttranscriptional event and that the thi box is an element indispensable for the expression of thiCOGE. We also show that the region surrounding the Shine-Dalgarno (S-D) sequence and the first 443 nt of the coding region participate in the regulation of thiCOGE expression by thiamin. We propose that the thi box is a conserved RNA regulatory element that exerts its function by folding into a secondary structure whose existence is supported by a covariational and phylogenetic analysis.
Materials and Methods
Bacterial Strains, Plasmids, and Culturing.
R. etli strain CE3 was used for all of the experiments and was grown at 30°C in minimal medium (MM) with 10 mM NH4Cl as the nitrogen source and 10 mM succinate as the carbon source or in peptone yeast extract medium as described (15). Thiamin hydrochloride (100 μg/ml) was added as indicated. Plasmids were conjugated into R. etli by biparental mating using the E. coli S17.1 strain previously transformed with the plasmid of interest (16). Culturing for studying the effect of added thiamin was done as follows: cells were grown to an OD600 of 0.6 in 100 ml of MM with or without thiamin as indicated. From this culture 30 ml were centrifuged and used to inoculate 50 ml of MM with or without thiamin. This culture was grown to an OD600 of 0.7, and the cells were harvested for β-galactosidase measurements or RNA preparations.
Construction of thiC-lacZ Fusions.
All of the deletion fragments were obtained by PCR amplification using plasmid pJMR01 as template (17). This plasmid contains an EcoRI fragment of 2.5 kb containing the regulatory region of thiC and 540 bp of thiC coding region. The amplified fragments were ligated to plasmids pBlueScript-SK (Stratagene) or pCAPs (18) and were sequenced on both strands. After purification and restriction, the fragments were ligated to vector pMP220 digested with BglII and XbaI or BglII and PstI. The vector pMP220 contains the E. coli lacZ gene without a promoter generating transcriptional lacZ gene fusions (19). The following pairs of oligonucleotides were used in the amplification reactions (the sequences of all PCR primers are available on request): A-B, A-B1, A-B2, and A-C2 to construct plasmids pMM7, pMM7.1, pMM7.2, and pMM7.3, respectively. To obtain construct pMM7.4, two fragments were amplified by using oligonucleotides A-C1 and B-D1. First, the A-C1 fragment was cloned into pCAPS BglII–EcoRI. Then the B-D1 fragment was cloned into the recombinant clone carrying the A-C1 fragment in the EcoRI and XbaI sites. Finally, the BglII–XbaI fragment was excised, purified, and cloned into pMP220 digested with BglII–XbaI (Fig. 1B).
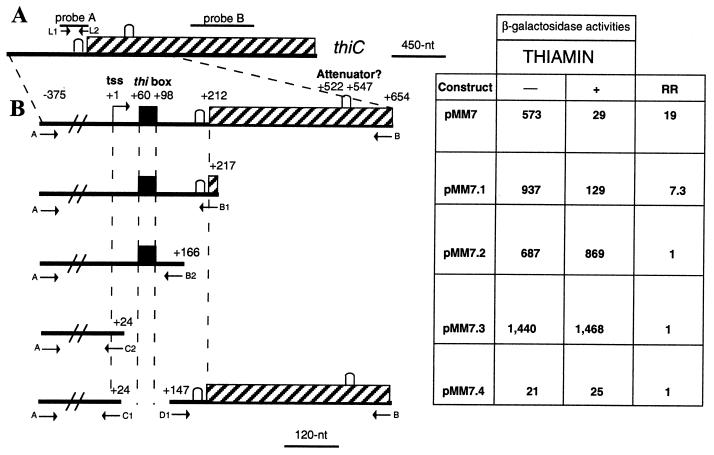
Figure 1.
(A) Schematic map of thiC gene of R. etli. The stippled box represents the thiC coding sequence. The lines above represent the location of the probes used in the Northern experiment. (B) Deletion constructions fused to lacZ. The fragments obtained by PCR were cloned into pMP220 vector to generate lacZ transcriptional fusions for determination of β-galactosidase activities from cultures grown in MM with or without thiamin as indicated. The letters alongside the arrows represent the names of the oligonucleotides used to obtain the different PCR fragments. The nucleotide positions are given with reference to the tss (position +1) of thiC. The black box represents the thi box sequence, and the potential stem-loop structures in the transcript that comprises the S-D sequence and the attenuator are marked by an inverted U. β-galactosidase activity is expressed as nmol of o-nitrophenol produced min−1⋅mg protein−1. Representative results of three experiments are shown. RR is the repression ratio, the β-galactosidase activity observed in media without thiamin divided by the activity observed in media supplemented with thiamin.
