Abstract
Transcription factors BABY BOOM (BBM), WUSCHEL (WUS), BSD, LEAFY COTYLEDON (LEC), LEAFY COTYLEDON LIKE (LIL), VIVIPAROUS1 (VP1), CUP SHAPED COTYLEDONS (CUC), BOLITA (BOL), and AGAMOUS LIKE (AGL) play a crucial role in somatic embryogenesis. In this study, we identified eighteen genes of these nine transcription factors families from the banana genome database. All genes were analyzed for their structural features, subcellular, and chromosomal localization. Protein sequence analysis indicated the presence of characteristic conserved domains in these transcription factors. Phylogenetic analysis revealed close evolutionary relationship among most transcription factors of various monocots. The expression patterns of eighteen genes in embryogenic callus containing somatic embryos (precisely isolated by Laser Capture Microdissection), non-embryogenic callus, and cell suspension cultures of banana cultivar Grand Naine were analyzed. The application of 2, 4-dichlorophenoxyacetic acid (2, 4-D) in the callus induction medium enhanced the expression of MaBBM1, MaBBM2, MaWUS2, and MaVP1 in the embryogenic callus. It suggested 2, 4-D acts as an inducer for the expression of these genes. The higher expression of MaBBM2 and MaWUS2 in embryogenic cell suspension (ECS) as compared to non-embryogenic cells suspension (NECS), suggested that these genes may play a crucial role in banana somatic embryogenesis. MaVP1 showed higher expression in both ECS and NECS, whereas MaLEC2 expression was significantly higher in NECS. It suggests that MaLEC2 has a role in the development of non-embryogenic cells. We postulate that MaBBM2 and MaWUS2 can be served as promising molecular markers for the embryogencity in banana.
Introduction
Banana is an important staple food fruit crop in several developing countries. Being a vegetatively propagated plant, its multiplication index through sucker is very low [1]. Somatic embryogenesis (SE) is considered to be an important route for mass production [2–4] and development of transgenic [5–6] banana plants. SE through embryogenic cell suspension (ECS) has been reported in various banana cultivars [7–9]. The process includes different developmental stages viz., callus, ECS, regeneration, germination, rooting and acclimatization (Fig 1). The occurrence of somaclonal variation in SE through callus culture has been demonstrated in several plant species [10–12]. A callus phase in tissue culture, the use of growth regulators, the number and duration of subcultures, stress and the genotype are the factors that enhance the somaclonal variation and restrict the potential use of SE [11–12]. Therefore, the molecular factors which are known to regulate SE in other plants can provide a lead to understand the mechanism of SE in banana.
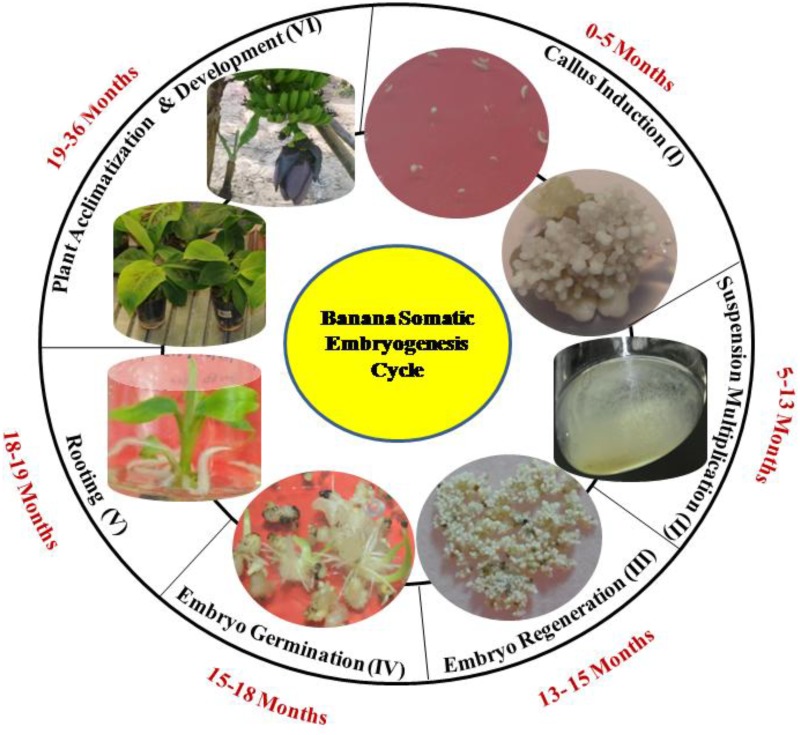
Fig 1. Banana somatic embryogenesis developmental stages.
Different developmental stages of banana somatic embryogenesis cycle are following: (I) Callus Induction (0–5 months) (II) Suspension and Multiplication (5–13 months) (III) Embryo Regeneration (13–15 months) (IV) Embryo Germination (15–18 months) (V) Rooting (18–19 months) (VI) Plant Acclimatization and development (19–36 months).
Transcription factors (TFs) are the evolutionary conserved regulatory proteins, which play a significant role in transcriptional regulation of gene expression. Plant TFs reported to have a higher rate of lineage-specific expansion [13–14]. They are imperative molecules in the regulation of different biological processes, including SE in plants. Certain TFs such as BABYBOOM (BBM) [15–19], WUSCHEL (WUS) [20–23], BSD [24–25], LEAFY COTYLEDON (LEC) [26–27], LEAFY COTYLEDON LIKE (LIL) [28–29], VIVIPAROUS1 (VP1) [30–31], BOLITA (BOL) [32] and AGAMOUS-LIKE (AGL) [33] are highly specific to plant lineage, suggesting their importance in plant-specific processes. These TFs are restricted to plant lineage and characterized by the presence of conserved domain in their proteins. The study directing towards understanding the role of these TFs in banana could be of great interest and has the potential for improvement of SE. For instance, BBM expression patterns in cacao tissue reported as a biomarker for embryogenesis [19]. WUS play a vital role in SE by promoting the vegetative to embryogenic transition in arabidopsis [21]. BSD is known to be associated with cell proliferation during the SE [24–25, 34]. LEC [27] and L1L [28] contain HAP3 subunit and reported for their role in embryo development, morphogenesis, and cellular differentiation. The role of AGL has been demonstrated in SE of arabidopsis and radish plants [35–36].
Several other TFs are known to be involved in the process of only organogenesis or both SE and organogenesis. For example, VP1 regulates seed dormancy and involved in organogenesis and SE [30, 37–38]. However, CUC plays a role only in the organogenesis [39–40]. CUC is also known to promote adventitious root formation in calli [41]. Another TF like BOL is known to be involved in the regulation of the cell expansion and proliferation [32].
Earlier, efforts have been made to characterize different TFs for their role in SE in several plant species [15, 20, 24, 28], but no report is available in banana. In the present study, 18 genes belonging to the nine TF families were identified in the banana genome. Phylogenetic tree analyses of these genes in land plants including monocot, dicot, and gymnosperm were conducted for their molecular evolution. In addition, the expression patterns of these genes were characterized during the critical steps of in-vitro cultures of cv. Grand Naine (AAA).
Materials and methods
Identification, chromosome distribution and exon-intron prediction of TFs in banana
Nine TF families (BBM, WUS, BSD, LEC, L1L, VP1, CUC, BOL and AGL) were selected for the in-silico study. Homologs of these TFs were identified in arabidopsis, maize and rice (TAIR, http://www.arabidopsis.org; http://bioinformatics.psb.ugent.be/plaza; TIGR, http://rice.plantbiology.msu.edu) and used as a query sequence in BLASTP search to retrieve the sequences from the banana genome (http://banana-genome.cirad.fr/). The putative protein sequences resulting from each blast search (E- value 10−5) in banana were collected and redundant sequences were removed. Gene models were refined and genomic coding sequences (CDS) were retrieved from the banana genome hub database [42, http://banana-genome.cirad.fr/]. For chromosomal localization, genomic coordinates were recovered from the banana genome. Genomic sequences of the selected banana TFs were downloaded from Gramene database [43, http://www.gramene.org/]. Genomic and CDS sequences of all homologs were aligned using multialin software [44, multalin.toulouse.inra.fr/multalin/]. The number of exons and introns were calculated manually.
