Abstract
Short interfering RNAs (siRNAs) are double-stranded RNAs of ≈21–25 nucleotides that have been shown to function as key intermediaries in triggering sequence-specific RNA degradation during posttranscriptional gene silencing in plants and RNA interference in invertebrates. siRNAs have a characteristic structure, with 5′-phosphate/3′-hydroxyl ends and a 2-base 3′ overhang on each strand of the duplex. In this study, we present data that synthetic siRNAs can induce gene-specific inhibition of expression in Caenorhabditis elegans and in cell lines from humans and mice. In each case, the interference by siRNAs was superior to the inhibition of gene expression mediated by single-stranded antisense oligonucleotides. The siRNAs seem to avoid the well documented nonspecific effects triggered by longer double-stranded RNAs in mammalian cells. These observations may open a path toward the use of siRNAs as a reverse genetic and therapeutic tool in mammalian cells.
Mechanisms that silence unwanted gene expression are critical for normal cellular function. Characterized gene silencing mechanisms include a variety of transcriptional and posttranscriptional surveillance processes (1–3). Double-stranded RNA (dsRNA) has been shown to trigger one of these posttranscriptional surveillance processes, in which gene silencing involves the degradation of single-stranded RNA (ssRNA) targets complementary to the dsRNA trigger (4). RNA interference (RNAi) effects triggered by dsRNA have been demonstrated in a number of organisms including plants, protozoa, nematodes, and insects (5). RNAi may play a role in the silencing of mobile elements in Caenorhabditis elegans and Drosophila (6–9). Similar posttranscriptional gene silencing (PTGS) effects have been implicated as an anti-viral response in plants. PTGS/RNAi seems to be a multistep pathway requiring the processing of the trigger, a facilitated interaction with, and degradation of, the target mRNA. In some cases, these processes may also involve physical amplification of the trigger RNA and long-term maintenance of gene silencing (10, 11).
A key finding from recent work has shown the generation of small (≈21–25 nucleotides) dsRNAs from the input dsRNA during PTGS and RNAi (12–16). These small dsRNAs have been detected in plants, Drosophila, and C. elegans and have been suggested to serve as guide RNAs for target recognition. In Drosophila extracts subjected to RNAi, these small dsRNAs [called short interfering (siRNAs)] resemble breakdown products of an RNase III-like digestion (17). In particular, each strand of the siRNAs carry 5′ phosphate and 3′ hydroxyl termini and 2- or 3-nt 3′ overhangs. siRNAs of 21–22 nucleotides can induce specific degradation when added to Drosophila cell extracts (17). Further, a Drosophila dsRNA-specific RNase has been identified that can degrade large dsRNA (200 and 500 bp) to small dsRNAs of ≈22 nucleotides. RNAi-triggered inhibition of this ribonuclease significantly reduces the effectiveness of RNAi in Drosophila S2 cells (18).
As yet, clear evidence for the generality of an RNAi-like mechanism in vertebrate cells is lacking. Several studies have reported evidence for dsRNA-triggered silencing in particular certain vertebrate systems, early embryos of mice, zebrafish, and Xenopus, as well as Chinese hamster ovary cells (19–25). At the same time, numerous reports have described failures to observe gene-specific RNAi effects in different vertebrate systems, demonstrating instead nonspecific effects of dsRNA on gene expression (26–29). These nonspecific effects have not been surprising as there is an extensive literature describing a variety of nonspecific responses induced by dsRNAs in mammalian cells. A major component of the mammalian nonspecific response to dsRNA is mediated by the dsRNA-dependent protein kinase, PKR, which phosphorylates and inactivates the translation factor eIF2α, leading to a generalized suppression of protein synthesis and cell death via both nonapoptotic and apoptotic pathways (30). PKR may be one of several kinases in mammalian cells that can mediate this response (31). A second dsRNA-response pathway involving the dsRNA-induced synthesis of 2′-5′ polyadenylic acid and a consequent activation of a sequence-non-specific RNase (RNaseL) has also been demonstrated (32). These nonspecific responses to dsRNA, however, do not necessarily preclude the presence of an RNAi-like mechanism in mammalian cells. The activation of PKR by dsRNA has been shown to be length-dependent; dsRNAs of less than 30 nucleotides are unable to activate PKR, and full activation requires ≈80 nucleotides (33, 34). Given the observations that (i) 21–25-nt dsRNAs with a characteristic structure can mediate RNAi in cell extracts and that (ii) dsRNAs of less than 30 bp do not activate PKR, we set out to determine whether short dsRNAs with an RNase III cleavage structure could trigger a gene-specific RNAi response in model invertebrates and mammalian cells.
Methods
Nucleic Acids.
