Summary
Drought represents a key limiting factor of global crop distribution. Receptor‐like kinases play major roles in plant development and defence responses against stresses such as drought. In this study, LRK2, which encodes a leucine‐rich receptor‐like kinase, was cloned and characterized and found to be localized on the plasma membrane in rice. Promoter–GUS analysis revealed strong expression in tiller buds, roots, nodes and anthers. Transgenic plants overexpressing LRK2 exhibited enhanced tolerance to drought stress due to an increased number of lateral roots compared with the wild type at the vegetative stage. Moreover, ectopic expression of LRK2 seedlings resulted in increased tiller development. Yeast two‐hybrid screening and bimolecular fluorescence complementation (BiFC) indicated a possible interaction between LRK2 and elongation factor 1 alpha (OsEF1A) in vitro. These results suggest that LRK2 functions as a positive regulator of the drought stress response and tiller development via increased branch development in rice. These findings will aid our understanding of branch regulation in other grasses and support improvements in rice genetics.
Keywords: rice, LRK2, drought stress, tiller
Introduction
Rice (Oryza sativa) is one of the most important crops, feeding more than one‐third of the world's population. This major staple requires large amounts of water during growth and is therefore susceptible to drought stress. At each stage of growth, drought hinders crop development and decreases yield. Molecular genetic tools aimed at improving rice drought tolerance and maintaining production, while expanding development into regions with limited water resources is therefore important (Fernie et al., 2006; Pennisi, 2008).
Leucine‐rich repeat receptor‐like kinase (LRK), which belongs to the largest subfamily of kinases in plants, contains a leucine‐rich extracellular domain, a transmembrane domain and a C‐terminal intracellular kinase domain (Shiu et al., 2004; Walker, 1993). At least 223 LRKs are known in Arabidopsis and more than 300 in rice (Shiu et al., 2001). LRKs function in a number of developmental processes and defence responses such as meristem maintenance (Clark et al., 1997), cellular proliferation (Matsubayashi et al., 2002), brassinosteroid signalling (Li and Chory, 1997), floral organ abscission (Jinn et al., 2000; Taylor et al., 2016) and defence bacterial flagellin (Zipfel et al., 2006). However, known functions in rice remain limited to, for example, ERECTA (Shen et al., 2015), Xa21 (Jiang et al., 2013; Song et al., 1997), OsSIK1 (Ouyang et al., 2010) and FON1 (Feng et al., 2014), with the majority of genes yet to be elucidated.
Eukaryotic elongation factor (eEF) proteins can be divided into eEF1 and eEF2. eEF1 proteins, which can further be divided into eEF1A, eEF1Bα, eEF1Bβ and eEF1Bγ subunits, are highly conserved in a number of species (Browning, 1996). eEF1‐mediated ammonia acyl‐tRNA has also been shown to bind to ribosomes (Riis et al., 1990). Moreover, recent studies have shown that eEF1A is not only important for translation, but is also an important multifunctional protein (Ejiri, 2002; Sasikumar et al., 2012), playing a role in processes such as cell proliferation (Pecorari et al., 2009; Sanders et al., 1992), cell apoptosis (Byun et al., 2009; Shepherd et al., 1989; Zhang et al., 2015), cell morphogenesis (Gross and Kinzy, 2005) and signal transduction (Numata et al., 2000). However, most of these studies have focused on animals and humans, with few findings in plants. In Arabidopsis, eEF1B is associated with cell wall biosynthesis and plant development (Hossain et al., 2012). Moreover, the AtEF2 gene has been shown to be involved in low temperature signalling (Guo et al., 2002). Since 1998, four EF1A genes have also been cloned in rice, but their functions have yet to be reported (Kidou and Ejiri, 1998).
Previously, an eight leucine‐rich LRK gene (LRK1‐LRK8) cluster, which has been shown to increase grain yield, was cloned from the rice quantitative trait locus (QTL) qGY2‐1 (Li et al., 2002a). Haplotype divergence of the LRK locus was subsequently found to be associated with the origin and differentiation of cultivated rice. Moreover, LRK2 was found to be highly expressed in Oryza sativa L. ssp. indica var. 9311, but was undetectable by RT‐PCR in Oryza sativa L. ssp. japonica cv. Nipponbare (He et al., 2006). In the present study, we cloned and characterized LRK2 and revealed an increase in branch number and drought tolerance in LRK2‐overexpressing transgenic lines. pLRK2::GUS was largely expressed in tiller buds, nodes, roots and anthers. Furthermore, yeast two‐hybrid screening and bimolecular fluorescence complementation (BiFC) analysis suggested an interaction between LRK2 and OsEF1A. The molecular mechanisms underlying the response of rice to drought was also discussed with the aim of improving crop growth under potentially adverse conditions.
Results
LRK2 encodes a leucine‐rich receptor‐like kinase
LRK2 contains extracellular LRR motifs, a transmembrane domain (TM) and a cytoplasmic kinase domain (Figure 1a). To investigate the relationships between LRK2 (GenBank accession no. AY756174.4 GI:54306232) and LRK members from other plant species, phylogenetic analysis of LRKs from rice and Arabidopsis thaliana was performed using Clustalx1.83 and MEGA 6. Analysis involved 1000 bootstrap replicates with the following sequences: OsCLVATA: EAY84170; AtCLVATA: AAB58929; AtHAESA: XP_002869498; AtERECTA: XP_002880777; AtFLS2: AAO41929.1; AtEFR: AAL77697.1; OsX21: NC_008395.2; LRK2: AY756174.4; AtBRI1: XP_002866847.1; OsBRI1: AAK52544.1; AtBAK1: NP_567920.1; OsBAK1: EEC82980.1; and AtPSKR1:At2g02220. Based on a comparison of homologous amino acid sequences, LRK2 was found to share a close genetic relationship with phytosulfokine receptor 1 (PSKR1) from A. thaliana (Hartmann et al., 2014; Matsubayashi et al., 2002, 2006; Figure 1b). In addition, the function of LRK2 as a novel leucine‐rich repeat receptor‐like kinase (LRR‐RLK) was reported for the first time.
Figure 1.
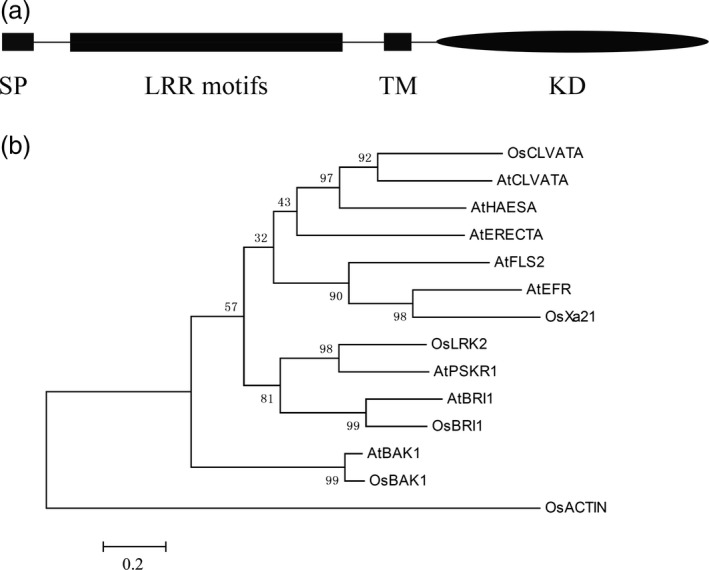
LRK2 encodes a leucine‐rich receptor‐like kinase. (a) Schematic diagram of the LRK2 protein. SP, signal peptide; LRR motif, leucine‐rich repeat region; TM, transmembrane domain; KD, intercellular kinase domain. (b) Phylogenetic analysis of deduced amino acid sequences of LRK2 compared with other homologous sequences.
Features of LRK2 in rice
To confirm the subcellular localization of LRK2, LRK2 with an enhanced GFP was constructed under control of the cauliflower mosaic virus 35S promoter (35S::LRK2:eGFP), then infiltrated into tobacco (Nicotiana benthamiana) leaves by Agrobacterium‐mediated transient transformation. The resulting construct, 35S::LRK2:eGFP, exhibited eGFP expression in the plasma membrane compared with the control, 35S::eGFP, which showed expression throughout the cell (Figure 2A). In addition, to determine expression patterns, the 2‐kb promoter region of the LRK2 gene was cloned into the expression vector pBIN121 with the β‐glucuronidase (GUS) reporter gene (Figure S1A). The construct pLRK2:GUS was subsequently transformed into rice (Figure S1B). A GUS staining assay of T2 transgenic lines revealed expression in the tiller buds, nodes, roots and anthers (Figure 2B).
Figure 2.