β-Galactosidase Measurements.
The level of β-galactosidase was determined as described (20), by measuring the rate of hydrolysis of o-nitrophenyl-β-d-galactopyranoside in cells permeabilized with SDS-CHCl3. All assays were performed in triplicate.
RNA Slot-Blot Hybridization.
Total bacterial RNA was isolated with the High Pure RNA Isolation Kit of Roche Molecular Biochemicals. Ten micrograms of RNA was slot-blotted into Hybond N+ membranes, hybridized, washed, and exposed to film as described by the manufacturer (Amersham Pharmacia). Probe A was obtained by using oligonucleotides L1-L2 and plasmid pJMR01 as template in a PCR amplification reaction. Probe B is a 453-bp XhoI fragment derived from the central part of the thiC coding region contained in plasmid pJMR5 (from nucleotide +1044 to +1494) (14). The 16S probe is a 500-bp HindIII fragment containing the 16S rRNA gene from E. coli (21). All probes were radioactively labeled by using 32P-dCTP and the random priming kit from Amersham Pharmacia.
Search of thi Box Sequences in GenBank.
Homologs to the thi box sequence were searched by using the blast program as implemented in the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) with the R. etli thi box as query (14). The GenBank entries of the sequences showing partial matches to the thi box were retrieved and were pairwise-aligned to the R. etli thi box by using the gap program of the Wisconsin package (GCG). In all of the cases analyzed, thiamin biosynthetic genes were present downstream of the newly found thi box sequences.
Results
Location of the thi Box Sequence in the E. coli Genome.
To determine the possible role of the thi box in gene expression, the E. coli genome was searched for the thi box sequence as detailed in Materials and Methods. Interestingly, the thi box sequence was found only in front of the three thiamin biosynthetic operons (thiCEFSGH, thiMD, and thiBPQ), whose expression in S. enterica has been shown to be negatively regulated by TPP (4, 12, 13). This correlation suggests that the thi box is a regulatory element involved in repression of gene expression by thiamin.
Effect of Deletions of the thiCOGE Regulatory Region on Expression.
As mentioned previously, an untranslated RNA leader of 211 nt containing the thi box is present in the 5′ region of the thiCOGE operon of R. etli. To determine whether the thi box and/or other sequences of this leader region participate in thiamin gene regulation, a series of transcriptional lacZ fusions to 3′-deletion derivatives were made.
Fig. 1B shows that the construct pMM7 with its fusion junction at +654 (all coordinates relative to thiC tss) exhibited 19-fold repression by thiamin. This construct contains 444 nt of the coding region of thiC encoding 148 aa of the N-terminal part of ThiC. The deletion derivative pMM7.1 with endpoint at +217 (carrying only 6 nt of the coding region of thiC) displayed a reduced degree of repression (repression ratio of 7.3). This result suggests that an element that plays a negative role in thiamin control lies in the region deleted (from +218 to +654).
3′ Deletion to +166 (pMM7.2) resulted in a complete loss of regulation. In this construct, the translation start codon of thiC is not present. This result suggests that in the region between +167 to +217 an element important in making thiCOGE expression repressible by thiamin is located.
Role of the thi Box Sequence.
Further 3′ deletion of the untranslated leader removing the thi box resulted in high constitutive expression (pMM7.3, Fig. 1). In accordance with this result, a primer extension analysis of cells grown in the presence or absence of thiamin showed the same tss (data not shown). Therefore, to study the role of the thi box in gene expression, the effect of deletion of the thi box was examined in a lacZ fusion (construct pMM7.4, Fig. 1B). Removal of a region from +24 to +147 containing the thi box resulted in extremely low levels of expression. This result shows that the thi box is an element indispensable for thiCOGE expression in the presence of the rest of the thiC transcript.
thi Box Secondary Structure.