Phylogenetic analysis, motif identification and sequence analysis
Protein sequences of the selected TFs of banana and their homologs from angiosperm (monocot and dicot) and gymnosperm were used for phylogeny analyses. The phylogenetic tree was constructed using the neighbor-joining method with MEGA 6.0 software [45]. The unrooted tree was generated through 1000 bootstrap values for the reliability of the tree. Multiple sequence alignment was carried out by using CLC genomic workbench (QIAGEN Denmark). The amino acid sequence corresponding to TF families in banana were studied for conserved domains analysis using conserved domain database [46]. Theoretical isoelectric point (pI) and molecular weight (MW) were predicted using the Compute pI/MW tool on the ExPASy server [47, http://web.expasy.org/compute_pi/]. Transmembrane helix (TMH) was analyzed on the TMHMM server v 2.0 [48, http://www.cbs.dtu.dk/services/TMHMM-2.0/] and the subcellular localization of deduced proteins was predicted on the Target P 1.1 server [49, http://www.cbs.dtu.dk/services/TargetP/].
Callus development
Total seventy immature male flower buds (source of explant) of cv. Grand Naine were collected from the experimental field of National Agri-Food Biotechnology Institute (NABI), Mohali, Punjab, India (310 m above sea level; Latitude 30° 47’ North; Longitude 76° 41’ East). Immature male flowers (explants) of rank (1–15) adjacent to the floral apex were isolated and inoculated on callus induction medium for callus formation [7, 50]. Six hundred explants derived from forty flower buds were inoculated on 2, 4-dichlorophenoxyacetic acid (2, 4-D) containing medium, while the four hundred fifty explants prepared from thirty buds were cultured on 2, 4-D -free MS basal medium. All cultures were incubated under the dark condition at 27°C for the emergence of somatic embryos in plant tissue culture chambers (Percival, USA). After 12 Week (W) of incubation, embryogenic calli (with proembryos) and non-embryogenic calli were identified under the upright microscope (Leica Microsystems, Germany), collected, frozen immediately in the liquid nitrogen and stored at −80°C until further use.
ECS establishment
Nearly six months (24W) old embryogenic calli comprised with somatic embryos and non-embryogenic calli were inoculated in suspension medium containing 2, 4-D and zeatin [7]. Suspension culture was kept in dark at 27°C with agitation at 90 rotations per minute (Kuhner, Switzerland) and sub-cultured weekly. ECS samples were collected at 0W, 1W, and 24W intervals while the NECS collected at 0W, and 1W for further study.
Histological analysis
Samples collected from callus induction medium with 2, 4-D and without 2, 4-D were used for histological analysis. Samples were preserved in 70% ethanol at 4°C for one day [51] and then fixed in blocks using freezing solution (Leica Biosystems, Germany). Ten to twelve μm thin sections were prepared by using cryomicrotome (Leica Biosystems, Germany) and observed under the microscope (Leica Microsystems, Germany). For suspension culture analysis, cells were placed on the slide along with suspension medium and cover slip. The embryogenic and non-embryogenic cells were stained with iodine to confirm the presence of starch granules. These cells were observed directly under the upright microscope (Leica Microsystems, Germany).
Laser capture microdissection (LCM) of embryogenic cells
RNAase free conditions (tools, solutions, handling) were followed throughout the experiment. The experiment was performed as per previously described protocol [52]. In brief, banana callus (24W) containing embryos were fixed at -23°C under vacuum using tissue freezing medium (Leica biosystems, Germany). Tissue blocks were fixed with the holding clamp of cryomicrotome (Leica biosystems, Germany) and 10–12 μm thin sections were prepared. These sections were taken on slides, air-dried at room temperature and observed under LCM microscope (Zeiss, Germany).
For microdissection, embryos were identified and marked using PALM (Zeiss, Germany) tool. Tissues were snipped-off with a laser beam along with marking in RNase-free tubes and stored at -80°C for RNA isolation.
RNA isolation and cDNA synthesis
Total RNA was isolated from different samples using RNA extraction kit (Sigma-Aldrich, USA). Isolated RNA was treated with DNase I kit (AmbionThermo Scientific, USA) to eliminate DNA contamination. Total RNA was analyzed by agarose gel electrophoresis for size and integrity. The quantification of total RNA was done with a NanoQuant (Infinite 200 PRO NanoQuant, Austria). DNA-free RNA was used for cDNA first strand synthesis by using revert aid first strand cDNA synthesis kit (Thermo Scientific, USA) as per manufacturer’s protocol. Oligo dT primer was used for cDNA preparation.
Quantitative real-time PCR (qPCR)
The qPCR study was performed using ABI 7700 Sequence Detector (Applied Biosystems, USA). Housekeeping gene Actin1 (GenBank Accession No. AF246288) was used to normalize variant expression of selected genes [53–54]. The primers were tested for single band amplification using conventional end-point PCR. Melting curve study was carried out using qPCR. The total volume of each reaction was 10 μl and consisted of 1X SYBR Green Master mix (Applied Biosystems, USA) 5 pmol of each primer, 0.5 μl cDNA template and sterile distilled H2O. PCR conditions followed during real-time PCR experiment were: step (1) 50°C 2 min, step (2) 95°C 10 min, step (3) (95°C 0.15 min, 60°C 1 min) x 40 cycles, followed by the thermal dissociation curve. The relative expression level was analyzed using the 2-ΔΔCt method [55], where ΔΔCt = (Ct target—Ct actin) time x−(Ct target—Ct actin) time 0. Primer details are mentioned in S1 Table. All experiments were performed in biological triplicates and each experiment consisted of three technical replicates. The t-test was carried out for assessment of statistical significance of data.
Results
Identification and sequence analysis of SE related TFs in Musa acuminata
The BLAST search was carried out for identification of BBM, WUS, BSD, LEC, L1L, VP1, CUC, BOL, and AGL protein sequences in banana by using query sequences of arabidopsis (27), rice (8), and maize (13). A total of 18 sequences belonging to nine TF families were retrieved from the banana genome. All deduced protein sequences contain conserved domains of their respective TF families (S1, S2, S3, S4, S5, S6, S7 and S8 Figs). Protein sequence identity between banana TFs and their homologs in arabidopsis, maize, and rice ranges from 24% to 83%, 25% to 74%, and 45% to 74%, respectively (S2 Table). Various features such as locus ids, chromosomal coordinates, predicted protein MW, pI, subcellular localization and TMH of banana sequences were summarized in Table 1. Protein length and MW of all TFs ranged from 63 to 678 amino acids and 7.20 to 75.04 kDa, respectively. Most of the TFs (11) showed pI in acidic range (4.3–6.83), while others had an alkaline range (8.6–10.14). However, only MaLEC1 had shown neutral pI. MaBBM1, MaBBM2, and MaLIL1 were localized in the chloroplast, whereas MaBSD3 and MaAGL1 in mitochondria. Subcellular localization of other TFs was not available. No transmembrane domain was predicted in any TF.
Table 1. Structural features of SE related transcription factors in Musa acuminata.
