Abstract
There is epidemiological evidence that H. pylori might predispose to Alzheimer’s disease. To understand the cellular processes potentially linking such unrelated events, we incubated the human gastric cells MNK-28 with the H. pylori peptide Hp(2-20). We then monitored the activated genes by global gene expression. The peptide modulated 77 genes, of which 65 are listed in the AlzBase database and include the hallmarks of Alzheimer’s disease: APP, APOE, PSEN1, and PSEN2. A large fraction of modulated genes (30 out of 77) belong to the inflammation pathway. Remarkably, the pathways dis-regulated in Alzheimer’s and Leasch-Nyhan diseases result dis-regulated also in this study. The unsuspected links between such different diseases – though still awaiting formal validation – suggest new directions for the study of neurological diseases.
Introduction
Alzheimer disease (AD) is a progressive, age-influenced neurodegenerative disease. AD can display an early or late onset depending upon the genome, diet and lifestyle of the patient1,2. The hallmark of both these forms of AD is the presence of neurofibrillary tangles (NFTs) of the phosphorylated protein tau and insoluble fibrils and plaques of the amyloid-β peptide (Aβ42)3. Early onset AD (EOAD) is a rare form of AD with a prevalence of 5.3 × 105 people at risk4. About 85% of the patients affected by EOAD display rare mutations in the amyloid precursor protein (APP) or the presenilin (PSEN1, PSEN2) loci4. Copy number variants (CNVs) have been detected in 21 unrelated EOAD patients with no mutations at the main APP or PSEN loci4. The more frequent late onset form of AD (LOAD) is associated with mutations of the apolipoprotein E (APOE) gene. The APOE-ε4 allele displays dosage effect: the proportion of affected subjects is 47% for heterozygotes (2/4 or 3/4) and 91% for homozygotes (4/4)5. The APOE-ε4 allele is a risk factor also for EOAD6. More recent studies have described 19 genes (11 of which are new) associated (P < 5 × 10−8) with LOAD7.
Helicobacter pylori (H. pylori) infection is limited to the human stomach8. This Gram-negative bacterium causes gastritis, peptic ulcer and more rarely gastric cancer. The life-time risks of developing ulcers or gastric carcinoma are 10–20% and < 1%, respectively8,9. H. pylori infection is also associated with non-gastric diseases: AD, Parkinson’s disease, atherosclerosis, and cardiovascular ischemia10–12. In the case of AD, two genetic association studies – both carried out on small numbers of patients of European ancestry – report an association between AD and H. pylori infection13,14. However, a larger study – carried out on Japanese patients - did not confirm the association15.
Recently, two of our patients with H. pylori infection16 manifested symptoms of AD. This observation stimulated the present study, aimed at detecting a potential biological link between H. pylori infection and AD. Case-control studies suffer from low replication17,18, resulting from confounding factors such as genetic heterogeneity19, pleiotropy20, population stratification21, or epistasis22. To test our hypothesis, we therefore opted to use the RNA sequencing technology (RNA-seq) that has become particularly attractive for gene expression studies because highly reproducible23. In addition, being independent of assumptions about the genes involved, RNA-seq can lead to the identification of new gene products or pathways.
The Hp(2-20) peptide - derived from the H. pylori ribosomal protein L124 - is a ligand of the formyl peptide receptors (FPRs) FPR1, FPRL1, and FPRL225. FPRs are seven transmembrane G protein-coupled receptors which regulate inflammation, a critical player in AD26. The gastric mucosal cell line MKN-28 expresses both the FPRL1 and FPRL2 proteins25. The Aβ42 peptide - the dominant component of amyloid plaques found in the brains of AD patients3 – is also a ligand for FPRL127. Strikingly, synthetic and secreted humanin peptides protect neural cells by inhibiting the access of Aβ42 to FPRL128. The FPRL1 ligands - in addition to Aβ42 and humanin- also include the host-derived agonists annexin A1 and lipoxin A427, which display strong anti-inflammatory activity and promote apoptosis and phagocytosis at the site of inflammation27. Notably, FPRL1 displays copy number variants associated with extreme forms of AD4.
Hp(2-20) upregulates the VEGF-A pathway expression at the mRNA and protein levels25 and activates the ERK and Akt pathways that in turn cooperate with the VEGF-A pathway29. VEGF-A plays a crucial role in mitigating neural injury and promoting neurogenesis and brain repair in AD patients29. The astrocytes from AD patients display increased VEGF-A immunoreactivity, which is interpreted as a compensatory mechanism countering the reduced vascularity occurring in AD patients30. At the same time, single nucleotide polymorphisms of the VEGF-A promoter that predispose to AD are also known31. Cumulatively, these traits make the MKN-28 cells and the Hp(2-20) peptide both well suited for tracing a potential thread connecting H. pylori infection with AD. Thus, MKN-28 cells were incubated with the peptide Hp(2-20) and the differentially expressed genes analyzed for known transcriptional associations with AD. The Hp(2-20) peptide induced the transcription of 5911 genes, of which 77 are listed in the AlzBase database.
Results
Identification of genes with altered expression levels
To understand the cellular processes potentially linking H. pylori infection and AD, we incubated the human gastric cell line MKN-28 with the peptide Hp(2-20) alone (condition A), the H. pylori growth broth alone (condition B), or with both the peptide and the growth broth (condition C). We then monitored the genes activated under these conditions by RNA-seq.
First, we performed a time course experiment to determine the optimal exposure time of MKN-28 cells to the conditions A, B, and C (Fig. 1). This preliminary pilot test was limited to some genes with a role in AD: FPR1, FPRL1, FPRL2, and CTSG. The latter codes for the cathepsin G, a protease cleaving the Aβ42 peptide from the APP precursor protein32 and a ligand for FPR133. From these experiments, we concluded that 1 h incubation time of MKN-28 cells is the optimal: it induces the expression of AD-related genes, mimicking this disorder at the cellular level without being cytotoxic (Fig. 2).
Figure 1.
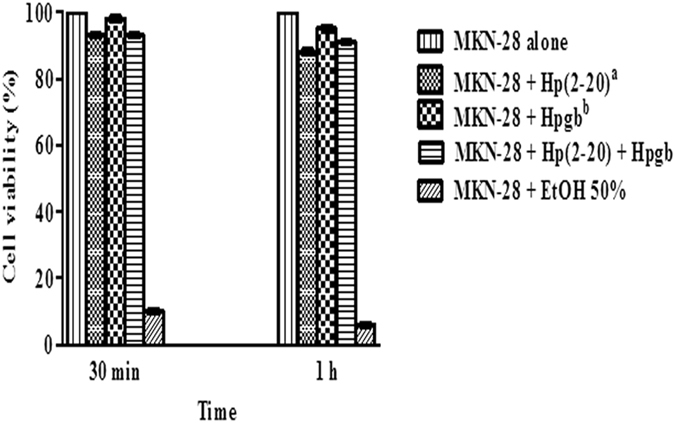
Analysis of MKN-28 cells viability. Results are representative of three independent experiments. Each value is the mean ± SD of three replicas. Statistical analysis was carried out with the GraphPad Prism version 5.03 (GraphPad, La Jolla, CA, USA). Cell viability was statistically significant (P < 0.001) in each case; aHp(2-20) (2 × 10–5 M); bHpgb = H. pylori growth broth (140 µl/well).
Figure 2.
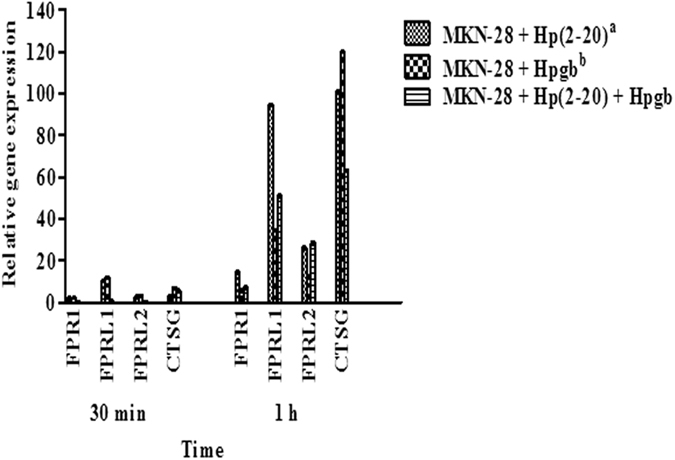
MKN-28 mRNAs levels of FPR1, FPRL1, FPRL2 and CTSG. Results are representative of three independent experiments. Each value is the mean ± SD of three replicas. Expression values were normalized against the human glyceraldehydes-3-phosphate dehydrogenase (GAPDH) gene. Stability assay, carried out using the BestKeeper tool, indicated that GAPDH was more stable then ACT-β at 30 (1.03 vs 2.89) and 60 min (1.89 vs 3.29). Statistical analysis was carried out with the GraphPad Prism version 5.03 (GraphPad, La Jolla, CA, USA). Differences in expression levels between 30 min and 1 h are all significantly different (P < 0.001); aHp(2-20) concentration was 2 × 10−5 M; bHpgb = H. pylori growth broth (140 µl/well).
