Abstract
To develop biomimetic three-dimensional (3D) tissue constructs for drug screening and biological studies, engineered blood vessels should be integrated into the constructs to mimic the drug administration process in vivo. The development of perfusable vascularized 3D tissue constructs for studying the drug administration process through an engineered endothelial layer remains an area of intensive research. Here, we report the development of a simple 3D vascularized liver tissue model to study drug toxicity through the incorporation of an engineered endothelial layer. Using a sacrificial bioprinting technique, a hollow microchannel was successfully fabricated in the 3D liver tissue construct created with HepG2/C3A cells encapsulated in a gelatin methacryloyl hydrogel. After seeding human umbilical vein endothelial cells (HUVECs) into the microchannel, we obtained a vascularized tissue construct containing a uniformly coated HUVEC layer within the hollow microchannel. The inclusion of the HUVEC layer into the scaffold resulted in delayed permeability of biomolecules into the 3D liver construct. In addition, the vascularized construct containing the HUVEC layer showed an increased viability of the HepG2/C3A cells within the 3D scaffold compared to that of the 3D liver constructs without the HUVEC layer, demonstrating a protective role of the introduced endothelial cell layer. The 3D vascularized liver model presented in this study is anticipated to provide a better and more accurate in vitro liver model system for future drug toxicity testing.
INTRODUCTION
Current drug discovery and development strategies face a high rate of failed clinical trials due to the use of inadequate animal models to anticipate the efficacy and safety of novel drugs in humans.1 Thus, there is an urgent demand to create advanced preclinical models that can supplement the current drug discovery process.2,3 Engineered in vitro organ models have received great attention as promising drug testing platforms in recent years.2,4–7 These three-dimensional (3D) models better mimic the complex human biology compared to two-dimensional (2D) models due to their enhanced cell-cell interactions and improved cellular functions.8–10 Particularly, 3D structures provide an improved tissue model by creating zones of differential proliferation,11 showing phenotypic heterogeneity,12 and delivering a more realistic drug response compared to their 2D counterparts.13 Furthermore, to assess the functionality of tissues, organs-on-a-chip platforms, where 3D organ models are cultured under continuous perfusion of medium, require continual monitoring of multiple physiological and biochemical parameters relevant to the cell responses to drug treatments.14
To create biomimetic native organ models, the development of a vascularized platform is needed for engineered 3D tissue constructs, since embedding vessels into the tissue constructs will allow transport of nutrients, oxygen, and drugs to preserve the metabolic activity of cells. In previous studies, vascular models including an artery model platform,15 in vitro microvessels to study thrombosis and angiogenesis,16 devices that vascularize multi-organ-on-chips,17 microfluidic platforms for microcirculation,18 and vascularized perfusable tissues19 have been developed. In recent years, bioprinting has been used to create a perfusable 3D engineered tissue construct with an endothelialized vasculature. Miller et al. introduced one such technology using 3D printed biocompatible sacrificial filaments.20 In this work, carbohydrate glass filaments were printed into a rat primary hepatocyte-laden 3D hydrogel and then the sacrificial filaments were dissolved to create vasculature. In another paper, Lee et al. also created a bioprinted vessel by using a gelatin precursor and human umbilical vein endothelial cells (HUVECs) in a 3D collagen matrix. By applying a layer-by-layer approach, a channel was created to study angiogenic cell invasion of the hydrogel.21 This layer-by-layer method also allows for the creation of dual cell type-printed tissue to create complex structures.22 Another model by Kolesky et al. created bioprinted 3D thick vascularized tissues that were perfused with growth factors for differentiation of hMSCs towards osteogenic fate.19 Although these methods provide a facile way to fabricate pre-vascularized tissue constructs, carbohydrate glass filaments exhibit a short lifespan, cell-laden gelatin printing has shown a low resolution only for thick constructs (>1 cm) fabrication, and pluronic multilayer systems are complex requiring larger amounts of bioprinting material. Despite the recent advances, efforts are still focused on developing more realistic in vitro vascularized organ platforms that can better mimic the in vivo cell behavior and overall tissue architecture.
In this paper, we created a 3D vascularized liver tissue model for mimicking physiological drug diffusion and toxicity testing. A HepG2/C3A cell-encapsulated gelatin methacryloyl (GelMA) hydrogel was used as a scaffold to engineer the 3D liver tissue construct with a central vessel. To create a smooth and reproducible hollow microchannel in the cell-laden GelMA hydrogel, we draw upon a technique previously developed by our group to create perfusable vascularized structures using a sacrificial agarose fiber in photocrosslinkable hydrogels.23 This bioprinting technique using agarose has several advantages over previously developed methods, such as the possibility to create reproducible microchannels with a wide range of diameters (100–1000 μm), a straightforward removal of bioprinted agarose fiber from the cell-laden hydrogel without additional process of dissolving sacrificial materials, and the creation of smooth surface of hollow microchannel for formation of a monolayer of endothelial cells. By utilizing this technique, we created a hollow capillary, and then by seeding cells, we created a channel covered by a monolayer of endothelial cells where nutrients, oxygen, media, and drugs can be introduced into the tissue constructs through the endothelialized channel. To evaluate the role of the endothelial layer, the vascularized liver tissue construct was loaded into a bioreactor and the diffusion properties were characterized by the perfusion of fluorescence dye as we modified channel diameters and flow rates. Subsequently, the vascularized 3D liver tissue constructs were incubated under continuous perfusion flow and cellular metabolic activity and viability under acetaminophen (APAP) treatment were assessed.