| Gene Name | Gene ID | Chromosome Position | Coordinates | Protein Length | Predicted pI | MW (kDa) | Subcellular Localization | Trans Membrane Helix (TMH) |
|---|---|---|---|---|---|---|---|---|
| MaBBM1 | GSMUA_Achr3P21460_001 | 3 | 22424668:22429036 | 612 | 6.30 | 66.8 | Chloroplast | 0 |
| MaBBM2 | GSMUA_Achr2P05880_001 | 2 | 10811480:10815136 | 572 | 6.83 | 62.8 | Chloroplast | 0 |
| MaWUS1 | GSMUA_Achr8P02040_001 | 8 | 1508245:1509831 | 128 | 9.8 | 15.02 | - | 0 |
| MaWUS2 | GSMUA_Achr10P26570_001 | 10 | 30070543:30072984 | 233 | 8.9 | 25.7 | - | 0 |
| MaBSD1 | GSMUA_Achr6P00640_001 | 6 | 420148:428830 | 421 | 4.3 | 46.1 | - | 0 |
| MaBSD2 | GSMUA_Achr8P25810_001 | 8 | 29568544:29574681 | 295 | 4.8 | 33.1 | - | 0 |
| MaBSD3 | GSMUA_Achr2P17230_001 | 2 | 17835096:17843511 | 496 | 4.3 | 54.9 | Mitochondria | 0 |
| MaLEC1 | GSMUA_Achr10P12560_001 | 10 | 21559530:21570493 | 482 | 7.0 | 54.6 | - | 0 |
| MaLEC2 | GSMUA_Achr3P23760_001 | 3 | 24489817:24495395 | 368 | 6.4 | 41.82 | - | 0 |
| MaLIL1 | GSMUA_Achr4P26330_001 | 4 | 25686876:25690660 | 269 | 5.3 | 30.2 | Chloroplast | 0 |
| MaLIL2 | GSMUA_Achr11P06580_001 | 11 | 5174935:5181575 | 205 | 5.6 | 25.8 | - | 0 |
| MaVP1 | GSMUA_AchrUn_randomP18520_001 | random | 88731276:88735269 | 678 | 6.2 | 75.04 | - | 0 |
| MaCUC1 | GSMUA_Achr9P20090_001 | 9 | 23156505:23159553 | 317 | 5.3 | 35.7 | - | 0 |
| MaCUC2 | GSMUA_Achr9P00570_001 | 9 | 481453:483257 | 168 | 9.3 | 19.5 | - | 0 |
| MaCUC3 | GSMUA_Achr10P22350_001 | 10 | 27580397:27583307 | 355 | 8.6 | 38.4 | - | 0 |
| MaBOL | GSMUA_Achr6P19570_001 | 6 | 13852244:13854193 | 89 | 6.4 | 9.7 | - | 0 |
| MaAGL1 | GSMUA_Achr5P20280_001 | 5 | 22027677:22029271 | 63 | 10.14 | 7.2 | Mitochondria | 0 |
| MaAGL2 | GSMUA_Achr8P07230_001 | 8 | 4776954:4778864 | 236 | 9.4 | 25.8 | - | 0 |
Chromosome localization and exon-intron composition
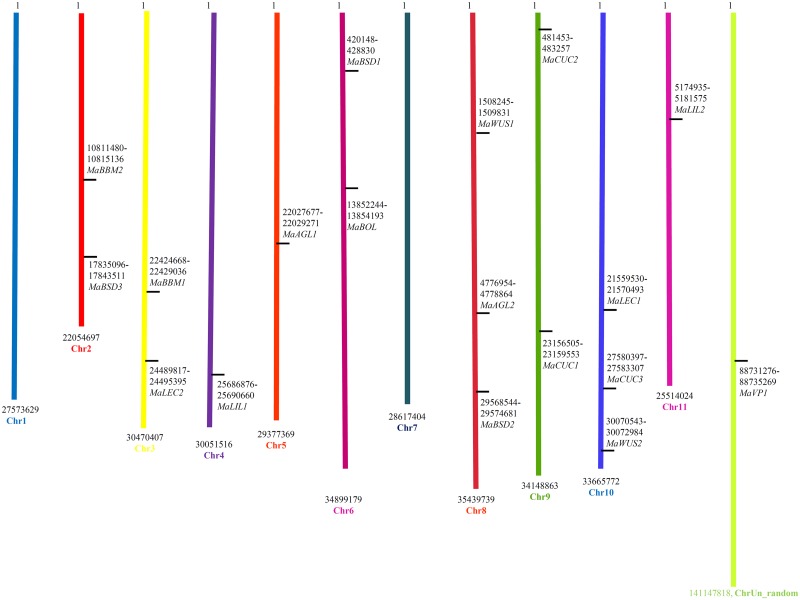
All TFs except one (MaVP1) that was found to be associated with un_random chromosome could be mapped on banana chromosomes (Fig 2). All the mapped TFs were distributed over nine chromosomes {MaAGL1 (Chr5), MaAGL2 (Chr8), MaBBM1 (Chr3), MaBBM2 (Chr2), MaBSD1 (Chr6), MaBSD2 (Chr8), MaBSD3 (Chr2), MaL1L1 (Chr4), MaL1L2 (Chr11), MaWUS1 (Chr8), MaWUS2 (Chr10), MaCUC1 (Chr9), MaCUC2 (Chr9), MaCUC3 (Chr10), MaLEC1 (Chr10), MaLEC2 (Chr3) and MaBOL (Chr6)}.
Fig 2. Localization of transcription factors and genes on banana chromosomes.
The black vertical lines on the chromosomes indicate the positions of the respective genes. Numbers represent nucleotide base pair positions with the name of the respective gene.
MaAGL1, MaAGL2, and MaWUS1 were intron-free genes, while other TFs have shown the presence of introns varying from 1 to 11. MaBBM1contained the highest number of introns (11) while its homolog in arabidopsis has only 8 introns. Except for BOL and CUC, no other TFs were found to have its homologs in rice (S3 Table).
Phylogenetic study and conserved motif analysis
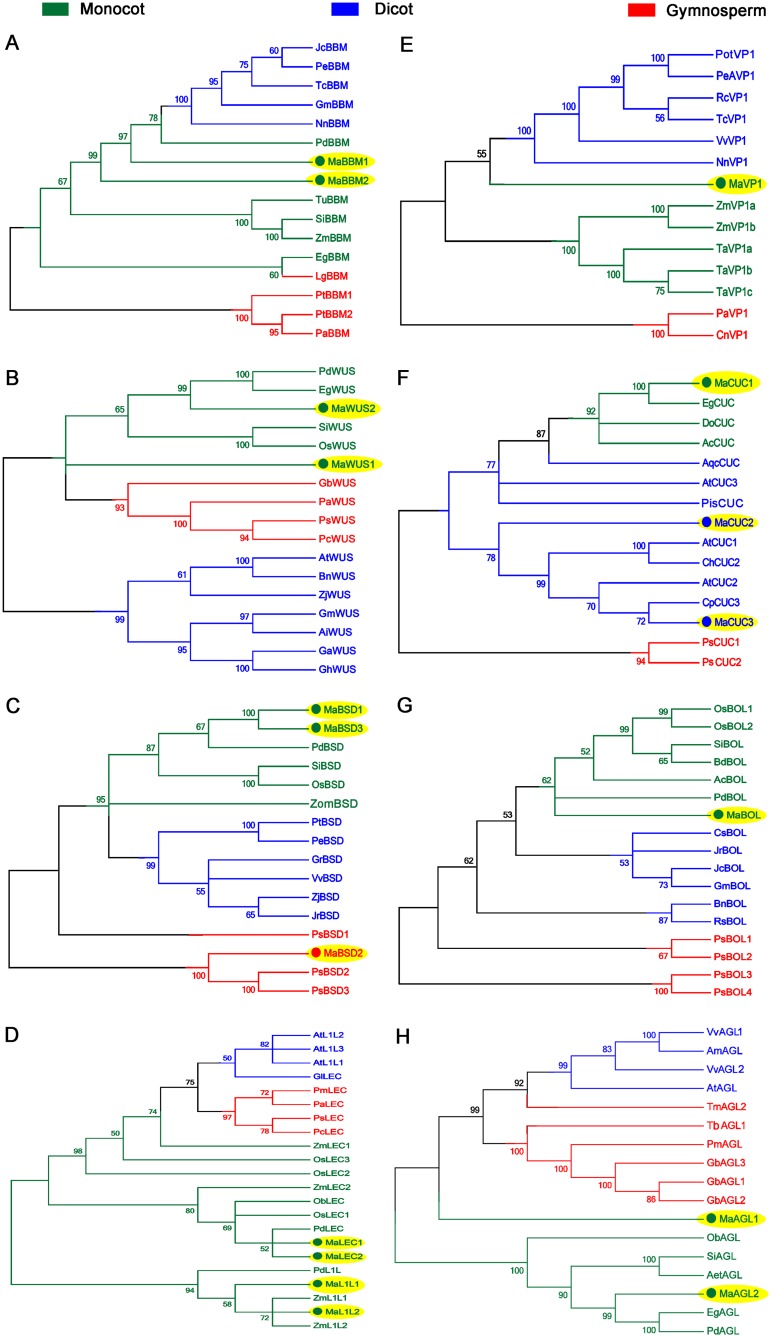
The TF sequences from the gymnosperm, monocot and dicot were included in the phylogenetic tree construction. As expected, all the homologs of banana TFs were clustered with the monocot clade except MaBSD2, MaCUC2, and MaCUC3. MaBSD2 grouped with gymnosperm, whereas MaCUC2 and MaCUC3 clustered with dicot (Fig 3).
Fig 3. The phylogenetic trees of transcription factors are showing clustering of sequences with different taxonomic groups of plants.
