Abstract
There is currently a pandemic of pseudorabies virus (PRV) variant strains in China. Despite extensive research on PRV variant strains in the past two years, few studies have investigated PRV pathogenicity-related genes. To determine which gene(s) is/are linked to PRV virulence, ten putative virulence genes were knocked out using clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 technology. The pathogenicity of these mutants was evaluated in a mouse model. Our results demonstrated that of the ten tested genes, the thymidine kinase (TK) and glycoprotein M (gM) knockout mutants displayed significantly reduced virulence. However, mutants of other putative virulence genes, such as glycoprotein E (gE), glycoprotein I (gI), Us2, Us9, Us3, glycoprotein G (gG), glycoprotein N (gN) and early protein 0 (EP0), did not exhibit significantly reduced virulence compared to that of the wild-type PRV. To our knowledge, this study is the first to compare virulence genes from the current pandemic PRV variant strain. This study will provide a valuable reference for scientists to design effective live attenuated vaccines in the future.
Introduction
Pseudorabies virus (PRV) is a swine alpha-herpesvirus that causes considerable economic losses to the swine industry1. In China, PRV has been well controlled for decades by the Bartha-K61 vaccine, and it was thought that this disease would be eradicated in the foreseeable future. Despite great efforts to promote PRV vaccination, an unprecedented large-scale outbreak of PRV variants in China has caused great economic losses to the Chinese swine industry2, 3. These re-emerging pseudorabies variants belong to genotype II, and their sequences exhibit significant differences from genotype I4, 5. Compared to classic virulent PRV strains, these variant strains exhibit increased virulence, with an earlier onset of clinical signs and higher mortality in swine5, 6. Although several studies have focused on PRV variant strains in the past two years, few studies have investigated PRV pathogenicity-related genes. This information will be valuable for controlling this re-emerging pathogen.
Because of its large genome, PRV has traditionally been genetically manipulated using bacterial artificial chromosome (BAC) techniques7. However, BAC mutagenesis is available only for virus isolates for which a useful BAC has been produced. In addition, during BAC system construction, the insertion of selection markers or parts of BAC plasmids into the viral genome may affect viral function. The clustered regularly interspaced palindromic repeats (CRISPR)/associated (Cas9) system was a revolutionary development in gene-editing technology8, 9. CRISPR/Cas9 can specifically break the targeted DNA with high efficiency and thereby cause indels in the target region via nonhomologous end joining (NHEJ) DNA damage repair or via homologous directed repair (HDR) in the presence of a homologous DNA donor8, 10. This method is simple, requiring only the design of an effective single-guide RNA (sgRNA) that is specific to a given target gene. Large genomic DNA viruses, such as PRV, can be edited easily, and the only delay involves determining the targeted sequence11–15.
In this study, we knocked out ten putative pathogenicity-related genes (glycoprotein E (gE), glycoprotein I (gI), Us2, Us9, thymidine kinase (TK), Us3, early protein 0 (EP0), glycoprotein M (gM), glycoprotein G (gG) and glycoprotein N (gN)) and compared the virulence of different mutants in a sensitive mouse model. We chose gE, gI, Us2 and Us9 because a classical live attenuated vaccine, Bartha-K, featured a large deletion that included these four genes16, 17. Additionally, TK, Us3, and EP0 have been reported to be correlated with classical PRV virulence18–21. We chose the envelope proteins gM, gG and gN because these genes have potential in the DIVA (differentiating infected from vaccinated animals) strategy. This study uncovered pathogenicity-related genes of re-emerging PRV variants for the first time and will provide a valuable reference for the control of PRV variants.
Materials and Methods
Cell lines and viruses
Vero cells were cultured in DMEM with 10% FBS and streptomycin/penicillin. The PRV HeN1 strain was the first PRV variant strain isolated in China and was isolated in our laboratory (GenBank accession number: KP098534.1). The properties of the HeN1 strain have been described previously2, 4. The Us2, Us3, Us9 and gE/gI knockout PRV strains were described in our previous reports12, 13.