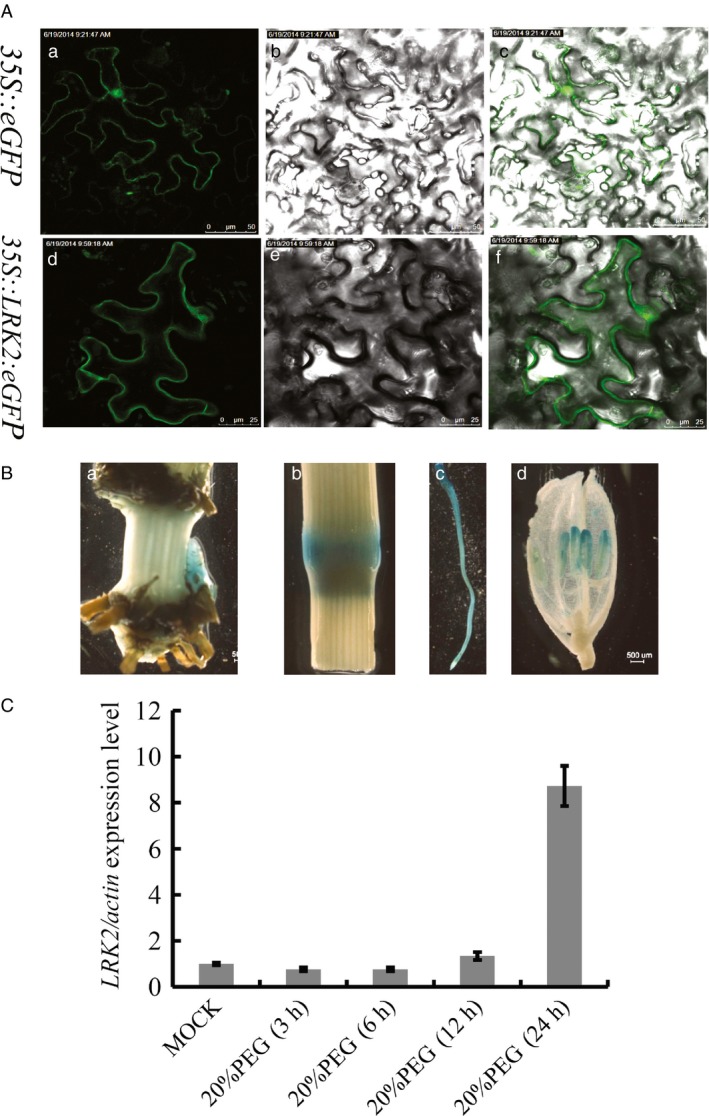
Expression patterns and subcellular localization of LRK2 in rice. (A) Subcellular localization of LRK2. a,d: under the laser (430 nm); b,e: bright field; c,f: merged image; a,b,c: sections for 35S::eGFP; d,e,f: sections for LRK2 construction. Bars = 25 μm. (B) GUS staining of different organs in the overexpressing line: a, tiller bud; b, node; c, root; d, anther. (C) Expression analysis of LRK2 in Nipponbare rice seedlings under drought treatment using real‐time PCR. Each column represents an average of three replicates and bars indicate the SD.
To examine whether LRK2 expression was regulated by drought stress, 7‐day‐old rice seedlings were subjected to 20% PEG6000 then harvested for RNA extraction at different time points and the transcripts of whole plants were quantified by real‐time PCR. As a result, transcript levels were found to be rapidly and strongly induced by drought (Figure 2C).
Expression of LRK2 in transgenic rice lines
To obtain further insight into LRK2, 2X35S::LRK2 and 2X35S::antiLRK2 (Figure 3a), plasmids containing the entire LRK2 gene were introduced into Nipponbare, and transformants selected on medium containing hygromycin. The transgenic plants were simultaneously examined by PCR using genomic DNA as a template with specific primers. Eleven and eight independent transformants (T0) were regenerated from hygromycin‐resistant calli of the 2X35S::LRK2 and 2X35S::antiLRK2 lines, respectively (Figure S2). Four T2 transgenic lines plus the wild type were subsequently selected for expression analysis (M2 and M6 to represent 2X35S::LRK2, and AM5 and AM8 for 2X35S::antiLRK2). Expression levels during the three‐leaf stage were determined by semiquantitative and RT‐PCR. LRK2 was strongly expressed in M2 and M6, but reduced in AM5 and AM8 (Figure 3b,c), suggesting that the 2X35S::LRK2 and 2X35S::antiLRK2 plasmids were genetically transmitted to the next generation.
Figure 3.
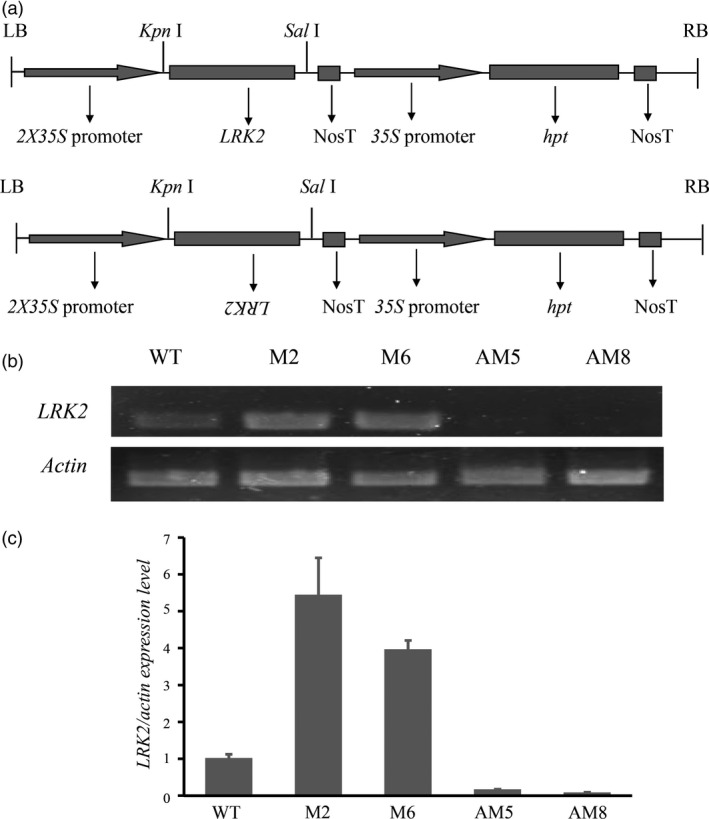
Expression of LRK2 in transgenic rice. (a) Schematic diagram of the plant expression vectors pCAMBIA1300‐2X35S::LRK2 and pCAMBIA1300‐2X35S::antiLRK2 used for LRK2 overexpression and decreased expression in transgenic plants. (b, c) Expression analysis of LRK2 in transgenic plants using semiquantitative PCR and real‐time PCR with gene‐specific primers.
Overexpression of LRK2 increases drought tolerance in rice
LRKs reportedly regulate a number of stress responses, including drought, salinity and low temperature (Ouyang et al., 2010; Shen et al., 2015; Yang et al., 2014). The performance of LRK2‐overexpressing, LRK2‐antisense and wild‐type plants under drought stress conditions was therefore examined (Figure 4). Under normal conditions, all plants grew well. After 12 days, plants were cultured in 20% PEG6000 solution and then 5 days later, phenotypic changes in wild‐type and transgenic plants were observed. Leaf rolling and wilting was delayed in the M2 and M6 transgenic lines compared with the wild type, while in AM5 and AM8 symptoms of drought stress were severe. After 8‐day treatment, the M2 and M6 lines showed increased tolerance to drought stress compared with the wild type, while AM5 and AM8 remained sensitive.
Figure 4.

Performance of LRK2 transgenic plants under drought stress. 0 day: Wild‐type, M2 and M6 seedlings grown for 12 days under normal conditions. 5, 8 days: Performance of wild‐type, M2 and M6 seedlings treated with 20% PEG6000 for 5 and 8 days, respectively. 1, 3, 11 days: Performance of wild‐type, AM5 and AM8 seedlings grown under drought stress for 1, 3 and 11 days, respectively. Recovery 20 days: Recovery of the treated wild‐type, M2 and M6 seedlings for 20 days. Recovery 8 days: Recovery of the treated wild‐type, AM5 and AM8 seedlings for 8 days.
Following drought treatment, the plants were watered to induce recovery, and then, the growth status was examined. M2 and M6 recovered and grew more vigorously than the wild type, with survival rates of 73.5% and 65.5%, respectively, compared with 49.2%. In contrast, only 12.2% of the AM5 and 10.2% of the AM8 plants recovered, compared with 28.6% of the wild type. All values were significantly different (P < 0.05, t‐test; Figure 5A), suggesting that LRK2 positively regulates the drought stress response in rice.
Figure 5.