Next, using RNA-seq, we measured the changes in gene expression of MKN-28 cells upon 1 h exposure to conditions A, B, and C. We identified 958 genes whose expression was affected by all the tested conditions. Hereafter we refer to these genes as “common”, while we name “unique” those whose expression changed exclusively upon exposure to one of the conditions. Specifically, we found 2066 genes unique to condition A, 2641 unique to condition B and 109 unique to condition C (Fig. 3). RNA-seq analysis was carried out on three biological replicates for each condition. Controls were untreated MKN-28 cells. Differential expression across the A, B, and C conditions, involved analysis of an average of 30 million reads for each sample; 90% of them mapped uniquely on the human reference genome. The total number of genes differentially expressed in conditions A, B and C were about 5900, 6500 and 1800, respectively (Fig. 3).
Figure 3.
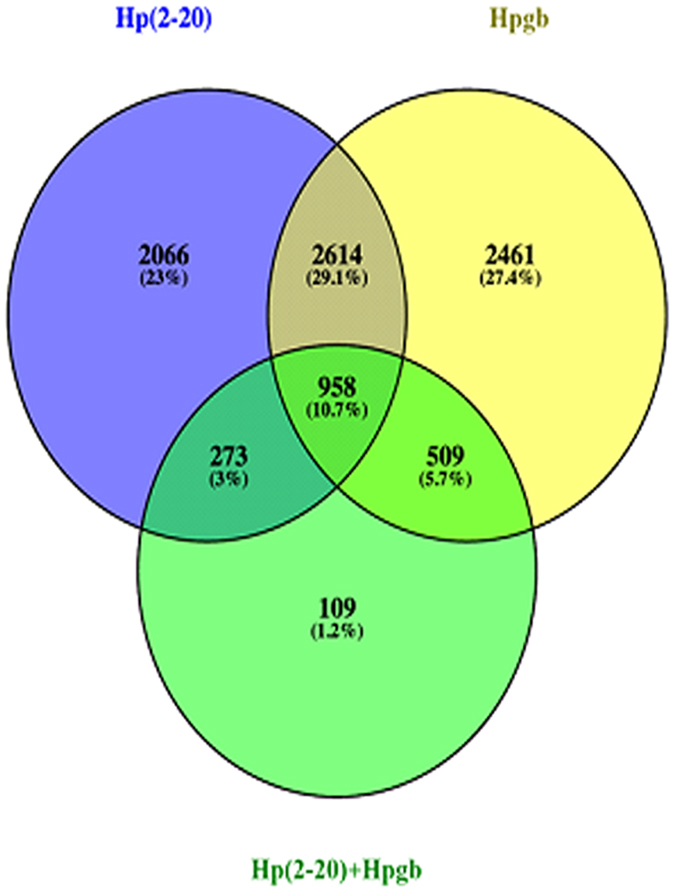
Venn diagram presentation of unique and common genes dysregulates in the presence of Hp(2-20) (A), the H. pylori growth broth (B), or both (C).
Common genes with altered gene expression
We first analyzed the common genes with MultiExperiment viewer (MeV) and the QT CLUST tools. The former displays single gene expressions under the three conditions (Fig. 4). The latter divides genes into six clusters on the basis of their similar trends in at least two conditions. In particular, the genes of clusters 3 and 4 are upregulated across conditions A, B, and C, while the genes of clusters 1 and 5 instead are downregulated across the same conditions. The genes of cluster 2 are downregulated in A and upregulated in B and C. On the contrary, the genes of cluster 6 are upregulated in A and downregulated in B and C (Fig. 5). The gene ontologies associated with each cluster are reported in Supplementary Table S1.
Figure 4.
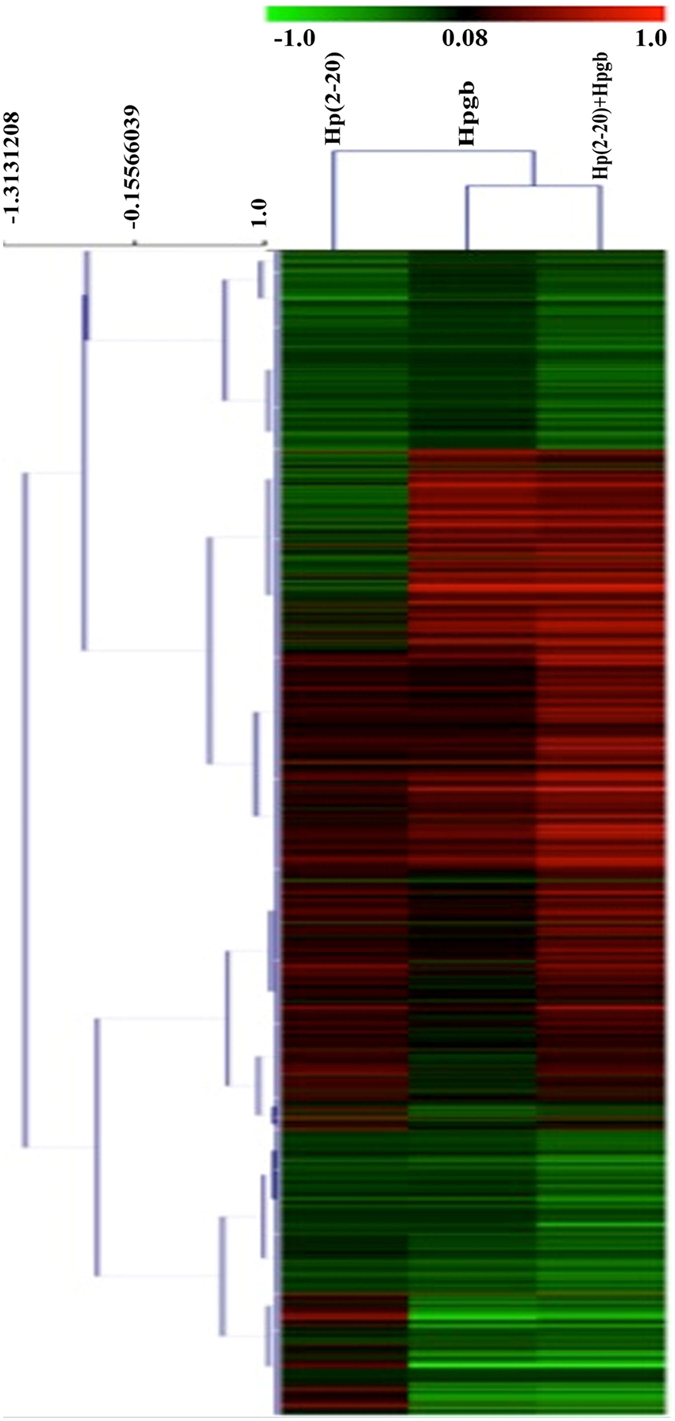
Heat map of the 958 common genes. Columns represent differences in expression levels (from green (down-regulated) to red (upregulated) in the presence of Hp(2-20) (A), the H. pylori growth broth (B), or both (C). Heat map and hierarchical clustering were obtained based on log2 fold-change.
Figure 5.
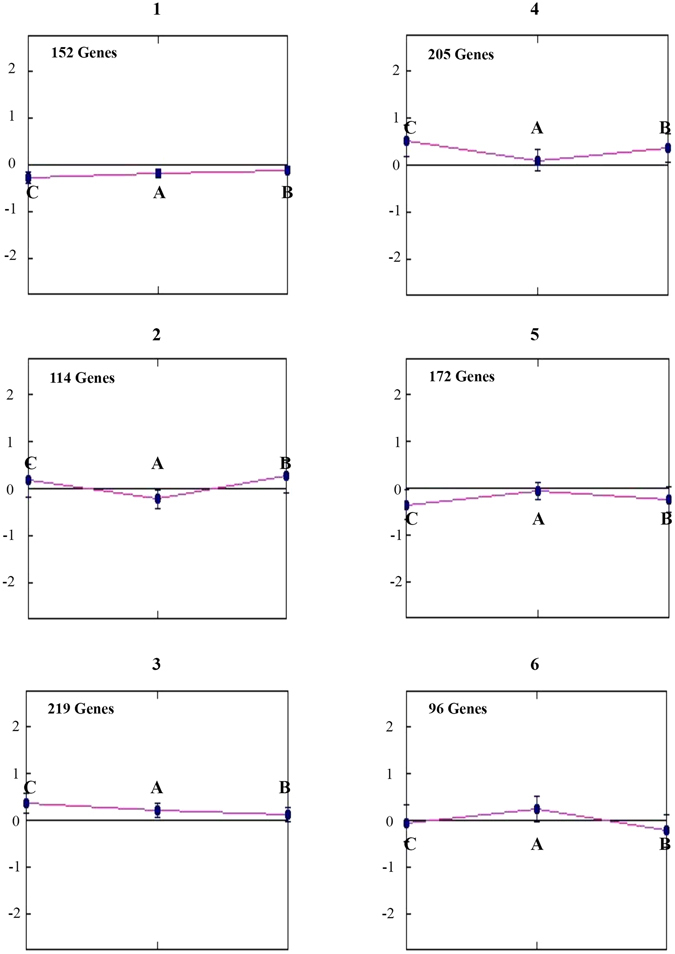
Classification of 958 common genes according to the QT-CLUST tool. Genes displaying similar trends in at least two conditions are clustered together. The letters A, B, and C indicate gene expressions in the presence of Hp(2-20), the H. pylori growth broth, or both, respectively.