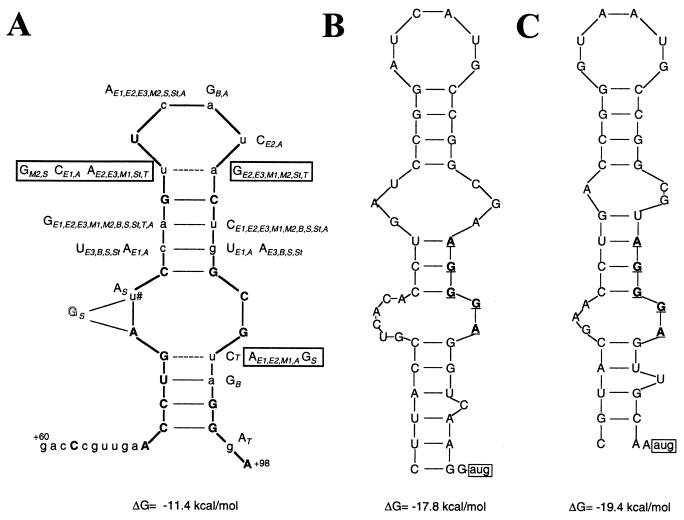
Previous work showed that the R. etli thi box is part of a 211-nt untranslated RNA leader (14). The tss of the three E. coli thiamin biosynthetic operons have not been mapped. However, in the three cases a −35,−10 promoter could be identified at around 200 bp upstream from the translation start codon, suggesting that the thi box sequence is part of long untranslated RNA leaders also in E. coli. As the function of many large single-stranded RNAs is thought to be determined, at least in part, by their structure, we investigated in silico whether the thi box sequence has the potential to form a hairpin structure by using the m-fold program (22, 23). A structure with a ΔG = −11.4 kcal/mol was found and is shown in Fig. 2A. The previously identified thi boxes from E. coli, Bacillus subtilis, M. tuberculosis, Synechocystis sp., S. enterica, Thiocystis violacea, and Aeromonas hydrophila (14) also could form a similar structure. In fact, if one superimposes the structures thus generated, two important facts are revealed. The first is that of the 18 bases conserved in all of the thi box sequences analyzed, nine bases are part of the two helixes that can be formed (Fig. 2A). The second is that in the case of nonconserved bases that form part of the two helixes the changes in base composition on one side of a helix are compensated by matching changes in base composition on the opposite side of the same helix (Fig. 2A). These results strongly suggest that the putative hairpin structures identified are formed in vivo and are conserved in diverse bacterial species.
Figure 2.
(A) Proposed secondary structure for the thi box sequence. Capital bold letters represent conserved bases in all of the thi box sequences analyzed. The dashed lines represent hydrogen bonds that can be formed only in the R. etli thi box. The capital letters alongside the R. etli thi box represent the changes in sequence observed in the other thi box sequences. The boxed capital letters represent changes that would not permit the formation of base pairs. Subscripts represent the thi box sequences in front of genes: E1, E2, and E3, thiC, thiB and thiM genes of E. coli; M1 and M2, thiO and thiC genes of M. tuberculosis; B1, thiA gene of B. subtilis; S, Synechocystis; St, Salmonella enterica; T, Thiocystis violacea; A, Aeromonas hydrophila. The u marked with a # is absent in M2 thi box (see C). The G in outlined style is inserted between the As in S thi box. (B and C) Hairpin structures formed by the thi box sequences in front of the thiO and thiC genes of M. tuberculosis, respectively. The sequence AGGGA that is highly conserved in the 3′ end of the thi box sequences analyzed is represented in bold, underlined letters. The translation start codon of both genes is boxed.
Previously we showed that in some organisms, like M. tuberculosis, the thi box is near the translation start codon whereas in other organisms, like R. etli or E. coli, the thi box is around 100 nt upstream. In Fig. 2 B and C, the hairpin structures formed by the thi box elements present in front of the thiO and thiC genes of M. tuberculosis are shown, respectively. The two presumptive S-D sequences are part of the thi box, which is located 7 and 8 nt upstream from the start codon of the thiO and thiC genes.
Other Putative RNA Secondary Structures Important for Regulation.
