Abstract
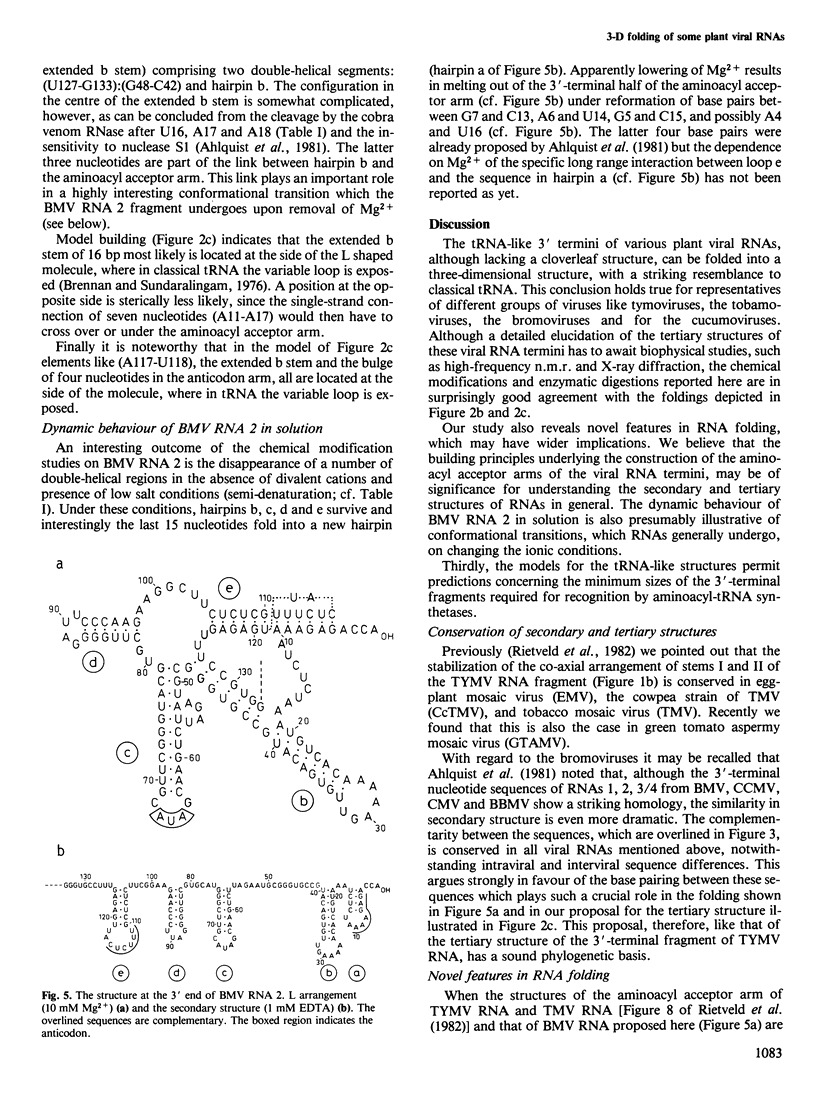
Various plant viral RNAs possess a 3' terminus with tRNA-like properties. These viral RNAs are charged with an amino acid upon incubation with the cognate aminoacyl-tRNA synthetase and ATP. We have studied the structure of end-labelled 3'-terminal fragments of turnip yellow mosaic virus RNA and brome mosaic virus RNA 2 with chemical modifications of the adenosine and cytidine residues and with enzymatic digestions using RNase T1, nuclease S1 and the double-strand-specific ribonuclease from cobra venom. The data indicate that the 3' termini of these plant viral RNAs lack a cloverleaf structure as found in classical tRNA. The three-dimensional folding, however, reveals a striking resemblance with classical tRNA. The models proposed are supported by phylogenetic data. Apparently distinct three-dimensional solutions have evolved to meet the requirements for faithful recognition by tRNA-specific enzymes. The way in which the aminoacyl acceptor arms of these tRNA-like structures are constructed reveal novel features in RNA folding which may have a bearing on the secondary and tertiary structures of RNA in general. The dynamic behaviour of brome mosaic virus RNA 2 in solution presumably is illustrative of conformational transitions, which RNAs generally undergo on changing the ionic conditions.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahlquist P., Dasgupta R., Kaesberg P. Near identity of 3- RNA secondary structure in bromoviruses and cucumber mosaic virus. Cell. 1981 Jan;23(1):183–189. doi: 10.1016/0092-8674(81)90283-x. [DOI] [PubMed] [Google Scholar]
- Auron P. E., Weber L. D., Rich A. Comparison of transfer ribonucleic acid structures using cobra venom and S1 endonucleases. Biochemistry. 1982 Sep 14;21(19):4700–4706. doi: 10.1021/bi00262a028. [DOI] [PubMed] [Google Scholar]