Single-stranded, gene-specific sense and antisense RNA oligomers were synthesized by using 2′-O-(tri-isopropyl) silyloxymethyl chemistry by Xeragon AG (Zurich, Switzerland). We have previously shown RNAs produced by this methodology are highly pure and efficiently form RNA duplexes (16, 27). For studies conducted in C. elegans, RNA oligomers were annealed and injected into adults at a concentration of 5 mg/ml as described (16). For experiments conducted using mammalian cells, dsRNA molecules were generated by mixing sense and antisense ssRNA oligomers (100 μg each) in 10 mM Tris⋅Cl (pH 7.0), 20 mM NaCl (total volume 300 μl), heating to 95°C, and cooling slowly (18 h) to room temperature. The dsRNAs were ethanol-precipitated and resuspended in water at ≈0.5 mg/ml. The integrity and the dsRNA character of the annealed RNAs were confirmed by gel electrophoresis. The sequences of the RNA oligonucleotides used are shown in Table 2, which is published as supplemental data on the PNAS web site, www.pnas.org; the cat 22 and 23 ssRNA oligomers were HPLC-purified. Plasmid pEGFP-N3 (CLONTECH) expresses a mammalian-enhanced version of green fluorescent protein (GFP) and neomycin phosphotransferase (neo). Plasmid pcDNA3.CAT (Invitrogen) expresses chloramphenicol acetyl transferase (CAT) and neo.
Cell Culture and Nucleic Acid Transfections.
All mammalian cells were grown in DMEM (Life Technologies, Rockville, MD) supplemented with 10% (vol/vol) FBS (Gemini Biological Products, Calabasas, CA). Primary mouse embryonic fibroblasts (MEFs) from wild-type I129 mouse embryos (a gift of J. Bell, Univ. of Ottawa, Ontario, Canada; ref. 31) were expanded to generate a more homogenous cell line and were used at passages 20–50 (35). 293 is a human embryonic kidney cell line (36); HeLa is a human epithelial cell line derived from a cervical adenocarcinoma [American Type Culture Collection (ATTC) no. CCL-2]. Plasmid/RNA cotransfection of mammalian cells was mediated by using the cationic lipid Lipofectamine (GIBCO) and the propriety plus reagent (Life Technologies). Cells were seeded ≈18 h before transfection and were transfected at ≈70–80% confluency. Plasmid DNA was complexed with the plus reagent (4–6 μl/2 μg DNA) in DMEM for ≈15 min. RNAs were added 5–10 min into the plasmid/plus reagent incubation. Lipofectamine diluted in DMEM was added to the plasmid/plus reagent/RNA mixture, and complexation was continued for an additional 15 min. The amount of Lipofectamine added (8–15 μg) was based on the total weight of nucleic acid (DNA and RNA) used and a weight to weight ratio of nucleic acid to lipid of 1:4. The amount of RNA used was adjusted to account for the variations in the sizes of RNA. For small RNAs (21–27 nucleotides), 70 pmols of ssRNA and dsRNA was used, corresponding to ≈0.5 μg of a 22-nt ssRNA and 1 μg of 22-nt dsRNA. For the larger RNAs (78–81 nucleotides), ≈30 pmols of RNA was used (0.85 μg of ssRNA and 1.7 μg of dsRNA). Three hours after initiation of transfection, DMEM supplemented with 20% (vol/vol) FBS was added to the cells.
Analysis of Gene Expression.
The C. elegans unc-22 gene encodes an abundant striated muscle component that results in a characteristic twitching phenotype. Animals were scored for the twitching phenotype as described (16). GFP expression was assessed in mammalian cells by fluorescence-activated cell sorter (FACS; FacsCaliber, Becton Dickinson) by using pcDNA3.CAT-transfected cells to control for background fluorescence. CAT expression was assessed by using an ELISA-based assay (Roche Molecular Biochemicals). Total protein was determined by using the Bradford method as described (27). Poly(A)+ RNA was purified from MEFs by using GTC extraction, oligo(dT) cellulose chromatography, and DNase digestion to remove residual plasmid DNA. After electrophoresis [1.2% agarose/1 × 4-morpholinepropanesulfonic acid (Mops)/5.0% formaldehyde] and Northern blot transfer, filters were sequentially hybridized with random prime-labeled cDNA probes corresponding to egfp and neo. Hybridization intensities were measured by using a BAS150 PhosphorImager (Molecular Dynamics), and pixel densities were calculated by using IMAGE READER 1.4 and IMAGE GAUGE 3.0 (Fuji).
Cell Survival and in Vitro Kinase Assays.