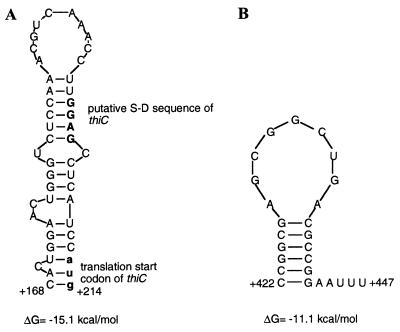
The results obtained with the deletion derivatives suggested that the regions where the S-D sequence is located and the proximal part of the coding region of thiC are involved in regulation. Upon further analysis of both regions, two putative hairpin structures were found and are shown in Fig. 3. One putative structure (from nucleotide +168 to +214) could mask the S-D sequence (see Fig. 3A). The other structure present in the coding region of thiC (from nucleotide +522 to +547) has characteristics of a Rho-independent terminator (a stem formed by six G-C pairs followed by a run of two As and three Us at its 3′ end) (see Fig. 3B).
Figure 3.
(A) Proposed hairpin structure in the region surrounding the S-D sequence of the thiC gene of R. etli. The presumptive S-D sequence is represented by bold, capital letters and the translation start of thiC by bold, lowercase letters. (B) Putative Rho-independent terminator found in the coding region of thiC.
Expression Levels of thiC mRNA Leader and Coding Regions.
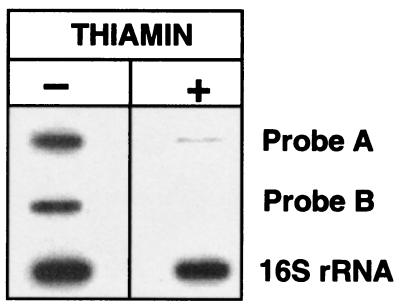
To determine whether the putative attenuator found in the coding region of thiC is functional, a Northern experiment was performed with RNAs obtained from thiamin-grown or thiamin-deprived cells. We used three different probes, one that hybridizes to the untranslated leader sequence (probe A, from nucleotide +6 to +217), another derived from the central part of the thiC gene (probe B, from nucleotide +1044 to +1494) (see Fig. 1A) and a third one derived from the 16S rRNA gene of E. coli that was used as loading control (21). With probes A and B, a signal was very evident in RNAs from thiamin-deprived cells. In contrast, a very faint signal was seen in RNAs from thiamin-grown cells only with probe A (see Fig. 4). The different pattern of hybridization observed with probes A and B suggests that the presumptive attenuator is functional.
Figure 4.
thiC gene expression. RNA from the CE3 strain grown in MM with (+) or without (−) thiamin was slot-blotted and hybridized with probe A (a DNA fragment from nucleotide +6 to +217), probe B (an intra thiC fragment from nucleotide +1044 to +1494), and with the 16S rRNA gene of E. coli.
Discussion
Although TPP is the cofactor of key enzymes of carbohydrate metabolism, little is known about the regulatory mechanisms involved in expression of its biosynthetic genes. Previous studies have shown that the thiCOGE operon of R. etli is transcribed with a 211-nt untranslated RNA leader (14). In this article we report a deletion analysis of reporter fusions to thiC that identified three regions of the transcript important for regulation by thiamin: the thi box located in the untranslated RNA leader, a second region located near the S-D site, and a third region located in the coding region of the thiC gene. In these three regions three putative RNA structures were identified in silico.