Generation of CRISPR/Cas9 sgRNA constructs
Specific gene-targeted sgRNAs were designed using an online CRISPR Design Tool (https://wwws.blueheronbio.com/external/tools/gRNASrc.jsp), and the target regions were primarily located downstream of the start codons of the coding regions of specific genes. Generally, four sgRNAs were designed for each gene, and we selected only the most effective sgRNA for further specific gene knockout. The effectiveness of the sgRNAs was screened with a firefly luciferase-tagged recombinant virus, as described in our previous report13. Using this virus, if the sgRNA targets the PRV genome, the genomic DNA will be broken by Cas9, and luciferase expression will thereby decrease accordingly. The most effective sgRNAs identified after screening are listed in Table 1.
Table 1.
sgRNAs used in this study.
| sgRNA-gG | 5′-CACCGCCTCGCCCTCGGGCTCCTCG-3′ |
| 5′-AAACCGAGGAGCCCGAGGGCGAGGC-3′ | |
| sgRNA-gM | 5′-CACCGGCAACGCCGAGGCCGTGAGC-3′ |
| 5′-AAACGCTCACGGCCTCGGCGTTGCC-3′ | |
| sgRNA-gN | 5′-CACCGCTCTTCCATAGTCTTTTCCG-3′ |
| 5′-AAACCGGAAAAGACTATGGAAGAGC-3′ | |
| sgRNA-TK | 5′-CACCGCATCAGCGCGGCGGCCTTCG-3′ |
| 5′-AAACCGAAGGCCGCCGCGCTGATGC-3′ | |
| sgRNA-EP0 | 5′-CACCGTCTGGACGTCGCGGCCACCG-3′ |
| 5′-AAACCGGTGGCCGCGACGTCCAGAC-3′ |
PRV gene knockout using CRISPR/Cas9 technology
The gene knockout procedure was similar to that described in our previous reports12, 13. First, Vero cells were seeded in 12-well plates and transiently transfected 12 h later with the indicated CRISPR/Cas9 plasmids (2 μg per well). Then, 12 h post-transfection, PRV HeN1 was inoculated at an MOI of 0.01. At 48 h post-infection (hpi), the supernatants were collected for plaque purification. Several plaques were selected randomly for viral DNA extraction, and gene knockout was confirmed via DNA sequencing and western blot. The western blot procedure was similar as described previously12. The PCR primers used to assess gene knockout are listed in Table 2.
Table 2.
PCR primers used in this study.
| gG-F | 5′-ACCGCTACGACACCAAGGTC-3′ |
| gG-R | 5′-GCCGCCGTCAAAGAACCAG-3′ |
| gN-F | 5′-TACAATCGCCTGCACCTCGC-3′ |
| gN-R | 5′-AGGAGCCGTGGCCATCGTAG-3′ |
| gM-F | 5′-AAGAAGCTGGTCACGGTGGG-3′ |
| gM-R | 5′-AGCTGCGCGTTGATCGTGGC-3′ |
| TK-F | 5′-AAGCAGAACGGCAGCCTGAGCG-3′ |
| TK-R | 5′-GGGCACGGCAAACTTTATTGGGAT-3′ |
| EP0-F | 5′-CGCAGCGCCGCT TTCAGACCCA-3′ |
| EP0-R | 5′-GGAGCATGGCC TCGGTCAC-3′ |
Plaque purification
Vero cells were seeded in 60-mm dishes, and the indicated viruses were collected and subjected to serial 10-fold dilutions for the plaque-forming assay. At 2 hpi, the cells were washed three times with PBS and overlaid with 2% low-melting-point agarose (Lonza, USA) in DMEM medium containing 2% FBS. The dishes were further incubated at 37 °C for 3–5 days to allow plaques to form, and a single plaque was then purified.
In vitro growth properties of mutant viruses
Vero cells were infected with wild-type PRV HeN1 or the indicated gene knockout PRV mutants at an MOI of 0.01. The infected cells were collected at 12, 24, 36 and 48 hpi. Serial 10-fold dilutions of the indicated viruses were used to infect Vero cells. The viral titers at different times were recorded as the 50% tissue culture infection dose (TCID50).