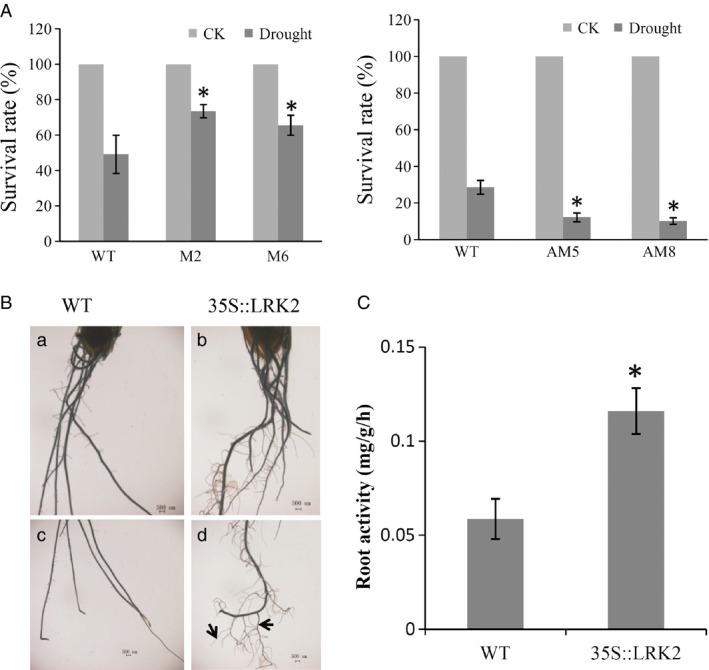
Performance of LRK2 transgenic plants under drought stress. (A) Survival rates of the wild‐type and transgenic plants after recovery. (B) (a–d) Root characters of the plants under drought stress. Arrows indicate the large and small lateral roots. Scale bars: 500 μm. (C) Root activity of the wild‐type and LRK2‐overexpressing lines (LRK2). *, P < 0.05, t‐test.
Large root systems are known to be more conducive to extraction of water from deep soil layers compared with small root systems. The root structures of transgenic and wild‐type plants after drought treatment were therefore examined under a stereoscopic microscope. The transgenic plants had significantly more lateral roots and a larger overall root system than the wild type (Figure 5B). Root activity in rice at different stages directly affects plant growth. In this study, root activity appeared higher in LRK2‐overexpressing lines compared with the wild type (Figure 5C). These results suggest that increased root activity in the LRK2‐overexpressing lines may be one reason for the increase in lateral root development.
To analyse the expression of genes involved in drought tolerance, expression levels of 9‐cis‐epoxycarotenoid dioxygenase 1 (NCED1), NCED2, plasma membrane intrinsic protein 2;3 (PIP2;3) and ABA‐responsive element binding factor (ABF) were detected using real‐time PCR in wild‐type and transgenic seedlings under drought (20% PEG6000) for 12 h (Hatmi et al., 2015; Redillas et al., 2012; Shi et al., 2015; Wu et al., 2015; Yoshida et al., 2010). The data showed that PIP2;3 was strongly expressed in LRK2 overexpression lines (Figure S3).
Effect of LRK2 on rice tiller development
To investigate the effect of LRK2 on yield traits, 14 transgenic lines were analysed (M1 to M6, AM1 to AM8; 20 individual plants per line). After cultivating transgenic lines and Nipponbare wild‐type plants under identical conditions, the following yield components were examined: numbers of tillers per plant, grains on the main panicle, grains per panicle and grains per plant (Table 1). The tiller is a specialized grain‐bearing branch that forms on unelongated basal internodes. At the tillering stage, the LRK2‐overexpressing lines exhibited increased tiller development (Figure S4A). At maturity, M2 and M6 produced 36% and 32% more panicles than the wild type, respectively, while AM5 and AM8 showed a decrease in panicles of 29.7% and 44.9% compared with the wild type, respectively (Table 1, Figures 6, S4B). The number of grains on the main panicle increased in overexpressing lines compared with the wild type (M2: 8.57%; M6: 5.46%), while the number of grains per panicle showed a slight decrease (M2: 10.13%; M6: 18.79%). In contrast, the number of grains per plant increased in the overexpressing lines (M2: 29.9%; M6; 25.3%) compared with the wild type (Table 1, Figure S4C). The number of grains per panicle is affected by the number of primary, secondary and sometimes higher‐order panicle branches. The number of primary and secondary panicle branches was therefore counted. The overexpressing lines showed a slight increase in the number of secondary branches on the main panicle compared with the wild type (M2: 7.86%; M6: 6.46%); however, the number of primary branches did not significantly differ (Table 2, Figure S4C). These results suggest that LRK2 improves rice tiller number and the number of grains per plant.
Table 1.
Yield components of wild‐type (WT) and transgenic plants M2, M6, AM5 and AM8
| Plant line | Number of tillers per plant | Number of grains on the main panicle | Number of grains per panicle | Number of grains per plant |
|---|---|---|---|---|
| WT | 16.38 ± 3.57 | 85.91 ± 4.9 | 76.89 ± 6.37 | 1036.63 ± 115.01 |
| M2 | 22.28 ± 4.79 | 93.27 ± 5.4 | 69.10 ± 4.58 | 1347.5 ± 156.81 |
| M6 | 21.34 ± 5.73 | 90.6 ± 5.64 | 62.44 ± 8.26 | 1298.53 ± 104.28 |
| AM5 | 11.5 ± 2.56 | 77.56 ± 3.3 | 52.56 ± 8.3 | 752.23 ± 82.02 |
| AM8 | 9.02 ± 1.41 | 75.13 ± 4.5 | 50.13 ± 7.5 | 612.58 ± 98.3 |
| P value | P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 |
Data were obtained from random samples at maturity and represent the mean ± SD of 20 individuals.
Figure 6.
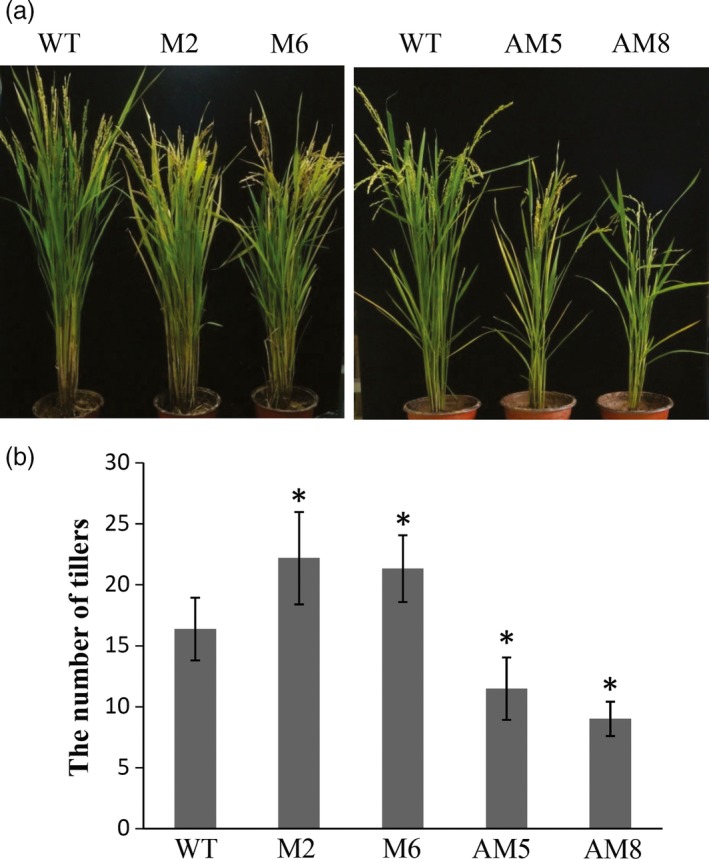
Effects of LRK2 in transgenic plants. (a) Comparison of wild‐type and transgenic lines at the flowering stage. (b) Number of panicles per plant in the wild‐type and transgenic plants. *, P < 0.05, t‐test.
Table 2.
Number of branches on the main panicle of wild‐type (WT) and transgenic lines M2 and M6
| Plant lines | Primary branches | Secondary branches |
|---|---|---|
| WT | 7.45 ± 0.82 | 15.64 ± 3.44 |
| M2 | 7.93 ± 1.1 | 16.87 ± 2.33a |
| M6 | 7.54 ± 0.9 | 16.65 ± 2.28a |
Data were obtained from random samples at maturity and represent the mean ±SD of 20 individuals.
P < 0.05, t‐test.