Inspection of Fig. 5 shows that gene modulation induced by the Hp(2-20) peptide is clearly different when used alone (1, 3, 4, 5) or in combination with H. pylori growth broth (2, 6). On practical grounds, the above results clearly indicate that the concurrent use of two preconditioning factors might interfere with the correct understanding of single genes expression.
Unique genes displaying altered expression in the presence of the H. pylori growth broth
When the preconditioning factor was the H. pylori growth broth, the majority of the genes with modulated expression were associated with H. pylori infection (data not shown). The relatively few genes associated with AD represented either the Alzheimer’s disease-amyloid or the Alzheimer-disease presenilin pathways. However, these genes displayed limited interaction (Fig. 6). We then analyzed the data from cells preconditioned with Hp(2-20) alone.
Figure 6.
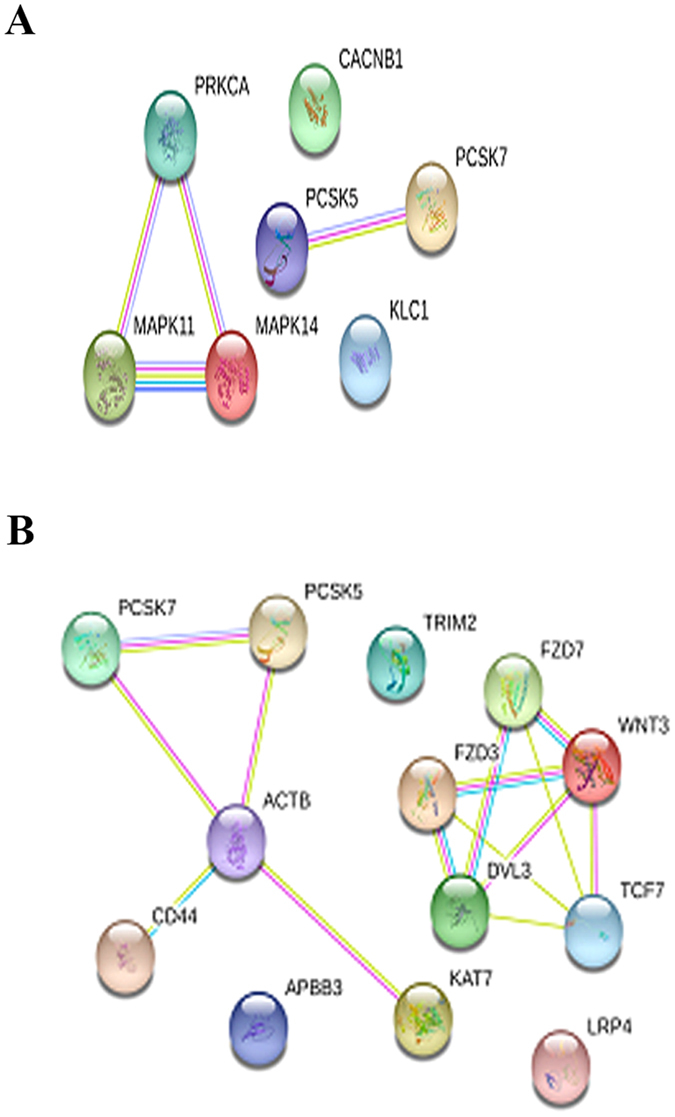
Network analysis of Alzheimer disease-amyloid secretase (A) and of Alzheimer-disease presenilin (B) genes catalogued in the Panther database and activated by the H. pylori growth broth. Lines indicate interactions between proteins (nodes).
Genes displaying altered expression in the presence of the Hp2-20 peptide
We identified 2066 genes which, following preconditioning with Hp(2-20), displayed a change in expression levels (data not shown). The list includes genes from 131 signaling pathways (Supplementary Table S2), some of which are indirectly relevant to AD (inflammation, angiogenesis, VEGF and Wnt signaling) (Table 1) and some more directly relevant (Alzheimer disease-amyloid secretase, and Alzheimer disease-presenilin) (Table 2). These pathways include 77 genes among which are the following hallmarks of AD: WNT10B, DKK1, and FZD5 (up-regulated); TCF7L2 and LRP6 (down-regulated); ANXA1, PSEN-1, PSEN-2, APOE, CTNNB1 (up-regulated); MTRNR2L2 (down-regulated).
Table 1.
Pathways represented among the 77 genes associated with AD and differentially expressed upon activation with Hp(2-20).
| Pathways | Genes |
|---|---|
| Alzheimer disease-amyloid secretase pathway (P00003) | PRKCI, MAPK8, MAPK13, ADAM9, MAPK3, ADAM17, PRKCQ, PKN2, APP, PSEN1, PSEN2, PRKACA |
| Alzheimer disease-presenilin pathway (P00004) | RBPJ, ACTL8, APBB2, FZD5, WNT10B, LRP6, ADAM17, NECTIN1, TCF7L2, PSEN1, PSEN2, NOTCH1, CTNNB1, CTNNA1, APP |
| Angiogenesis (P00005) | PRKCI, FRS2, RBPJ, RASA1, FZD5, MAPK8, PIK3CB, NRAS, WNT10B, SOS2, PRR5, PLA2G4A, MAP3K1, MAPK3, MAP2K4, SRC, PTPN, PLD2, PLCG2, RHOC, BRAF, TCF7L2, PRKCQ, PRKACA, NOTCH1, CTNNB1 |
| VEGF signaling pathway (P00056) | PRKCI, PIK3CB, NRAS, PRR5, PLA2G4A, MAPK3, PLCG2, BRAF, PRKCQ, PRKACA |
| EGF receptor signaling pathway (P00018) | PPP4C, PRKCI, PPP2CB, RASA1, RHOG, YWHAH, MAPK8, PIK3CB, NRAS, GAB1, SOS2, SPRY4, NEK1, MAPK13, MAP3K1, PPP6, MAPK3, MAP2K4, RASA2, PLCG2, BRAF, PRKCQ, CBLB, ERBB3, PRKACA |
| Wnt signaling pathway (P00057) | LRP6, PRKCI, PPP2CB, PRKACA, PRKCQ, ARRB1, ACTL8, GNAQ, FZD5, TCF7L2, WNT10B, CTNNB1, CTNNA1 |
| Inflammation mediated by chemokine and cytokine signaling pathway (P00031) | ARRB1, PRKACA, ACTL8, RHOG, GNAQ, REL, PIK3CB, NRAS, IFNAR1, NFKB1, ARPC1B, PDPK1, IKBKE, PLA2G4A, ROCK1, PRKX, GRK6, MAPK3, ARPC4, PLCG2, RELA, SOCS6, RHOC, BRAF, PTEN, JUNB, FPR1, FPR2, FPR3, ANXA1 |
*The FDR value of listed genes was <0.05.
Table 2.
Genes of Alzheimer disease-amyloid secretase and Alzheimer disease-presenilin pathways catalogued in the Panther database and differently expressed upon activation with Hp(2-20).
| Pathways name | Gene ID | Gene name | FDR | log2FC |
|---|---|---|---|---|
| Alzheimer disease-amyloid secretase pathway | PRKCI | Protein kinase C iota type | 3.11E-02 | −0.11 |
| MAPK8 | Mitogen-activated protein kinase 8 | 4.24E-02 | −0.15 | |
| MAPK13 | Mitogen-activated protein kinase 13 | 3.51E-02 | 0.10 | |
| ADAM9 | Disintegrin and metalloproteinase domain-containing protein 9 | 2.51E-02 | −0.08 | |
| MAPK3 | Mitogen-activated protein kinase 3 | 1.52E-02 | 0.23 | |
| ADAM17 | Disintegrin and metalloproteinase domain-containing protein 17 | 2.55E-02 | −0.26 | |
| PRKCQ | Protein kinase C theta type | 3.03E-02 | −0.22 | |
| PKN2 | Serine/threonine-protein kinase N2 | 1.73E-02 | −0.26 | |
| APP | Amyloid Beta Precursor Protein | 2.94E-02 | 0.04 | |
| PSEN1 | Presenilin 1 | 2.77E-02 | 0.35 | |
| PSEN2 | Presenilin 2 | 1.83E-02 | 0.49 | |
| PRKACA | Protein Kinase CAMP-Activated Catalytic Subunit Alpha | 3.50E-02 | 0.14 | |
| Alzheimer disease-presenilin pathway | RBPJ | Recombining binding protein suppressor of hairless | 2.48E-02 | −0.21 |
| ACTL8 | Actin-like protein 8 | 3.14E-02 | −0.48 | |
| APBB2 | Amyloid beta A4 precursor protein-binding family B member 2 | 2.99E-02 | −0.18 | |
| FZD5 | Frizzled-5 | 4.99E-02 | 0.10 | |
| WNT10B | Protein Wnt-10b | 2.72E-02 | 0.30 | |
| LRP6 | Low-density lipoprotein receptor-related protein 6 | 2.48E-02 | −0.22 | |
| ADAM17 | Disintegrin and metalloproteinase domain-containing protein 17 | 2.55E-02 | −0.26 | |
| NECTIN1 | Nectin-1 | 3.86E-02 | 0.10 | |
| PSEN1 | Presenilin 1 | 2.77E-02 | 0.35 | |
| PSEN2 | Presenilin 2 | 1.83E-02 | 0.49 | |
| NOTCH1 | Notch 1 | 1.21E-002 | 0.36 | |
| CTNNB1 | Catenin Beta 1 | 8.98E-03 | −0.18 | |
| CTNNA1 | Catenin Alpha 1 | 1.87E-02 | 0.30 | |
| APP | Amyloid Beta Precursor Protein | 2.94E-02 | 0.04 | |
| TCF7L2 | Transcription factor 7-like 2 | 3.14E-02 | −0.18 |
WNT10B activates the canonical WNTs/β-catenin signaling pathway34, while DKK1 - preventing LRP6 from interacting with WNTs35 – down-regulates the WNTs/β-catenin pathway. Attenuation of this pathway is known to favor the development of AD36. TCF7L2 codes for a key transcription factor of the WNT signaling pathway37. FZD5 is the receptor for the WNT5A ligand and participates in the β-catenin pathway induction38,39 (Fig. 7).