RESULTS AND DISCUSSION
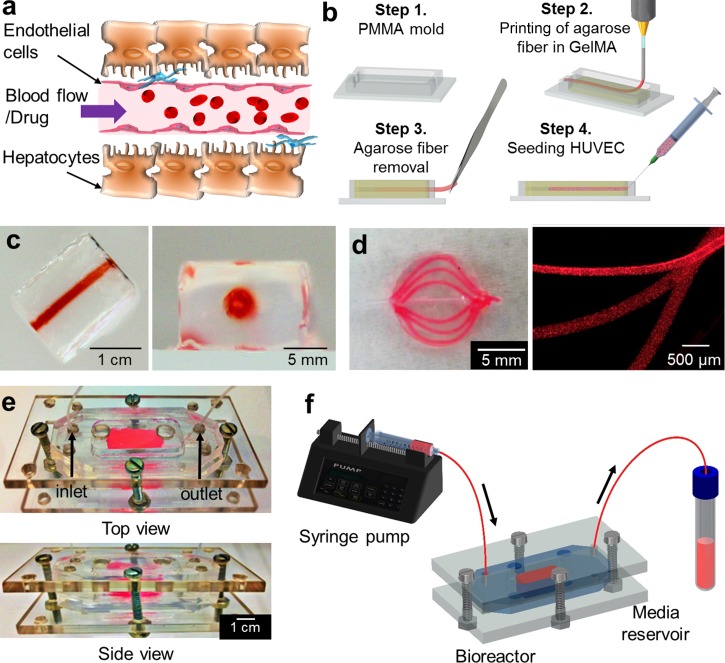
Figure 1(a) shows an illustration of the perfusable vascularized liver construct directly exposed to injected drug via blood vessel. Figure 1(b) shows the fabrication steps of the 3D liver construct with the perfusable vasculature using a sacrificial bioprinting technique.24 HepG2/C3A was selected as a reliable alternative cell source, especially for use in proof-of-concept studies in 3D vascularized tissue model platforms because HepG2/C3A serves as a stable, easy-to-handle, and readily available source of human hepatocytes for in vitro studies compared to human primary hepatocytes which have a limited lifespan, a high batch-to-batch variability, and difficulty in obtaining a large amount of cells.7,25 A HepG2/C3A cell-laden GelMA prepolymer solution was poured into a polymethyl methacrylate (PMMA) mold. An agarose solution was aspirated by a glass capillary connected to a bioprinter and then exposed to a 4 °C chamber for 10 s to make agarose fiber in a glass capillary. The agarose fiber was then directly printed to the center of cell-laden prepolymer solution, and the whole construct was exposed to UV light for 20 s to crosslink the HepG2/C3A cell-laden GelMA prepolymer solution. The obtained hydrogel construct was removed from the PMMA mold, and the agarose fiber was then manually removed to create a hollow microchannel in the cell-laden hydrogel construct. The agarose fiber could be easily removed from the construct due to the weak adhesion between the GelMA hydrogel and the agarose fiber.24 We selected HUVECs to create an endothelial layer and to provide a stable, easy-to-handle, and readily available source of human endothelial cells for in vitro study in combination with HepG2/C3A cells. Subsequently, green fluorescent protein (GFP)-expressing HUVECs (GFP-HUVECs) were seeded inside the hollow channel and incubated for 48 h in HUVEC media to create an endothelial layer on the inner surface of microchannel.
FIG. 1.
(a) Schematic of vascularized liver structure. (b) Fabrication of a 3D engineered liver tissue construct with a perfusable blood vessel structure. (c) Photographic images of the fabricated hydrogel construct with a central hollow microchannel structure and (d) curved microchannels embedded inside a circular GelMA structure. Red dye solution was perfused into the hydrogel construct after the molding and crosslinking process. (e) Photographs of the resealable bioreactor. (f) Illustration of the bioreactor connected to the perfusion module.
Figures 1(c) and 1(d) show the photographic images of the fabricated hydrogel construct having a central hollow microchannel structure and curved microchannels embedded inside a circular GelMA structure. The integrity of the hollow channel structures was assessed by the perfusion of a red dye solution. The construct was placed into a polydimethylsiloxane (PDMS) bioreactor containing HepG2/C3A media [Fig. 1(e)]. The bioreactor was sealed using a set of screws and nuts, providing hydraulic tightness and an easy access to the hydrogel scaffold-containing bioreactor making the construct's analysis rapid and straightforward.7,26–29 The bioreactor was connected to a syringe pump, and inlet/outlet ports were integrated to perfuse the system [Fig. 1(f)]. The tubes were gently connected to the inlet and outlet of the inner hydrogel channel through orifices in the PMMA and inside the chamber. During the operation, the HepG2/C3A hydrogel construct was surrounded by HepG2/C3A static media while HUVEC media flowed through the central channel to mimic the blood flow.