To assay cell survival, MEFs were plated in 96-well plates ≈18 h before transfection and were transfected at ≈70–80% confluency by using Lipofactamine as a carrier. RNA transfections were conducted as above, except for the omission of the plus complexation step, and using 1/10th the amount of RNA and lipid and 1/10th the volume of medium. Cell viability was determined by using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) labeling reagent as described by the manufacturer (Roche Molecular Biochemicals) 48 h after initiation of transfection. In vitro kinase reactions were conducted in a final volume of 12.5 ml by using 100 mM [γ-32P]ATP (specific activity 1 Ci/mM, Amersham Pharmacia), 100 mM ATP (Sigma) in 20 mM Hepes (pH 7.5), 90 mM KCl, 5 mM MgOAc, 1 mM DTT, and an equal amount of cell lysate prepared from 1 × 106 human Jurkat T lymphocytes treated with 100 units/ml of rhIFN-β for 24 h before lysis (lysis buffer: 20 mM Hepes/120 mM KCl/5 mM MgOAc/1 mM benzamidine/1 mM DTT/1% Nonidet P-40). dsRNA (1 μg/ml) was added to each reaction mixture, and the reactions were incubated for 10 min at 30°C. Reactions were quenched by addition of an equal volume of 2 times sample buffer (2 times sample buffer: 62.5 mM Tris⋅Cl, pH 6.8/10% glycerol/2% SDS/0.0125% bromophenol blue/5% β-mercaptoethanol), boiled for 2 min, and subjected to electrophoresis [10% (vol/vol) SDS/PAGE]. Labeled proteins were visualized by autoradiography of dried gels.
Results
Short RNase III-Like Products Can Induce Inhibition of Gene Expression in Invertebrate Cells.
A series of dsRNAs with characteristics of siRNAs (5′ phosphate, 3′ hydroxyl, and 2 base 3′ overhangs on each strand) were generated from chemically synthesized ssRNAs. The siRNAs varied from 21–27 nucleotides and had sequences that matched three different target RNAs, unc-22, cat, and egfp (for sequences see Table 2).
To determine whether siRNAs can be used directly to inhibit gene expression we first assessed interference in C. elegans by using siRNAs corresponding to C. elegans unc-22 (Table 1). unc-22 provides a sensitive and specific assay for genetic interference as this is the only gene in the C. elegans genome that can mutate by loss of function to give a twitching phenotype. unc-22 siRNAs induced a decrease in unc-22 gene expression as measured by the presence of the twitching phenotype in the progeny of injected adults. Small dsRNAs of 23, 24, and 25 nucleotides produced interference with the 25-nt unc-22 siRNA inducing the highest fraction of animals with an affected phenotype (16.3%). As a control, siRNAs directed against an unrelated sequence (egfp) induced no phenotypic changes (Table 1).
Table 1.
Short RNase III-like products can induce specific interference in C. elegans
| Injection | Fraction affected (number scored) |
|---|---|
| unc-22siRNA 23 nts | 1.4% (145) |
| unc-22siRNA 24 nts | 3.6% (279) |
| unc-22siRNA 25 nts | 16.3% (768) |
| unc-22 sense ssRNA 25 nts | 0% (>1100) |
| unc-22 antisense ssRNA 25 nts | 0% (>600) |
| unc-22dsRNA 81 nts | 88.9% (180) |
| egfpsiRNA 22 nts | 0% (>300) |
| egfpsiRNA 23 nts | 0% (>300) |
| egfpsiRNA 24 nts | 0% (>300) |
| egfpsiRNA 25 nts | 0% (>300) |
| No injection | 0% (>300) |
dsRNA molecules were formed with each strand carrying a 5′-PO4, 3′-OH, and 2-base 3′ overhangs. These were injected into adult C. elegans as described in Methods. Percentages shown denote portion of progeny broods that show a specific decrease in unc-22 function as evidenced by twitching behavior in 330 μM levamisole. Numbers in parenthesis are total numbers of animals scored. nts, nucleotides.
21–23-nt dsRNAs Inhibit Expression in MEFs.
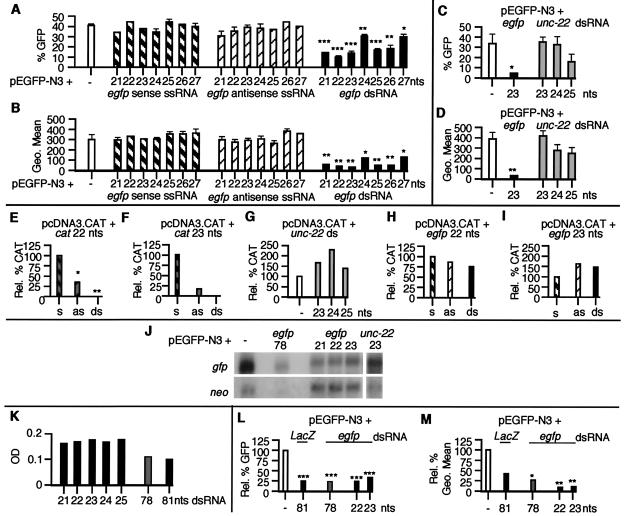
To test whether small dsRNA molecules can specifically inhibit gene expression in vertebrate cells, we cotransfected MEFs with expression plasmids encoding GFP (pEGFP-N3) and CAT (pcDNA3.CAT), and synthetic siRNAs corresponding to egfp, cat, or unc-22 (Fig. 1). The egfp dsRNAs (21–27 nucleotides) all inhibited GFP expression in MEFs. The 22- and 23-nt egfp siRNAs (20 and 21 nucleotides base-paired with 2-nt 3′ overhangs) showed the greatest degree of inhibition, both with respect to the total number of cells expressing GFP (Fig. 1A) and the fluorescence intensity of the GFP expression observed in GFP-positive cells (Fig. 1B). In contrast, unc-22 dsRNAs of 23–25 nucleotides had no significant effect on GFP expression (Fig. 1 C and D).