Figure 7.
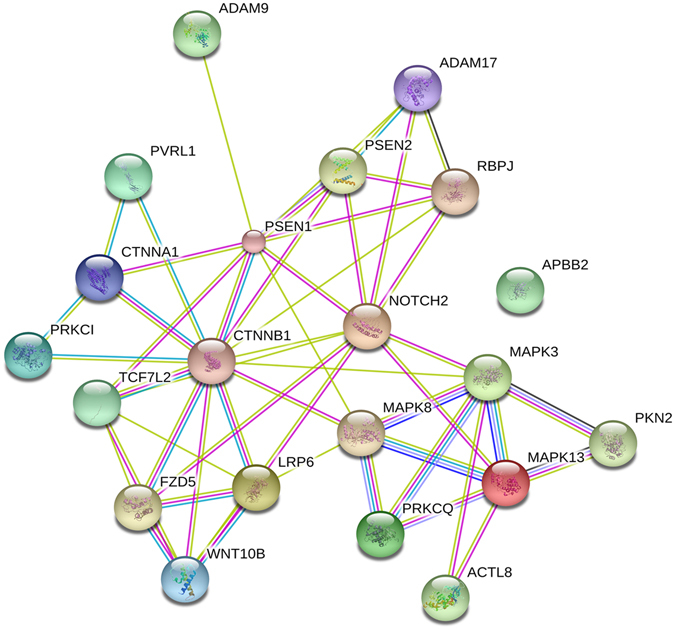
Network analysis of Alzheimer disease-amyloid secretase and of Alzheimer-disease presenilin genes catalogued in the Panther database and activated by Hp(2-20). Lines indicate interactions between proteins (or nodes).
ANXA1 exerts a strong local anti inflammatory activity40. APP, PSEN-1, and PSEN-2 mutations account for about 85% of EOAD cases4. The APOE-ε4 allele is a major risk factor for LOAD5. CTNNB1 is present in several key biological pathways highly relevant to AD41. MTRNR2L2 codes for the neuroprotective humanin protein, which in this study functions as a hub molecule for 17 molecules of the AD transcriptome (Fig. 8). FPR1 and FPRL1 are part of the AlzBase database and are both up-regulated in this study.
Figure 8.
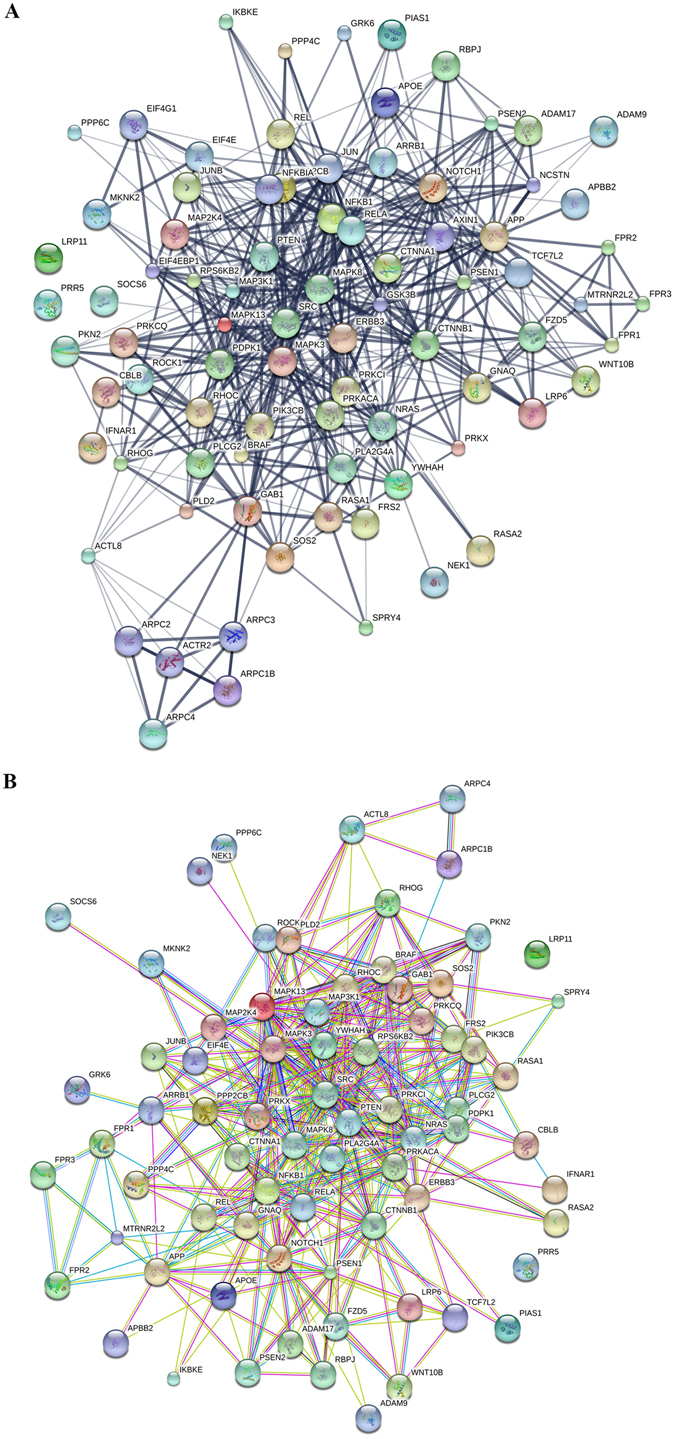
Network analysis of the gene pathways* catalogued in the Panther database, which - directly or indirectly - are connected with AD upon activation with Hp(2-20). Protein interactions were analyzed with the STITCH (A) and STRING (B) tools. The former catalogues known or predicted interactions between chemicals and proteins; the latter known or predicted protein-protein interactions. *List of the pathways included in the figure: Alzheimer disease-amyloid secretase pathway, Alzheimer disease-presenilin pathway, Angiogenesis, VEG-F signaling pathway, and Inflammation mediated by chemokine and cytokine signaling pathway.
Finally, unique and total genes activated with Hp(2-20) - the most interesting category of genes – were studied by the ingenuity pathway analysis (IPA). We found that the unique – but not the total genes - target the amyloid processing pathway among the top five. This finding confirms that the peptide preferentially targets AD genes. Instead, total and unique genes do not display major differences with respect to the “top diseases and bio functions category”, suggesting that the peptide can induce H. pylori infection genes as well (Table 3).
Table 3.
Pathways and functions activated by unique and total Hp(2-20) genes detected by IPA software.
| Hp(2-20) peptide | |||||
|---|---|---|---|---|---|
| Total | Unique | ||||
| Top Canonical Pathways | Top Canonical Pathways | ||||
| Name | p-value | Overlap | Name | p-value | Overlap |
| EIF2 Signaling | 5.05E-19 | 52.0% 115/221 | Insulin Receptor Signaling | 1.34E-06 | 22.0% 31/141 |
| Regulation of eIF4 and p70S6K Signaling | 1.22E-14 | 52.9% 83/157 | Tight Junction Signaling | 2.70E2.70E-06 | 20.4% 34/167 |
| mTOR Signaling | 6.16E-14 | 48.7% 97/199 | Amyloid Processing | 4.18E-06 | 31.40% 16/51 |
| Protein Ubiquitination Pathway | 2.82E-13 | 45.1% 115/255 | IGF-1 Signaling | 1.20E-05 | 22.6% 24/106 |
| Glucocorticoid Receptor Signaling | 3.35E-12 | 42.9% 123/287 | RAR Activation | 1.96E-05 | 18.4% 35/190 |
| Top Diseases and Bio Functions | Top Diseases and Bio Functions | ||||
| Diseases and Disorders | Diseases and Disorders | ||||
| Name | p-value | #Molecules | Name | p-value | #Molecules |
| Cancer | 1.50E-06-5.74E-101 | 4703 | Cancer | 7.67E-03-4.82E-28 | 1683 |
| Organismal Injury and Abnormalities | 1.67E-06-5.74E-101 | 4746 | Organismal Injury and Abnormalities | 7.67E-03-4.82E-28 | 1695 |
| Gastrointestinal Disease | 1.30E-06-1.75E-69 | 3977 | Gastrointestinal Disease | 7.67E-03-8.77E-17 | 1402 |
| Infectious Diseases | 1.93E-08-7.62E-31 | 796 | Infectious Diseases | 7.67E-03-1.08E-07 | 273 |
| Developmental Disorder | 8.60E-07-5.97E-23 | 644 | Hematological Disease | 7.67E-03-5.04E-05 | 151 |
Modulation of the D-proline pathway genes
Microbial metabolites can reach the brain through direct interaction with enteric neurons42. D-proline, a metabolite of H. pylori43, ranks first among the 20 metabolites associated with AD44.