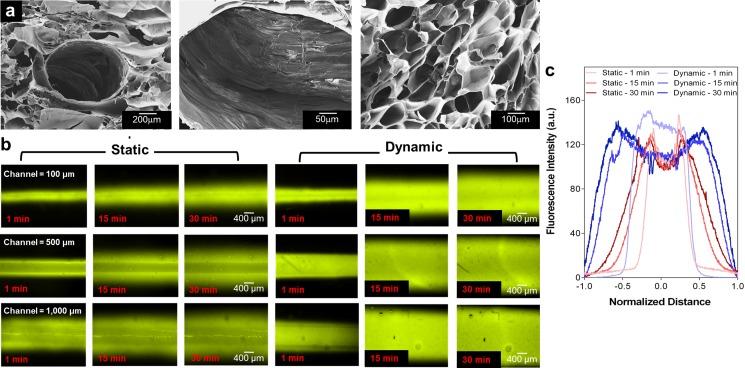
To create a perfusable hollow microchannel within the GelMA hydrogel, it is critical to have a hydrogel scaffold that can handle the weakened mechanical properties due to the main channel. To optimize the concentration of agarose and GelMA pre-polymer solutions, we fabricated agarose fiber encapsulated GelMA hydrogels using a bioprinter with three different concentrations of agarose solutions (4%, 6%, and 8%) and three different concentrations of GelMA pre-polymer solutions (5%, 8%, and 10%). In this case, we selected 6% agarose fiber rather than 4% and 8% agarose fibers due to the reason that 4% was too soft and 8% was too stiff. This made it difficult to completely remove the printed agarose fiber from the GelMA hydrogel. Additionally, to optimize the concentration of GelMA and UV exposure times, we evaluated the HepG2/C3A growth in the GelMA hydrogel as well as HUVECs' attachment and proliferation on the surface of the GelMA hydrogel. In addition, the concentration of the GelMA hydrogel and the UV exposure time were further optimized to create a robust hydrogel construct with integrated channels that has appropriate HepG2/C3A growth with high cell viability in the GelMA hydrogel while supporting the attachment and proliferation of GFP-HUVECs on the surface of the microchannel. Furthermore, this process was optimized to mimic the mechanical property of healthy human liver tissue and resulted in appropriate cell growth compared with the concentrations used before.30 Finally, we chose the 8% GelMA hydrogel with a 20-s UV crosslinking time. Although the Young's Modulus of the 8% GelMA hydrogel (12.1 ± 1.1 kPa) as shown in Fig. S1 (supplementary material) is still higher than that of a healthy human liver (5.5 ± 1.6 kPa),31 this is the minimum concentration of the GelMA hydrogel and UV exposure time that showed appropriate cell growth compared to other conditions that were tried during optimization. Hence, the concentration of the GelMA hydrogel (8%) and agarose solution (6%) was found to be optimum conditions to successfully create a scaffold with microchannels having diameters ranging from 100 μm to 1000 μm. Figure 2(a) shows typical scanning electron microscopy (SEM) images of the porous GelMA hydrogel construct with a microchannel. The average pore size of the GelMA hydrogel was measured to be 143.2 ± 6.4 μm. The porous characteristic of the GelMA hydrogel allows the diffusion and exchange of nutrients, oxygen, and drug molecules from the microchannel into the construct. To evaluate the variations in the diffusion patterns due to the size of the microchannels, we prepared hydrogel constructs having microchannels which are 100, 500, and 1000 μm in diameter. After injecting 40 nM of fluorescent dye (Rhodamine 6G) into the microchannels, we then evaluated the diffusive gradient of the dye under static and dynamic (50 μl/h) conditions at pre-defined time points (1, 15, and 30 min). Fluorescence intensity quantified across the central portion of the channel displayed the profile of the dye gradient and the normalized average distance the dye travelled [Figs. 2(b) and 2(c)]. This microchannel provided a uniform gradient across the hydrogel scaffold that offered a means to study how cell function is regulated by a specific gradient. Under dynamic conditions, the dye was found to diffuse throughout the channel at a faster speed compared with that of static condition.
FIG. 2.
(a) SEM images of a freeze dried porous GelMA hydrogel and central channel structure after agarose fiber removal. (b) Diffusion profiles with different channel diameters (100, 500, and 1000 μm) and time points (1, 15, and 30 min) under static and dynamic conditions. (c) Fluorescence intensity measured in 500 μm channels at several time points (1, 15, and 30 min). The diffusion profile was normalized to the distance from the center of the channel.
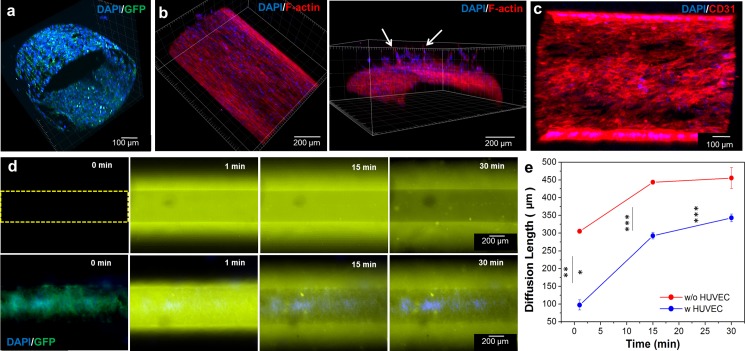
To evaluate the effect of the endothelial barrier layer on the diffusion of molecules, we engineered an endothelial layer by seeding HUVECs inside the hollow microchannel of the hydrogel construct. Channels with different diameters (100, 500, and 1000 μm) were used to observe and evaluate the formation of the endothelial layer along different microchannels. To create a uniform monolayer of cells, the whole construct was manually rotated every 1.5 h after the GFP-HUVECs were injected into the channel. Endothelial cells were found to clog the channels for the 100 μm channels upon cell injection preventing the perfusion flow while for the 1000 μm channels we found that it would take the cells up to 5 days to cover the periphery of the channel surface. As seen in the previous work, the ideal channel diameter for similar cell seeding techniques ranged from 280 μm to 1270 μm for a 7 day culture.32 After 48 h, the endothelial cells proliferated and uniformly covered the microchannel with a 500 μm diameter, as shown in the confocal microscopy images [Figs. 3(a) and 3(b)]. The endothelial cells were aligned along the longitudinal direction. Furthermore, 3D endothelial cell invasion and sprouting into the surrounding GelMA hydrogel was evident as determined by F-actin/nuclei staining [Fig. 3(b), arrow in the right side image]. To access the intercellular junction of HUVECs, we immunostained the construct for the CD31 marker. As shown in Fig. 3(c), immunostaining of the HUVEC layer displayed strong CD31 signals throughout the entire construct.
FIG. 3.
Confocal microscopy images of (a) the integrity of the GFP-HUVEC layer in the construct with nuclei staining after 2 days and (b) top and cross-sectional views of F-actin/nuclei staining after 7 days which showed endothelial cell invasion and sprouting into the surrounding GelMA hydrogel (arrows). (c) Top view of a HUVEC layer immunostained for CD31 (red) and 4′,6-diamidino-2-phenylindole″ (DAPI) (blue) after 2 days in culture. (d) Diffusion profiles of Rhodamine 6G into the microchannel endothelialized without (upper row) and with (lower row) GFP-HUVECs at several time points. The channel's diameter was 500 μm. (e) Diffusion length of Rhodamine 6 G with and without the endothelial cell barrier. Fluorescence intensity was measured at the center of the channel. The barrier delayed the diffusion of the dye (*** p < 0.001).