The thi box is highly conserved in the 5′ region of thiamin biosynthetic and transport genes from Gram-negative and Gram-positive organisms and cyanobacteria (14). Based on its evolutionary conservation and its presence in E. coli genes whose expression is regulated by TPP, it is thought to be involved in thiamin regulation. Further search for the thi box element in other bacterial genomes showed that it is present in Thermotoga maritima, Streptomyces coelicolor, Mycobacterium leprae, Vibrio cholera, Sinorhizobium meliloti, Pseudomonas fluorescens and in one species from Archae, Thermoplasma volcanium (GenBank accession nos. AE001747, AL133219, AL583918, AE004097, AF070520, AY007258, AP000994, respectively). These data show that the thi box sequence plays a fundamental role in an ancient mechanism regulating thiamin gene expression in bacteria. Phylogenetic analysis is the most accurate method for RNA secondary structure prediction. Some structures determined by this method include tRNAs (24), ribosomal RNAs (25–27), group I and group II introns (28, 29), RNAs from small nuclear ribonucleoproteins (30), hammerhead catalytic RNA (31), and RNase P RNAs (32). Further studies have confirmed these structures. A phylogenetic comparative analysis showed that all of the thi box sequences considered can be folded into a similar hairpin structure formed by two helixes, one interior loop, and a hairpin loop. From the 18 conserved bases in all of the thi box sequences analyzed, eight are involved in forming G-C pairs and two form an U-A pair (U-G in B. subtilis thi box) in the two helixes. One conserved base is present in the hairpin loop and four more lie in the interior loop. The other three conserved bases are outside of the hairpin structure. It is possible that the eight conserved bases that do not form part of the helixes are specifically recognized by a regulatory factor. The other nonconserved bases that form part of the helixes show compensatory base changes that ensure base pair formation. The high conservation of sequence and structure of the thi boxes analyzed, the presence of the thi box in the 5′ region of thiamin biosynthetic and transport genes in different bacterial genomes, and the fact that the thi box associated with the R. etli thiC gene is transcribed and is absolutely necessary for thiC expression show that the thi box is a novel RNA regulatory element.
Concerning the second RNA structure identified in silico, a deletion with endpoint at +166 displayed unregulated high activities. This result suggests that in the region between +167 and +217 an element involved in repression of thiC expression by thiamin is present. A putative hairpin structure with a ΔG = −15.1 kcal/mol was predicted in this region. This structure could mask the S-D sequence and the translation start codon of thiC. It is tempting to speculate that repression of thiC expression by thiamin depends on the formation of this structure. Mutagenesis of this structure will be important to determine its role in regulation. Interestingly, in M. tuberculosis, the first two RNA structures (the thi box and the hairpin structure) overlap (Fig. 2, B and C). In this bacterial species, the stabilization of the thi box structure in the presence of thiamin could occlude the ribosomal binding site, preventing translation of the thiC or thiO genes.
Finally, regarding the third RNA structure, deletion from nucleotides +218 to +654 revealed the existence of an element that plays a negative role in thiamin-dependent control. Such an element was found in the region from nucleotide +522 to +547. This sequence has the potential to form a hairpin structure with characteristics of a Rho-independent terminator. A Northern blot experiment using two probes, one derived from the leader region and the other from the central part of the coding sequence of thiC, downstream from the putative attenuator, showed that only the leader region-derived probe hybridized, giving a very faint signal, suggesting an effect on RNA stability. These results are consistent with the idea of a message halting at +547.
As mentioned previously, the polypurine sequence AGGGA, which is highly conserved in the 3′ end of the thi box sequences analyzed, is situated at a distance that would allow it to function as the S-D sequence of the thiC and thiO genes of M. tuberculosis. It is possible that all of the thi box sequences would be able to interact with the 30S ribosomal subunit. This interaction would be responsible for the protective effect of the thi box on thiC expression when the putative attenuator located in the coding region of thiC is also present. Examples of interactions between a S-D sequence and ribosomes leading to an increase in expression are documented for the genes ermC and cryIIIA of Bacillus subtilis and Bacillus thuringiensis, respectively. The expression of the ermC gene (which confers resistance to erythromycin) is increased upon addition of the antibiotic to the growth medium (33). The increase in expression results from the protection from degradation delivered by the ribosomes stalled on the ermC message (33). In B. thuringiensis it has been shown that a S-D sequence designated as STAB-SD is a determinant of stability of toxin cryIIIA mRNA (34). This STAB-SD is located 116 nt upstream of the start codon of the cryIIIA gene and is not involved in translation initiation (34). The STAB-SD is thought to enhance stability by interacting with the 30S ribosomal subunit.
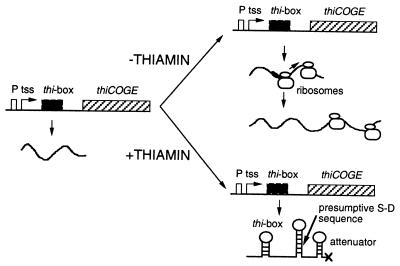
A possible model of thi gene regulation by thiamin that can be drawn from the results reported here is shown in Fig. 5. As was stated before, transcription from the thiC promoter is constitutive. In the absence of thiamin, the thi box would not be folded and the sequence AGGGA could interact with the ribosomes. The interaction between the thi box and the ribosomes would impede the degradation of the message or, alternatively, the ribosomes so recruited by the thi box can be passed on to the S-D region where translation would start. The ribosomes translating the message would not allow the formation of the attenuator and transcription would proceed. In contrast, in the presence of TPP, the thi box would be folded into a hairpin structure and would not interact with the ribosomes, the hairpin structure in the S-D region will form and the ribosomes would not bind to the message. As a consequence, when transcription reaches the putative terminator, this sequence would be folded into a secondary structure and transcription would be prematurely terminated. The untranslated message then will be degraded.