- Bastin M., Hall T. C. Interaction of elongation factor 1 with aminoacylated brome mosaic virus and tRNA's. J Virol. 1976 Oct;20(1):117–122. doi: 10.1128/jvi.20.1.117-122.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan T., Sundaralingam M. Structlre of transfer RNA molecules containing the long variable loop. Nucleic Acids Res. 1976 Nov;3(11):3235–3250. doi: 10.1093/nar/3.11.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briand J. P., Jonard G., Guilley H., Richards K., Hirth L. Nucleotide sequence (n=159) of the amino-acid-accepting 3'-OH extremity of turnip-yellow-mosaic-virus RNA and the last portion of its coat-protein cistron. Eur J Biochem. 1977 Feb;72(3):453–463. doi: 10.1111/j.1432-1033.1977.tb11269.x. [DOI] [PubMed] [Google Scholar]
- Florentz C., Briand J. P., Romby P., Hirth L., Ebel J. P., Glegé R. The tRNA-like structure of turnip yellow mosaic virus RNA: structural organization of the last 159 nucleotides from the 3' OH terminus. EMBO J. 1982;1(2):269–276. doi: 10.1002/j.1460-2075.1982.tb01158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenni A. L., Joshi S., Chapeville F. tRNA-like structures in the genomes of RNA viruses. Prog Nucleic Acid Res Mol Biol. 1982;27:85–104. doi: 10.1016/s0079-6603(08)60598-x. [DOI] [PubMed] [Google Scholar]
- Hall T. C. Transfer RNA-like structures in viral genomes. Int Rev Cytol. 1979;60:1–26. doi: 10.1016/s0074-7696(08)61257-7. [DOI] [PubMed] [Google Scholar]
- Joshi S., Chapeville F., Haenni A. L. Length requirements for tRNA-specific enzymes and cleavage specificity at the 3' end of turnip yellow mosaic virus RNA. Nucleic Acids Res. 1982 Mar 25;10(6):1947–1962. doi: 10.1093/nar/10.6.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Moras D., Comarmond M. B., Fischer J., Weiss R., Thierry J. C., Ebel J. P., Giegé R. Crystal structure of yeast tRNAAsp. Nature. 1980 Dec 25;288(5792):669–674. doi: 10.1038/288669a0. [DOI] [PubMed] [Google Scholar]