To evaluate the effect of the endothelial cell layer on the diffusion of the small molecules, 40 nM of Rhodamine 6G in Dulbecco's phosphate-buffered saline (DPBS) was perfused into the microchannel under static conditions and the diffusion profiles were analyzed [Fig. 3(d)]. The average fluorescence intensity was measured in the 500 μm channels after the initial perfusion at different time points (1, 15, and 30 min). Differences in fluorescence intensity were observed inside the channels with and without the HUVEC layer. Since the porosity of the HepG2/C3A-laden GelMA with and without the HUVEC layer was the same throughout the construct, the diffusion coefficient of molecules inside the scaffold remained unaffected. However, the presence of the HUVEC layer was the main player affecting the diffusion of the dye into the hydrogel scaffold, as shown in Fig. 3(e). The presence of the HUVEC layer delayed the diffusion of the dye into the scaffold, thus reducing the fluid flux across the layer.33 This behavior was observed in a previously published study where the permeability of fluorescein isothiocyanate (FITC)-dextran decreased in the presence of a endothelial layer.19
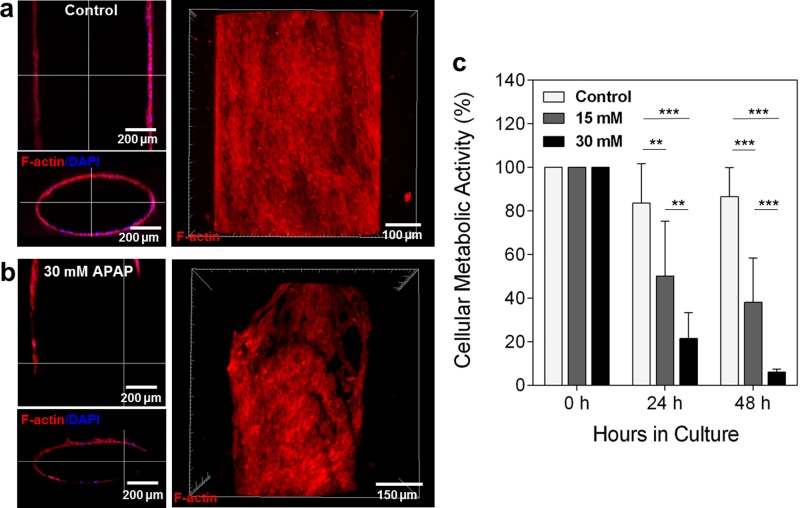
APAP, which is a leading cause of acute liver failure, is known for having direct action on endothelial cells at the liver sinusoid.34,35 To evaluate the direct pharmacological damage to the engineered endothelial barrier, we continuously perfused the scaffold with 30 mM of APAP added into the HUVEC media for 48 h. Inside the bioreactor, the hydrogel scaffold was situated so that it was in direct contact with the bottom and top of the PMMA chamber and the sides of the hydrogel were in contact with the HepG2/C3A media. This design forced the perfused media to go through the microchannel. Figures 4(a) and 4(b) show the confocal microscopy images of the HUVEC layer with and without the APAP treatment, respectively. The discontinuous or non-uniform F-actin/nuclei staining was observed in the cross-sectional and top view of the confocal images of the HUVEC layer after APAP treatment compared to that of the control. To confirm that the HUVEC layer was damaged by the APAP and the destruction of the HUVEC layer was attributed to the APAP treatment instead of the continuous flow along the channel, HUVECs were treated with different doses of APAP (15 and 30 mM) at two time points (24 and 48 h) under static conditions. As shown in Fig. 4(c), the cellular metabolic activity of HUVECs decreased in all the conditions studied, showing that APAP damaged the HUVECs in the endothelial layer regardless of the flow.36 Viability is presumed to be lower at 24 h compared to 0 h as a result of cell contact inhibition by high confluency.
FIG. 4.
Confocal microscopy images of the engineered blood vessel layer after 48 h perfusion (a) without APAP and (b) with 30 mM of APAP. Media only perfusion (control) showed that the HUVEC layer retained its integrity throughout the channel while the scaffold treated with 30 mM APAP displayed a damaged HUVEC layer. (c) Cellular metabolic activity of HUVECs in static drug studies of 15 and 30 mM APAP.
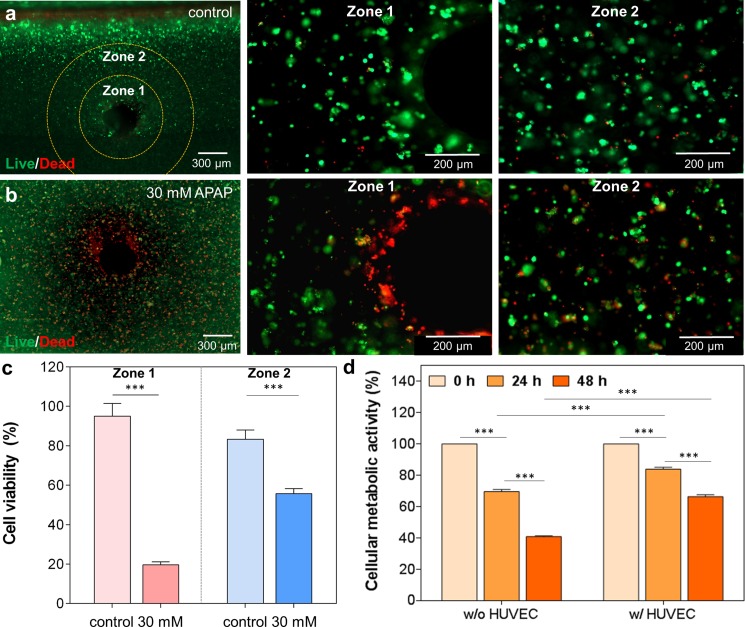
To evaluate the effect of the endothelial layer of vascularized liver tissue construct during the drug treatment, an endothelialized microchannel was incorporated into a 3D liver tissue construct which was created by encapsulating HepG2/C3A cells inside a GelMA hydrogel. 30 mM of APAP added to HUVEC media was perfused continuously into the endothelialized microchannel for 48 h. Two distinctive zones were found in the hydrogel scaffold which was classified according to cell death after 48 h perfusion of 30 mM of APAP into the engineered HUVEC layer (Fig. 5). “Zone 1” was defined as the region closest to the channel, while “Zone 2” was the region further away from the channel and more into the hydrogel. In the absence of any drug treatment, both Zone 1 and Zone 2 exhibited higher cell viability (more than 80%) even for 3D liver tissue constructs that were thick (total thickness: ∼3 mm) [Fig. 5(a)]. The higher cell viability might be attributed to the proper diffusion of nutrients and oxygen into the deeper regions of the 3D liver tissue from perfusion of the channels with HUVEC media. In contrast, the 30 mM APAP treatment resulted in cell death near the channel where the HUVEC layer was located [Fig. 5(b)]. However, the encapsulated HepG2/C3A cells located in the vicinity of the channel within the GelMA hydrogel showed higher viability than HUVECs in the same Zone. This observation indicated that the APAP toxicity was more profound in HUVECs because HUVECs may possess a significant ability to metabolize APAP prior to any evidence of HepG2/C3A cell injury that is consistent with previously reported in vitro and in vivo studies.37,38 After performing quantification analysis, it was found that the cells with no drug treatment (control) in Zone 1 remained viable whereas their viability decreased to 20% after a 48 h exposure to APAP [Fig. 5(c)]. Additionally, in the APAP treated constructs, the cells in Zone 2 were less affected by the drug compared to those in Zone 1. The use of APAP toxicity in this platform gives information on cell disruption and necrosis in the hydrogel similar to what ex vivo human models have shown.39 Additionally, the HUVEC layer is also shown to be disrupted, making this a suitable system for drug testing and assessment of cell death within the hydrogel.