Figure 5.
Model for R. etli thiCOGE expression. (Left) The structural and functional organization of the regulatory and coding regions of the genes thiCOGE is represented. Transcription is constitutive from the thiC promoter. Depending on the availability of thiamin in the growth medium, this transcript will be elongated up to the end of the thiE gene (−thiamin condition) or would be prematurely terminated at the putative attenuator located at position +522 to +547 (+thiamin condition). The hairpin structures that can be formed by the thi box, the S-D sequence, and the putative attenuator are marked. thiCOGE genes are not drawn to scale. For details see the text.
In our model, a thiamin-dependent regulatory element could be involved in the interaction with the thi box RNA structure. However, no mutants affected in such a regulatory element have been isolated so far (10, 12). Also, titration of this putative regulator by the overexpression of the untranslated RNA leader failed to reveal its presence (data not shown). Thus, an interesting possibility is that in this case the direct binding of TPP to the thi box RNA structure could modulate the formation of the RNA structures identified in this work. Interactions between RNAs and small molecules like the one proposed here have been observed in the case of RNA aptamers that bind theophylline (35) and chloramphenicol (36) with very high affinity and specificity.
Regulation of thiCOGE expression shows striking similarities to repression of expression of the btuB gene and the cob operon by vitamin B12 or cyanocobalamin (37–39). In E. coli and S. enterica, these genes code for an outer membrane cobalamin transport protein and cobalamin biosynthetic enzymes, respectively. These genes are transcribed with unusually long leaders of 241 nt (btuB) or 468 nt (cob operon). Several mutations that led to decreased repression have been mapped to these leaders (40–42). The B12 box, a 25-nt element that is conserved in the leaders of the btuB and cbiA genes, is required for the expression of btuB (41, 43), as the thi box is required for thiamin-regulated expression of thi genes. In addition, a potential Rho-independent terminator structure that could serve as a transcriptional attenuator lies in the proximal portion of the coding sequence of btuB, as in the case of thiC (43). The primary control of btuB expression by adenosyl-cobalamin (the effector molecule) is exerted at the level of translation initiation where a hairpin structure sequesters the S-D sequence (44, 45). Similar to the case for thiamin modulation of gene expression, no trans-acting factors involved in cobalamin repression have been found (46). Recently, it has been demonstrated that adenosyl-cobalamin inhibits the binding of 30S ribosomal subunits to btuB RNA and that a ribosome-associated, RNase-sensitive factor is needed to unfold the hairpin structure that masks the S-D sequence to allow ribosome binding (47). Furthermore, a cis-acting translational enhancer located upstream of the S-D sequence of the cob mRNA of S. typhimurium is required to unfold the inhibitory ribosome binding site hairpin in the absence of cobalamin (48). Also, it was shown that the untranslated leader of the cob mRNA is highly structured and that secondary and tertiary folding is required for normal repression by cobalamin (48). Other vitamin biosynthetic genes that seem to be regulated by a similar mechanism are the riboflavin genes, for which a conserved RNA structure element has been identified (49). Therefore, regulatory mechanisms similar to the one proposed here for thiamin could operate in the regulation of expression of other vitamin biosynthesis genes in bacteria.
Acknowledgments
We thank C. Morera, M. L. Tabche, and O. López for technical assistance and E. Morett for reviewing the manuscript. Funding was provided by Consejo Nacional de Ciencia y Tecnología (Conacyt, México) Grants 31561-N and 0028, and Dirección General de Asuntos del Personal Académico Grant IN204697.
Abbreviations
- TPP
thiamin pyrophosphate
- tss
transcription start site
- S-D
Shine-Dalgarno
- MM
minimal medium
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 9465.