(A) Integration of BBM TFs within AP2/ERF family members from the different taxonomic group of plants. (B) Integration of WUS TFs with other WOX gene family members from the different taxonomic group of plants. (C) Integration of BSD TFs with its other family members from the different taxonomic group of plants. (D) Integration of LEC/L1L TFs with other CBF family members from the different taxonomic group of plants. (E) Integration of VP1 TFs with its other homologs (ARF family) from a different taxonomic group of plants. (F) Integration of CUC TFs within NAC family members from the different taxonomic group of plants. (G) Integration of BOL TFs with its other homologs (BOL A family) from a different taxonomic group of plants. (H) Integration of AGL TFs with its other homologs (MADS family) from a different taxonomic group of plants.
The amino acid sequences corresponding to 9 TFs in banana were studied for conserved domains analysis (S1, S2, S3, S4, S5, S6, S7 and S8 Figs). MaBBM was found to have DNA binding sites within two conserved AP2 domains. It is required for transcription regulation of developmental processes, whereas MaWUS had DNA binding site along with specific DNA base contact site in the homeobox conserved domain. It is known to play a key role in plant development. MaBSD protein containing signature BSD domain is synapse-associated protein and is not much explored. LEC/L1L was noticed to contain a conserved HAP3 domain that played an important role in signal transduction and light harvesting. MaVP1 contained one acidic amino-terminal region (A1) and three basic regions (B1, B2, B3), having a role in seed development and auxin transport. MaBOL possessed BolA region that is involved in callus induction. MaCUC a member of NAC gene family is known to be related with organogenesis. The presence of these domains in the selected TFs confirmed their belongings to the respective families.
Development of somatic embryogenesis
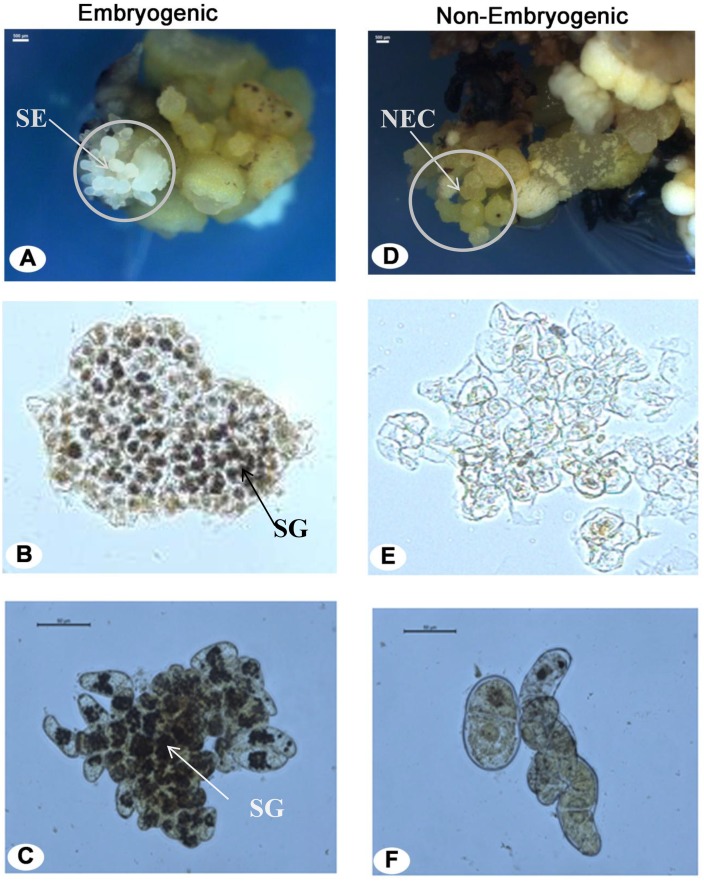
Total eighteen explants responded on the 2, 4-D containing medium with 3% frequency of embryogenic callus formation. On the other side, none of the explants responded for embryogenecity on MS basal medium (without 2, 4-D). Nearly after 3–6 months (12W-24W) of incubation one or few somatic embryos emerged on the surface of callus. Microscopic visualization of callus consisted of somatic embryos, non-embryogenic callus, and their respective suspension cells are shown in Fig 4. The somatic embryos were globular and translucent in nature and present on the surface of the callus (Fig 4A). Embryogenic region beneath the somatic embryos contained many numbers of friable embryogenic cells and suitable for initiation of ECS. The non-embryogenic callus was yellow, compact, hard and lacked embryo-like structure on the surface (Fig 4D). Embryogenic as well as non-embryogenic cells were inoculated in the suspension medium. Suspension cultures of independent embryogenic and non-embryogenic lines were kept separately in the shaker. Embryogenic cell aggregates multiplied and formed many lobed structures from peripheries of which new aggregates were released. Non-embryogenic cells did not divide and eventually led to cell death after 1W. The responded ECS were sub-cultured weekly and maintained for more than 20 months with the high efficiency of embryogenic response.
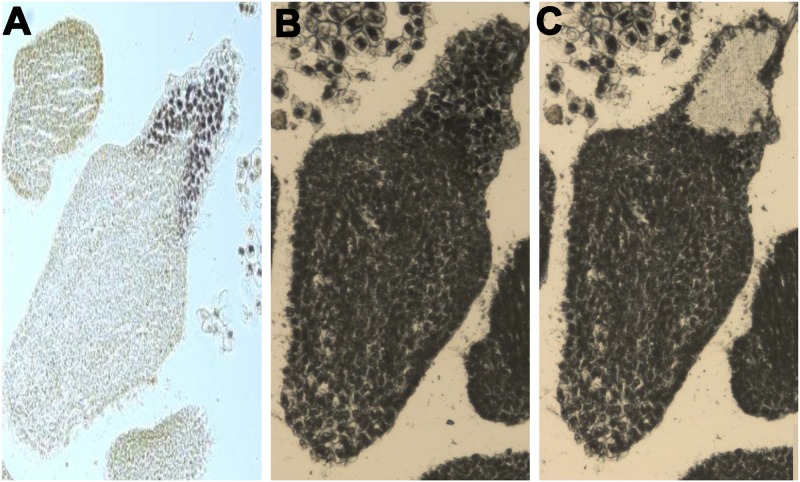
Fig 4. Histological analysis of callus and cell suspension cultures under the microscope.
(A) Somatic embryos in callus. Iodine-stained (B) embryogenic cells and (C) ECS with packed starch granules in deep black color. Iodine-stained (D) non-embryogenic callus and(E) NECS. SE, NEC, and SG in the figure represent somatic embryos, non-embryogenic callus, and starch granules, respectively.
Embryogenic cells have dense cytoplasm with small vacuole and abundant starch granules at borders (Fig 4B). These cells were mostly isodiametric and spherical in shape. The cells of non-embryogenic nature were highly vacuolated, contained fewer starch granules and abnormal in shape and size (Fig 4E). These observations are in agreement with previous studies [2, 7, 56]. The darkly stained cell aggregates have also been confirmed the presence of dense cytoplasm with abundant starch granules in ECS (Fig 4C). The similar observation was reported in the other studies [2, 57]. These embryogenic cells were spherical in shape and multiplied at a higher rate. The cells in NECS were the irregular shape, highly vacuolated and hardly contained any starch granules (Fig 4F). Apart from this, the potent embryogenic cells were snipped off using LCM from a mature embryo, which is used as 0W ECS for the expression study (Fig 5).
Fig 5. Representative image of LCM-derived tissue for RNA isolation.
(A) Iodine-stained embryogenic cells for LCM. (B) Somatic embryo before LCM-derived tissue extraction. (C) Somatic embryo after embryogenic cells snipped off.
