Abstract
Predicting antibody pair performance in a sandwich format streamlines development of antibody-based diagnostics and laboratory research tools, such as enzyme-linked immunosorbent assays (ELISAs) and lateral flow immunoassays (LFAs). We have evaluated panels of monoclonal antibodies against the malarial parasite biomarker Plasmodium falciparum histidine rich protein 2 (HRP2), including 9 new monoclonal antibodies, using biolayer interferometry (BLI) and screened antibody pairs in a checkerboard ELISA. This study showed BLI predicts antibody pair ELISA performance for HRP2. Pairs that included capture antibodies with low off-rate constants and detection antibodies with high on-rate constants performed best in an ELISA format.
Keywords: Biolayer interferometry, ELISA, Malaria, Histidine-rich protein 2, Binding kinetics
Graphical abstract
Highlights
-
•
Kinetic parameters of 15 anti-HRP2 antibodies are measured by biolayer interferometry.
-
•
Kinetic constants are compared to a checkerboard ELISA of 225 antibody pairs.
-
•
Biolayer interferometry predicts antibody pair performance for HRP2 ELISA.
-
•
Capture mAbs with low koff and detection mAbs with high kon are best in HRP2 ELISA.
Antibody-based diagnostics for disease-specific biomarkers are invaluable tools for patient care, disease surveillance, and intervention management. Lateral flow immunoassays (LFAs), which leverage high-affinity and specific antibody-antigen interactions in a sandwich immunoassay format, make infectious disease diagnosis possible in low-resource settings, where laboratory infrastructure is unavailable. For example, approximately 314 million antibody-based LFA rapid diagnostic tests for malaria were sold in 2014. Most of these tests were deployed in sub-Saharan Africa, where the disease burden is high and good-quality, high-throughput microscopy is not readily available [1].
While LFAs have made malaria diagnosis accessible and affordable, they lack the sensitivity needed to detect low parasite densities. Submicroscopic and asymptomatic cases are often missed by these tests and left untreated, remaining a reservoir for malaria transmission [2], [3], [4], [5]. Recent models suggest that diagnostics with detection limits of 200 parasites/μl only detect 55% of the infectious reservoir, depending on the setting [6]. These data highlight the need not only for more sensitive malaria rapid diagnostic tests, but also for laboratory tools with limits of detection capable of defining the clinically relevant protein biomarker concentrations required to accurately diagnose asymptomatic infections.
The primary protein biomarker used in malaria rapid diagnostic tests for Plasmodium falciparum infection is Plasmodium falciparum histidine-rich protein 2 (HRP2), which is expressed by one out of five species of malaria parasites known to infect humans. HRP2 is a unique biomarker because it lacks native tertiary structure, and its sequence is 30% histidine, consisting largely of AHHAHHAAD and AHHAAD repeat motifs [7]. Expression of HRP2 varies over the erythrocytic life cycle of the parasite, though the function of the protein remains unconfirmed [8], [9].
Sensitive antibody-based diagnostics and laboratory tools for HRP2 detection at extremely low parasite densities will depend on high-affinity molecular recognition events for both capture and detection of the biomarker. Biolayer interferometry (BLI) is a label-free bioanalytical technique used to quantify the strength of antibody-antigen interactions, allowing for measurement of kinetic parameters such as the dissociation constant (KD), on-rate constant (kon), and off-rate constant (koff) [10], [11]. This optical technique allows for real-time monitoring of the interference pattern of white light reflected from two surfaces within fiber optic sensors that are immersed in biomolecule solutions. This experimental set-up is advantageous over evanescent (eg: surface plasmon resonance) or acoustic label-free systems, which typically require microfluidics to deliver the sample to the sensing surface. These systems are prone to clogging when complex sample matrices are used. Further, since BLI detection occurs at the biosensor tip surface, matrix effects, such as those from unbound proteins in solution, are minimized [11].
The crucial need for improved malaria point-of-care diagnostics and laboratory assays and the reliance of these tools on the strength of antibody-antigen interactions highlight the importance of selecting the best monoclonal antibodies (mAbs) for capture and detection. The goal of this work is to assess biolayer interferometry as a tool for predicting antibody pair performance in an enzyme-linked immunosorbent assay (ELISA) for HRP2. The kinetic parameters of 9 novel mAbs as well as 6 commercially available clones (Table S1) are determined and compared to antibody pair performance in a sandwich ELISA format.