Figure 1.
Gene-specific inhibition of expression in MEFs by siRNAs. MEFs transfected with plasmid DNA, ssRNAs, and dsRNAs were harvested 48 h after transfection and were assayed for (A–D) GFP expression by FACS analysis (each transfection was assayed in triplicate and data are shown as mean ± SEM). A and C show the percentage of GFP-positive cells and B and D show the fluorescence intensity (Geo Mean) of GFP-positive cells. (E–I) CAT expression (each transfection condition was assayed in triplicate; data in E are normalized to the amount of CAT pg/μg of protein observed in pcDNA3.CAT-transfected cells; data in F–I are normalized to the amount of CAT pg/μg of protein in plasmid and sense ssRNA-transfected cells. s, sense ssRNA; as, antisense ssRNA). (J) egfp and neo RNA levels by Northern analysis of poly(A)+ mRNA. (K) Cell survival (assayed in duplicate and shown as a mean OD560–650; dsRNAs of 21–25 and 78 nucleotides correspond to egfp; the dsRNA of 81 nucleotides corresponds to LacZ). (L and M) GFP expression by FACS analysis (data are shown as relative percentage normalized to pEGFP-N3-transfected cells). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To further assess the efficacy and specificity of the inhibition mediated by siRNAs in mammalian cells, we used a second reporter, CAT (Fig. 1 E–I). cat siRNAs of 22 and 23 nucleotides completely inhibited CAT expression (Fig. 1 E and F), whereas unc-22 and egfp dsRNAs had no little or no effect on CAT expression (Fig. 1 G–I). Although no antisense effect had been seen by using GFP as a reporter, the cat ssRNA antisense oligomers partially inhibited CAT expression. However, the siRNA-mediated inhibition was more potent (≈1.5-fold), suggesting that the gene silencing mediated by the small dsRNAs can be distinguished from a purely antisense-based mechanism.
To analyze this inhibition of egfp expression at an RNA level, poly(A)+ RNA was purified from transfected MEFs and subjected to Northern analysis by using cDNA probes corresponding to egfp and neo, both encoded by the pEGFP-N3 plasmid (Fig. 1J). Quantitative PhosphorImager analysis showed a decrease in the levels of the egfp mRNA obtained from cells cotransfected with the pEGFP-N3 plasmid and the 21-, 22-, and 23-nt egfp siRNAs, compared with cells transfected only with the GFP plasmid. The percentage decrease was ≈60% for all three egfp siRNAs when compared with the levels of egfp mRNA in cells transfected only with plasmid. Importantly, no effect was seen on the levels of the neo transcript compared with plasmid-only transfected cells, indicating that the inhibition induced by the small egfp dsRNAs was sequence-specific. Consistent with this hypothesis, the 23-nt dsRNA corresponding to the C. elegans unc-22 gene had no effect on either egfp or neo expression.
To follow the fate of cells transfected with siRNAs and larger dsRNAs, we assayed MEF cell survival (Fig. 1K). Longer dsRNAs (78 or 81 nucleotides with flush ends) induced a substantial degree of cell death (up to 50%) in a 48-h period, whereas the smaller dsRNAs had a minimal effect on the growth of cells. By examining the effect of the larger dsRNAs on gene expression, we observed that the larger dsRNAs (78 or 81 nucleotides) induced a sequence nonspecific decrease of 75% in the percentage of cells expressing GFP (Fig. 1L) and in CAT protein levels (data not shown), compared with plasmid controls. This nonspecific decrease in gene expression is consistent with previous data from numerous mammalian systems and contrasts with the specific gene silencing the 78-nt egfp dsRNA induces in Drosophila S2 cells (27). However, it should be noted that the decrease in transgene expression after siRNA transfection could be distinguished from the nonspecific inhibition by examination of the GFP fluorescence intensity seen in viable cells. The fluorescence intensity of GFP expression best illustrates a change in the total amount of GFP made by a live cell and therefore is less influenced by nonspecific cell death. Although some decrease (≈60%) in the fluorescence intensity was seen by using the larger ≈80-nt dsRNA molecules (irrespective of sequence), the egfp siRNAs of 22 and 23 nucleotides consistently reduced the intensity of the GFP signal (by ≈90%) to near background levels (Fig. 1M). The difference in specificity between the longer dsRNAs and siRNAs could also be seen at an RNA level were the 78-nt egfp dsRNA induced a significant decrease in both the egfp and neo transcripts, whereas the siRNAs inhibited only egfp (Fig. 1J).