Expression study of TFs in response to 2, 4-D treatment in callus
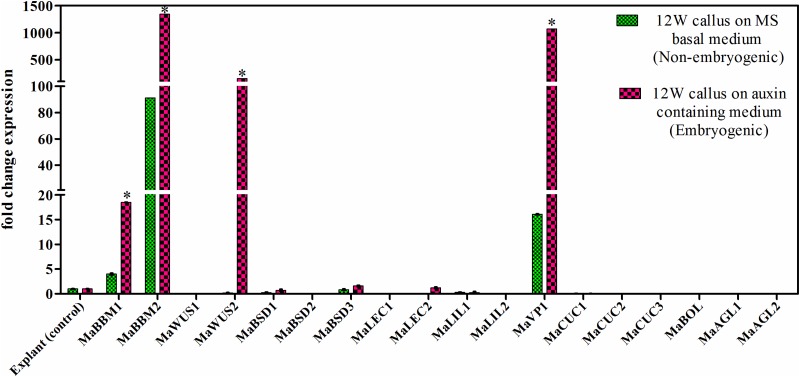
Banana male flowers were inoculated on callus induction medium without 2, 4-D and with 2, 4-D treatment. After 12W, the callus was harvested and further used for gene expression study (Fig 6). The explant (male flower) was used as a control for the study. The expression of MaBBM1, MaBBM2, and MaVP1 was upregulated on 2, 4-D free medium with respect to the explant. The significantly higher expression of MaWUS2 along with MaBBM1, MaBBM2, and MaVP1 was observed on 2, 4-D supplemented medium. The expression of MaBBM1, MaBBM2, and MaVP1 was increased to 4.6, 14.7, and 66.9 fold respectively, in 2, 4-D supplemented medium as compared to 2, 4-D free medium (Fig 6). MaWUS1, MaBSD1, MaBSD2, MaBSD3, MaLEC1, MaLEC2, MaL1L1, and MaCUC1 showed no significant change (p<0.05) in their expression level in both conditions (with 2, 4-D and without 2, 4-D) as compared to the explant. The expression of MaCUC2, MaCUC3, MaBOL, MaAGL1, and MaAGL2 was not detected in the study.
Fig 6. Quantitative real-time PCR study of selected TFs in callus.
The differential expression of TFs in 12W callus is shown in response to 2, 4 –D treatment. The gene expression was normalized using Actin1 as an internal control. Bars denote mean fold expression as compared to the control (explant) ± SD. Statistical analysis was performed using student’s paired t-test. Statistical significance was checked at p<0.05 (*) with respect to the MS basal medium (without 2, 4 -D treatment).
Gene expression analysis of TFs at different developmental stages of ECS and NECS
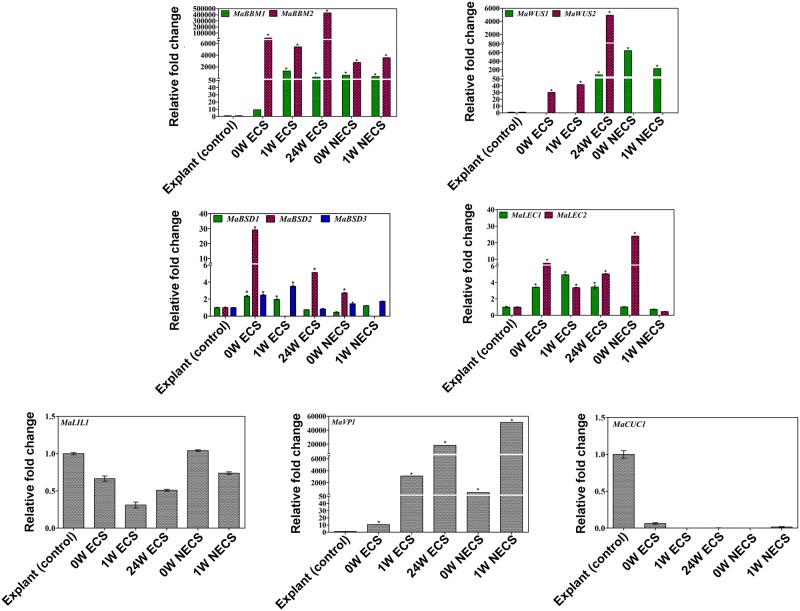
Differential expression of all 18 genes was observed at various developmental stages of ECS (0W, 1W, and 24W) and NECS (0W and 1W) cultures (Fig 7). The male flower (explant) was used as a control for the expression study. MaBBM1, MaBBM2, MaWUS1, MaLEC2, and MaVP1 were expressed differentially in 0W and 1W NECS. The higher expression of MaBBM (MaBBM1 and MaBBM2) was noticed in both ECS and NECS. However, MaBBM2 showed increased transcript level in comparison to MaBBM1 in both ECS and NECS (Fig 7). The expression of MaWUS2 was gradually increased in different stages of ECS, but it was undetected in NECS. In contrast, MaWUS1 was highly expressed in different stages of NECS, but in the case of ECS, its expression was detected only at 24W. Out of three paralogs of MaBSD, the expression of MaBSD2 was significantly upregulated at early (0W) and late (24W) stages of ECS, while MaBSD3 showed higher expression at 1W ECS. MaBSD2 and MaBSD3 were expressed only at 0W and 1W NECS, respectively.
Fig 7. Quantitative real-time PCR study of the selected transcription factors in ECS.
Differential expression of selected TFs in different development stages of ECS (0W, 1W, and 24W) and NECS (0W and 1W). The gene expression was normalized using Actin1 as an internal control. Bars denote mean fold expression as compared to the control (explant) ± SD. The color bars indicate the homolog of the respective gene. (* indicates p<0.05).
The higher expression of MaLEC2 was noticed in all stages of ECS and 0W NECS. On the other side, MaLEC1 showed higher expression at different stages of ECS but not NECS. MaLEC2 was highly expressed at 0W NECS. MaVP1 in banana was significantly upregulated at different development stages of ECS and NECS. MaL1L1 and MaCUC1 did not show any significant upregulation in expression level as compared to the explant. The expression of MaL1L2, MaCUC2, MaCUC3, MaAGL, and MaBOL was not detected in the qPCR study.
Discussion
SE is widely utilized for micropropagation and genetic transformation, but the basic molecular mechanism behind it is not well understood [58–59]. Recently, the proteome approach has been reported to identify the differentially expressed proteins during the SE in banana [2]. Gene expression study of SE related TFs could be the significant approach for understanding their role in in-vitro developmental biology. In banana potency of explant to develop embryogenic callus is very low [59]. Moreover, the prolonged culturing in the callus induction medium could also lead to a somaclonal variation [11]. Therefore, it is important to find the molecular regulators that can be explored to enhance the SE potential in banana. In this study, in-silico characterization and expression analysis of the 18 genes of 9 TF families were studied for their role in SE of banana. Homologs of most of these genes in other plant species have already been reported for their role in SE [30–31, 15, 38, 60, 29, 25, 61]. The presence of multiple homologs of TFs in banana may be the result of gene duplication events during evolution, which may have significance for functional divergence [53].
The differential expression patterns of 18 genes in embryogenic and non-embryogenic tissues, and cell suspension cultures with respect to the explant of cv. Grand Naine were determined. MaBBM2 is highly expressed in all stages of ECS. The expression of MaBBM2 in ECS was increased to 22.95 fold at 24W as compared to 0W. However, the lower expression of MaBBM2 was noticed in NECS as compared to ECS. BBM role in the conversion of vegetative tissue to embryogenic culture has been reported in Brassica napus [15]. In arabidopsis, overexpression of GmBBM1 resulted in somatic embryos emergence in transgenic lines [62]. Similarly, our expression study also revealed that MaBBM2 may play an important role in the conversion of explant to embryogenic callus and ECS.
WUS contains a homeodomain that is involved in regulation of developmental processes [21, 63]. WUS role in promoting vegetative to embryogenic transition and stem cell maintenance has been reported in arabidopsis [21]. Among the two homologs of WUS under study, MaWUS2 was found to be highly expressed in the late stage of ECS that suggests its potential role in ECS maintenance. Recently, BBM and WUS have also been reported to improve transformation efficiency in monocots i.e., sorghum, sugarcane and rice [64].
BSD domain containing genes are known to be a basal TF which are reported for their association with cell proliferation during SE [34]. Here, we identified the three homologs of BSD (MaBSD1, MaBSD2, and MaBSD3) in banana. MaBSD homologs shared two distinct clades in phylogenetic tree analysis. MaBSD2 clustered with gymnosperm, while MaBSD1 and MaBSD3 were grouped in a clade with monocot. This indicates their divergence during the evolution. MaBSD2 was differentially expressed in ECS, whereas MaBDS1 did not show any significant change in expression level. These results suggested that gene duplication during the evolution process may hamper the functionality of MaBSD homologs.
LEC plays role in controlling embryogenesis [65]. The present study showed high accumulation of MaLEC2 transcript in 0W NECS. It suggests that MaLEC2 could lead to the cell necrosis via non-embryogenic callus in banana. However, LEC1 was expressed in embryogenic cells as compared to non-embryogenic cells in carrot [66]. The L1L has close structural resemblance with LEC and known to be a regulator of embryo development. In our study, the homologs of MaL1L did not show any significant change in the expression.