Interaction between LRK2 and eukaryotic elongation factor 1A
To identify interactors of LRK2, LRK2 kinase domain (LRK2D) was used as bait in yeast two‐hybrid analysis. As a result, six potential interacting molecules were identified. Nucleotide sequence analysis further revealed one cDNA fragment encoding the eukaryotic elongation factor OsEF1A (GenBank Accession no. GQ848073.1), which encodes a protein involved in protein synthesis and cell proliferation. To confirm the direct interaction between LRK2 and OsEF1A, a yeast two‐hybrid assay was performed via cotransformation of the pGBKT7‐LRK2D bait construct and full‐length pGADT7‐OsEF1A. As expected, only yeast cells harbouring both LRK2D and OsEF1A grew vigorously on both SD/Leu‐Trp‐ and SD/Leu‐Trp‐His‐/50 mm 3‐amino‐1,2,4‐triazole (3‐AT) plus X‐gal media. In contrast, the wild type grew well on SD/Leu‐Trp‐ but not SD/Leu‐Trp‐His‐/50 mm 3‐AT/ X‐gal medium (Figure 7A). pGBKT7‐LRK2D and a truncated version of OsEF1A were also cotransformed with pGADT7, revealing that the C‐terminal region alone is sufficient, and indeed necessary, for binding with the LRK2 kinase domain (Figure 7B). To further verify the interaction between LRK2D and OsEF1A, BiFC analysis of Nicotiana benthamiana leaves was performed (Figure 7C). Under 513 nm illumination, cells harbouring LRK2D:YEP CE and OsEF1A:YEP NE emitted yellow fluorescence, confirming the interaction between OsEF1A and LRK2.
Figure 7.
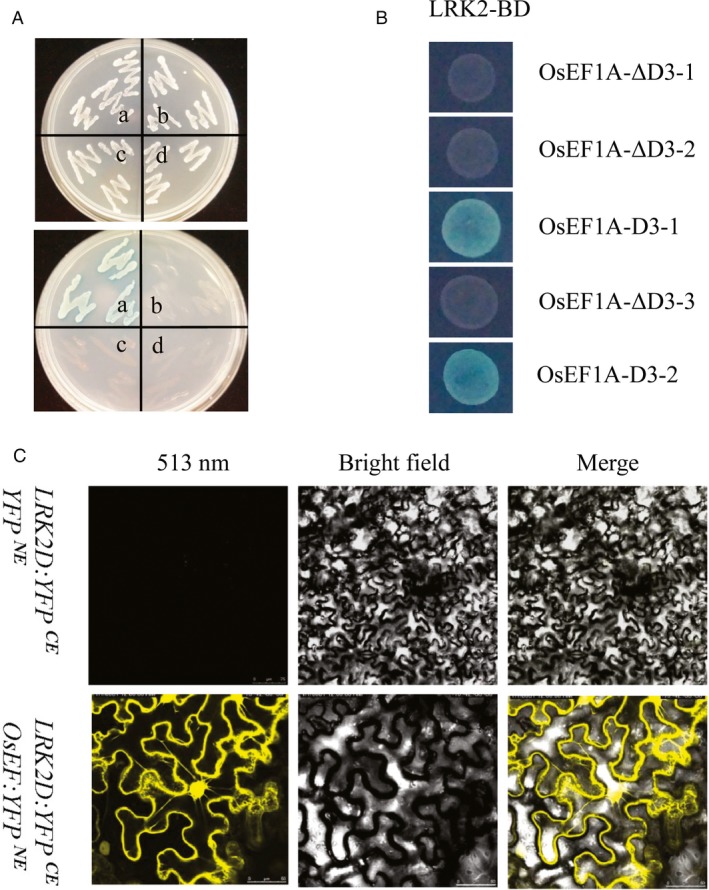
Interaction between LRK2D and OsEF1A in vitro. (A) Interaction between OsEF1A and the LRK2 kinase domain as bait in a yeast two‐hybrid system. Photograph shows the growth behaviour of transformants on SD/Leu‐Trp medium (upper) and SD/Leu‐Trp‐His‐/50 mM3‐AT medium (lower). The yeast cells harboured various pairs of plasmids: a. pGBKT7‐LRK2D with pGADT7‐OsEF1A; b. pGBKT7 with pGADT7‐OsEF1A; c. pGBKT7‐LRK2D with pGADT7; d. pGBKT7 with pGADT7. (B) The carboxyl domain of OsEF1A was found sufficient, and necessary, for interaction with the LRK2 kinase domain in yeast. Residues of OsEF1A present in the constructs were 1 to 230aa (OsEF1A‐ΔD3‐1), 231 to 320aa (OsEF1A‐ΔD3‐2), 321 to 447aa (OsEF1A‐D3‐1), 1 to 320aa (OsEF1A‐ΔD3‐3), 231 to 447aa (OsEF1A‐D3‐2). (C) Interaction between LRK2D and OsEF1A as examined by BiFC. The indicated constructs were transformed into leaves of Nicotiana benthamiana. Scale bars in upper images: 20 mm. Scale bars in lower images: 50 mm.
Discussion
It is predicted that global climate change will increase temperatures, alter geographical patterns of rainfall and increase the frequency of extreme climatic events (Harrison et al., 2014). Drought is a major constraint of crop development and production worldwide. Accordingly, a number of studies have suggested that overexpression of stress‐related genes may help improve drought tolerance in cereal crops (Cheng et al., 2016; Uga et al., 2013). In this study, a new rice leucine‐rich repeat receptor‐like kinase, LRK2, was found to have the ability to increase drought stress by promoting root growth and significantly increasing tiller number, while reducing plant height (Figures 6 and S4; Table S1). The data indicated that the LRK2 gene encodes a protein localized to the plasma membrane, and expressed in tiller buds, nodes, roots and anthers (Figure 2). These results suggest that the LRK2 gene is essential for stable and adequate crop production in drought‐prone areas.
The mature rice fibrous root system is composed of adventitious and lateral roots. Adventitious root branching results in both large and small lateral roots (Coudert et al., 2010), which are essential for water and nutrient uptake and critical for increased yield under stress (Atkinson et al., 2014; Coudert et al., 2010). In rice, DRO1 was previously found to be negatively regulated by auxin and involved in cell elongation, resulting in an enlarged root system and increased drought avoidance (Uga et al., 2013). Moreover, rice OsAHP1 and OsAHP2 knockdown plants were previously found to exhibit phenotypes representative of a deficiency in cytokinin signalling, including enhanced lateral root growth and resistant to osmotic stress compared with wild‐type plants (Sun et al., 2014). Moreover, ectopic OCI expression was found to increase the lateral root density and drought tolerance in Arabidopsis and soya bean (Quain et al., 2014). In the present study, LRK2 overexpression resulted in an improved root system with an increased number of both large and small lateral roots compared with the wild type. This is one reason for the increased drought tolerance in LRK2‐overexpressing lines. The 2,3,5‐triphenyltetrazolium chloride (TTC) test was used here to determine root activity in the LRK2‐overexpressing plants (Hu et al., 2016; Steponkus and Lanphear, 1967). Roots of the transgenic plants were significantly more active than those of the wild‐type plants (Figure 5C), further contributing to increased drought resistance in the LRK2‐expressing lines. PIP2;3, one of the aquaporins, plays a crucial role in response to drought stress (Yu et al., 2006). SIRK1, a member of the LRK family in Arabidopsis, was shown to interact with and activate PIP2;3 by phosphorylation (Wu et al., 2013). Experiments should be undertaken to evaluate whether LRK2 can specifically phosphorylate PIP2;3.
Plant architecture, such as the structure of the roots, shoots and inflorescences, is affected by branching. Shoot branches in rice are referred to as tillers (Tanaka et al., 2015). Root branches are located underground, while tiller and inflorescence structures in rice undergo lateral branching above ground during the vegetative and reproductive stages, respectively. Tiller number is generally regarded as the determining factor of yield as tillers are specialized panicle‐bearing branches (Grillo et al., 2009). Several genes related to tiller development have been characterized; for example, MOC1 is important in both tiller bud formation and outgrowth, while OsTB1 negatively regulates axillary bud outgrowth (Li et al., 2003; Takeda et al., 2003). Moreover, both MOC1 and OsTB1 expression was found in the axillary meristem and tiller buds. MOC1 was also detected at the leaf axils. The LRK2 gene studied here was expressed in tiller buds and nodes, suggesting a role in tiller formation and outgrowth. In line with this, LRK1 was previously found to increase both tiller and grain number (Zha et al., 2009). In the current study, overexpression of LRK2 resulted in an increase in tiller number, but no significant change in the number of grains per panicle. Similarly, both LRK1‐ and LRK2‐overexpressing plants exhibited an increase in tiller number. These results suggest that LRK2 and LRK1 are expressed in different tissues, despite belonging to the same gene cluster. However, it is also possible that the diverse phenotypes of the transgenic lines were partly due to the different genetic backgrounds of the rice cultivars used for transformation (i.e. 9311 and Nipponbare).