The strength of mAb-rcHRP2 interactions was measured using BLI. For each anti-HRP2 IgG mAb, the antibody was biotinylated, loaded onto streptavidin biosensors, and HRP2 was allowed to associate and dissociate. Specific experimental conditions can be found in Supplemental Information. The binding profiles, pseudo-first order fit curves, and residuals are plotted in Tables S2 and S3. Kinetic parameters for all anti-HRP2 mAbs are listed in Table S4. Notably, 11 out of 15 anti-HRP2 antibodies had off-rate constants below the limit of detection of the OctetRed96 instrument (<1 × 10−7 1/s). Although experiments were optimized such that one-to-one fits could provide accurate estimations of mAb-HRP2 affinities, it is likely that repeated motifs throughout the protein allowed for re-binding of HRP2 to the mAb-functionalized sensors during the dissociation phase, resulting in no net dissociation.
While the off-rate constants make distinguishing the anti-HRP2 mAbs by koff or KD difficult, the measured association rate constants varied over three orders of magnitude. The 3 mAbs with the highest on-rate constants were custom-made IgG from Precision Antibody (10F5, 10C1, 6C8). Three commercial antibodies had the lowest kon values, two of which were IgM (MPFM-55A and PTL3). This was not surprising, since IgM generally have low-affinity, high-avidity interactions with their targets [12].
To identify the best-performing antibody pairs for an HRP2 sandwich immunoassay, all 225 possible pairs of anti-HRP2 mAbs (15 × 15 matrix) were screened in a checkerboard format. The average signal-to-noise ratio (S/N) for each pair was determined by dividing the average A450 at 34.1 pM of rcHRP2 by the average absorbance of the blank (Fig. S1). Several mAbs, such as MPFG, C1-13, 4D6, 8D3, 11H7, and 12F12 performed poorly as capture, while MPFM, PTL3, 2g6 and 0445 performed well as capture elements. Additionally, numerous custom mAbs were successful as detection components, including 4D6, 6C8, 10C1, 10F5, 11E10, 12D4 and 12F12.
With the quantification of individual mAb-HRP2 interactions by BLI and the relative ranking of anti-HRP2 mAb pairs in a checkerboard ELISA screening comes the question: can BLI be used to predict antibody pair performance in a traditional plate ELISA format?
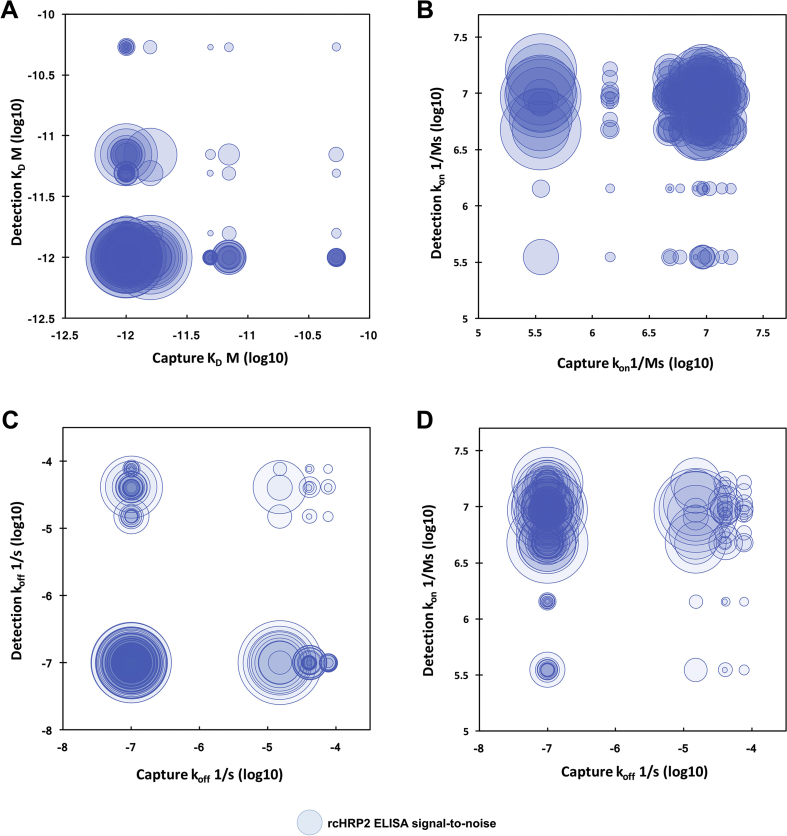
Fig. 1 relates the measured kinetic parameters of anti-HRP2 capture and detection antibodies to the S/N measured for each pair in the checkerboard ELISA. In these plots, a kinetic parameter for the capture antibody is plotted on the abscissa, and a kinetic parameter for the detection antibody is plotted on the ordinate. The size of the circle at the ordinate pair (capture, detection) represents the relative S/N for that pair measured by ELISA. Larger circles indicate better-performing ELISA antibody pairs, while small circles indicate poor antibody pairs. The best-performing mAb pairs in the ELISA format are associated with capture and detection antibodies with low KD values, since the largest circles are concentrated in the lower left-hand corner of the plot in Fig. 1A. This result is anticipated, as one would expect stronger interactions to lead to better ELISA performance. However, when the ELISA data is compared to kon and koff values of the capture and detection antibodies, interesting trends emerge. Fig. 1B plots kon values for capture and detection mAbs versus ELISA S/N. In this visualization, it becomes apparent that no trend relates ELISA S/N to kon of capture antibodies, since the sizes of the circles neither increase nor decrease along the x-axis. However, there is a dependence of ELISA S/N on the kon of the detection antibody along the y-axis; greater detection antibody kon results in better ELISA antibody pair performance for anti-HRP2 mAbs. For koff (Fig. 1C), the trends are less clear, since there were fewer discrete koff values for the anti-HRP2 mAbs compared to kon values. However, Fig. 1D shows that the highest concentration of high-performing anti-HRP2 ELISA antibody pairs occurs when capture antibody koff is low and detection antibody kon is high.