VP1 an auxin-inducible gene that encodes a TF involved in ABA signaling [67–68]. We noticed that MaVP1 was highly expressed in callus, ECS, and NECS. Expression of VP1 in ECS of arabidopsis has been correlated with its role in embryo development [38]. In Secale cereale, VP1 has been reported to have a negative effect on the development of embryogenic callus [69].
CUC is known to induce adventitious shoots in arabidopsis [70–72]. It is utilized as a predictive marker for root and shoot organogenesis [39, 73]. Hence, we selected CUC gene from banana as a negative control for the expression study. As expected CUC homologs were either absent or having basal expression level in different development stages of SE in banana. The expression of MaAGL and MaBOL was not detected. Previous reports showed AGL15 accumulation in tissues derived from double fertilization and its participation in the early stages of zygotic embryo development [33]. The significantly higher expression of MaLEC2 at 0W NECS has suggested their role towards non-embryogenecity in banana.
Based on the differential expression patterns, we anticipate that MaBBM2 and MaWUS2 are the promising candidates for the embryogenicity in banana. Besides, it would be needed to confirm the functional role of MaLEC2 in the development of non-embryogenic callus. Therefore, future studies will be directed to functionally assess the role of MaBBM2, MaWUS2, and MaLEC2 for the better understanding of SE in banana.
Supporting information
Amino acid sequences with two repeats of the conserved AP2 domain are highlighted. Homologs selected for the study, BnBBM1 (Brassica napus accession no. AF317904), BnBBM2 (Brassica napus accession no. AF317905), AtBBM (Arabidopsis thaliana accession no. NP_197245), GmBBM (Glycine max accession no. HM775856).
(TIF)
Amino acid sequences with conserved Homeodomain are highlighted. Homologs selected for the study, VvWUS (Vitus vinifera accession no. XP_002266323.1), AtWUS1 (Arabidopsis thaliana accession no.NP_565429.1), OsWUS (Oryzasativa accession no.AB218894), PaWUS (Picea abies accession no. JX512364), AtWUS2 (Arabidopsis thaliana accession no. NM_125325).
(TIF)
Amino acid sequences with the conserved domain (BSD) are highlighted. Homologs selected for the study, PtBSD (Populus trichocarpa accession no. XP_002310888), VvBSD (Vitis vinifera accession no. XP_010647076), ZomBSD (Zostera marina accession no. KMZ65018), PsBSD (Picea sitchensis accession no. ABK24663).
(TIF)
Amino acid sequences with the conserved HAP3-B domain are highlighted. Homologs selected for the study, GlLEC (Glycine latifolia accession no. ABW7151), ZmLEC (Zea mays accession no. AF410176), PcLEC (Pinus contorta accession no. HM852975) AtLEC (Arabidopsis thaliana accession no. NP_173616), PdL1L (Phoenix dactylifera accession no.XP_008786198), ZmL1L (Zea mays accession no. NP_001167647), PsL1L (Picea sitchensis accession no. ABK25387).
(TIF)
Amino acid sequences with the conserved domain (acidic and basic regions) are highlighted. Homologs selected for the study, NnVP1 (Nelumbo nucifera accession no. XP_010245692), VvVP1 (Vitis vinifera accession no. XP_003632397), ZmVP1 (Zea mays accession no. NM_001112070), PaVP1 (Picea abies accession no. AAG22585).
(TIF)
Amino acid sequences with the conserved domain (NAC) are highlighted. Homologs selected for the study, EgCUC (Elaeis guineens accession no. HM62227) PsCUC (Picea sitchensis accession no. ABR16679), CpCUC (Carica papaya accession no. BK007973), PaCUC (Picea abies accession no. ADQ47506), PsCUC (Picea sitchensis accession no. ABR16679).
(TIF)
Amino acid sequences with the conserved domain (BOLA) are highlighted. JrBOL (Juglans regia accession no. XP_018860422), CsBOL (Cucumis sativus accession no. XP_004147354), PdBOL (Phoenix dactylifera accession no.XP_008785827), PsBOL (Picea sitchensis accession no. ABK22779).
(TIF)
Amino acid sequences with the conserved domain (MADS domain) are highlighted. Homologs selected for the study, AtAGL (Arabidopsis thaliana accession no. AAB3897), VvAGL (Vitis vinifera accession no. AF373603), GbAGL (Ginkgo biloba accession no. AAM76208), EgAGL (Elaeis guineensis accession no. XP_01090993).
(TIF)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The research was supported by National Agri-Food Biotechnology Institute (NABI), Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India for grant and research facility. Authors are thankful to Biotechnology Industry Research Assistance Council (BIRAC) for banana biofortification project grant to NABI. S and NK are thankful to Department of Biotechnology, Panjab University Chandigarh for Ph.D. registration.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The research was supported by National Agri-Food Biotechnology Institute (NABI), Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India for research facility. This study was funded by NABI CORE Research Grant and supported by BIRAC for Banana Biofortification Project to Dr Siddharth Tiwari. The funders did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Singh H, Uma S, Selvarajan R, Karihaloo J. Micropropagation for production of quality banana planting material in Asia-Pacific. Asia-Pacific Consortium on Agricultural Biotechnology (APCoAB). 2011;New Delhi, India 92.
- 2.Kumaravel M, Uma S, Backiyarani S, Saraswathi MS, Vaganan MM, Muthusamy M, Sajith KP. Differential proteome analysis during early somatic embryogenesis in Musa spp. AAA cv. Grand Naine. Plant Cell Reps. 2017;36:163–78. [DOI] [PubMed] [Google Scholar]
- 3.Hussein S, Ibrahim R, Ling A, Kiong P. Somatic embryogenesis: an alternative method for in vitro micropropagation. Iran J Biotechnol. 2006;4:156–161. [Google Scholar]
- 4.Wirakarnain S, Hossain ABMS, Chandran S. Plantlet production through development of competent multiple meristem cultures from male inflorescence of banana, Musa acuminata cv. ‘Pisang Mas’ (AA). Am J Biochem Biotechnol. 2008;4:325–328. doi: 10.3844/ajbbsp.2008.325.328 [Google Scholar]
- 5.Tripathi JN, Oduor RO, Tripathi L. A high-throughput regeneration and transformation platform for production of genetically modified banana. Front. Plant Sci. 2015;6 doi: 10.3389/fpls.2015.01025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganapathi TR, Higgs NS, Balint-Kurti PJ, Arntzen CJ, May GD, Van Eck JM. Agrobacterium-mediated transformation of embryogenic cell suspensions of the banana cultivar Rasthali (AAB). Plant cell Reps. 2001;20:157–62. [DOI] [PubMed] [Google Scholar]
- 7.Côte FX, Domergue R, Monmarson S, Schwendiman J, Teisson C, Escalant JV. Embryogenic cell suspensions from the male flower of Musa AAA cv. Grand nain. Physiol Plant. 1996;97:285–290. doi: 10.1034/j.1399-3054.1996.970211.x [Google Scholar]
- 8.Grapin A, Schwendiman J, Teisson C. Somatic embryogenesis in plantain banana. In Vitro-Plant. 1996;32:66–71. [Google Scholar]
- 9.Khalil SM and Elbanna AAM. Highly efficient somatic embryogenesis and plant regeneration via suspension cultures of banana (Musa spp.). Arab J Biotech. 2004;7: 99–110. [Google Scholar]
- 10.Abdellatif KF, Hegazy AE, Aboshama HM, Emara HA, El-Shahed AA. Morphological and molecular characterization of somaclonal variations in tissue culture-derived banana plants. J Genet Eng Biotechnol. 2012;30:47–53. [Google Scholar]
- 11.Bairu MW, Aremu AO, Van Staden J. Somaclonal variation in plants: causes and detection methods. Plant Growth Regul. 2011; 63: 147–173 [Google Scholar]
- 12.Dey T, Saha S, Ghosh PD. Somaclonal variation among somatic embryo derived plants-evaluation of agronomically important somaclones and detection of genetic changes by RAPD in Cymbopogon winterianus. S Afr J Bot. 2015;96:112–21. [Google Scholar]