References
- 1.White R L, Spencer I D. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. 2nd Ed. Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 680–686. [Google Scholar]
- 2.Begley T P, Downs D M, Ealick S E, McLafferty F W, Van Loon A P G M, Taylor S, Campobasso N, Chiu H-J, Kinsland C, Reddick J J, et al. Arch Microbiol. 1999;171:293–300. doi: 10.1007/s002030050713. [DOI] [PubMed] [Google Scholar]
- 3.van der Horn P B, Backstrom A D, Stewart V, Begley T P. J Bacteriol. 1993;175:982–992. doi: 10.1128/jb.175.4.982-992.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petersen L A, Downs D M. J Bacteriol. 1997;179:4894–4900. doi: 10.1128/jb.179.15.4894-4900.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sprenger G A, Schorken U, Wiegert T, Grolle S, de Graaf A A, Taylor S V, Begley T P, Bringer-Meyer S, Sahm H. Proc Natl Acad Sci USA. 1997;94:12857–12862. doi: 10.1073/pnas.94.24.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lois L M, Campos N, Putra S R, Danielson K, Rohmer M, Boronat A. Proc Natl Acad Sci USA. 1998;95:2105–2110. doi: 10.1073/pnas.95.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor S V, Kelleher N L, Kinsland C, Chiu H-J, Costello C A, Backstrom A D, McLafferty F W, Begley T P. J Biol Chem. 1998;273:16555–16560. doi: 10.1074/jbc.273.26.16555. [DOI] [PubMed] [Google Scholar]
- 8.Webb E, Claas K, Downs D M. J Bacteriol. 1997;179:4399–4402. doi: 10.1128/jb.179.13.4399-4402.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Backstrom A D, Austin R, McMordie S, Begley T P. J Am Chem Soc. 1995;117:2351–2352. [Google Scholar]
- 10.Webb E, Downs D. J Biol Chem. 1997;272:15702–15707. doi: 10.1074/jbc.272.25.15702. [DOI] [PubMed] [Google Scholar]
- 11.Imamura N, Nakayama H. J Bacteriol. 1982;151:708–717. doi: 10.1128/jb.151.2.708-717.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webb E, Febres F, Downs D. J Bacteriol. 1996;178:2533–2538. doi: 10.1128/jb.178.9.2533-2538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webb E, Claas K, Downs D. J Biol Chem. 1998;273:8946–8950. doi: 10.1074/jbc.273.15.8946. [DOI] [PubMed] [Google Scholar]
- 14.Miranda-Ríos J, Morera C, Taboada H, Dávalos A, Encarnacion S, Mora J, Soberón M. J Bacteriol. 1997;179:6887–6893. doi: 10.1128/jb.179.22.6887-6893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noel K D, Sánchez A, Fernández L, Leemans J, Cevallos M A. J Bacteriol. 1984;158:148–155. doi: 10.1128/jb.158.1.148-155.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon R, Priefer U, Puhler A. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 17.Miranda J, Membrillo-Hernández J, Tabche M L, Soberón M. Appl Microbiol Biotechnol. 1996;45:182–188. [Google Scholar]
- 18.Schlieper D, von Wilcken-Bergman B, Schmidt M, Sobek H, Muller-Hill B. Anal Biochem. 1998;257:203–209. doi: 10.1006/abio.1997.2558. [DOI] [PubMed] [Google Scholar]
- 19.Spaink H P, Okker R J H, Wijffelman C A, Pees E, Lugtenberg B J J. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- 20.Reyes J D, Tabche M L, Morera C, Girard M L, Romero D, Krol E, Miranda J, Soberón M. Gene. 2000;250:149–157. doi: 10.1016/s0378-1119(00)00176-1. [DOI] [PubMed] [Google Scholar]
- 21.Brosius J, Dull T J, Sleeter D D, Noller H F. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 22.Zuker M, Mathews D H, Turner D H. In: RNA Biochemistry and Biotechnology. Barciszewski J, Clark B F C, editors. Kluwer, Dordrecht, The Netherlands: NATO ASI Series; 1999. pp. 11–43. [Google Scholar]
- 23.Mathews D H, Sabina J, Zuker M, Turner D H. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 24.Madison J T, Everett G A, Kung H. Science. 1966;153:531–534. doi: 10.1126/science.153.3735.531. [DOI] [PubMed] [Google Scholar]