Inhibition of Gene Expression in Human Somatic Cells.
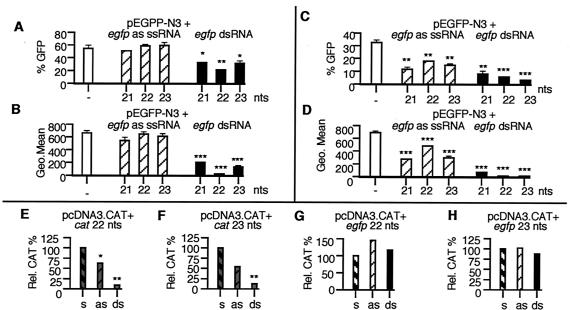
To date, there has been no evidence of an RNAi-like process occurring in human somatic cells. To determine whether siRNAs could also specifically inhibit gene expression in human cells, we cotransfected two commonly used human cell lines, the embryonic kidney cell line 293 and the epithelial carcinoma cell line HeLa, with plasmids and RNA (Fig. 2). All of the egfp siRNAs tested inhibited GFP gene expression in 293 (Fig. 2 A and B) and HeLa (Fig. 2 C and D) cells, with the 22- and 23-nt egfp siRNAs inducing the greatest decrease in GFP expression. In 293 cells cotransfected with pEGFP-N3 and the 22-nt egfp siRNA, the intensity of GFP expression was reduced to near background levels (Fig. 2B). Similar results were seen in HeLa cells cotransfected with pEGFP-N3 and the 22- or 23-nt egfp siRNAs (Fig. 2D). dsRNAs corresponding to unc-22 had no effect on GFP expression in these cells (data not shown). The siRNA-triggered inhibition of GFP expression was dose-dependent in that doubling the amount of dsRNA (from 70 to 140 pmols) decreased GFP intensity by an additional 25% for the egfp 22-nt siRNA and by 45% for the egfp 23-nt siRNA. CAT expression was also significantly inhibited by siRNAs corresponding to cat (Fig. 2 E and F) in HeLa cells. Again, the inhibition mediated by the siRNAs was significantly higher than that seen by using ssRNA antisense oligomers. Cotransfection of the pcDNA3.CAT plasmid and the egfp siRNAs of the same size and of similar GC/AT complexity had no effect on CAT expression (Fig. 2 G and H).
Figure 2.
siRNA-mediated gene silencing in human cells. (A and B) 293 and (C and D) HeLa cells transfected with pEGFP-N3 and antisense (as) ssRNAs and dsRNAs were harvested 48 h after transfection and were assayed for GFP expression by FACS analysis (assayed in triplicate; data are shown as mean ± SEM). A and C show the percentage of GFP-positive cells and B and D show the fluorescence intensity (Geo Mean) of GFP-positive cells. (E–H) HeLa cells transfected with pcDNA3-CAT, ssRNAs, and dsRNAs were harvested 48 h after transfection and assayed for CAT expression (assayed in triplicate and normalized to the amount of CAT pg/μg of protein observed in plasmid plus sense-transfected cells. s, sense ssRNA; as, antisense ssRNA). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
siRNA-Mediated Inhibition of Gene Expression Is Independent of Nonspecific Interference Pathways Activated by Larger dsRNAs.
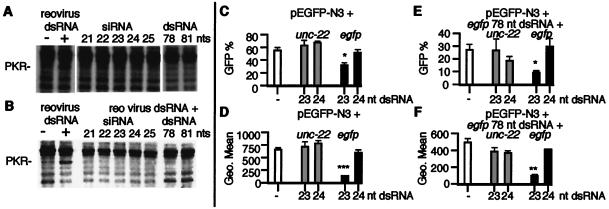
It has been reported that small blunt-ended dsRNAs of less than 30 bp do not activate PKR (34). Indeed, at high concentrations these short dsRNAs can competitively inhibit activation of PKR by larger dsRNAs. Similarly, the synthetic siRNAs used in this study did not activate PKR (Fig. 3A) and inhibited the activation of PKR by a large viral dsRNA (Fig. 3B). Interestingly, in this assay we were unable to detect activation of PKR by the 78- and 81-nt dsRNAs, despite observing a substantial level of cell death, suggesting that other dsRNA-dependent kinases or other pathways may be contributing in MEFs to the decrease in gene expression and cell death observed with these RNAs.