Plants have evolved a number of morphophysiological and biochemical strategies at both the cellular and molecular levels to allow them to adapt to biotic and abiotic stresses. When plants encounter abiotic stresses, membrane‐localized receptors rapidly sense environmental signals and transmit them downstream, thereby activating stress‐related responses. During these processes, LRKs play important roles as both sensors and transducers (Lease et al., 1998; Shiu et al., 2004; Torii, 2004). Meanwhile, RPK1 (receptor‐like protein kinase 1) is required for embryonic pattern formation and enhances both water and oxidative stress tolerance in Arabidopsis (Mandel et al., 2014; Masle et al., 2005; Meng et al., 2012; Shen et al., 2015; Shpak et al., 2004; Van Zanten et al., 2009). Similarly, BAK1 (BRI1‐associated receptor kinase 1) was found to play a role in a number of diverse processes, including brassinosteroid signalling, the phytosulfokine (PSK) signal pathway, light responses, cell death and plant innate immunity (Chinchilla et al., 2007, 2009; Ingram, 2007; Ladwig et al., 2015; Li et al., 2002b; Nam and Li, 2002; Sun et al., 2013). In this study, the novel LRK gene LRK2 was found to function in drought tolerance and tiller development. Identification and further elucidation of the molecular mechanisms of such genes would be valuable in rice production management and genetic improvement studies.
EF1A was first shown to function in protein synthesis, and since then has been implicated in a number of biochemical processes such as interactions with the cytoskeleton (Gross et al., 2005), apoptosis (Byun et al., 2009; Zhang et al., 2015) and cell proliferation (Sanders et al., 1992). EF1A also interacts with phospho‐Akt in breast cancer cells and regulates their proliferation, survival and motility (Pecorari et al., 2009), and is expressed ubiquitously in humans. Although a highly abundant cellular protein associated with the cytoskeleton, the function of EF1A in rice remains unknown. Phylogenetic analysis revealed a close genetic relationship between LRK2 and PSKR1 from A. thaliana. PSKR1 is a PSK receptor that stimulates plant growth and differentiation (Igarashi et al., 2012). In this study, the LRK2 intracellular domain was also found to directly interact with OsEF1A in double molecule fluorescence analysis and a yeast two‐hybrid assay. The findings further suggest that LRK2 interacts with OsEF1A to regulate plant developmental processes such as cell proliferation, thereby increasing plant branching.
Based on the results of this study, a model was proposed to describe the functions of rice LRK2 in regulating tiller size and the drought stress response (Figure 8). By interacting with other molecules such as the eukaryotic translation elongation factor, LRK2 is thought to regulate cellular proliferation, promote branch development and subsequently increase tiller number. The larger root system subsequently contributes to an increase in drought tolerance. As rice is an important food crop and drought one of the main factors affecting crop growth and yield, methods aimed at increasing yield and creating drought‐resistant varieties are crucial. The findings of this study demonstrate the potential of LRK2 as a useful tool for crop improvement, particularly with regard to drought tolerance, helping enhance agronomically useful traits such as tiller number, yield and the number of grains per plant. Future research will facilitate further improvements in abiotic stress tolerance in crops through genetic manipulation, thereby paving the way for a new green revolution.
Figure 8.
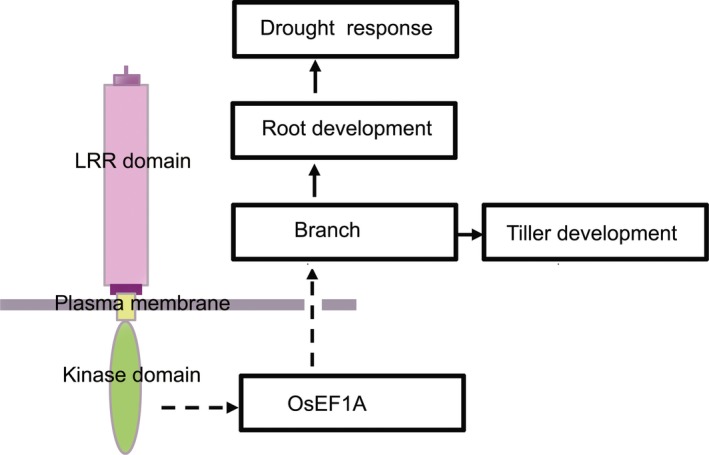
Proposed model of the role of the LRK2 gene in drought tolerance and tiller development in rice. LRR: leucine‐rich repeat.
Experimental procedures
Phylogenetic analysis
Phylogenetic analysis of LRK2 and other LRK proteins was carried out using MEGA 6.0 and a phylogenetic tree constructed using ClustalX 1.83 and UltraEdit21 software.
Generation of transgenic rice
Full‐length cDNA of LRK2 was amplified from the rice cultivar Nipponbare, and the confirmed sense and antisense sequences inserted into the pCAMBIA 1300‐2 × 35S vector under control of the cauliflower mosaic virus 35S promoter to produce LRK2‐overexpressing and knockdown lines. The primers 5′‐GTC GGTACCATGCAGCCACCTCATTCTTCATGCAAC‐3′ and 5′‐CAGGTCGAC TCAGTCGGAGCCTACACTGTCCAG‐3′ were used to construct 2 × 35S::LRK2, and primers 5′‐GTCGGTACCTTATATCTTTATTTCAGTGCCTATACTGTC‐3′ and 5′‐CAGGTCGACATGCAGCTACTTCATTACAAGAAACACAG‐3′ to construct 2 × 35S::antiLRK2. The constructs were transformed into Nipponbare mediated by Agrobacterium‐mediated transformation as described previously (Attia et al., 2005). The primers 5′‐CAAGACCTGCCTGAAACCGAACTG‐3′ and 5′‐GCGCGTCTGCTGCTCCATACA‐3′ were used to confirm the transgenic plants.
Subcellular localization of rice LRK2
To investigate the subcellular localization of LRK2, the LRK2 coding sequence was amplified with the cDNA clone as the template, using primers 5′‐CGGGGTACCATG CAG CCACCTCATTCTTCATGCA‐3′ and 5′‐TCCCCCGGGGTCGGA GCCTACACT GTCCAGGCAG‐3′. The PCR product was used to construct a 35S::LRK2‐eGFP fusion plasmid, which was transformed into tobacco leaves by Agrobacterium‐mediated transformation, as described previously (Yang et al., 2000). The transformed plants were cultured at 22 °C under 16‐h light for 24–48 h. The GFP image was subsequently obtained using a Leica TCS SP5 AOBS confocal laser microscope.
Promoter–GUS analysis
The LRK2 promoter, an approximately 2000‐bp DNA fragment upstream of the translation start site, was amplified using primers 5′‐TTGAAGCTTCCTCCCACCTCCAAGTGTTCAAC‐3′ and 5′‐CCAGGATCCGGTTTTCTGGTGATACTAGCATGGAAG‐3′ from 9311. The DNA fragment was then cloned into the pBI121 expression vector between the HindIII and BamHI sites using the above transformation method. The primers 5′‐CTGGATCCGTAGATCTGAGGAACCGACGA‐3′ and 5′‐GAGGACGTCTCACACGTGGTGGTGGTGGT‐3′ were used to confirm the transgenic plants. A GUS assay was performed at various developmental stages as described previously (Jefferson et al., 1987).
Total RNA isolation, RT‐PCR and real‐time quantitative PCR analysis (qRT‐PCR)
Total RNA extraction was performed using an RNeasy Plant Mini Kit following the manufacturer's instructions (Qiagen, Germany). Translation of RNA into cDNA was performed using a ReverTra Ace qPCR‐RT Kit following the manufacturer's instructions (TOYOBO, Japan). Amplification of genes from the cDNA template was performed using specific primers with high‐fidelity primeSTAR HS DNA Polymerase according the user's manual (TaKaRa, Japan). Diluted reaction products were used as templates for RT‐PCR and real‐time quantitative PCR analysis. The following LRK2‐specific primers were used for RT‐PCR: 5′‐GTCGGTACCATGCAGCCACCTCATTCTTCATGCAAC‐3′ and 5′‐CAGGTCGACTCAGTCGGAGCCTACACTGTCCAG‐3′, and the following specific primers for qRT‐PCR analysis: 5′‐TCAGCATCCAAAAAACAGTTGAAC‐3′ and 5′‐CTCTGGATCAGAGGTGAACGAAC‐3′. Each data point represents three replicates, and each experiment was repeated twice.
Growth conditions
Rice seeds (O. sativa ssp. japonica cv. Nipponbare) and transgenic plants were sown in pots after 3 day of germination at 37 °C. All plants were grown under 28 °C/16‐h light and 25 °C/8‐h dark conditions at 75% relative humidity in a glasshouse. For gene expression analysis, the roots of Nipponbare rice seedlings at the three‐leaf stage were immersed in PEG6000 (20%) or water for 3, 6, 12 and 24 h. After treatment, the seedlings were harvested and used for total RNA isolation.