Fig. 1.
Relationship between anti-HRP2 (A) KD, (B) kon, and (C) koff in the capture (x-axis) and detection (y-axis) positions as measured by BLI and ELISA signal-to-noise ratios (size of circles). (D) Plot of capture koff (x-axis) vs. detection kon (y-axis) vs. ELISA signal-to-noise (circles). Larger circles represent higher S/N. Note: only anti-HRP2 IgG pairs are plotted.
Using these BLI parameters, it would be predicted that pairs with 10F5, 10C1, and 6C8 in the detection position paired with capture mAbs with low off-rate constants, such as the two IgM mAbs (MPFM-55A, PTL-3) or an IgG such as 0445, would perform well in an ELISA format. In fact, these pairs are some of the top-performers in the ELISA checkerboard screening, despite the low kon values for the capture mAbs (Fig. S1). Capture antibodies with low koff may be favored due to the high number of washes and additional incubation steps the capture mAb-HRP2 complex is subjected to during the ELISA protocol. In contrast, kon may be most important for detection antibodies, since the time scale of this interaction is lower. These emperical results agree with a previously reported model [13]. Notably, kinetic parameters were determined by BLI in under 35 min for each mAb, highlighting the advantage of this technique as a rapid screening tool.
These results demonstrate that BLI can be used as a rapid, predictive tool for development of an HRP2 ELISA. Because HRP2 lacks structure and contains a series of repeated epitopes for mAb binding, it is important to ask whether trends observed for HRP2 are generalizable to protein biomarkers with defined tertiary structure and no or few repeat motifs in the sequence. For HRP2, if a capture and detection antibody target the same epitope, both may be able to bind to the protein due to the repeat motifs in the amino acid sequence. However, for targets lacking repeated epitopes, if two antibodies both target the same epitope, it is likely that little-to-no ELISA signal would be observed.
The utility of BLI for quantifying the strength of antibody-antigen interactions is clear, and this work demonstrates that BLI is also useful for predicting the ELISA performance of antibody pairs when an antigen contains many repeats in its sequence. There are many targets of interest that could benefit from building a sensitive ELISA from the bottom-up using BLI as a predictive tool. For example, the circulating anodic antigen biomarker for Schistosoma infections is decorated with repeating polysaccharide motifs [14], [15]. Further, BLI may be useful for developing ELISAs for viral capsids containing many oligomeric subunits or multimeric proteins with identical subunits.
Conflict of interest
PATH funded the development of the antibodies and entered into a licensing agreement with A&G to make the antibodies commercially available. PATH and its affiliated authors do not have any financial interest in A&G nor receive any royalties under the license agreement.
Acknowledgements
This work was supported by the Bill and Melinda Gates Foundation (Grant OPP1135840) and PATH: Diagnostics for malaria elimination toward eradication (1758-00-06-00). This material is based on work supported by the National Science Foundation Graduate Research Fellowship Program under Grant #1445197 (C.F.M). The authors thank M. F. Richards for critical comments in the preparation of this manuscript.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ab.2017.07.010.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.World Health Organization, “Malaria rapid diagnostic test performance: results of WHO product testing of malaria RDTs: round 6 (2014-2015)”.