D-proline occurs at significantly higher levels (17.4 vs 2.4 nm/ml) in the gastric juice of patients with H. pylori infection compared to healthy controls43. While predisposing to AD43,44, D-proline improves the cognitive decline of AD patients44. These results open the possibility of using bacteria producing D-proline to curb the cognitive decline of AD patients. We found that both the Hp(2-20) peptide and H. pylori culture broth dis-regulate 14 and 12 of the genes participating to the synthesis of D-proline; besides, 70% of these genes are up-regulated (Supplementary Table S3). Thus, D-proline shows that the same metabolite can influence two different traits of the same disease in opposite directions; this unique trait of D-proline can permit to understand how this can occur.
Genes dis-regulated in the Lesch-Nyhan and Alzheimer diseases
Several genes of the canonical WNT signaling, Alzheimer’s disease-presenilin, and Alzheimer disease-amyloid pathways are dis-regulated in both AD and Lesch-Nyhan disease45–47. Lesch-Nyhan is an incurable neurological disease caused by mutations of the hypoxanthine guanine phosphorybosyltransferase gene (HPRT)45. The canonical Wnt signaling pathway controls several aspects of vertebrate development (stem cell self-renewal, neurogenesis, and tumorigenesis)48 and has been associated with AD49. Dis-regulation of the Alzheimer’s disease-presenilin, and Alzheimer disease-amyloid pathways characterizes AD3,4. The former pathway also interferes with neural differentiation by stabilizing the β-catenin transcription48. Four of the 10 genes dis-regulated in Lesch-Nyhan disease and AD45 are also dis-regulated in the present study (Table 4). The lack of concordance in the transcriptional levels between the two studies very likely reflect differences in time-course.
Table 4.
Genes dis-regulated in LND1 and AD. A comparison between two studies.
| Gene Symbol | LND2 | AD3 | FDR | log2FC |
|---|---|---|---|---|
| APOE | Up | Up | 2.56E-02 | 0.78 |
| ADAM9 | Up | Down | 2.51E-02 | −0.08 |
| LRP11 | Up | Up | 4.31E-02 | 0.11 |
| PCLG2 | Up | Down | 3.71E-02 | −0.09 |
| CDK5R1 | Up | Nr4 | Nr4 | Nr4 |
| CAPN6 | Up | Nr | Nr4 | Nr4 |
| ADAMTS4 | Up | Nr | Nr4 | Nr4 |
| TNFRSF19 | Up | Nr | Nr4 | Nr4 |
| BACE2 | Up | Nr | Nr4 | Nr4 |
| PLCL2 | Down | Nr | Nr4 | Nr4 |
1Leasch-Nyhan disease; 2Reference 64; 3This study; 4Nr = normo-regulated.
Discussion
There is evidence – mainly inferred from apparently conflicting results between independent case-controls studies13–15 – that H. pylori infection might predispose to AD. To find a biologically plausible basis to this claim, we preconditioned the MNK-28 gastric cells with the Hp(2-20) peptide, the H. pylori culture broth or both and then interrogated the transcriptomes separately. To exclude possible artifacts deriving from interactions between bacterial components (please see the Results section), here we only discuss the data obtained with Hp (2-20) alone.
Why did we use the MNK-28 cells and Hp(2-20) peptide in the first place? First - to avoid confounding factors due to differences in immune response genes between hosts50,51 and the extreme differences existing between H. pylori strains52 - we opted for a less complex system: the transcriptome analysis of a human gastric cell line challenged with an H. pylori synthetic peptide. This experimental approach was also thought to facilitate replication of our results by independent workers. This when reproducibility is formally demanded in biomedical research53. Second, inflammation alters the permeability of the blood-brain barrier54,55 causing accumulation of the A42 peptide in the microglia cells (the macrophages residing in the brain)3,56. A42 is a regular resident in the brain where it exerts antimicrobial activity57,58, but – when in excess - stimulates inflammation and neuron apoptosis59. Thus, by activating the expression of the FPRL1 receptor on the MNK-28 cells with the Hp(2-20) ligand, we expected to probe two hallmarks of AD: inflammation and the Aβ42 plaque-forming process26,60.
How can H. pylori from the stomach affect the brain? There is credible evidence that the human microbiota communicates with the central nervous system through neural, endocrine, and immune pathways61. The proposed association of trimethylamine N-oxide (TMAO), succinic acid, mannitol44, and D-proline43 with AD is a demonstration that microbial metabolites might provoke AD. Also, it has been proposed that the immune response to H. pylori causes apoptosis and neural cell destruction by releasing pro-inflammatory molecules and inducing reactive oxygen metabolites51,62. There is evidence that H. pylori damages the brain-blood barrier63,64 and the gut metabolites reaches the brain through direct interaction with enteric neurons42. Microbial metabolites can also influence the peripheral immune response, which in sequence affects the blood- brain barrier64.
The peptide alone modulated 77 AD genes, of which 65 are listed in the AlzBase database (Table 5). This result excludes an experimental bias and proves the efficiency of the adopted RNA-seq approach. A large fraction of the modulated genes (30 out of 77) belong to the inflammation pathway (Table 1). This finding confirms what was anticipated above. Of the individual proteins that the 64 genes code for, here we discuss those deeply involved with AD and consequently more congruent with the objective of the present study.
Table 5.
Profiles of 65 AD genes catalogued in the AlzBase database.
| Gene | AlzBase | This study | Gene | AlzBase | This study | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sum | Up | Dn | Pe | FDR | Log2FC | Sum | Up | Dn | Pe | FDR | Log2FC | ||
| ADAM17 | 4 | 3 | 1 | 0 | 2.55E-02 | −0.26 | PDPK1 | 6 | 4 | 2 | 0 | 4.15E-02 | −0.13 |
| ADAM9 | 2 | 0 | 1 | 1 | 2.51E-02 | −0.08 | PIAS1 | 3 | 2 | 1 | 0 | 3.16E-02 | −0.18 |
| ANXA1 | 9 | 9 | 0 | 0 | 3.21E-02 | 0.13 | PIK3CB | 3 | 3 | 0 | 0 | 3.24E-02 | −0.12 |
| APBB2 | 3 | 0 | 3 | 0 | 2.99E-02 | −0.18 | PKN2 | 9 | 7 | 1 | 1 | 1.73E-02 | −0.26 |
| APOE | 4 | 4 | 0 | 0 | 2.56E-02 | 0.78 | PLA2G4A | 2 | 2 | 0 | 0 | 3.12E-02 | −0.17 |
| APP | 7 | 0 | 6 | 1 | 2.94E-02 | 0.04 | PLCG2 | 4 | 4 | 0 | 0 | 4.27E-02 | −0.2 |
| ARPC1B | 6 | 6 | 0 | 0 | 1.85E-02 | 0.18 | PLD2 | 5 | 2 | 3 | 0 | 2.83E-02 | 0.15 |
| ARPC4 | 3 | 0 | 3 | 0 | 2.86E-02 | 0.11 | PPP2CB | 4 | 0 | 4 | 0 | 3.79E-02 | −0.08 |
| ARRB1 | 9 | 5 | 3 | 1 | 2.29E-02 | 0.33 | PRKACA | 3 | 0 | 3 | 0 | 3.50E-02 | 0.14 |
| BRAF | 1 | 0 | 1 | 0 | 2.44E-02 | −0.34 | PRKCI | 4 | 4 | 0 | 0 | 3.11E-02 | −0.11 |
| CBLB | 8 | 6 | 1 | 1 | 3.19E-02 | −0.29 | PRKCQ | 3 | 0 | 3 | 0 | 3.03E-02 | −0.22 |
| CTNNA1 | 6 | 4 | 2 | 0 | 1.87E-02 | 0.3 | PRKX | 10 | 9 | 0 | 1 | 3.11E-02 | −0.2 |
| CTNNB1 | 1 | 0 | 1 | 0 | 8.98E-03 | −0.18 | PRR5 | 2 | 1 | 1 | 0 | 2.88E-02 | 0.27 |
| EIF4E | 5 | 0 | 5 | 0 | 2.05E-02 | −0.24 | PSEN1 | 2 | 1 | 1 | 0 | 2.77E-02 | 0.35 |
| ERBB3 | 7 | 6 | 1 | 0 | 4.24E-02 | −0.08 | PSEN2 | 6 | 0 | 4 | 2 | 1.83E-02 | 0.49 |
| FPR1 | 8 | 6 | 2 | 0 | 3.06E-02 | 0.12 | PTEN | 1 | 1 | 0 | 0 | 3.26E-02 | −0.11 |
| FPR2 | 4 | 2 | 2 | 0 | 2.93E-02 | 0.59 | RASA1 | 4 | 0 | 4 | 0 | 2.43E-02 | −0.19 |
| FZD5 | 3 | 3 | 0 | 0 | 4.99E-02 | 0.1 | RASA2 | 5 | 1 | 4 | 0 | 3.11E-02 | −0.28 |
| GAB1 | 8 | 5 | 3 | 0 | 2.48E-02 | −0.28 | RBPJ | 1 | 0 | 1 | 0 | 2.48E-02 | −0.21 |
| GNAQ | 4 | 4 | 0 | 0 | 2.53E-02 | −0.21 | REL | 2 | 1 | 1 | 0 | 2.42E-02 | −0.54 |
| JUNB | 7 | 7 | 0 | 0 | 1.95E-02 | 0.34 | RELA | 8 | 7 | 0 | 1 | 4.35E-02 | −0.09 |