FIG. 5.
Live/dead staining images of cross sectional views of the 3D vascularized liver tissue construct (a) without and (b) with 30 mM APAP treatment under dynamic conditions. Zone 1 was defined as the perfusion territory in close proximity to the channel (∼300 μm) and Zone 2 was defined as the perfusion zone furthest away (∼600 μm) from the channel. (c) Quantification of cell viability after 48 h perfusion of cell culture media (control) and 30 mM APAP treatment. Media only perfusion (control) showed good cell viability in both zones, while 30 mM APAP perfusion exhibited a higher cell death accentuated in Zone 1 compared to Zone 2. (d) Cellular metabolic activity in the 3D liver tissue constructs with and without HUVEC layer after 24 and 48 h 30 mM APAP treatment. HepG2/C3A only constructs showed a lower metabolic activity compared to the HUVEC-HepG2/C3A constructs suggesting a protective action of the HUVECs during drug treatment compared to the other construct.
To assess the functionality of the engineered liver constructs and evaluate the endothelial cell barrier effects on the drug administration, Presto Blue assays were performed. Figure 5(d) indicates that constructs with only HepG2/C3A encapsulated cells showed a lower metabolic activity compared to the vascularized constructs having HUVECs surrounded by HepG2/C3A encapsulated scaffolds suggesting a protective action of the HUVECs during drug treatment compared to the constructs without HUVECs. Consequently, HepG2/C3As exhibited higher resistance to APAP toxicity when co-cultured with the HUVEC layer. We believe that the HUVECs could possibly be (i) delaying the diffusion of drugs as they form a barrier, or (ii) consuming/metabolizing some of the drugs lowering their concentration, or a (iii) combination of (i) and (ii). This protective effect of the endothelial cells when co-cultured with hepatocytes while the cells were exposed to APAP treatments has been previously reported.36,40 The incorporation of vascular components into the in vitro tissue platforms plays a key role in more accurately reproducing the in vivo behavior which is suitable in creating a realistic drug response from in vitro constructs similar to in vivo drug tests. Therefore, the 3D vascularized liver model presented in this study, while still preliminary, is anticipated to provide a better in vitro liver model system for more systematic analyses of drug toxicity. The vascularized liver construct placed inside a bioreactor can be readily used for drug toxicity studies and would possibly better represent in vivo conditions by providing a more physiologically relevant in vitro tool of co-culture systems utilizing human primary hepatocytes and liver endothelial cells. In addition, the study can be conveniently expanded to other organ-on-a-chip platforms for drug screening studies such as the heart, kidney, and bone.
MATERIALS AND METHODS
GelMA synthesis
GelMA was synthesized as described in previous reports.41 Briefly, DPBS (Sigma-Aldrich) was used to dissolve 10% (w/v) gelatin (Sigma-Aldrich) at 50 °C. Then, at a constant rate of 0.5 ml/min, 8% (v/v) of methacrylic anhydride (Sigma-Aldrich) was added to the mixture and left to react for 2 h. After adding warm DPBS (∼40 °C), the solution mixture was dialyzed at 40 °C for 7 days using 12–14 kDa dialysis membranes. The mixture was then placed at −80 °C for 2 days and lyophilized for the following 5 days. GelMA foam obtained was kept at room temperature.
Cell culture
HepG2/C3A cells (ATCC) were cultured at 37 °C with a 5% CO2 atmosphere in low glucose Dulbecco's Modified Eagle Medium (DMEM, Gibco) with 1% penicillin-streptomycin and 10% fetal bovine serum (FBS) (Gibco). Media was substituted every 48 h. GFP-tagged HUVECs (donated by Dr. Yunzhi Peter Yang at Stanford University) were cultured in Endothelial Cell Growth Medium (EGM™ BulletKit™, Lonza) until 70% confluency was reached.
Fabrication of vascularized liver tissue construct in GelMA
The GelMA prepolymer solution was prepared by dissolving 8% (w/v) GelMA in DPBS with 0.5% (w/v) photoinitiator (2-hydroxy-4′-(2-hydroxyethoxy)‐2-methylpropiophenone, Sigma-Aldrich). With a final concentration of 5 × 106 cells/ml, HepG2/C3A cells were trypsinized and dispersed into the GelMA prepolymer solution. 1 ml of the cell-dispersed solution was poured into a 2 × 1 cm2 PMMA mold and then placed below the dispensing tip of the 3D bioprinter (NovoGen MMX bioprinter, Organovo, San Diego, CA, USA). Subsequently, 6% agarose (Sigma-Aldrich) in DPBS was printed as a fiber into the GelMA solution using a microextrusion technique by solidifying the liquid agarose at 4 °C for 10 s. The agarose fiber-embedded GelMA solution was then crosslinked under UV light with a light intensity of 850 mW for 20 s with a working distance of 8.5 cm. After the photocrosslinking process, the agarose fiber was manually removed to create a hollow channel. GFP-HUVECs were seeded inside the channel at 2 × 105 cells/ml cell density.