- 13.Shiu SH, Shih MC, Li WH. Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiol. 2005;139:18–26. doi: 10.1104/pp.105.065110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmermann R, Werr W. Pattern formation in the monocot embryo as revealed by NAM and CUC3 orthologues from Zea mays L. Plant Mol Biol. 2005; 58:669–85 doi: 10.1007/s11103-005-7702-x [DOI] [PubMed] [Google Scholar]
- 15.Boutilier K. Ectopic Expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell. 2002;14:1737–1749. doi: 10.1105/tpc.001941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srinivasan C, Liu Z, Heidmann I, Supena ED, Fukuoka H, Joosen R, et al. Heterologous expression of the BABY BOOM AP2/ERF transcription factor enhances the regeneration capacity of tobacco (Nicotiana tabacum L.). Planta. 2007;225:341–351. doi: 10.1007/s00425-006-0358-1 [DOI] [PubMed] [Google Scholar]
- 17.Passarinho P, Ketelaar T, Xing M, van Arkel J, Maliepaard C, Hendriks MW, et al. BABY BOOM target genes provide diverse entry points into cell proliferation and cell growth pathways. Plant Mol Biol. 2008;68:225–237. doi: 10.1007/s11103-008-9364-y [DOI] [PubMed] [Google Scholar]
- 18.Heidmann I, De Lange B, Lambalk J, Angenent GC, Boutilier K. Efficient sweet pepper transformation mediated by the BABY BOOM transcription factor. Plant Cell Rep. 2011;30:1107–1115. doi: 10.1007/s00299-011-1018-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Florez SL, Erwin RL, Maximova SN, Guiltinan MJ, Curtis WR. Enhanced somatic embryogenesis in Theobroma cacao using the homologous BABY BOOM transcription factor. BMC Plant Biol. 2015;15:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laux T, Mayer KF, Berger J, Jürgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in arabidopsis. Development. 1996;122:87–96 [DOI] [PubMed] [Google Scholar]
- 21.Zuo J, Niu QW, Frugis G, Chua NH. The WUSCHEL gene promotes a vegetative-to-embryonic transition in arabidopsis. Plant J. 2002;30:349–359. doi: 10.1046/j.1365-313X.2002.01289.x [DOI] [PubMed] [Google Scholar]
- 22.Xu YY, Wang XM, Li J, Li JH, Wu JS, Walker JC, et al. Activation of the WUS gene induces ectopic initiation of floral meristems on mature stem surface in Arabidopsis thaliana. Plant Mol Biol. 2005;57:773–784. doi: 10.1007/s11103-005-0952-9 [DOI] [PubMed] [Google Scholar]
- 23.Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV. WUSCHEL protein movement mediates stem cell homeostasis in the arabidopsis shoot apex. Genes Dev. 2011;25:2025–2030. doi: 10.1101/gad.17258511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doerks T, Huber S, Buchner E, Bork P. BSD: A novel domain in transcription factors and synapse-associated proteins. Trends Biochem Sci. 2002;27:168–170. doi: 10.1016/S0968-0004(01)02042-4 [DOI] [PubMed] [Google Scholar]
- 25.Park J, Kim MJ, Jung SJ, Suh MC. Identification of a novel transcription factor AtBSD1 containing a BSD domain in Arabidopsis thaliana. J Plant Biol. 2009;52:141–146. doi: 10.1007/s12374-009-9015-0 [Google Scholar]
- 26.Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, et al. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA. 2001;98:11806–11811. doi: 10.1073/pnas.201413498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaj MD, Zhang S, Harada JJ, Lemaux PG. Leafy cotyledon genes are essential for induction of somatic embryogenesis of arabidopsis. Planta. 2005;222:977–988. doi: 10.1007/s00425-005-0041-y [DOI] [PubMed] [Google Scholar]
- 28.Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, et al. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell. 2003;15:5–18. doi: 10.1105/tpc.006973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alemanno L, Devic M, Niemenak N, Sanier C, Guilleminot J, Rio M, et al. Characterization of LEAFY COTYLEDON1-LIKE during embryogenesis in Theobroma cacao L. Planta. 2008;227:853–866. doi: 10.1007/s00425-007-0662-4 [DOI] [PubMed] [Google Scholar]
- 30.Jones HD, Kurup S, Peters NCB, Holdsworth MJ. Identification and analysis of proteins that interact with the Avena fatua homologue of the maize transcription factor VIVIPAROUS 1. Plant J. 2000;21:133–142. doi: 10.1046/j.1365-313X.2000.00662.x [DOI] [PubMed] [Google Scholar]
- 31.Su YH, Zhao XY, Liu YB, Zhang CL, O’Neill SD, Zhang XS. Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in arabidopsis. Plant J. 2009;59:448–460. doi: 10.1111/j.1365-313X.2009.03880.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marsch-Martinez N, Greco R, Becker JD, Dixit S, Bergervoet JH, Karaba A, et al. BOLITA an arabidopsis AP2/ERF-like transcription factor that affects cell expansion and proliferation/differentiation pathways. Plant Mol Biol. 2006;62:825–843. doi: 10.1007/s11103-006-9059-1 [DOI] [PubMed] [Google Scholar]
- 33.Perry SE, Lehti MD, Fernandez DE. The MADS-domain protein AGAMOUS-like 15 accumulates in embryonic tissues with diverse origins. Plant Physiol. 1999;120:121–130. doi: 10.1104/pp.120.1.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maldonado-Borges JI, Ku-Cauich JR, Escobedo-Graciamedrano RM. Annotation of differentially expressed genes in the somatic embryogenesis of Musa and their location in the banana genome. Sci World J. 2013;2013:1–8. doi: 10.1155/2013/535737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thakare D, Tang W, Hill K, Perry SE. The MADS-domain transcriptional regulator AGAMOUS-LIKE15 promotes somatic embryo development in arabidopsis and soybean. Plant Physiol. 2008;146:1663–1672. doi: 10.1104/pp.108.115832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhai L, Xu L, Wang Y, Zhu X, Feng H, Li C, et al. Transcriptional identification and characterization of differentially expressed genes associated with embryogenesis in radish (Raphanus sativus L.). Sci Rep. 2016;6:21652 doi: 10.1038/srep21652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikeda-Iwai M, Satoh S, Kamada H. Establishment of a reproducible tissue culture system for the induction of arabidopsis somatic embryos. J Exp Bot. 2002;53:1575–1580. doi: 10.1093/jxb/erf006 [DOI] [PubMed] [Google Scholar]
- 38.Footitt S, Ingouff M, Clapham D, Von Arnold S. Expression of the viviparous 1 (Pavp1) and p34cdc2 protein kinase (cdc2Pa) genes during somatic embryogenesis in Norway spruce (Picea abies [L.]Karst). J Exp Bot. 2003;54:1711–1719. doi: 10.1093/jxb/erg178 [DOI] [PubMed] [Google Scholar]
- 39.Ikeda Y, Banno H, Niu QW, Howell SH, Chua NH. The enhancer of shoot regeneration 2 gene in arabidopsis regulates CUP-SHAPED COTYLEDON 1 at the transcriptional level and controls cotyledon development. Plant Cell Physiol. 2006;47:1443–1456. doi: 10.1093/pcp/pcl023 [DOI] [PubMed] [Google Scholar]
- 40.Hibara K, Karim MR, Takada S, Taoka KI, Furutani M, Aida M, et al. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell. 2006;18:2946–2957. doi: 10.1105/tpc.106.045716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daimon Y, Takabe K, Tasaka M. The CUP-SHAPED COTYLEDON genes promote adventitious shoot formation on calli. Plant Cell Physiol. 2003;44:113–121. doi: 10.1093/pcp/pcg038 [DOI] [PubMed] [Google Scholar]
- 42.D’Hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O, et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012;488:213–218. doi: 10.1038/nature11241 [DOI] [PubMed] [Google Scholar]
- 43.Jaiswal P. Gramene database: a hub for comparative plant genomics. Plant Reverse Genetics: Methods and Protoco. 2011;678:247–275 [DOI] [PubMed] [Google Scholar]
- 44.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: A conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bjellqvist B, Basse B, Olsen E, Celis JE. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis. 1994;15:529–539. [DOI] [PubMed] [Google Scholar]
- 48.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 49.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131 [DOI] [PubMed] [Google Scholar]
- 50.Ganapathi TR, Suprasanna P, Bapat VA, Kulkarni VM, Rao PS. Somatic embryogenesis and plant regeneration from male flower buds in banana. Curr Sci. 1999;76:1228–1231. [Google Scholar]