Drought stress treatment
For drought stress treatment, 12‐day‐old 2 × 35S::LRK2, 2 × 35S::antiLRK2 and wild‐type plants were grown in pots under the indicated conditions then treated with 20% PEG6000, respectively, until the leaves of the wild type were rolled as a result of drought stress. Plants were then rewatered. The phenotypes of the plants were subsequently observed and photographed at various time points. After recovery, survival rates were calculated by counting plants with green healthy young leaves.
Measurements of root activity
Root activity in terms of TTC reduction was measured as described previously (Hu et al., 2016; Steponkus and Lanphear, 1967). Fresh roots from wild‐type and transgenic plants were placed in six test tubes, each containing 2 mg sodium thiosulfate and 5 mL of various concentrations of TTC or distilled water, followed by the addition of 5 mL phosphate buffer (pH 7.5). The roots were incubated at 37 °C for 2 h after which 2 mL 1 m sulphuric acid was added to terminate the reaction. The roots were removed and ground in a mortar containing 3 mL ethyl acetate to extract the triphenylformazan (TTCH). TTCH was measured based on the absorbance of the supernatant at 485 nm. A standard curve was constructed with TTCH on the x‐axis and OD on the y‐axis. The root activity was determined by TTCH concentration for each fresh root as follows: root activity (mg g−1 h−1) = TTCH reduction (TTCH mg)/fresh root weight (FW g)/time (h). For each root activity measurement, data points represent the average of three replicates.
Yeast two‐hybridization analysis
Yeast two‐hybrid library construction and screening was performed using BD Matchmaker library construction and screening kits (Clontech). The LRK2D coding region was fused in‐frame with the GAL4 DNA binding domain in the pGBKT7 vector to generate the bait vector. Primers included LRK2D‐F (5′‐CTG CATATG CTTTTCTCGCTCAGGGATGC‐3′) and LRK2D‐R (5′‐CGC GTCGAC GTCGGAGCCTACACTGTCCAG‐3′). Rice ds‐cDNA, the vector pGADT7‐Rec2 and bait construct pGBKT7‐LRK2D were then cotransformed into yeast strain AH109, which was subsequently plated directly onto SD/ Leu‐Trp‐His‐/50 mm 3‐AT medium followed by incubation at 30 °C for 4 days. Positive clones were screened according to the manufacturer's protocol. The OsEF1A coding region was cloned into pGADT7 and yeast two‐hybrid analysis performed using primers OsEF1A‐F (5′‐CCAGAATTCATGGGTAAGGAGAAGACGCACATCA‐3′), OsEF1A‐R (5′‐TTT GGATCCTTATTTCTTCTTGGCGGCAGCCTTG‐3′), OsEF1A D1‐AD‐EcoRI‐F (5′‐CCAGAATTCATTGTGGTCATTGGCCACG‐3′), OsEF1A D1‐AD‐BamHI‐R (5′‐TTTGGATCCGGGCTCGTTGATCTGGTCAAG‐3′), OsEF1A D2‐AD‐EcoRI‐F (5′‐CCAGAATTCGACAAGCCCCTACGTCTTCCC‐3′), OsEF1A D2‐AD‐BamHI‐R (5′‐TTTGGATCCGTCATCCTTGGAGTTGGAGGC‐3′), OsEF1A D3‐AD‐EcoRI‐F (5′‐CCAGAATTCGAGGCTGCCAGCTTCACCTC‐3′) and OsEF1AD3‐AD‐BamHI‐R (5′‐TTTGGATCCGATGACGCCAACAGCCACC‐3′). Yeast cells harbouring pGBKT7‐LRK2D+ pGADT7‐OsEF1A, pGBKT7+ pGADT7‐OsEF1A, pGBKT7‐LRK2D+ pGADT7, pGBKT7+ pGBKT7, pGBKT7‐LRK2D+ OsEF1A‐ΔD3‐1, pGBKT7‐LRK2D+ OsEF1A‐ΔD3‐2, pGBKT7‐LRK2D+ OsEF1A‐D3‐1, pGBKT7‐LRK2D+ OsEF1A‐ΔD3‐3, pGBKT7‐LRK2D+ OsEF1A‐D3‐2 were selected on SD/Leu‐Trp‐ and SD/Leu‐Trp‐His‐/50 mm 3‐AT with X‐gal plates.
Bimolecular fluorescence complementation assay
Bimolecular fluorescence complementation in tobacco was carried out as described previously (Schweiger and Schwenkert, 2014). LRK2D and OsEF1A cDNA fragments were amplified with the following primers OsEF1A‐pSPYNE‐F (5′‐CACACTAGTAT GGGTAAGGAGAAGACGCACAT‐3′), OsEF1A‐pSPYNE‐R (5′‐CGACCCGGGTT TCTTCTTGGCGGCAGCC‐3′), LRK2D‐pSPYCE‐F (5′‐CACTCTCGAATGCT TTTCTCGCTCAGGGATG‐3′) and LRK2D‐pSPYCE‐R (5′‐CGAGTCGACGTC GGAGCCTACACTGTCC‐3′). They were then cloned into the following split‐YFP vectors with nonoverlapping coding regions: 35S::SPYNE and 35S::SPYCE (Walter et al., 2004). The constructs were verified by sequencing and transformed, respectively, into Agrobacterium. Agrobacterium were grown at 28 °C to a final OD600 of 0.8 for agroinfiltration. Next, we mixed equal volumes of Agrobacterium culture carrying the constructs OsEF1A‐pSPYNE and LRK2D‐pSPYCE, selected the leaves of 3‐week‐old tobacco plants, and infiltrated the Agrobacterium suspension carefully into the tobacco leaves by pressing a syringe without a needle. Plants were watered and cultured at 22 °C under 16‐h light for 2 days. Images were collected using a Leica TCS SP5 AOBS confocal laser microscope.
Conflict of interest
The authors declare no conflict of interests.
Supporting information
Figure S1 LRK2 promoter expression in transgenic rice.
Figure S2 PCR analysis to confirm the transgenic lines.
Figure S3 Gene expression levels in rice seedlings in response to drought treatment.
Figure S4 Overexpression of LRK2 increased the tiller number in rice.
Table S1 Plant heights of control and transgenic lines.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31000741, 31671650), the Natural Science Foundation of Zhejiang Province, China (Grant No. LY13C130009), the Zhejiang Province University Students’ Science and Technology Innovation Program (Grant No. 2015R404005) and the Open Funds for Key Modern Agricultural Biotechnology and Crop Disease Prevention and Control Program of Zhejiang Province, China (Grant No. 2012KFJJ0014).