Analysis of the virulence of mutant viruses
Six- to eight-week-old female SPF BALB/c mice were divided into 12 groups (5 mice per group), and each group was infected with HeN1 or the indicated PRV mutant via subcutaneous injection with 2 × 104 PFUs of virus or DMEM (100 μL). The results from two independent experiments are presented as two experiments together. All animal experiments were approved by the Animal Ethics Committee of Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences and were performed in accordance with animal use ethical guidelines and approved protocols.
Statistical analysis
Statistical analyses were performed using an ANOVA, as implemented in SPSS version 19.0. P < 0.05 was considered statistically significant.
Results
Knockout and replicative properties of ten putative virulence genes
In this study, we evaluated ten putative pathogenicity-related genes in mice. To create the indicated mutants, we applied CRISPR/Cas9 technology to rapidly knock out putative virulence gene(s). First, we designed sgRNAs and screened for effective sgRNAs, as described previously12–14. Second, we used the most effective sgRNA to knock out the corresponding gene. Finally, the sgRNA-treated PRV was purified via several rounds of plaque purification. The PRV mutations were confirmed via DNA sequencing (Fig. S), and the CRISPR/Cas9 knockout efficiency for all ten genes was determined, as shown in Table S. Western blot further confirmed that gE, gI, Us3, and EP0 were successfully knocked out (Fig. S (F to H)). For the TK knockout, it was previously reported that acyclovir could inhibit PRV containing an intact TK gene. Therefore, in this study, we used acyclovir to evaluate whether TK was successfully knocked out. As shown in Fig. S (I), acyclovir inhibited wild-type viral replication and had no effect on the triple-mutant TK HeN1 strain. For other genes, no antibody was available, so we were unable to examine these mutants at the protein level. However, it is known that CRISPR/Cas9 targeting of the 3′ region of an ORF will potentially result in the expression of truncated products, whereas targeting of the 5′ ORF will create alternative ATG translation start sites with the potential to produce protein products. In our study, we designed sgRNAs targeting the 5′ ORF. When we further analyzed alternative ATG translation start sites with the potential to produce protein products, we found only very small ORFs, which we do not believe could produce functional proteins. After the desired mutants were successfully prepared, we tested whether the replication of these mutants was influenced by the knockout of the indicated gene(s) in vitro. Our results indicated that the TK and Us3 null mutants had replication kinetics similar to wild-type PRV; however, other mutations affected viral replication to different degrees (Fig. 1). Of all the investigated mutants, the EP0-negative mutant replicated at the slowest rate (Fig. 1). This finding indicates that EP0 gene products are very important for viral replication.
Figure 1.
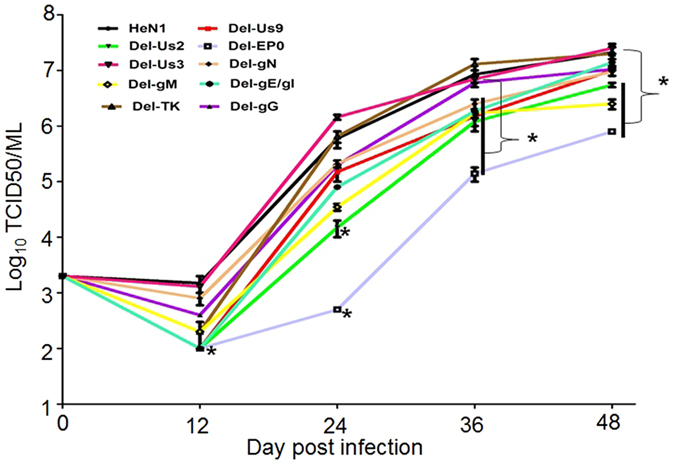
Replication kinetics of knockout viruses. Vero cells were infected with wild-type PRV HeN1 or the indicated gene knockout PRV mutants at an MOI of 0.01. The infected cells were collected at 12, 24, 36 and 48 hpi. The viral titers at different times were recorded as the 50% tissue culture infection dose (TCID50).