- 51.Zeller R. Fixation embedding and sectioning of tissues embryos and single cells. Curr Protoc Mol Bio. 2001;l7:14.1. [DOI] [PubMed] [Google Scholar]
- 52.Gautam V, Singh A, Singh S, Sarkar AK. An efficient LCM-based method for tissue specific expression analysis of genes and miRNAs. Sci Rep. 2016;6:21577 doi: 10.1038/srep21577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pandey A, Misra P, Alok A, Kaur N, Sharma S, Lakhwani D, et al. Genome-wide identification and expression analysis of homeodomain leucine zipper subfamily iv (HDZ IV) gene family from Musa accuminata. Front Plant Sci. 2016;7:20 doi: 10.3389/fpls.2016.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaur N, Pandey A, S, Kumar P, Pandey P, Kesarwani AK, et al. Regulation of banana Phytoene Synthase (MaPSY) expression, characterization and there modulation under various abiotic stress conditions. Front Plant Sci. 2017; 8:462 doi: 10.3389/fpls.2017.00462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmittgen T.D, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- 56.Magnard JL, Le Deunff E, Domenech J, Rogowsky PM, Testillano PS, Rougier M, et al. Genes normally expressed in the endosperm are expressed at early stages of microspore embryogenesis in maize. Plant Mol Biol. 2000;44:559–574. doi: 10.1023/A:1026521506952 [DOI] [PubMed] [Google Scholar]
- 57.Ghosh A, Ganapathi TR, Nath P, Bapat VA. Establishment of embryogenic cell suspension cultures and Agrobacterium-mediated transformation in an important Cavendish banana cv. Robusta (AAA). Plant Cell Tiss Org 2009;97:131–9. [Google Scholar]
- 58.Joshi R and Kumar P. Regulation of somatic embryogenesis in crops. Agri. Reviews. 2013;34:1–20. [Google Scholar]
- 59.Xu CX, Zou R, Pan X, Chen HB. Somatic embryogenesis in banana (Musa spp.). Int J Plant Dev Biol. 2008;2:52–58. [Google Scholar]
- 60.Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, et al. Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc Natl Acad Sci USA. 2006;103:3468–73. doi: 10.1073/pnas.0511331103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng Q, Zheng Y, Perry SE. AGAMOUS-Like15 promotes somatic embryogenesis in arabidopsis and soybean in part by the control of ethylene biosynthesis and response. Plant Physiol. 2013;161:2113–27. doi: 10.1104/pp.113.216275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ouakfaoui S El, Schnell J, Abdeen A, Colville A, Labbé H, Han S, et al. Control of somatic embryogenesis and embryo development by AP2 transcription factors. Plant Mol Biol. 2010;74:313–326. doi: 10.1007/s11103-010-9674-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van der Graff E, Laux T, Rensing SA. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009;10:248 doi: 10.1186/gb-2009-10-12-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho MJ, et al. Morphogenic regulators BABY BOOM and WUSCHEL improve monocot transformation. Plant Cell. 2016; 28:1998–2015. doi: 10.1105/tpc.16.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mahdavi-Darvari F, Noor NM, Ismanizan I. Epigenetic regulation and gene markers as signals of early somatic embryogenesis. Plant Cell Tissue Organ Cult. 2014;120:407–422. doi: 10.1007/s11240-014-0615-0 [Google Scholar]
- 66.Yazawa K, Takahata K, Kamada H. Isolation of the gene encoding carrot LEAFY COTYLEDON1 and expression analysis during somatic and zygotic embryogenesis. Plant Physiol Biochem. 2004;42:215–223. doi: 10.1016/j.plaphy.2003.12.003 [DOI] [PubMed] [Google Scholar]
- 67.Suzuki M, Kao CY, Cocciolone S, McCarty DR. Maize VP1 complements arabidopsis ABI3 and confers a novel ABA/auxin interaction in roots. Plant J. 2001;28:409–418. doi: 10.1046/j.1365-313X.2001.01165.x [DOI] [PubMed] [Google Scholar]
- 68.Brady SM, Sarkar SF, Bonetta D, McCourt P. The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in arabidopsis. Plant J. 2002;34:67–75. doi: 10.1046/j.1365-313X.2003.01707.x [DOI] [PubMed] [Google Scholar]
- 69.Gruszczyńska A, Rakoczy-Trojanowska M. Expression analysis of somatic embryogenesis-related SERK LEC1 VP1 and NiRortologues in rye (Secale cereale L). J Appl Genet. 2011;52:1–8. doi: 10.1007/s13353-010-0015-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takada S, Hibara K, Ishida T, Tasaka M. The CUP-SHAPED COTYLEDON1 gene of arabidopsis regulates shoot apical meristem formation. Development. 2001;128:1127–1135. [DOI] [PubMed] [Google Scholar]
- 71.Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in arabidopsis. Plant Cell. 2003;15:1563–77. doi: 10.1105/tpc.012203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Larsson E, Sundström JF, Sitbon F, Von Arnold S. Expression of PaNAC01 a Picea abies CUP-SHAPED COTYLEDON orthologue is regulated by polar auxin transport and associated with differentiation of the shoot apical meristem and formation of separated cotyledons. Ann Bot. 2012;110:923–934. doi: 10.1093/aob/mcs151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Motte H, Verstraeten I, Werbrouck S, Geelen D. CUC2 as an early marker for regeneration competence in arabidopsis root explants. J Plant Physiol. 2011;168:1598–1601. doi: 10.1016/j.jplph.2011.02.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequences with two repeats of the conserved AP2 domain are highlighted. Homologs selected for the study, BnBBM1 (Brassica napus accession no. AF317904), BnBBM2 (Brassica napus accession no. AF317905), AtBBM (Arabidopsis thaliana accession no. NP_197245), GmBBM (Glycine max accession no. HM775856).
(TIF)
Amino acid sequences with conserved Homeodomain are highlighted. Homologs selected for the study, VvWUS (Vitus vinifera accession no. XP_002266323.1), AtWUS1 (Arabidopsis thaliana accession no.NP_565429.1), OsWUS (Oryzasativa accession no.AB218894), PaWUS (Picea abies accession no. JX512364), AtWUS2 (Arabidopsis thaliana accession no. NM_125325).
(TIF)
Amino acid sequences with the conserved domain (BSD) are highlighted. Homologs selected for the study, PtBSD (Populus trichocarpa accession no. XP_002310888), VvBSD (Vitis vinifera accession no. XP_010647076), ZomBSD (Zostera marina accession no. KMZ65018), PsBSD (Picea sitchensis accession no. ABK24663).
(TIF)
Amino acid sequences with the conserved HAP3-B domain are highlighted. Homologs selected for the study, GlLEC (Glycine latifolia accession no. ABW7151), ZmLEC (Zea mays accession no. AF410176), PcLEC (Pinus contorta accession no. HM852975) AtLEC (Arabidopsis thaliana accession no. NP_173616), PdL1L (Phoenix dactylifera accession no.XP_008786198), ZmL1L (Zea mays accession no. NP_001167647), PsL1L (Picea sitchensis accession no. ABK25387).
(TIF)
Amino acid sequences with the conserved domain (acidic and basic regions) are highlighted. Homologs selected for the study, NnVP1 (Nelumbo nucifera accession no. XP_010245692), VvVP1 (Vitis vinifera accession no. XP_003632397), ZmVP1 (Zea mays accession no. NM_001112070), PaVP1 (Picea abies accession no. AAG22585).
(TIF)
Amino acid sequences with the conserved domain (NAC) are highlighted. Homologs selected for the study, EgCUC (Elaeis guineens accession no. HM62227) PsCUC (Picea sitchensis accession no. ABR16679), CpCUC (Carica papaya accession no. BK007973), PaCUC (Picea abies accession no. ADQ47506), PsCUC (Picea sitchensis accession no. ABR16679).
(TIF)
Amino acid sequences with the conserved domain (BOLA) are highlighted. JrBOL (Juglans regia accession no. XP_018860422), CsBOL (Cucumis sativus accession no. XP_004147354), PdBOL (Phoenix dactylifera accession no.XP_008785827), PsBOL (Picea sitchensis accession no. ABK22779).
(TIF)
Amino acid sequences with the conserved domain (MADS domain) are highlighted. Homologs selected for the study, AtAGL (Arabidopsis thaliana accession no. AAB3897), VvAGL (Vitis vinifera accession no. AF373603), GbAGL (Ginkgo biloba accession no. AAM76208), EgAGL (Elaeis guineensis accession no. XP_01090993).
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.