References
- Atkinson, J.A. , Rasmussen, A. , Traini, R. , Voss, U. , Sturrock, C. , Mooney, S.J. , Wells, D.M. et al (2014) Branching out in roots: uncovering form, function, and regulation. Plant Physiol. 166, 538–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attia, K. , Li, K.G. , Wei, C. , He, G.M. , Su, W. and Yang, J.S. (2005) Transformation and functional expression of the rFCA‐RRM2 gene in rice. J. Integr. Plant Biol. 47, 823–830. [Google Scholar]
- Browning, K.S. (1996) The plant translational apparatus. Plant Mol. Biol. 32, 107–144. [DOI] [PubMed] [Google Scholar]
- Byun, H.O. , Han, N.K. , Lee, H.J. , Kim, K.B. , Ko, Y.G. , Yoon, G. , Lee, Y.S. et al (2009) Cathepsin D and eukaryotic translation elongation factor 1 as promising markers of cellular senescence. Cancer Res. 69, 4638–4647. [DOI] [PubMed] [Google Scholar]
- Cheng, S. , Zhou, D.X. and Zhao, Y. (2016) WUSCHEL‐related homeobox gene WOX11 increases rice drought resistance by controlling root hair formation and root system development. Plant Signal. Behav. 11, e1130198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla, D. , Zipfel, C. , Robatzek, S. , Kemmerling, B. , Nurnberger, T. , Jones, J.D. , Felix, G. et al (2007) A flagellin‐induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature, 448, 497–500. [DOI] [PubMed] [Google Scholar]
- Chinchilla, D. , Shan, L. , He, P. , de Vries, S. and Kemmerling, B. (2009) One for all: the receptor‐associated kinase BAK1. Trends Plant Sci. 14, 535–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S.E. , Williams, R.W. and Meyerowitz, E.M. (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell, 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Coudert, Y. , Perin, C. , Courtois, B. , Khong, N.G. and Gantet, P. (2010) Genetic control of root development in rice, the model cereal. Trends Plant Sci. 15, 219–226. [DOI] [PubMed] [Google Scholar]
- Ejiri, S. (2002) Moonlighting functions of polypeptide elongation factor 1: from actin bundling to zinc finger protein R1‐associated nuclear localization. Biosci. Biotechnol. Biochem. 66, 1–21. [DOI] [PubMed] [Google Scholar]
- Feng, L. , Gao, Z. , Xiao, G. , Huang, R. and Zhang, H. (2014) Leucine‐rich repeat receptor‐like kinase FON1 regulates drought stress and seed germination by activating the expression of ABA‐responsive genes in rice. Plant Mol. Biol. Rep. 32, 1158–1168. [Google Scholar]
- Fernie, A.R. , Tadmor, Y. and Zamir, D. (2006) Natural genetic variation for improving crop quality. Curr. Opin. Plant Biol. 9, 196–202. [DOI] [PubMed] [Google Scholar]
- Grillo, M.A. , Li, C. , Fowlkes, A.M. , Briggeman, T.M. , Zhou, A. , Schemske, D.W. and Sang, T. (2009) Genetic architecture for the adaptive origin of annual wild rice, oryza nivara. Evolution, 63, 870–883. [DOI] [PubMed] [Google Scholar]
- Gross, S.R. and Kinzy, T.G. (2005) Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat. Struct. Mol. Biol. 12, 772–778. [DOI] [PubMed] [Google Scholar]
- Guo, Y. , Xiong, L. , Ishitani, M. and Zhu, J.K. (2002) An Arabidopsis mutation in translation elongation factor 2 causes superinduction of CBF/DREB1 transcription factor genes but blocks the induction of their downstream targets under low temperatures. Proc. Natl Acad. Sci. USA, 99, 7786–7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, M.T. , Tardieu, F. , Dong, Z. , Messina, C.D. and Hammer, G.L. (2014) Characterizing drought stress and trait influence on maize yield under current and future conditions. Glob. Chang. Biol. 20, 867–878. [DOI] [PubMed] [Google Scholar]
- Hartmann, J. , Fischer, C. , Dietrich, P. and Sauter, M. (2014) Kinase activity and calmodulin binding are essential for growth signaling by the phytosulfokine receptor PSKR1. Plant J. 78, 192–202. [DOI] [PubMed] [Google Scholar]
- Hatmi, S. , Gruau, C. , Trotel‐Aziz, P. , Villaume, S. , Rabenoelina, F. , Baillieul, F. , Eullaffroy, P. et al (2015) Drought stress tolerance in grapevine involves activation of polyamine oxidation contributing to improved immune response and low susceptibility to Botrytis cinerea. J. Exp. Bot. 66, 775–787. [DOI] [PubMed] [Google Scholar]
- He, G. , Luo, X. , Tian, F. , Li, K. , Zhu, Z. , Su, W. , Qian, X. et al (2006) Haplotype variation in structure and expression of a gene cluster associated with a quantitative trait locus for improved yield in rice. Genome Res. 16, 618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain, Z. , Amyot, L. , McGarvey, B. , Gruber, M. , Jung, J. and Hannoufa, A. (2012) The translation elongation factor eEF‐1Bbeta1 is involved in cell wall biosynthesis and plant development in Arabidopsis thaliana . PLoS ONE, 7, e30425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y. , Xia, S. , Su, Y. , Wang, H. , Luo, W. , Su, S. and Xiao, L. (2016) Brassinolide increases potato root growth in vitro in a dose‐dependent way and alleviates salinity stress. Biomed Res. Int. 3, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi, D. , Tsuda, K. and Katagiri, F. (2012) The peptide growth factor, phytosulfokine, attenuates pattern‐triggered immunity. Plant J. 71, 194–204. [DOI] [PubMed] [Google Scholar]
- Ingram, G.C. (2007) Cell signalling: the merry lives of BAK1. Curr. Biol. 17, R603–R605. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. , Kavanagh, T.A. and Bevan, M.W. (1987) GUS fusions: beta‐glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y. , Chen, X. , Ding, X. , Wang, Y. , Chen, Q. and Song, W.Y. (2013) The XA21 binding protein XB25 is required for maintaining XA21‐mediated disease resistance. Plant J. 73, 814–823. [DOI] [PubMed] [Google Scholar]
- Jinn, T.L. , Stone, J.M. and Walker, J.C. (2000) HAESA, an Arabidopsis leucine‐rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14, 108–117. [PMC free article] [PubMed] [Google Scholar]
- Kidou, S. and Ejiri, S. (1998) Isolation, characterization and mRNA expression of four cDNAs encoding translation elongation factor 1A from rice (Oryza sativa L.). Plant Mol. Biol. 36, 137–148. [DOI] [PubMed] [Google Scholar]
- Ladwig, F. , Dahlke, R.I. , Stuhrwohldt, N. , Hartmann, J. , Harter, K. and Sauter, M. (2015) Phytosulfokine regulates growth in Arabidopsis through a response module at the plasma membrane that includes CYCLIC NUCLEOTIDE‐GATED CHANNEL17, H+‐ATPase, and BAK1. Plant Cell, 27, 1718–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease, K. , Ingham, E. and Walker, J.C. (1998) Challenges in understanding RLK function. Curr. Opin. Plant Biol. 1, 388–392. [DOI] [PubMed] [Google Scholar]
- Li, J. and Chory, J. (1997) A putative leucine‐rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell, 90, 929–938. [DOI] [PubMed] [Google Scholar]
- Li, D.J. , Sun, C.Q. , Fu, Y.C. , Li, C. , Zhu, Z.F. , Chen, L. , Cai, H.W. et al (2002a) Identification and mapping of genes for improving yield from Chinese common wild rice (O. rufipogon Griff.) using advanced backcross QTL analysis. Chin. Sci. Bull. 47, 1533–1537. [Google Scholar]
- Li, J. , Wen, J. , Lease, K.A. , Doke, J.T. , Tax, F.E. and Walker, J.C. (2002b) BAK1, an Arabidopsis LRR receptor‐like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell, 110, 213–222. [DOI] [PubMed] [Google Scholar]
- Li, X.Y. , Qian, Q. , Fu, Z.M. , Wang, Y.H. , Xiong, G.S. , Zeng, D.L. , Wang, X.Q. et al (2003) Control of tillering in rice. Nature, 422, 618–621. [DOI] [PubMed] [Google Scholar]
- Mandel, T. , Moreau, F. , Kutsher, Y. , Fletcher, J.C. , Carles, C.C. and Eshed Williams, L. (2014) The ERECTA receptor kinase regulates Arabidopsis shoot apical meristem size, phyllotaxy and floral meristem identity. Development, 141, 830–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masle, J. , Gilmore, S.R. and Farquhar, G.D. (2005) The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature, 436, 866–870. [DOI] [PubMed] [Google Scholar]
- Matsubayashi, Y. , Ogawa, M. , Morita, A. and Sakagami, Y. (2002) An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science, 296, 1470–1472. [DOI] [PubMed] [Google Scholar]
- Matsubayashi, Y. , Ogawa, M. , Kihara, H. , Niwa, M. and Sakagami, Y. (2006) Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiol. 142, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X. , Wang, H. , He, Y. , Liu, Y. , Walker, J.C. , Torii, K.U. and Zhang, S. (2012) A MAPK cascade downstream of ERECTA receptor‐like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell, 24, 4948–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, K.H. and Li, J.M. (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell, 110, 203–212. [DOI] [PubMed] [Google Scholar]
- Numata, O. , Kurasawa, Y. , Gonda, K. and Watanabe, Y. (2000) Tetrahymena elongation factor‐1 alpha is localized with calmodulin in the division furrow. J. Biochem. 127, 51–56. [DOI] [PubMed] [Google Scholar]