Figure 3.
siRNAs and mammalian dsRNA-dependent pathways. To detect PKR autophosphorylation, we performed in vitro kinase assays as described in Methods. (A) In vitro kinase reactions were performed without exogenous RNA (−) or with 1 μg/ml of reovirus dsRNA or 1 μg/ml of siRNA (21–25 nucleotides), or 1 μg/ml of 78- or 81-nt dsRNA. (B) In vitro kinase competition assays were performed by using si- and dsRNAs. Reactions were performed without exogenous RNA (−) or 1 μg/ml of reovirus RNA, or 75-fold excess siRNA (21–25 nucleotides) or 78- or 81-nt dsRNA, plus reovirus dsRNA (1 μg/ml). siRNAs of 21–25 nucleotides and dsRNA of 78 nucleotides corresponded to egfp (the 81-nt dsRNA corresponds to LacZ). (C and D) 293 cells transfected with pEGFP-N3 and unc-22 or egfp siRNAs, and (E and F) 293 cells transfected with pEGFP-N3 and 78 egfp dsRNA and unc-22 or egfp siRNAs were assayed for GFP expression by FACS analysis 48 h after transfection (each transfection was assayed in triplicate; data are shown as mean ± SEM). B and D show the percentage of GFP-positive cells and C and E show the fluorescence intensity (Geo Mean) of GFP-positive cells. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To see whether the small dsRNAs could block the toxic effect of the larger dsRNAs in cells, we cotransfected 293 cells with the pEGFP-N3 plasmid, the egfp 78-nt dsRNA, and the unc-22 23- or 24-nt siRNAs or the egfp 23- or 24-nt siRNAs (Fig. 3 C–F). The cell death induced by the 78-nt egfp dsRNA was not inhibited by the unc-22 or egfp siRNAs (Fig. 3 C and D vs. E and F) but importantly the 78-nt egfp dsRNA did not block the specific inhibition of GFP expression mediated by the 23-nt egfp siRNA. This result suggests that the siRNA-mediated gene silencing mechanism is independent of nonspecific responses of mammalian cells to dsRNA.
Discussion
A consistent observation of PTGS and RNAi in several species has been the detection of small dsRNAs (≈21–25 nucleotides) and siRNAs derived from the triggering dsRNA. These small dsRNAs have been observed irrespective of whether the initiating dsRNA is delivered directly, is derived from a viral RNA, or is produced from a transgene (12–17). These findings and further biochemical analysis (18) have suggested that the generation of siRNAs represents a critical step in the RNAi/PTGS mechanism. We now present evidence that these siRNAs can have direct effects on gene expression in C. elegans and mammalian cell culture in vivo. Our results in mammalian cells are particularly striking in that previous attempts to assay RNAi effects in vertebrate somatic cells have encountered effects that were predominately gene-nonspecific (26–29). We propose that the small size of the siRNAs avoids the induction of the nonspecific responses of mammalian cells to dsRNA.
Several models have been put forward to explain RNAi, in particular the mechanisms by which siRNAs interact with the target mRNA and thus facilitate its degradation (12–15, 17, 37). It has been proposed that the siRNAs act as a guide for the enzymatic complex required for the sequence-specific cleavage of the target mRNA. Evidence for the role of siRNA as a guide includes cleavage of the target mRNA at regular intervals of ≈21–23 nucleotides in the region corresponding to the input dsRNA (13), with the exact cleavage sites corresponding to the middles of sequences covered by individual 21- or 22-nt siRNAs (17). Although mammals and lower organisms seem to share dsRNA-triggered responses that involve a related intermediate (siRNAs), it is likely that there will be differences as well as similarities in the underlying mechanism.
Several of the proteins shown to play key roles in RNA-triggered gene silencing in plants and invertebrates share homology with potential coding regions from the human or other vertebrate genomes. These include putative RNA-dependent polymerases (RdRp; refs. 38–41), the RDE-1/Argonaute family (8), and a variety of putative helicases and nucleases (9, 18, 42–44). Mammalian homologs of the RNAi-associated Drosophila RNase III have been identified (45, 46). Importantly, one of these putative RNases has been shown to generate small dsRNA molecules of ≈22 nucleotides from larger dsRNAs (18). However, even in invertebrate systems, the precise role of these factors in RNAi remains to be elucidated. Because factors from each of these homology classes have identified roles in normal physiology and development (i.e., beyond genome surveillance), a full analysis of the reaction mechanisms in the different biological systems may be needed before a clear picture of the commonality between RNAi in these different systems will emerge.
Our experiments do not address possible differences in mechanism between invertebrate and vertebrate systems, although we observed some variation between the different assay systems in the optimal size and effectiveness of the inhibiting dsRNA. These differences could be gene-, species-, cell type-, or assay-specific; it will be particularly interesting to determine whether there are species-dependent differences in the length or structure of natural siRNAs. It is not yet clear what roles RNAi/PTGS might play in mammalian systems. RNAi-related silencing mechanisms in plant and invertebrate systems have been implicated in the silencing of viruses and transposons. Mammalian genomes have a need to cope with a considerable load of viruses, selfish DNA, and aberrant transcription. RNAi-related mechanisms could certainly function as a part of the defense network for any or all of these genomic hazards. Alternatively, specific gene silencing by dsRNA could function in normal mammalian gene regulation, e.g., in imprinting or X inactivation (47).