- 25.Fox G, Woese C R. Nature (London) 1975;256:505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- 26.Woese C R, Magrum L J, Gupta R, Siegel R B, Stahl D A, Kop J, Crawford N, Brosius J, Gutell R, Hogan J J, et al. Nucleic Acids Res. 1980;8:2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noller H F, Kop J, Wheaton V, Brosius J, Gutell R R, Kopylov A M, Dohme F, Herr W, Stahl D A, Gupta R, et al. Nucleic Acids Res. 1981;9:6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies R W, Waring R B, Ray J A, Brown T A, Scazzochcio C. Nature (London) 1982;300:719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- 29.Michel F, Jacquier A, Dujon B. Biochimie. 1982;64:867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- 30.Siliciano P G, Jones M H, Guthrie C. Science. 1987;237:1484–1487. doi: 10.1126/science.3306922. [DOI] [PubMed] [Google Scholar]
- 31.Foster A C, Symons R H. Cell. 1987;49:211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- 32.Pace N R, Smith D K, Olsen G J, James B D. Gene. 1989;82:65–75. doi: 10.1016/0378-1119(89)90031-0. [DOI] [PubMed] [Google Scholar]
- 33.Hue K K, Bechoffer D H. J Bacteriol. 1991;173:3732–3740. doi: 10.1128/jb.173.12.3732-3740.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agaisse H, Lereclus D. Mol Microbiol. 1996;20:633–643. doi: 10.1046/j.1365-2958.1996.5401046.x. [DOI] [PubMed] [Google Scholar]
- 35.Jenison R D, Gill S C, Pardi A, Polisky B. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 36.Burke D H, Hoffman D C, Brown A, Hansen M, Pardi A, Gold L. Chem Biol. 1997;4:833–843. doi: 10.1016/s1074-5521(97)90116-2. [DOI] [PubMed] [Google Scholar]
- 37.Kadner R J. J Bacteriol. 1978;136:1050–1057. doi: 10.1128/jb.136.3.1050-1057.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei B-Y, Bradbeer C, Kadner R J. Res Microbiol. 1992;143:459–466. doi: 10.1016/0923-2508(92)90091-2. [DOI] [PubMed] [Google Scholar]
- 39.Escalante-Semerena J C, Roth J R. J Bacteriol. 1987;169:2251–2258. doi: 10.1128/jb.169.5.2251-2258.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lundrigan M D, Koster W, Kadner R J. Proc Natl Acad Sci USA. 1991;88:1479–1483. doi: 10.1073/pnas.88.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richter-Dahlfors A A, Andersson D I. Mol Microbiol. 1992;6:743–749. doi: 10.1111/j.1365-2958.1992.tb01524.x. [DOI] [PubMed] [Google Scholar]
- 42.Richter-Dahlfors A A, Ravnum S, Andersson D I. Mol Microbiol. 1994;13:541–553. doi: 10.1111/j.1365-2958.1994.tb00449.x. [DOI] [PubMed] [Google Scholar]
- 43.Franklund C V, Kadner R J. J Bacteriol. 1997;179:4039–4042. doi: 10.1128/jb.179.12.4039-4042.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ravnum S, Andersson D I. Mol Microbiol. 1997;23:35–42. doi: 10.1046/j.1365-2958.1997.1761543.x. [DOI] [PubMed] [Google Scholar]
- 45.Nou X, Kadner R J. J Bacteriol. 1998;180:6719–6728. doi: 10.1128/jb.180.24.6719-6728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roth J R, Lawrence J G, Bobick T A. Annu Rev Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 47.Nou X, Kadner R J. Proc Natl Acad Sci USA. 2000;97:7190–7195. doi: 10.1073/pnas.130013897. . (First Published June 13, 2000; 10.1073/pnas.130013897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ravnum S, Andersson D I. Mol Microbiol. 2001;39:1585–1594. doi: 10.1046/j.1365-2958.2001.02346.x. [DOI] [PubMed] [Google Scholar]
- 49.Gelfand M S, Mironov A A, Jomantas J, Kozlov Y I, Perumov D A. Trends Genet. 1999;15:439–442. doi: 10.1016/s0168-9525(99)01856-9. [DOI] [PubMed] [Google Scholar]