Bioreactor fabrication
A bioreactor was constructed by modifying our recently published protocols.7,28 Top and bottom layers of the bioreactor were fabricated using a laser cut PMMA mold. A center PDMS chamber was fabricated by casting PDMS around the PMMA mold. To prepare PDMS, the curing agent and elastomer were mixed at a 1:10 ratio and left in an oven at 80 °C for 1 h. After printing the vascularized liver construct, it was placed inside the PDMS chamber and cell culture medium was supplied. A polytetrafluoroethylene (PTFE) tube (#30 AWG thin wall tubing natural, Cole-Palmer, Veron Hills, IL, USA) was attached to the inlet and outlet of the internal HUVEC channel (Fig. S2, supplementary material) and was used to flow media to the reactor employing a syringe pump (Harvard Apparatus PHD 2000, Cambridge, MA, USA). The connections between the tubes and the hydrogel construct were further reinforced by the addition of 20% GelMA hydrogels. HUVEC media was perfused for 48 h at a 50 μl/h flow rate. The HUVEC media flowed through the channel having the endothelial monolayer where the outlet tube was connected to a 50 ml reservoir for media collection. Nuts and screws were used to attach the two PMMA outer layers together with the PDMS chamber to prevent possible leakage during the experiments.
Chemical diffusion along the engineered blood vessel structure
40 nM of Rhodamine 6G (Sigma-Aldrich) was injected with a syringe needle into the microchannels with and without the HUVEC monolayer. Fluorescent images were captured using a fluorescence microscope (Zeiss D1 system, Oberkochen, Germany) at 0, 1, 15, and 30 minutes after the injection of Rhodamine 6G. Fluorescence-based diffusion profiles along a straight line across the entire construct were characterized using ImageJ (NIH). At least 6 region of interests (ROIs) for each time point were measured and normalized to the highest fluorescence value.
APAP treatment
APAP dose was adjusted for the static and dynamic conditions in order to evaluate the drug response of HUVECs. For static conditions, HUVECs were cultured in a 96-well plate at a cell density of 1 × 105 cells/well. After 24 h, cells were treated with APAP (0, 15, and 30 mM) in HUVEC media for 48 h (n = 3 for each condition). Media with APAP was changed every 24 h. Dynamic experiments were performed in separate bioreactors where HUVEC media containing 30 mM APAP was continuously perfused though the channel in the center of each construct for 48 h.
Metabolic activity assay and cell viability
The viability of cells inside the vascularized liver construct was assessed before and after APAP treatment by performing a calcein-AM/ethidium homodimer Live/Dead assay (Invitrogen). Presto Blue Reagent (Life Technologies) was used to assess the cellular metabolic activity. After 2 h of incubation at 37 °C, the reagent was removed and the fluorescence intensity was measured using a microplate reader (BioTek Synergy 2, Vermont, USA). Metabolic activity was measured every 24 h for both static and dynamic APAP conditions. Live/Dead images were taken using an inverted fluorescence microscope (Zeiss Axio Observer D1 Fluorescence Microscope, Carl Zeiss, Germany).
F-actin, CD31, and nuclei staining
Samples were washed with DPBS and fixed with 4% (v/v) paraformaldehyde for 15 min at room temperature for the F-actin and nuclei staining. Then, the washed samples were treated with 0.1% (v/v) Triton X-100 diluted in DPBS for 10 min. Alexa Fluor 488® phalloidin (Thermo Fisher Scientific) was diluted at a 1:50 ratio in DPBS and incubated with the construct for 1 h to stain for F-actin. Hydrogel scaffolds were then washed with DPBS, and a 300 nM DAPI (Thermo Fisher Scientific) solution was added to the sample. The sample with the DAPI solution was incubated for 15 min and washed 3 times with DPBS before imaging. CD31 antibody (Dako) was diluted at a 1:50 ratio in 1% (w/v) bovine serum albumin (BSA) in DPBS for 24 h at 4 °C. After washing with DPBS, the samples were incubated with Alexa Fluor 594-goat anti-mouse secondary antibodies (1:200) for 1 h at room temperature. Confocal images were taken using a Leica-SP5 Confocal Microscope (Leica, Germany) and reconstructed using Fiji42 and Imaris (Bitplane Scientific Software).
Statistical analysis
Data were shown as mean ± standard deviation. Using Graphpad Prism Software, statistical analyses were performed using a Tukey's post-hoc test and two-way analysis of variance (ANOVA). Statistical significance was calculated at 95% (*p < 0.05), 99% (**p < 0.01), and 99.9% (***p < 0.001) confidence intervals.
CONCLUSIONS
In conclusion, we engineered a vascularized 3D liver construct with a vessel layer for drug toxicity testing. By integrating hydrogel scaffolding, 3D bioprinting, and microfluidics, the novel platform could efficiently mimic the vessel layer in vitro. APAP was perfused into the engineered HUVEC layer construct for 48 h to identify the effects of the endothelial layer on the drug administration process. The barrier delayed the passage of the drugs from the channel to the hydrogel scaffold and protected the HepG2/C3A cells from the toxicity caused due to the drug treatment, showing that the engineered endothelial layer plays a crucial role in the drug administration process. By creating advanced in vitro platforms that aim to replicate biological functions and environments, this vascularized platform will help observe and predict drug toxicity mechanisms at a microcirculatory level. The integration of our bioprinted vascularized construct with a bioreactor contributes to the creation of a more realistic “liver-on-a-chip” platform narrowing the gap between in vitro and in vivo drug testing models.
SUPPLEMENTARY MATERIAL
See supplementary material for the mechanical property of GelMA hydrogels and a photograph showing that the construct was connected through the inlet and outlet with tubing.