Pathogenic evaluation of ten putative virulence gene knockout mutants in mice
To investigate whether these putative virulence genes were critical for PRV pathogenicity, we used a susceptible mouse model. Although the TK null mutant replicated similarly to the wild-type PRV in vitro, this mutant completely lost its pathogenicity (Fig. 2). No mice died, and no clinical signs were observed in the TK groups. The gM gene was also significantly associated with virulence: only two of ten mice infected with mutants for this gene died (Fig. 2). The two dead mice manifested clinical signs similar to those observed in mice infected with wild-type PRV, but the clinical signs were delayed compared to those in the wild-type PRV. Most of the mutants, such as the gE, gI, Us9, Us3, gG, gN and EP0 mutants, exhibited no significant virulence reduction compared to that of the wild-type PRV (Fig. 2). However, interestingly, the Us2 mutant caused a more rapid onset of symptoms and earlier death than the wild-type PRV (Fig. 2).
Figure 2.
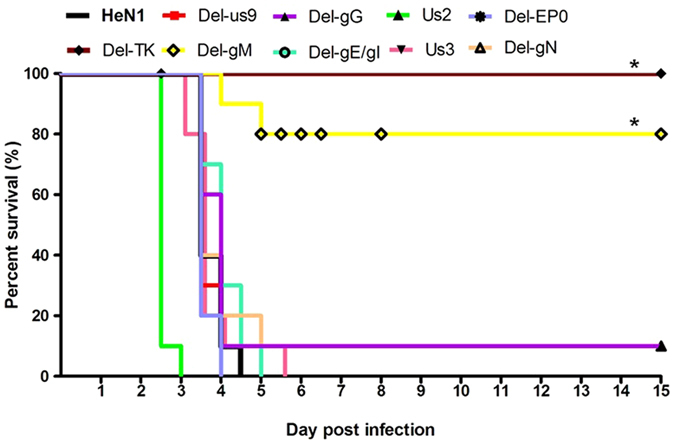
Pathogenicity of the indicated PRV mutants. Six- to eight-week-old female SPF BALB/c mice were divided into 11 groups, and each group was infected with HeN1 or the indicated PRV mutant via subcutaneous injection with 2 × 104 PFUs of each virus or DMEM (100 μL). Survival was recorded for 15 days.
Discussion
In this study, we chose sensitive mice to evaluate PRV pathogenesis. Among all PRV hosts, pigs are known to be the most insensitive to PRV infection, while mice and sheep are more sensitive to PRV infection. Thus, the use of pigs may not reveal a pathogenic difference. In another study that supports our opinions, mice infected with a gE/gI deletion mutant displayed high morbidity and mortality, whereas infected pigs remained clinically normal22. A similar result was achieved in a sheep experiment22. Another group in our laboratory also performed a pathogenicity experiment with gE/gI and TK deletion mutants. After being infected with these two mutants, all the pigs remained clinically normal, and no adverse reactions were observed (unpublished data). Therefore, we think that mice are more susceptible to PRV infection than pigs, and mice may be more suitable for evaluating the pathogenicity of PRV.
The TK gene is found only within Alphaherpesvirinae and Gammaherpesvirinae. This gene differs from cellular TK genes and plays a critical role in the synthesis of dTTP23. For classical PRV strains, TK-negative PRV mutants were highly attenuated in mice, rabbits and pigs and conferred protective immunity against PRV challenge in pigs18, 19. Consistent with classical PRV strains, the TK gene was also an important virulent gene in the re-emerging PRV variant. In mice sensitive to PRV infection, the TK knockout mutant completely lost its pathogenicity. This finding indicates that future vaccine development against PRV variants should better inactivate this gene.
gM is a nonessential glycoprotein that is conserved throughout the herpesvirus family. gM was found to inhibit PRV-induced membrane fusion by altering the membrane trafficking itineraries24, 25. gM and gN can form a disulfide-linked complex, and the gM mutant exhibited significantly decreased plaque size without impaired viral penetration26, 27. gM was also shown to be involved in different steps during virus secondary envelopment28. The gM mutant of the classical PRV strain exhibited significantly impaired viral virulence in piglets and conferred protection against a challenge infection29. Our study also showed that, among all the tested genes, the role of gM in virulence was second only to that of TK. More importantly, gM is an envelope protein, and the gM mutant can be used to differentiate infected from vaccinated animals, similar to gE-deleted vaccines. We propose that both the TK and gM knockout strains may represent promising candidate genetically marked vaccines for the future control of this re-emerging pathogen.