- Ouyang, S.Q. , Liu, Y.F. , Liu, P. , Lei, G. , He, S.J. , Ma, B. , Zhang, W.K. et al (2010) Receptor‐like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 62, 316–329. [DOI] [PubMed] [Google Scholar]
- Pecorari, L. , Marin, O. , Silvestri, C. , Candini, O. , Rossi, E. , Guerzoni, C. , Cattelani, S. et al (2009) Elongation Factor 1 alpha interacts with phospho‐Akt in breast cancer cells and regulates their proliferation, survival and motility. Mol. Cancer., 8, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi, E. (2008) Plant genetics. The blue revolution, drop by drop, gene by gene. Science, 320, 171–173. [DOI] [PubMed] [Google Scholar]
- Quain, M.D. , Makgopa, M.E. , Marquez‐Garcia, B. , Comadira, G. , Fernandez‐Garcia, N. , Olmos, E. , Schnaubelt, D. et al (2014) Ectopic phytocystatin expression leads to enhanced drought stress tolerance in soybean (Glycine max) and Arabidopsis thaliana through effects on strigolactone pathways and can also result in improved seed traits. Plant Biotechnol. J. 12, 903–913. [DOI] [PubMed] [Google Scholar]
- Redillas, M.C.F.R. , Jeong, J.S. , Kim, Y.S. , Jung, H. , Bang, S.W. , Choi, Y.D. , Ha, S.‐H. et al (2012) The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol. J. 10, 792–805. [DOI] [PubMed] [Google Scholar]
- Riis, B. , Rattan, S.I. , Clark, B.F. and Merrick, W.C. (1990) Eukaryotic protein elongation factors. Trends Biochem. Sci. 15, 420–424. [DOI] [PubMed] [Google Scholar]
- Sanders, J. , Maassen, J.A. and Moller, W. (1992) Elongation factor‐1 messenger‐RNA levels in cultured cells are high compared to tissue and are not drastically affected further by oncogenic transformation. Nucleic Acids Res. 20, 5907–5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasikumar, A.N. , Perez, W.B. and Kinzy, T.G. (2012) The many roles of the eukaryotic elongation factor 1 complex. Wiley Interdiscip. Rev. RNA, 3, 543–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger, R. and Schwenkert, S. (2014) Protein‐protein interactions visualized by bimolecular fluorescence complementation in tobacco protoplasts and leaves. J. Vis. Exp. 85, e51327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, H. , Zhong, X. , Zhao, F. , Wang, Y. , Yan, B. , Li, Q. , Chen, G. et al (2015) Overexpression of receptor‐like kinase ERECTA improves thermotolerance in rice and tomato. Nat. Biotechnol. 33, 996–1003. [DOI] [PubMed] [Google Scholar]
- Shepherd, J.C. , Walldorf, U. , Hug, P. and Gehring, W.J. (1989) Fruit flies with additional expression of the elongation factor EF‐1 alpha live longer. Proc. Natl Acad. Sci. USA, 86, 7520–7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, L. , Guo, M. , Ye, N. , Liu, Y. , Liu, R. , Xia, Y. , Cui, S. et al (2015) Reduced ABA accumulation in the root system is caused by ABA exudation in upland rice (Oryza sativa L. var. Gaoshan1) and this enhanced drought adaptation. Plant Cell Physiol. 56, 951–964. [DOI] [PubMed] [Google Scholar]
- Shiu, S.H. and Bleecker, A.B. (2001) Receptor‐like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proceedings of the National Academy of Sciences of the United States of America, 98, 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, S.H. , Karlowski, W.M. , Pan, R.S. , Tzeng, Y.H. , Mayer, K.F.X. and Li, W.H. (2004) Comparative analysis of the receptor‐like kinase family in Arabidopsis and rice. Plant Cell, 16, 1220–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak, E.D. , Berthiaume, C.T. , Hill, E.J. and Torii, K.U. (2004) Synergistic interaction of three ERECTA‐family receptor‐like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development, 131, 1491–1501. [DOI] [PubMed] [Google Scholar]
- Song, W.‐Y. , Pi, L.‐Y. , Wang, G.‐L. , Gardner, J. , HoIsten, T. and Ronald, P.C. (1997) Evolution of the rice Xa2I disease resistance gene family. Plant Cell 9, 1279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steponkus, P.L. and Lanphear, F.O. (1967) Refinement of the triphenyl tetrazolium chloride method of determining cold injury. Plant Physiol. 42, 1423–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. , Li, L. , Macho, A.P. , Han, Z. , Hu, Z. , Zipfel, C. , Zhou, J.M. et al (2013) Structural basis for flg22‐induced activation of the Arabidopsis FLS2‐BAK1 immune complex. Science, 342, 624–628. [DOI] [PubMed] [Google Scholar]
- Sun, L. , Zhang, Q. , Wu, J. , Zhang, L. , Jiao, X. , Zhang, S. , Zhang, Z. et al (2014) Two rice authentic histidine phosphotransfer proteins, OsAHP1 and OsAHP2, mediate cytokinin signaling and stress responses in rice. Plant Physiol. 165, 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, T. , Suwa, Y. , Suzuki, M. , Kitano, H. , Ueguchi‐Tanaka, M. , Ashikari, M. , Matsuoka, M. et al (2003) The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33, 513–520. [DOI] [PubMed] [Google Scholar]
- Tanaka, W. , Ohmori, Y. , Ushijima, T. , Matsusaka, H. , Matsushita, T. , Kumamaru, T. , Kawano, S. et al (2015) Axillary Meristem formation in rice requires the WUSCHEL Ortholog TILLERS ABSENT1. Plant Cell, 27, 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, I. , Wang, Y. , Seitz, K. , Baer, J. , Bennewitz, S. , Mooney, B.P. and Walker, J.C. (2016) Analysis of phosphorylation of the receptor‐like protein kinase HAESA during arabidopsis floral abscission. PLoS ONE, 11, e0147203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii, K.U. (2004) Leucine‐rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. Int. Rev. Cytol. 234, 1–46. [DOI] [PubMed] [Google Scholar]
- Uga, Y. , Sugimoto, K. , Ogawa, S. , Rane, J. , Ishitani, M. , Hara, N. , Kitomi, Y. et al (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102. [DOI] [PubMed] [Google Scholar]
- Van Zanten, M. , Snoek, L.B. , Proveniers, M.C. and Peeters, A.J. (2009) The many functions of ERECTA. Trends Plant Sci. 14, 214–218. [DOI] [PubMed] [Google Scholar]
- Walker, J.C. (1993) Receptor‐like protein kinase genes of Arabidopsis thaliana . Plant J. 3, 451–456. [DOI] [PubMed] [Google Scholar]
- Walter, M. , Chaban, C. , Schutze, K. , Batistic, O. , Weckermann, K. , Nake, C. , Blazevic, D. et al (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. [DOI] [PubMed] [Google Scholar]
- Wu, X.N. , Sanchez Rodriguez, C. , Pertl‐Obermeyer, H. , Obermeyer, G. and Schulze, W.X. (2013) Sucrose‐induced receptor kinase SIRK1 regulates a plasma membrane aquaporin in Arabidopsis. Mol. Cell Proteomics, 12, 2856–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F. , Sheng, P. , Tan, J. , Chen, X. , Lu, G. , Ma, W. , Heng, Y. et al (2015) Plasma membrane receptor‐like kinase leaf panicle 2 acts downstream of the DROUGHT AND SALT TOLERANCE transcription factor to regulate drought sensitivity in rice. J. Exp. Bot. 66, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Li, R. and Qi, M. (2000) In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 22, 543–551. [DOI] [PubMed] [Google Scholar]
- Yang, L. , Wu, K. , Gao, P. , Liu, X. , Li, G. and Wu, Z. (2014) GsLRPK, a novel cold‐activated leucine‐rich repeat receptor‐like protein kinase from Glycine soja, is a positive regulator to cold stress tolerance. Plant Sci. 215–216, 19–28. [DOI] [PubMed] [Google Scholar]
- Yoshida, T. , Fujita, Y. , Sayama, H. , Kidokoro, S. , Maruyama, K. , Mizoi, J. , Shinozaki, K. et al (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE‐dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685. [DOI] [PubMed] [Google Scholar]
- Yu, X. , Peng, Y.H. , Zhang, M.H. , Shao, Y.J. , Su, W.A. and Tang, Z.C. (2006) Water relations and an expression analysis of plasma membrane intrinsic proteins in sensitive and tolerant rice during chilling and recovery. Cell Res. 16, 599–608. [DOI] [PubMed] [Google Scholar]
- Zha, X. , Luo, X. , Qian, X. , He, G. , Yang, M. , Li, Y. and Yang, J. (2009) Over‐expression of the rice LRK1 gene improves quantitative yield components. Plant Biotechnol. J. 7, 611–620. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Lin, W. , Li, X. , Cao, H. , Wang, Y. and Zheng, S.J. (2015) Critical role of eukaryotic elongation factor 1 alpha 1 (EEF1A1) in avian reovirus sigma‐C‐induced apoptosis and inhibition of viral growth. Arch. Virol. 160, 1449–1461. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Kunze, G. , Chinchilla, D. , Caniard, A. , Jones, J.D. , Boller, T. and Felix, G. (2006) Perception of the bacterial PAMP EF‐Tu by the receptor EFR restricts Agrobacterium‐mediated transformation. Cell, 125, 749–760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 LRK2 promoter expression in transgenic rice.
Figure S2 PCR analysis to confirm the transgenic lines.
Figure S3 Gene expression levels in rice seedlings in response to drought treatment.
Figure S4 Overexpression of LRK2 increased the tiller number in rice.
Table S1 Plant heights of control and transgenic lines.