ACKNOWLEDGMENTS
The authors gratefully acknowledge funding from the Defense Threat Reduction Agency (DTRA) under Space and Naval Warfare Systems Center Pacific (SSC PACIFIC) Contract No. N66001-13-C-2027. The publication of this material does not constitute approval by the government of the findings or conclusions herein. The authors also acknowledge funding from the Institute for Soldier Nanotechnology, National Institutes of Health (HL092836, EB012597, AR057837, and HL099073), and the Office of Naval Research PECASE Award. Solange Massa is thankful for the support received by Universidad de los Andes (Chile) and Programa de Mejoramiento Institucional (PMI). Su Ryon Shin would like to recognize and thank Brigham and Women's Hospital President Betsy Nabel, MD, and the Reny family, for the Stepping Strong Innovator Award through their generous funding. Dr. Seo was partially supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2016R1A6A3A03006491). Andrea Arneri acknowledges the funding from the Italian Ministry of Health. Yu Shrike Zhang acknowledges the National Institutes of Health National Cancer Institute Pathway to Independence Award (K99CA201603).
The authors declare no conflict of interest.
References
- 1. van Midwoud P. M., Verpoorte E., and Groothuis G. M., Integr. Biol. 3, 509–521 (2011). 10.1039/c0ib00119h [DOI] [PubMed] [Google Scholar]
- 2. Bhatia S. N. and Ingber D. E., Nat. Biotechnol. 32, 760–772 (2014). 10.1038/nbt.2989 [DOI] [PubMed] [Google Scholar]
- 3. Ghaemmaghami A. M., Hancock M. J., Harrington H., Kaji H., and Khademhosseini A., Drug Discovery Today 17, 173–181 (2012). 10.1016/j.drudis.2011.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Toh Y.-C., Lim T. C., Tai D., Xiao G., van Noort D., and Yu H., Lab Chip 9, 2026–2035 (2009). 10.1039/b900912d [DOI] [PubMed] [Google Scholar]
- 5. Wagner I., Materne E.-M., Brincker S., Süßbier U., Frädrich C., Busek M., Sonntag F., Sakharov D. A., Trushkin E. V., and Tonevitsky A. G., Lab Chip 13, 3538–3547 (2013). 10.1039/c3lc50234a [DOI] [PubMed] [Google Scholar]
- 6. Sonntag F., Schilling N., Mader K., Gruchow M., Klotzbach U., Lindner G., Horland R., Wagner I., Lauster R., and Howitz S., J. Biotechnol. 148, 70–75 (2010). 10.1016/j.jbiotec.2010.02.001 [DOI] [PubMed] [Google Scholar]
- 7. Bhise N. S., Manoharan V., Massa S., Tamayol A., Ghaderi M., Miscuglio M., Lang Q., Shrike Zhang Y., Shin S. R., Calzone G., Annabi N., Shupe T. D., Bishop C. E., Atala A., Dokmeci M. R., and Khademhosseini A., Biofabrication 8, 014101 (2016). 10.1088/1758-5090/8/1/014101 [DOI] [PubMed] [Google Scholar]
- 8. Teixeira L. M., Leijten J., Sobral J., Jin R., Apeldoorn V. A., Feijen J., Blitterswijk V. C., Dijkstra P., and Karperien H., Eur. Cells Mater. 23, 387–399 (2012). 10.22203/eCM.v023a30 [DOI] [PubMed] [Google Scholar]
- 9. Seo J., Lee J. S., Lee K., Kim D., Yang K., Shin S., Mahata C., Jung H. B., Lee W., and Cho S. W., Adv. Mater. 26, 7043–7050 (2014). 10.1002/adma.201402273 [DOI] [PubMed] [Google Scholar]
- 10. Friedrich J., Seidel C., Ebner R., and Kunz-Schughart L. A., Nat. Protoc. 4, 309–324 (2009). 10.1038/nprot.2008.226 [DOI] [PubMed] [Google Scholar]
- 11. Lin R. Z. and Chang H. Y., Biotechnol. journal. 3, 1172–1184 (2008). 10.1002/biot.200700228 [DOI] [PubMed] [Google Scholar]
- 12. Lawrenson K., Sproul D., Grun B., Notaridou M., Benjamin E., Jacobs I. J., Dafou D., Sims A. H., and Gayther S. A., Carcinogenesis 32, 1540 (2011). 10.1093/carcin/bgr140 [DOI] [PubMed] [Google Scholar]
- 13. Perche F., Patel N. R., and Torchilin V. P., J. Controlled Release 164, 95–102 (2012). 10.1016/j.jconrel.2012.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wikswo J. P., Block F. E. III, Cliffel D. E., Goodwin C. R., Marasco C. C., Markov D. A., McLean D. L., McLean J. A., McKenzie J. R., Reiserer R. S., Samson P. C., Schaffer D. K., Seale K. T., and Sherrod S. D., IEEE Trans. Bio-Med. Eng. 60, 682–690 (2013). 10.1109/TBME.2013.2244891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yasotharan S., Pinto S., Sled J. G., Bolz S.-S., and Günther A., Lab Chip 15, 2660–2669 (2015). 10.1039/C5LC00021A [DOI] [PubMed] [Google Scholar]
- 16. Zheng Y., Chen J., Craven M., Choi N. W., Totorica S., Diaz-Santana A., Kermani P., Hempstead B., Fischbach-Teschl C., and López J. A., Proc. Natl. Acad. Sci. U.S.A. 109, 9342–9347 (2012). 10.1073/pnas.1201240109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schimek K., Busek M., Brincker S., Groth B., Hoffmann S., Lauster R., Lindner G., Lorenz A., Menzel U., and Sonntag F., Lab Chip 13, 3588–3598 (2013). 10.1039/c3lc50217a [DOI] [PubMed] [Google Scholar]