Since the late 1980s, the Bartha-K61 vaccine has been widely applied in China, resulting in effective control of PRV pandemics16. The Bartha-K61 vaccine was developed via multiple passages of a virulent field strain in cultured chicken cells and embryos23. Molecular and genetic analyses have identified a deletion of approximately 3 kb that encompasses Us8 (gE), Us9 and a large portion of Us7 (gI) and Us217, 30, 31. gE, gI and Us9 are associated with neurovirulence and are required for efficient anterograde spread in the nervous system32–35. The virulence of the Us9 null mutant was reduced in a rat model of eye infection32. However, in this study, the gE/gI mutant and the Us9 mutant manifested a virulence level similar to that of wild-type PRV (Fig. 2). This difference may be a result of the use of mice that were more sensitive to PRV infection. In a previous report, the virulence of a gE/gI mutant was evaluated in mice, sheep and pigs, and the gE/gI mutant was shown to be avirulent in pigs but virulent in mice and sheep22. From a biosafety perspective, the gE/gI deletion alone failed to generate a safe vaccine; the strain must be further attenuated. A limited number of studies have focused on Us2, which is a virion tegument component that is prenylated in infected cells36. Us2 binds to extracellular signal-regulated kinase (ERK), inhibiting the activation of ERK37. No previous reports have addressed the role of Us2 in virulence. Interestingly, we show for the first time that the Us2 null mutant caused an earlier onset of symptoms and earlier death than the wild-type virus. Why the Us2 null mutant exhibited increased pathogenicity needs to be explored further.
The PRV Us3 gene encodes a serine/threonine protein kinase that functions in multiple processes, including anti-apoptosis responses38, 39, cytoskeletal reorganization40, 41 and MHC-I downregulation42. Us3 is not required for growth in vitro, and the Us3 null mutant is slightly attenuated in mice, with a delayed onset of symptoms compared to that of the wild-type virus43. However, the Us3 null mutant exhibits strongly reduced virulence in pigs compared to that of the wild-type PRV20. In our report, we also obtained similar results in mice. Whether the Us3 null mutant exhibits reduced virulence in pigs must be explored further.
EP0 is expressed as an early protein in the PRV lifecycle, and this protein transactivates viral promoters, such as IE180, TK and gG44. In previous reports, an EP0 null mutant replicated slowly, exhibited significantly reduced virulence and elicited strong protective immunity against a lethal PRV challenge21, 45, 46. EP0 knockout attenuates PRV without affecting its immunogenicity, and such mutants were thought to be desirable vaccine candidates for PRV46. Although the EP0 mutant replicated slowly in vitro in previous reports, our results showed that the EP0 mutant replicated more slowly than other mutants, whereas the pathogenicity of the EP0 mutant was equivalent to that of wild-type PRV. This finding indicates that EP0 is not a critical virulence gene for this re-emerging PRV. Our result differed from previous reports, possibly because the PRV variant exhibited increased pathogenicity and the EP0 knockout alone may have a minimal effect on virulence in mice.
gG is the most abundant PRV protein found in the supernatant of PRV-infected cell cultures. gG binds to chemokines with high affinity, which in turn inhibits chemokine function, suggesting a role for gG in immune evasion47. In our study, we showed that gG is not a virulence gene in mice. However, because gG is involved in immune evasion, a candidate vaccine that contains a gG knockout may exhibit enhanced immunogenicity.
In conclusion, we evaluated ten putative virulence genes of the PRV variant and found that TK and gM were critical virulence genes in sensitive animals. The information obtained in this study will provide a valuable reference for scientists to effectively control PRV in the future.