Because of the efficacy and ease with which RNAi can be induced, RNAi has been rapidly exploited in C. elegans and Drosophila as a reverse genetics tool (48). Currently, the principal method used to reduce gene expression in mammalian cells utilizes antisense sequences in the form of single-stranded oligonucleotides and transcripts. The interaction of antisense sequences with mRNA through Watson–Crick base-pairing leads to a decrease in gene expression by several possible mechanisms, including the activation of RNaseH, which cleaves RNA/DNA duplexes, and the inhibition of RNA processing and/or translational blockade (49). Several issues have limited wider use of antisense technology. Problems have included a lack of suitable target sequences within a given mRNA caused by RNA secondary folding, which necessitates screening of multiple antisense sequences to identify those that mediate the greatest level of inhibition and inefficient delivery in vitro and in vivo. We have tested only a limited number of siRNAs in mammalian cells but as yet all of the siRNAs that were tested produced specific inhibition of gene expression, and the siRNAs seem to be very stable and thus may not require the extensive chemical modifications that ssRNA antisense oligonucleotides require to enhance the in vivo half-life. Our initial experiments suggest that siRNAs may be useful for triggering RNAi-like responses that could be used as functional genomics and therapeutic tools. Certain applications may be facilitated by the simple transfection protocols that we have used, whereas other applications may benefit from further optimization and additional exploration of the RNAi mechanism.
Supplementary Material
Acknowledgments
We thank Phil Sharp, Lisa Timmons, Patrick Weiss, Mario Mautino, and members of the Fire Laboratory for insightful discussions, and John Bell for MEFs. We thank the National Institutes of Health (AF R01-GM37706, T32-GM07321, and FI AI446962) and the Carnegie Institution for support. N.J.C. is a National Institutes of Health Fogarty Fellow.
Abbreviations
- dsRNA
double-stranded RNA
- ssRNA
single-stranded RNA
- siRNA
short interfering RNA
- RNAi
RNA interference
- PTGS
posttranscriptional gene silencing
- GFP
green fluorescence protein
- CAT
chloramphenicol acetyl transferase
- PKR
dsRNA-dependent protein kinase
- neo
neomycin phosphotransferase
- MEF
mouse embryonic fibroblast
- FACS
fluorescence-activated cell sorter
Note
A recent report by Elbashir et al. (50) describes a specific interference response in mammalian cells by using 21-nt siRNAs.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wolffe A P, Matzke M A. Science. 1999;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- 2.Frischmeyer P A, Dietz H C. Hum Mol Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell P, Tollervey D. Curr Opin Genet Dev. 2000;10:193–198. doi: 10.1016/s0959-437x(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 4.Fire A. Trends Genet. 1999;15:358–363. doi: 10.1016/s0168-9525(99)01818-1. [DOI] [PubMed] [Google Scholar]
- 5.Cogoni C, Macino G. Curr Opin Genet Dev. 2000;10:638–643. doi: 10.1016/s0959-437x(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 6.Kasschau K D, Carrington J C. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- 7.Llave C, Kasschau K D, Carrington J C. Proc Natl Acad Sci USA. 2000;97:13401–13406. doi: 10.1073/pnas.230334397. . (First Published November 14, 2000; 10.1073/pnas.230334397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tabara H, Sarkissian M, Kelly W G, Fleenor J, Grishok A, Timmons L, Fire A, Mello C C. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 9.Ketting R F, Haverkamp T H, van Luenen H G, Plasterk R H. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 10.Matzke M A, Matzke A J, Pruss G J, Vance V B. Curr Opin Genet Dev. 2001;11:221–227. doi: 10.1016/s0959-437x(00)00183-0. [DOI] [PubMed] [Google Scholar]
- 11.Carthew R W. Curr Opin Cell Biol. 2001;13:244–248. doi: 10.1016/s0955-0674(00)00204-0. [DOI] [PubMed] [Google Scholar]
- 12.Hamilton A J, Baulcombe D C. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 13.Zamore P D, Tuschl T, Sharp P A, Bartel D P. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 14.Hammond S M, Bernstein E, Beach D, Hannon G J. Nature (London) 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 15.Yang D, Lu H, Erickson J W. Curr Biol. 2000;10:1191–1200. doi: 10.1016/s0960-9822(00)00732-6. [DOI] [PubMed] [Google Scholar]