- 18. Sato M., Sasaki N., Ato M., Hirakawa S., Sato K., and Sato K., PloS One 10, e0137301 (2015). 10.1371/journal.pone.0137301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kolesky D. B., Homan K. A., Skylar-Scott M. A., and Lewis J. A., Proc. Natl. Acad. Sci. U.S.A. 113, 3179–3184 (2016). 10.1073/pnas.1521342113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miller J. S., Stevens K. R., Yang M. T., Baker B. M., Nguyen D.-H. T., Cohen D. M., Toro E., Chen A. A., Galie P. A., and Yu X., Nat. Mater. 11, 768–774 (2012). 10.1038/nmat3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee V. K., Kim D. Y., Ngo H., Lee Y., Seo L., Yoo S.-S., Vincent P. A., and Dai G., Biomaterials 35, 8092–8102 (2014). 10.1016/j.biomaterials.2014.05.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee J. S., Hong J. M., Jung J. W., Shim J. H., Oh J. H., and Cho D. W., Biofabrication 6, 024103 (2014). 10.1088/1758-5082/6/2/024103 [DOI] [PubMed] [Google Scholar]
- 23. Bertassoni L. E., Cardoso J. C., Manoharan V., Cristino A. L., Bhise N. S., Araujo W. A., Zorlutuna P., Vrana N. E., Ghaemmaghami A. M., and Dokmeci M. R., Biofabrication 6, 024105 (2014). 10.1088/1758-5082/6/2/024105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bertassoni L. E., Cecconi M., Manoharan V., Nikkhah M., Hjortnaes J., Cristino A. L., Barabaschi G., Demarchi D., Dokmeci M. R., Yang Y., and Khademhosseini A., Lab Chip 14, 2202–2211 (2014). 10.1039/C4LC00030G [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gomez-Lechon M. J., Donato M. T., Castell J. V., and Jover R., Curr. Drug Metab. 4, 292–312 (2003). 10.2174/1389200033489424 [DOI] [PubMed] [Google Scholar]
- 26. Zhang Y. S., Busignani F., Ribas J., Aleman J., Rodrigues T. N., Shaegh S. A. M., Massa S., Rossi C. B., Taurino I., and Shin S.-R., Sci. Rep. 6, 22237 (2016). 10.1038/srep22237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Riahi R., Shaegh S. A. M., Ghaderi M., Zhang Y. S., Shin S. R., Aleman J., Massa S., Kim D., Dokmeci M. R., and Khademhosseini A., Sci. Rep. 6, 24598 (2016). 10.1038/srep24598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y. S., Arneri A., Bersini S., Shin S. R., Zhu K., Goli-Malekabadi Z., Aleman J., Colosi C., Busignani F., Dell'Erba V., Bishop C., Shupe T., Demarchi D., Moretti M., Rasponi M., Dokmeci M. R., Atala A., and Khademhosseini A., Biomaterials 110, 45–59 (2016). 10.1016/j.biomaterials.2016.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shin S. R., Zhang Y. S., Kim D. J., Manbohi A., Avci H., Silvestri A., Aleman J., Hu N., Kilic T., Keung W., Righi M., Assawes P., Alhadrami H. A., Li R. A., Dokmeci M. R., and Khademhosseini A., Anal. Chem. 88, 10019–10027 (2016). 10.1021/acs.analchem.6b02028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shin S. R., Bae H., Cha J. M., Mun J. Y., Chen Y.-C., Tekin H., Shin H., Farshchi S., Dokmeci M. R., Tang X., and Khademhosseini A., ACS Nano 6, 362–372 (2012). 10.1021/nn203711s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roulot D., Czernichow S., Clesiau H. L., Costes J. L., Vergnaud A. C., and Beaugrand M., J. Hepatol. 48, 606–613 (2008). 10.1016/j.jhep.2007.11.020 [DOI] [PubMed] [Google Scholar]
- 32. Zhang W., Zhang Y. S., Bakht S. M., Aleman J., Shin S. R., Yue K., Sica M., Ribas J., Duchamp M., and Ju J., Lab Chip 16, 1579–1586 (2016). 10.1039/C6LC00001K [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brogioli D. and Vailati A., Phys. Rev. E 63, 012105 (2000). 10.1103/PhysRevE.63.012105 [DOI] [PubMed] [Google Scholar]
- 34. Deleve L. D., Wang X., Kaplowitz N., Shulman H. M., Bart J. A., and Van Der Hoek A., Biochem. Pharmacol. 53, 1339–1345 (1997). 10.1016/S0006-2952(97)00048-8 [DOI] [PubMed] [Google Scholar]
- 35. Jollow D., Thorgeirsson S., Potter W., Hashimoto M., and Mitchell J., Pharmacology 12, 251–271 (1974). 10.1159/000136547 [DOI] [PubMed] [Google Scholar]
- 36. Nelson L. J., Navarro M., Treskes P., Samuel K., Tura-Ceide O., Morley S. D., Hayes P. C., and Plevris J. N., Sci. Rep. 5, 17455 (2015). 10.1038/srep17455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Walker R. M., Racz W. J., and McElligott T. F., Hepatology 5, 233–240 (1985). 10.1002/hep.1840050213 [DOI] [PubMed] [Google Scholar]
- 38. Ito Y., Bethea N. W., Abril E. R., and McCuskey R. S., Microcirculation 10, 391–400 (2003). 10.1038/sj.mn.7800204 [DOI] [PubMed] [Google Scholar]
- 39. Schreiter T., Sowa J. P., Schlattjan M., Treckmann J., Paul A., Strucksberg K. H., Baba H. A., Odenthal M., Gieseler R. K., Gerken G., Arteel G. E., and Canbay A., Sci. Rep. 6, 31916 (2016). 10.1038/srep31916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Navarro M., Nelson L., Treskes P., Hayes P., Samuel K., and Plevris J., J. Hepatol. 60, S179 (2014). 10.1016/S0168-8278(14)60499-2 [DOI] [Google Scholar]
- 41. Akbari M., Tamayol A., Laforte V., Annabi N., Najafabadi A. H., Khademhosseini A., and Juncker D., Adv. Funct. Mater. 24, 4060–4067 (2014). 10.1002/adfm.201303655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., and Schmid B., Nat. Methods 9, 676–682 (2012). 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for the mechanical property of GelMA hydrogels and a photograph showing that the construct was connected through the inlet and outlet with tubing.