Electronic supplementary material
Acknowledgements
This study was supported by grants from the National Program on Key Research Project (2016YFD0500100). We thank professor Chang-Jiang Weng of Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences for providing the EP0 antibody.
Author Contributions
Yan-Dong Tang, Zhi-Jun Tian and Xue-Hui Cai designed and wrote the text of the main manuscript, and Ji-Ting Liu, Tong-Yun Wang, Ming-Xia Sun prepared Figures 1 and 2. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-08269-3
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mettenleiter TC. Molecular biology of pseudorabies (Aujeszky’s disease) virus. Comparative immunology, microbiology and infectious diseases. 1991;14:151–163. doi: 10.1016/0147-9571(91)90128-Z. [DOI] [PubMed] [Google Scholar]
- 2.An TQ, et al. Pseudorabies virus variant in Bartha-K61-vaccinated pigs, China, 2012. Emerging infectious diseases. 2013;19:1749–1755. doi: 10.3201/eid1911.130177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu X, et al. Pathogenic pseudorabies virus, China, 2012. Emerging infectious diseases. 2014;20:102–104. doi: 10.3201/eid2001.130531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye C, et al. Genomic characterization of emergent pseudorabies virus in China reveals marked sequence divergence: Evidence for the existence of two major genotypes. Virology. 2015;483:32–43. doi: 10.1016/j.virol.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 5.Luo Y, et al. Pathogenicity and genomic characterization of a pseudorabies virus variant isolated from Bartha-K61-vaccinated swine population in China. Veterinary microbiology. 2014;174:107–115. doi: 10.1016/j.vetmic.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Tong W, et al. Emergence of a Pseudorabies virus variant with increased virulence to piglets. Veterinary microbiology. 2015;181:236–240. doi: 10.1016/j.vetmic.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 7.Zhang C, et al. Construction of a triple gene-deleted Chinese Pseudorabies virus variant and its efficacy study as a vaccine candidate on suckling piglets. Vaccine. 2015;33:2432–2437. doi: 10.1016/j.vaccine.2015.03.094. [DOI] [PubMed] [Google Scholar]
- 8.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pellagatti A, Dolatshad H, Valletta S, Boultwood J. Application of CRISPR/Cas9 genome editing to the study and treatment of disease. Archives of toxicology. 2015;89:1023–1034. doi: 10.1007/s00204-015-1504-y. [DOI] [PubMed] [Google Scholar]
- 11.Xu A, et al. A simple and rapid approach to manipulate pseudorabies virus genome by CRISPR/Cas9 system. Biotechnol. Lett. 2015;37:1265–1272. doi: 10.1007/s10529-015-1796-2. [DOI] [PubMed] [Google Scholar]
- 12.Tang YD, et al. Live attenuated pseudorabies virus developed using the CRISPR/Cas9 system. Virus research. 2016;225:33–39. doi: 10.1016/j.virusres.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Tang YD, et al. Recombinant Pseudorabies Virus (PRV) Expressing Firefly Luciferase Effectively Screened for CRISPR/Cas9 Single Guide RNAs and Antiviral Compounds. Viruses. 2016;8:90. doi: 10.3390/v8040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo JC, et al. Highly Efficient CRISPR/Cas9-Mediated Homologous Recombination Promotes the Rapid Generation of Bacterial Artificial Chromosomes of Pseudorabies Virus. Frontiers in microbiology. 2016;7:2110. doi: 10.3389/fmicb.2016.02110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang X, et al. A CRISPR/Cas9 and Cre/Lox system-based express vaccine development strategy against re-emerging Pseudorabies virus. Scientific reports. 2016;6:19176. doi: 10.1038/srep19176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y, et al. Control of swine pseudorabies in China: Opportunities and limitations. Veterinary microbiology. 2016;183:119–124. doi: 10.1016/j.vetmic.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Lomniczi B, Watanabe S, Ben-Porat T, Kaplan AS. Genome location and identification of functions defective in the Bartha vaccine strain of pseudorabies virus. Journal of virology. 1987;61:796–801. doi: 10.1128/jvi.61.3.796-801.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kit S, Kit M, Pirtle EC. Attenuated properties of thymidine kinase-negative deletion mutant of pseudorabies virus. American journal of veterinary research. 1985;46:1359–1367. [PubMed] [Google Scholar]