- 16.Parrish S, Fleenor J, Xu S, Mello C, Fire A. Mol Cell. 2000;6:1077–1087. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- 17.Elbashir S M, Lendeckel W, Tuschl T. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernstein E, Caudy A A, Hammond S M, Hannon G J. Nature (London) 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 19.Wianny F, Zernicka-Goetz M. Nat Cell Biol. 2000;2:70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- 20.Svoboda P, Stein P, Hayashi H, Schultz R M. Development (Cambridge, UK) 2000;127:4147–4156. doi: 10.1242/dev.127.19.4147. [DOI] [PubMed] [Google Scholar]
- 21.Wargelius A, Ellingsen S, Fjose A. Biochem Biophys Res Commun. 1999;263:156–161. doi: 10.1006/bbrc.1999.1343. [DOI] [PubMed] [Google Scholar]
- 22.Li Y X, Farrell M J, Liu R, Mohanty N, Kirby M L. Dev Biol. 2000;217:394–405. doi: 10.1006/dbio.1999.9540. [DOI] [PubMed] [Google Scholar]
- 23.Nakano H, Amemiya S, Shiokawa K, Taira M. Biochem Biophys Res Commun. 2000;274:434–439. doi: 10.1006/bbrc.2000.3178. [DOI] [PubMed] [Google Scholar]
- 24.Ui-Tei K, Zenno S, Miyata Y, Saigo K. FEBS Lett. 2000;479:79–82. doi: 10.1016/s0014-5793(00)01883-4. [DOI] [PubMed] [Google Scholar]
- 25.Oelgeschlager M, Larrain J, Geissert D, De Robertis E M. Nature (London) 2000;405:757–763. doi: 10.1038/35015500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tuschl T, Zamore P D, Lehmann R, Bartel D P, Sharp P A. Genes Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caplen N J, Fleenor J, Fire A, Morgan R A. Gene. 2000;252:95–105. doi: 10.1016/s0378-1119(00)00224-9. [DOI] [PubMed] [Google Scholar]
- 28.Oates A C, Bruce A E, Ho R K. Dev Biol. 2000;224:20–28. doi: 10.1006/dbio.2000.9761. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Z, Cao Y, Li M, Meng A. Dev Biol. 2001;229:215–223. doi: 10.1006/dbio.2000.9982. [DOI] [PubMed] [Google Scholar]
- 30.Clemens M J, Elia A. J Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 31.Abraham N, Stojdl D F, Duncan P I, Methot N, Ishii T, Dube M, Vanderhyden B C, Atkins H L, Gray D A, McBurney M W, et al. J Biol Chem. 1999;274:5953–5962. doi: 10.1074/jbc.274.9.5953. [DOI] [PubMed] [Google Scholar]
- 32.Player M R, Torrence P F. Pharmacol Ther. 1998;78:55–113. doi: 10.1016/S0163-7258(97)00167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minks M A, West D K, Benvin S, Baglioni C. J Biol Chem. 1979;254:10180–10183. [PubMed] [Google Scholar]
- 34.Manche L, Green S R, Schmedt C, Mathews M B. Mol Cell Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Todaro G J, Green H. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graham F L, Smiley J, Russell W L, Nairn R. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 37.Bass B L. Cell. 2000;101:235–238. doi: 10.1016/s0092-8674(02)71133-1. [DOI] [PubMed] [Google Scholar]
- 38.Cogoni C, Macino G. Nature (London) 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- 39.Smardon A, Spoerke J M, Stacey S C, Klein M E, Mackin N, Maine E M. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- 40.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe D C. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 41.Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel J B, Jouette D, Lacombe A M, Nikic S, Picault N, et al. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 42.Wu-Scharf D, Jeong B, Zhang C, Cerutti H. Science. 2000;290:1159–1162. doi: 10.1126/science.290.5494.1159. [DOI] [PubMed] [Google Scholar]
- 43.Dernburg A F, Zalevsky J, Colaiacovo M P, Villeneuve A M. Genes Dev. 2000;14:1578–1583. [PMC free article] [PubMed] [Google Scholar]
- 44.Domeier M E, Morse D P, Knight S W, Portereiko M, Bass B L, Mango S E. Science. 2000;289:1928–1931. doi: 10.1126/science.289.5486.1928. [DOI] [PubMed] [Google Scholar]
- 45.Matsuda S, Ichigotani Y, Okuda T, Irimura T, Nakatsugawa S, Hamaguchi M. Biochim Biophys Acta. 2000;1490:163–169. doi: 10.1016/s0167-4781(99)00221-3. [DOI] [PubMed] [Google Scholar]
- 46.Wu H, Xu H, Miraglia L J, Crooke S T. J Biol Chem. 2000;275:36957–36965. doi: 10.1074/jbc.M005494200. [DOI] [PubMed] [Google Scholar]
- 47.Sharp P A. Genes Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- 48.Kuwabara P E, Coulson A. Parasitol Today. 2000;16:347–349. doi: 10.1016/s0169-4758(00)01677-x. [DOI] [PubMed] [Google Scholar]
- 49.Crooke S T. Biochim Biophys Acta. 1999;1489:31–44. doi: 10.1016/s0167-4781(99)00148-7. [DOI] [PubMed] [Google Scholar]
- 50.Elbashir S M, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature (London) 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.