- Peattie D. A., Gilbert W. Chemical probes for higher-order structure in RNA. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A., RajBhandary U. L. Transfer RNA: molecular structure, sequence, and properties. Annu Rev Biochem. 1976;45:805–860. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
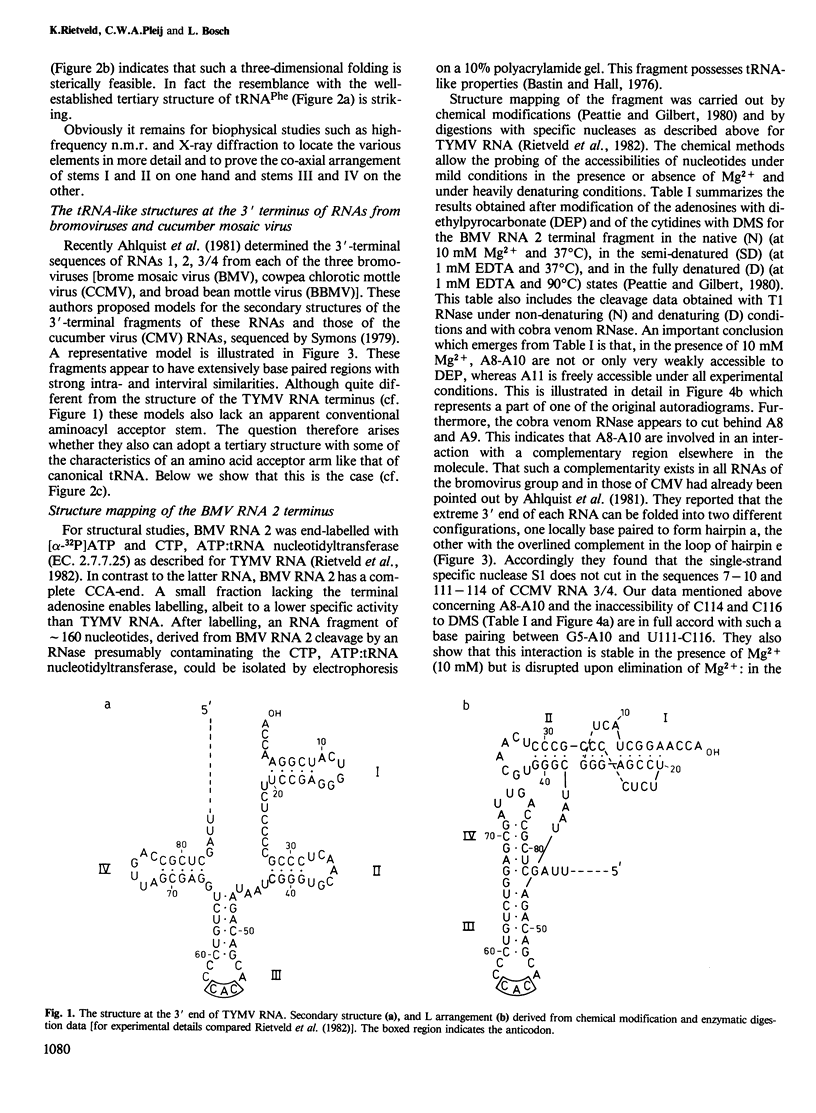
- Rietveld K., Van Poelgeest R., Pleij C. W., Van Boom J. H., Bosch L. The tRNA-like structure at the 3' terminus of turnip yellow mosaic virus RNA. Differences and similarities with canonical tRNA. Nucleic Acids Res. 1982 Mar 25;10(6):1929–1946. doi: 10.1093/nar/10.6.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974 Aug 16;250(467):546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Silberklang M., Prochiantz A., Haenni A. L., Rajbhandary U. L. Studies on the sequence of the 3'-terminal region of turnip-yellow-mosaic-virus RNA. Eur J Biochem. 1977 Feb;72(3):465–478. doi: 10.1111/j.1432-1033.1977.tb11270.x. [DOI] [PubMed] [Google Scholar]
- Symons R. H. Extensive sequence homology at the 3'-termini of the four RNAs of cucumber mosaic virus. Nucleic Acids Res. 1979 Oct 25;7(4):825–837. doi: 10.1093/nar/7.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo N. H., Roe B. A., Rich A. Three-dimensional structure of Escherichia coli initiator tRNAfMet. Nature. 1980 Jul 24;286(5771):346–351. doi: 10.1038/286346a0. [DOI] [PubMed] [Google Scholar]
- Wrede P., Wurst R., Vournakis J., Rich A. Conformational changes of yeast tRNAPhe and E. coli tRNA2Glu as indicated by different nuclease digestion patterns. J Biol Chem. 1979 Oct 10;254(19):9608–9616. [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]