- 19.McGregor S, Easterday BC, Kaplan AS, Ben-Porat T. Vaccination of swine with thymidine kinase-deficient mutants of pseudorabies virus. American journal of veterinary research. 1985;46:1494–1497. [PubMed] [Google Scholar]
- 20.Kimman TG, De Wind N, De Bruin T, de Visser Y, Voermans J. Inactivation of glycoprotein gE and thymidine kinase or the US3-encoded protein kinase synergistically decreases in vivo replication of pseudorabies virus and the induction of protective immunity. Virology. 1994;205:511–518. doi: 10.1006/viro.1994.1672. [DOI] [PubMed] [Google Scholar]
- 21.Boldogkoi Z, Braun A, Fodor I. Replication and virulence of early protein 0 and long latency transcript deficient mutants of the Aujeszky’s disease (pseudorabies) virus. Microbes and infection. 2000;2:1321–1328. doi: 10.1016/S1286-4579(00)01285-5. [DOI] [PubMed] [Google Scholar]
- 22.Cong X, et al. Pathogenicity and immunogenicity of a gE/gI/TK gene-deleted pseudorabies virus variant in susceptible animals. Veterinary microbiology. 2016;182:170–177. doi: 10.1016/j.vetmic.2015.11.022. [DOI] [PubMed] [Google Scholar]
- 23.Pomeranz LE, Reynolds AE, Hengartner CJ. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiology and molecular biology reviews: MMBR. 2005;69:462–500. doi: 10.1128/MMBR.69.3.462-500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klupp BG, Nixdorf R, Mettenleiter TC. Pseudorabies virus glycoprotein M inhibits membrane fusion. Journal of virology. 2000;74:6760–6768. doi: 10.1128/JVI.74.15.6760-6768.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crump CM, et al. Alphaherpesvirus glycoprotein M causes the relocalization of plasma membrane proteins. The Journal of general virology. 2004;85:3517–3527. doi: 10.1099/vir.0.80361-0. [DOI] [PubMed] [Google Scholar]
- 26.Jons A, Dijkstra JM, Mettenleiter TC. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. Journal of virology. 1998;72:550–557. doi: 10.1128/jvi.72.1.550-557.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brack AR, Dijkstra JM, Granzow H, Klupp BG, Mettenleiter TC. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. Journal of virology. 1999;73:5364–5372. doi: 10.1128/jvi.73.7.5364-5372.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopp M, Granzow H, Fuchs W, Klupp B, Mettenleiter TC. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. Journal of virology. 2004;78:3024–3034. doi: 10.1128/JVI.78.6.3024-3034.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dijkstra JM, Gerdts V, Klupp BG, Mettenleiter TC. Deletion of glycoprotein gM of pseudorabies virus results in attenuation for the natural host. The Journal of general virology. 1997;78(Pt 9):2147–2151. doi: 10.1099/0022-1317-78-9-2147. [DOI] [PubMed] [Google Scholar]
- 30.Mettenleiter TC, Lukacs N, Rziha HJ. Pseudorabies virus avirulent strains fail to express a major glycoprotein. Journal of virology. 1985;56:307–311. doi: 10.1128/jvi.56.1.307-311.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrovskis EA, Timmins JG, Gierman TM, Post LE. Deletions in vaccine strains of pseudorabies virus and their effect on synthesis of glycoprotein gp63. Journal of virology. 1986;60:1166–1169. doi: 10.1128/jvi.60.3.1166-1169.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brideau AD, Card JP, Enquist LW. Role of pseudorabies virus Us9, a type II membrane protein, in infection of tissue culture cells and the rat nervous system. Journal of virology. 2000;74:834–845. doi: 10.1128/JVI.74.2.834-845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Card JP, Whealy ME, Robbins AK, Enquist LW. Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat visual system. Journal of virology. 