| LRP11 | 5 | 3 | 2 | 0 | 4.31E-02 | 0.11 | RHOC | 6 | 6 | 0 | 0 | 2.82E-02 | 0.21 |
| LRP6 | 4 | 2 | 2 | 0 | 2.48E-02 | −0.22 | RHOG | 7 | 7 | 0 | 0 | 4.91E-02 | 0.11 |
| MAP2K4 | 11 | 1 | 10 | 0 | 2.48E-02 | −0.26 | ROCK1 | 5 | 5 | 0 | 0 | 3.11E-02 | −0.14 |
| MAP3K1 | 3 | 3 | 0 | 0 | 2.17E-02 | 0.23 | RPS6KB2 | 2 | 2 | 0 | 0 | 2.84E-02 | 0.15 |
| MAPK13 | 3 | 2 | 1 | 0 | 3.51E-02 | 0.1 | SOCS6 | 2 | 0 | 2 | 0 | 4.28E-02 | −0.16 |
| MAPK3 | 2 | 0 | 2 | 0 | 1.52E-02 | 0.23 | SOS2 | 3 | 3 | 0 | 0 | 2.41E-02 | −0.29 |
| MAPK8 | 9 | 2 | 6 | 1 | 4.24E-02 | −0.15 | SPRY4 | 6 | 1 | 5 | 0 | 2.85E-02 | 0.32 |
| MKNK2 | 10 | 10 | 0 | 0 | 3.03E-02 | 0.13 | SRC | 3 | 2 | 1 | 0 | 2.39E-02 | −0.18 |
| NEK1 | 5 | 2 | 3 | 0 | 2.73E-02 | −0.3 | TCF7L2 | 3 | 3 | 0 | 0 | 3.14E-02 | −0.18 |
| NFKB1 | 9 | 8 | 0 | 1 | 2.35E-02 | −0.15 | WNT10B | 1 | 1 | 0 | 0 | 2.72E-02 | 0.3 |
| NOTCH1 | 14 | 14 | 0 | 0 | 1.21E-02 | 0.36 | YWHAH | 13 | 1 | 12 | 0 | 3.52E-02 | 0.08 |
| NRAS | 3 | 0 | 3 | 0 | 2.44E-02 | −0.15 | |||||||
Sum: Total number of differential expression from all transcriptome studies of Alzheimer’s disease (AD). Up: Total number of up regulation from all transcriptome studies of AD. Dn: Total number of down regulation from all transcriptome studies of AD. Pe: Total number of dys-regulation with unknown direction from all transcriptome studies of AD.
Because of its multiple roles in AD, we discuss first (and in more detail) ANXA1. This molecule is an endogenous ligand of the FPRL1 receptor40 but - at the transcriptional level - is directly connected with all three FPRs (Fig. 8). ANXA1 exerts a strong anti-inflammatory activity by promoting the removal of apoptotic neurons without inducing pro-inflammatory molecules (TNF-α, IL-6, and NO), a condition that limits local inflammation and spares healthy neurons65,66. ANXA1 accomplishes this task by bridging microglia cells through the FPRL1 receptor and apoptotic neurons by recognizing on their surface the phosphotidylserine, the “eat me” signal40. ANXA1 thus supports the protective role of phagocytosis (removal of apoptotic neurons) and at the same time curbs the negative effects of phagocytosis (loss of healthy neurons). Thus, the upregulation of ANXA1 by the Hp(2-20) peptide (observed in this study) and the ANXA1 accumulation in the brain of AD patients40 might be interpreted as compensatory mechanisms -occurring in vitro and in vivo - aimed to attenuate the side effects of inflammation. These results - along with concurrent ones40 - suggest a potential use of ANXA1 for the treatment of AD. In a mouse model of AD, the human recombinant ANXA1 (hrANXA1) has already displayed the property of repairing the blood–brain barrier integrity damaged by the Aβ42 peptide67.
MTRNR2L2 and ARRB1 are two more neuroprotective proteins. In this study, the former is down-regulated and the latter up-regulated. The humanin (MTRNR2L2) protein is directly connected to seven proteins and indirectly to ten more, all part of the AD transcriptome (Fig. 8). MTRNR2L2 is a 24 aminoacid polypeptide expressed in the occipital area of the brain; it recruits microglia cells to the site of inflammation to clear activated neutrophils68; induces Ca++ mobilization; and exerts neuroprotective activity, presumably by competing with the neurotoxic Aβ42 peptide for the FPRL1 receptor68. The down-regulation of MTRNR2L2 observed in this study possibly reflects the characteristic of this gene to be highly expressed only in testis, kidney, skeletal muscles, and heart69. ARRB1 (β arrestin1) also displays neuroprotective activity during cerebral ischemic stress by regulating the BECN-dependent autophagosome formation70. In transgenic AD mice, ablation of the ARRB1 gene reduces brain damage71.
Given the strict relationship between microbiome and neurodegenerative diseases including AD, we would like to explain why we did not attempt to identify H. pylori in the gut microbiome of AD patients. The gut microbiome generally is used to identify bacterial communities. The large difference in the number of H. pylori positive samples among healthy individuals reported in gastric microbiome72 and gut microbiome73 studies argues that faecal samples might be inadequate for H. pylori identification. Thus, a negative result of the gut microbiome assay would not necessarily exclude the presence of the pathogen. Further, it is well known that the human microbiome influences distant organs such as the brain through chemical signalling44,74. Excluded the use of biopsies for ethical reasons, we attempted to trace the link between H. pylori and AD indirectly, looking how the pathogen and its metabolites affect genes associated with AD.
AD and Lesch-Nyhan disease share several dis-regulated genes from the canonical Wnt signaling, Alzheimer-amyloid, and Alzheimer-presenilin pathways (Table 4). These results have been interpreted as evidence on a link between the two diseases. More recent data have shown that alternative splicing generates nine isoforms from the single copy APP gene75. PCR and sequencing techniques have demonstrated that the isoforms contain deletions that could affect the stability and function of the APP protein75. Given that APP is one of the proteins implicated in both AD and Lesch-Nyhan diseases, it has been speculated that the genetic diversity originated by the alternative splicing mechanism could potentially explain the clinical diversity and complexity of these diseases.
The link connecting H. pylori and AD emerged clearly only when the Hp(2-20) was used alone. In combination with the H. pylori growth broth, the genes hallmarks of AD (APP, APOE, PSEN1, PSEN2, ANXA1, MTRNR2L2) remained silent. If the Hp(2-20) silencing here observed occurs also in vivo, then the risk of AD attributable to H. pylori infection is expected to be one of the factors contributing to this multifactorial disease76.
In conclusion, here we identified 77 genes, 65 of which are listed in the AlzBase database. Remarkably, the pathways that result dis-regulated in AD and Leasch-Nyhan diseases in one study68 are dis-regulated in this one as well. The above data lend biological plausibility to the hypothesis of a connection between H. pylori infection and AD. The FPRL1 receptor and its ligand Aβ42 are part of this connection. FPRL1 is expressed at high levels by the microglia cells infiltrating the brain tissue of AD patients77 and it has also been associated with AD4. The most difficult task will be to understand when inflammation and oxidant stress caused by the RPRL1- Aβ42 liaison is useful and when harmful to AD patients.
The unsuspected links between so different neurological diseases – though still awaiting formal validation – suggest new directions for these studies.
Materials and Methods
Bacteria and H. pylori growth broth
H. pylori strain ATCC 43504 was grown in 10 ml of liquid brain heart infusion medium (BHI; Oxoid, UK) supplemented with 10% foetal bovine serum (FBS; Oxoid), and incubated under microaerophilic condition generated by the CampyGen system (Oxoid) at 37 °C78. Bacteria were harvested at mid exponential phase, centrifuged (4.7 × 103 g; 5 min), filtered (0.22 µ) and added (140 µl/well; 30 min and 1 h) to growing MKN-28 cells.
Peptide
The cecropin-like peptide Hp(2-20) (sequence: NH2-AKKVFKRLEKLFSKIQNDK-COOH) corresponding to the amino-terminal part of the ribosomal protein L1 of H. pylori was synthesized by Innovagen (Lund, Sweden).