- 2.Maltha J., Gillet P., Jacobs J. Malaria rapid diagnostic tests in endemic settings. Clin. Microbiol. Infect. May 2013;19(5):399–407. doi: 10.1111/1469-0691.12151. [DOI] [PubMed] [Google Scholar]
- 3.Bousema T., Okell L., Felger I., Drakeley C. Asymptomatic malaria infections: detectability, transmissibility and public health relevance. Nat. Rev. Microbiol. Dec. 2014;12(12):833–840. doi: 10.1038/nrmicro3364. [DOI] [PubMed] [Google Scholar]
- 4.Baum E., Sattabongkot J., Sirichaisinthop J., Kiattibutr K., Davies D.H., Jain A., Lo E., Lee M.-C., Randall A.Z., Molina D.M., Liang X., Cui L., Felgner P.L., Yan G. Submicroscopic and asymptomatic Plasmodium falciparum and Plasmodium vivax infections are common in western Thailand - molecular and serological evidence. Malar. J. 2015;14:95. doi: 10.1186/s12936-015-0611-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babiker H.A., Gadalla A.A.H., Ranford-Cartwright L.C. The role of asymptomatic P. falciparum parasitaemia in the evolution of antimalarial drug resistance in areas of seasonal transmission. Drug Resist. Updat. Feb. 2013;16(1–2):1–9. doi: 10.1016/j.drup.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Slater H.C., Ross A., Ouédraogo A.L., White L.J., Nguon C., Walker P.G.T., Ngor P., Aguas R., Silal S.P., Dondorp A.M., La Barre P., Burton R., Sauerwein R.W., Drakeley C., Smith T.A., Bousema T., Ghani A.C. Assessing the impact of next-generation rapid diagnostic tests on Plasmodium falciparum malaria elimination strategies. Nature. Dec. 2015;528(7580):S94–S101. doi: 10.1038/nature16040. [DOI] [PubMed] [Google Scholar]
- 7.Baker J., McCarthy J., Gatton M., Kyle D.E., Belizario V., Luchavez J., Bell D., Cheng Q. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of PfHRP2-based rapid diagnostic tests. J. Infect. Dis. Sep. 2005;192(5):870–877. doi: 10.1086/432010. [DOI] [PubMed] [Google Scholar]
- 8.Baker J., Gatton M.L., Peters J., Ho M.-F., McCarthy J.S., Cheng Q. Transcription and expression of Plasmodium falciparum histidine-rich proteins in different stages and strains: implications for rapid diagnostic tests. PLoS One. Jul. 2011;6(7) doi: 10.1371/journal.pone.0022593. e22593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desakorn V., Dondorp A.M., Silamut K., Pongtavornpinyo W., Sahassananda D., Chotivanich K., Pitisuttithum P., Smithyman A.M., Day N.P.J., White N.J. Stage-dependent production and release of histidine-rich protein 2 by Plasmodium falciparum. Trans. R. Soc. Trop. Med. Hyg. Jul. 2005;99(7):517–524. doi: 10.1016/j.trstmh.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 10.Sultana A., Lee J.E. Current Protocols in Protein Science. John Wiley & Sons, Inc.; 2001. Measuring protein-protein and protein-nucleic acid interactions by biolayer interferometry. [DOI] [PubMed] [Google Scholar]
- 11.Concepcion J., Witte K., Wartchow C., Choo S., Yao D., Persson H., Wei J., Li P., Heidecker B., Ma W., Varma R., Zhao L.-S., Perillat D., Carricato G., Recknor M., Du K., Ho H., Ellis T., Gamez J., Howes M., Phi-Wilson J., Lockard S., Zuk R., Tan H. Label-free detection of biomolecular interactions using BioLayer interferometry for kinetic characterization. Comb. Chem. High. Throughput Screen. Sep. 2009;12(8):791–800. doi: 10.2174/138620709789104915. [DOI] [PubMed] [Google Scholar]
- 12.Delves P.J., Martin S.J., Burton D.R., Roitt I.M. John Wiley & Sons; 2017. Roitt's Essential Immunology. [Google Scholar]
- 13.Choi D.H., Katakura Y., Ninomiya K., Shioya S. Rational screening of antibodies and design of sandwich enzyme linked immunosorbant assay on the basis of a kinetic model. J. Biosci. Bioeng. Mar. 2008;105(3):261–272. doi: 10.1263/jbb.105.261. [DOI] [PubMed] [Google Scholar]
- 14.Bergwerff A.A., van Dam G.J., Rotmans J.P., Deelder A.M., Kamerling J.P., Vliegenthart J.F. The immunologically reactive part of immunopurified circulating anodic antigen from Schistosoma mansoni is a threonine-linked polysaccharide consisting of--> 6)-(beta-D-GlcpA-(1--> 3))-beta-D-GalpNAc-(1--> repeating units. J. Biol. Chem. Dec. 1994;269(50):31510–31517. [PubMed] [Google Scholar]
- 15.Van Dam G.J., Bergwerff A.A., Thomas-Oates J.E., Rotmans J.P., Kamerling J.P., Vliegenthart J.F.G., Deelder A.M. The immunologically reactive O-Linked polysaccharide chains derived from circulating cathodic antigen isolated from the human blood fluke Schistosoma Mansoni have lewis x as repeating unit. Eur. J. Biochem. Oct. 1994;225(1):467–482. doi: 10.1111/j.1432-1033.1994.00467.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.