1992;66:3032–3041. doi: 10.1128/jvi.66.5.3032-3041.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Husak PJ, Kuo T, Enquist LW. Pseudorabies virus membrane proteins gI and gE facilitate anterograde spread of infection in projection-specific neurons in the rat. Journal of virology. 2000;74:10975–10983. doi: 10.1128/JVI.74.23.10975-10983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kratchmarov R, Taylor MP, Enquist LW. Role of Us9 phosphorylation in axonal sorting and anterograde transport of pseudorabies virus. PloS one. 2013;8:e58776. doi: 10.1371/journal.pone.0058776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clase AC, et al. The pseudorabies virus Us2 protein, a virion tegument component, is prenylated in infected cells. Journal of virology. 2003;77:12285–12298. doi: 10.1128/JVI.77.22.12285-12298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang MH, Banfield BW. Pseudorabies virus tegument protein Us2 recruits the mitogen-activated protein kinase extracellular-regulated kinase (ERK) to membranes through interaction with the ERK common docking domain. Journal of virology. 2010;84:8398–8408. doi: 10.1128/JVI.00794-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deruelle MJ, De Corte N, Englebienne J, Nauwynck HJ, Favoreel HW. Pseudorabies virus US3-mediated inhibition of apoptosis does not affect infectious virus production. The Journal of general virology. 2010;91:1127–1132. doi: 10.1099/vir.0.015297-0. [DOI] [PubMed] [Google Scholar]
- 39.Geenen K, Favoreel HW, Olsen L, Enquist LW, Nauwynck HJ. The pseudorabies virus US3 protein kinase possesses anti-apoptotic activity that protects cells from apoptosis during infection and after treatment with sorbitol or staurosporine. Virology. 2005;331:144–150. doi: 10.1016/j.virol.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 40.Jacob T, Van den Broeke C, Van Waesberghe C, Van Troys L, Favoreel HW. Pseudorabies virus US3 triggers RhoA phosphorylation to reorganize the actin cytoskeleton. The Journal of general virology. 2015;96:2328–2335. doi: 10.1099/vir.0.000152. [DOI] [PubMed] [Google Scholar]
- 41.Van den Broeke C, et al. The kinase activity of pseudorabies virus US3 is required for modulation of the actin cytoskeleton. Virology. 2009;385:155–160. doi: 10.1016/j.virol.2008.11.050. [DOI] [PubMed] [Google Scholar]
- 42.Deruelle MJ, Van den Broeke C, Nauwynck HJ, Mettenleiter TC, Favoreel HW. Pseudorabies virus US3- and UL49.5-dependent and -independent downregulation of MHC I cell surface expression in different cell types. Virology. 2009;395:172–181. doi: 10.1016/j.virol.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 43.Olsen LM, Ch’ng TH, Card JP, Enquist LW. Role of pseudorabies virus Us3 protein kinase during neuronal infection. Journal of virology. 2006;80:6387–6398. doi: 10.1128/JVI.00352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe S, Ono E, Shimizu Y, Kida H. Pseudorabies virus early protein 0 transactivates the viral gene promoters. The Journal of general virology. 1995;76(Pt 11):2881–2885. doi: 10.1099/0022-1317-76-11-2881. [DOI] [PubMed] [Google Scholar]
- 45.Cheung AK, Fang J, Wesley RD. Characterization of a pseudorabies virus that is defective in the early protein 0 and latency genes. American journal of veterinary research. 1994;55:1710–1716. [PubMed] [Google Scholar]
- 46.Wesley RD, Cheung AK. A pseudorabies virus mutant with deletions in the latency and early protein O genes: replication, virulence, and immunity in neonatal piglets. Journal of veterinary diagnostic investigation: official publication of the American Association of Veterinary Laboratory Diagnosticians, Inc. 1996;8:21–24. doi: 10.1177/104063879600800104. [DOI] [PubMed] [Google Scholar]
- 47.Viejo-Borbolla A, Munoz A, Tabares E, Alcami A. Glycoprotein G from pseudorabies virus binds to chemokines with high affinity and inhibits their function. The Journal of general virology. 2010;91:23–31. doi: 10.1099/vir.0.011940-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