Cell culture
The human gastric adenocarcinoma MKN-28 cell line, (ATCC, MD, USA) was grown in RPMI medium (Gibco, Scotland) supplemented with 10% FBS, penicillin (100 IU/ml) and streptomycin (100 μg/ml) (both from Gibco, Paisley, Scotland) at 37 °C in a 5% CO2 atmosphere. The cells were then distributed in a 24-well plate (105 cells/well) (BD Falcon) and incubated (1 h) in the presence of the Hp(2-20) peptide (10−5 M)25, the H. pylori growth broth diluted 1:3 with serum-free medium, or both the peptide and the H. pylori growth broth.
RNA extraction and Quantitative Real-time PCR
Total RNA was extracted from individual wells according to the TRIzol reagent protocol (Gibco/BRL Life Technologies, Inc., Gaithersburg, MD) and then reverse-transcribed using the high-Capacity cDNA Reverse transcription kit (Applied Biosystem). Expression levels of the FPR1 (Hs04235426_s1), FPRL1 (Hs02759175_s1), FPRL2 (Hs00266666_s1), and CTSG (Hs01113415_g1) were measured by rt-PCR using the TaqMan PCR master 2X reagent (Applied Biosystem) and the Applied Biosystem iCycler according to the manufacturer’s protocol. PCR reactions were carried out in triplicate. The TaqMan assay probes were from Life Technologies (Monza, Italy). Expression values were normalized versus the untreated MKN-28 (control) cells. The reference gene was the housekeeping GAPDH. Stability assay was carried out using the BestKeeper tool79.
Transcriptome profiling with the RNA-Seq approach
Illumina reads were processed to remove adapter sequences and low quality bases (Phred score less than 25) by using Trimmomatic (version 0.33)80. The reads longer than 35 nt were retained for further analyses. Trimmed reads were then mapped against the human reference genome assembly (GRCh38.p3) from Ensembl version 82 with the program STAR (version 020201)81 The alignment files were filtered to retain the properly paired reads with a mapping quality higher than 30 by using SAMtools (version 1.2)82. Raw expression counts were then calculated by using featureCounts (version 1.4.6-p5)83. Raw counts were imported in R and, following TMM normalization, the lowly expressed genes were filtered out with the HTSFilter package84. Differential expression analysis of filtered genes was carried out with the NOISEQ package85. Gene Ontology Enrichment Analysis of differentially expressed genes was carried out using the human GO annotation from Ensembl version 82 and in-house-scripts. Significantly enriched GO categories were identified using the hyper geometric test.
Electronic supplementary material
Acknowledgements
The authors thank an anonymous referee for his insightful comments on the draft of the manuscript.
Author Contributions
R.C. conceived the experiments. A.F., F.Ca., F. Co. performed the experiments. F.Co., R.A.C., W.S., C.M. performed bioinformatic analysis. F.Ca., R.A.C. contributed reagents/materials. D.I., R.C., F.Co. wrote the paper. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Felice Contaldi and Federico Capuano contributed equally to this work.
A correction to this article is available online at https://doi.org/10.1038/s41598-018-23613-x.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-07532-x
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Domenico Iannelli, Email: iannelli@unina.it.
Rosanna Capparelli, Email: capparel@unina.it.
References
- 1.Lindsay J, et al. Risk factors for Alzheimer’s disease: a prospective analysis from the Canadian Study of Health and Aging. Am J Epidemiol. 2002;156(5):445–453. doi: 10.1093/aje/kwf074. [DOI] [PubMed] [Google Scholar]
- 2.Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430(7000):631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 4.Rovelet-Lecrux A, et al. A genome-wide study reveals rare CNVs exclusive to extreme phenotypes of Alzheimer disease. Eur J Hum Genet. 2012;20(6):613–617. doi: 10.1038/ejhg.2011.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corder EH, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 6.Chartier-Harlin MC, et al. Apolipoprotein E, epsilon 4 allele as a major risk factor for sporadic early and late-onset forms of Alzheimer’s disease: analysis of the 19q13.2 chromosomal region. Hum Mol Genet. 1994;3(4):569–574. doi: 10.1093/hmg/3.4.569. [DOI] [PubMed] [Google Scholar]
- 7.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45(12):1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19(3):449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kodaman N, et al. Human and Helicobacter pylori coevolution shapes the risk of gastric disease. Proc Natl Acad Sci USA. 2014;111(4):1455–1460. doi: 10.1073/pnas.1318093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan HJ, Goh KL. Extragastrointestinal manifestations of Helicobacter pylori infection: facts or myth? A critical review. J Dig Dis. 2012;13(7):342–349. doi: 10.1111/j.1751-2980.2012.00599.x. [DOI] [PubMed] [Google Scholar]
- 11.Tsang KW, Lam SK. Helicobacter pylori and extra-digestive diseases. J Gastroenterol Hepatol. 1999;14(9):844–850. doi: 10.1046/j.1440-1746.1999.01974.x. [DOI] [PubMed] [Google Scholar]
- 12.Wedi B, Kapp A. Helicobacter pylori infection in skin diseases: a critical appraisal. Am J Clin Dermatol. 2002;3(4):273–282. doi: 10.2165/00128071-200203040-00005. [DOI] [PubMed] [Google Scholar]
- 13.Kountouras J, et al. Relationship between Helicobacter pylori infection and Alzheimer disease. Neurology. 2006;66(6):938–940. doi: 10.1212/01.wnl.0000203644.68059.5f. [DOI] [PubMed] [Google Scholar]
- 14.Kountouras J, et al. Eradication of Helicobacter pylori may be beneficial in the management of Alzheimer’s disease. J Neurol. 2009;256(5):758–767. doi: 10.1007/s00415-009-5011-z. [DOI] [PubMed] [Google Scholar]
- 15.Shiota S, et al. The relationship between Helicobacter pylori infection and Alzheimer’s disease in Japan. J Neurol. 2011;258(8):1460–1463. doi: 10.1007/s00415-011-5957-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fulgione A, et al. Epistatic interaction between MyD88 and TIRAP against Helicobacter pylori. FEBS Lett. 2016;590(14):2127–37. doi: 10.1002/1873-3468.12252. [DOI] [PubMed] [Google Scholar]
- 17.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29(3):306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 18.Anonymous. Freely associating. Nat Genet22, 1–2 (1999). [DOI] [PubMed]
- 19.Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265(5181):2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 20.Fumagalli M, et al. Population genetics of IFIH1: ancient population structure, local selection, and implications for susceptibility to type 1 diabetes. Mol Biol Evol. 2010;27(11):2555–2566. doi: 10.1093/molbev/msq141. [DOI] [PubMed] [Google Scholar]
- 21.Zondervan KT, Cardon LR. Designing candidate gene and genome-wide case-control association studies. Nat Protoc. 2007;2(10):2492–2501. doi: 10.1038/nprot.2007.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Liu JS. Bayesian inference of epistatic interactions in case-control studies. Nat Genet. 2007;39(9):1167–1173. doi: 10.1038/ng2110. [DOI] [PubMed] [Google Scholar]
- 23.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18(9):1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pütsep K, Brändén CI, Boman HG, Normark S. Antibacterial peptide from H. pylori. Nature. 1999;398(6729):671–672. doi: 10.1038/19439. [DOI] [PubMed] [Google Scholar]
- 25.de Paulis A, et al. Helicobacter pylori Hp(2-20) promotes migration and proliferation of gastric epithelial cells by interacting with formyl peptide receptors in vitro and accelerates gastric mucosal healing in vivo. J Immunol. 2009;183(6):3761–3769. doi: 10.4049/jimmunol.0900863. [DOI] [PubMed] [Google Scholar]
- 26.Wyss-Coray T. Inflammation in Alzheimer disease: driving force, bystander or beneficial response? Nat Med. 2006;12(9):1005–1015. doi: 10.1038/nm1484. [DOI] [PubMed] [Google Scholar]
- 27.Dufton N, et al. Anti-inflammatory role of the murine formyl-peptide receptor 2: ligand-specific effects on leukocyte responses and experimental inflammation. J Immunol. 2010;184(5):2611–2619. doi: 10.4049/jimmunol.0903526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ying G, et al. Humanin, a newly identified neuroprotective factor, uses the G protein-coupled formylpeptide receptor-like-1 as a functional receptor. J Immunol. 2004;172(11):7078–7085. doi: 10.4049/jimmunol.172.11.7078. [DOI] [PubMed] [Google Scholar]
- 29.Greenberg DA, Jin K. From angiogenesis to neuropathology. Nature. 2005;438(7070):954–959. doi: 10.1038/nature04481. [DOI] [PubMed] [Google Scholar]
- 30.Kalaria RN, et al. Vascular endothelial growth factor in Alzheimer’s disease and experimental cerebral ischemia. Brain Res Mol Brain Res. 1998;62(1):101–105. doi: 10.1016/S0169-328X(98)00190-9. [DOI] [PubMed] [Google Scholar]
- 31.Del BR, et al. Vascular endothelial growth factor gene variability is associated with increased risk for AD. Ann Neurol. 2005;57(3):373–380. doi: 10.1002/ana.20390. [DOI] [PubMed] [Google Scholar]
- 32.Savage MJ, et al. Cathepsin G: localization in human cerebral cortex and generation of amyloidogenic fragments from the beta-amyloid precursor protein. Neuroscience. 1994;60(3):607–619. doi: 10.1016/0306-4522(94)90490-1. [DOI] [PubMed] [Google Scholar]
- 33.Vacchelli E, et al. Chemotherapy-induced antitumor immunity requires formyl peptide receptor 1. Science. 2015;350(6263):972–978. doi: 10.1126/science.aad0779. [DOI] [PubMed] [Google Scholar]
- 34.Luo J, et al. Wnt signaling and human diseases: what are the therapeutic implications? Lab Invest. 2007;87(2):97–103. doi: 10.1038/labinvest.3700509. [DOI] [PubMed] [Google Scholar]
- 35.Zorn AM. Wnt signaling: antagonistic Dickkopfs. Curr Biol. 2001;11(15):R592–R595. doi: 10.1016/S0960-9822(01)00360-8. [DOI] [PubMed] [Google Scholar]
- 36.Liu L, Wan W, Xia S, Kalionis B, Li Y. Dysfunctional Wnt/β-catenin signaling contributes to blood-brain barrier breakdown in Alzheimer’s disease. Neurochem Int. 2014;75:19–25. doi: 10.1016/j.neuint.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 37.Grant SF, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38(3):320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 38.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13(1):11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 39.Ring L, Neth P, Weber C, Steffens S, Faussner A. β-Catenin-dependent pathway activation by both promiscuous “canonical” WNT3a-, and specific “noncanonical” WNT4- and WNT5a-FZD receptor combinations with strong differences in LRP5 and LRP6 dependency. Cell Signal. 2014;26(2):260–267. doi: 10.1016/j.cellsig.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 40.McArthur S, et al. Annexin A1: a central player in the anti-inflammatory and neuroprotective role of microglia. J Immunol. 2010;185(10):6317–6328. doi: 10.4049/jimmunol.1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conejero-Goldberg C, et al. Molecular signatures in post-mortem brain tissue of younger individuals at high risk for Alzheimer’s disease as based on APOE genotype. Mol Psychiatry. 2011;16(8):836–847. doi: 10.1038/mp.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Kasper LH. The role of microbiome in central nervous system disorders. Brain Behav Immun. 2014;38:1–12. doi: 10.1016/j.bbi.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagata K, et al. L-Serine, D- and L-proline and alanine as respiratory substrates of Helicobacter pylori: correlation between in vitro and in vivo amino acid levels. Microbiology. 2003;149(Pt 8):2023–2030. doi: 10.1099/mic.0.26203-0. [DOI] [PubMed] [Google Scholar]
- 44.Xu R, Wang Q. Towards understanding brain-gut-microbiome connections in Alzheimer’s disease. BMC Syst Biol. 2016;10(Suppl 3):63. doi: 10.1186/s12918-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang TH, Friedmann T. Alzheimer’s disease shares gene expression aberrations with purinergic dysregulation of HPRT deficiency (Lesch-Nyhan disease) Neurosci Lett. 2015;590:35–39. doi: 10.1016/j.neulet.2015.01.042. [DOI] [PubMed] [Google Scholar]
- 46.Kang TH, Guibinga GH, Jinnah HA, Friedmann T. HPRT deficiency coordinately dysregulates canonical Wnt and presenilin-1 signaling: a neuro-developmental regulatory role for a housekeeping gene? PLoS One. 2011;6(1):e16572. doi: 10.1371/journal.pone.0016572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mastrangelo L, Kim JE, Miyanohara A, Kang TH, Friedmann T. Purinergic signaling in human pluripotent stem cells is regulated by the housekeeping gene encoding hypoxanthine guanine phosphoribosyltransferase. Proc Natl Acad Sci USA. 2012;109(9):3377–3382. doi: 10.1073/pnas.1118067109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet. 2004;5(9):691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 49.Inestrosa NC, Arenas E. Emerging roles of Wnts in the adult nervous system. Nat Rev Neurosci. 2010;11(2):77–86. doi: 10.1038/nrn2755. [DOI] [PubMed] [Google Scholar]
- 50.Fu S, et al. Increased expression and cellular localization of inducible nitric oxide synthase and cyclooxygenase 2 in Helicobacter pylori gastritis. Gastroenterology. 1999;116(6):1319–1329. doi: 10.1016/S0016-5085(99)70496-8. [DOI] [PubMed] [Google Scholar]
- 51.Peek RM, Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2(1):28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 52.Go MF, Kapur V, Graham DY, Musser JM. Population genetic analysis of Helicobacter pylori by multilocus enzyme electrophoresis: extensive allelic diversity and recombinational population structure. J Bacteriol. 1996;178(13):3934–3938. doi: 10.1128/jb.178.13.3934-3938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Munafò M. Metascience: Reproducibility blues. Nature. 2017;543(7647):619–620. doi: 10.1038/543619a. [DOI] [Google Scholar]
- 54.Abbott NJ, Rönnbäck L, Hansson E. Astrocyte-endothelial interactions at the blood-brain barrier. Nat Rev Neurosci. 2006;7(1):41–53. doi: 10.1038/nrn1824. [DOI] [PubMed] [Google Scholar]
- 55.Jaeger LB, et al. Lipopolysaccharide alters the blood-brain barrier transport of amyloid beta protein: a mechanism for inflammation in the progression of Alzheimer’s disease. Brain Behav Immun. 2009;23(4):507–517. doi: 10.1016/j.bbi.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deane R, et al. RAGE mediates amyloid-beta peptide transport across the blood-brain barrier and accumulation in brain. Nat Med. 2003;9(7):907–913. doi: 10.1038/nm890. [DOI] [PubMed] [Google Scholar]
- 57.Harris F, Dennison SR, Phoenix DA. Aberrant action of amyloidogenic host defense peptides: a new paradigm to investigate neurodegenerative disorders? FASEB J. 2012;26(5):1776–1781. doi: 10.1096/fj.11-199208. [DOI] [PubMed] [Google Scholar]
- 58.Soscia SJ, et al. The Alzheimer’s disease-associated amyloid beta-protein is an antimicrobial peptide. PLoS One. 2010;5(3):e9505. doi: 10.1371/journal.pone.0009505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rogers J, Luber-Narod J, Styren SD, Civin WH. Expression of immune system-associated antigens by cells of the human central nervous system: relationship to the pathology of Alzheimer’s disease. Neurobiol Aging. 1988;9(4):339–349. doi: 10.1016/S0197-4580(88)80079-4. [DOI] [PubMed] [Google Scholar]
- 60.Sastre M, Klockgether T, Heneka MT. Contribution of inflammatory processes to Alzheimer’s disease: molecular mechanisms. Int J Dev Neurosci. 2006;24(2-3):167–176. doi: 10.1016/j.ijdevneu.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 61.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13(10):701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 62.Kountouras J, et al. Alzheimer’s disease and Helicobacter pylori infection: Defective immune regulation and apoptosis as proposed common links. Med Hypotheses. 2007;68(2):378–388. doi: 10.1016/j.mehy.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 63.van de Haar HJ, et al. Blood-brain barrier impairment in dementia: current and future in vivo assessments. Neurosci Biobehav Rev. 2015;49:71–81. doi: 10.1016/j.neubiorev.2014.11.022. [DOI] [PubMed] [Google Scholar]
- 64.Main BS, Minter MR. Microbial Immuno-Communication in Neurodegenerative Diseases. Front Neurosci. 2017;11:151. doi: 10.3389/fnins.2017.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Coupade C, Ajuebor MN, Russo-Marie F, Perretti M, Solito E. Cytokine modulation of liver annexin 1 expression during experimental endotoxemia. Am. J. Pathol. 2001;159:1435–1443. doi: 10.1016/S0002-9440(10)62530-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Szabó C, Thiemermann C, Wu CC, Perretti M, Vane JR. Attenuation of the induction of nitric oxide synthase by endogenous glucocorticoids accounts for endotoxin tolerance in vivo. Proc. Natl. Acad. Sci. USA. 1994;91:271–275. doi: 10.1073/pnas.91.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park JC, et al. Annexin A1 restores Aβ1-42 -induced blood-brain barrier disruption through the inhibition of RhoA-ROCK signaling pathway. Aging Cell. 2016;16(1):149–161. doi: 10.1111/acel.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harada M, et al. N-Formylated humanin activates both formyl peptide receptor-like 1 and 2. Biochem. Biophys. Res. Commun. 2004;324:255–261. doi: 10.1016/j.bbrc.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 69.Bodzioch M, et al. Evidence for potential functionality of nuclearly-encoded humanin isoforms. Genomics. 2009;94(4):247–256. doi: 10.1016/j.ygeno.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 70.Wang P, et al. ARRB1/β-arrestin-1 mediates neuroprotection through coordination of BECN1-dependent autophagy in cerebral ischemia. Autophagy. 2014;10(9):1535–1548. doi: 10.4161/auto.29203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu X, et al. β-arrestin1 regulates γ-secretase complex assembly and modulates amyloid-β pathology. Cell Res. 2013;23:351–65. doi: 10.1038/cr.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He C, Yang Z, Lu N. Imbalance of Gastrointestinal Microbiota in the Pathogenesis of Helicobacter pylori-Associated Diseases. Helicobacter. 2016;21(5):337–348. doi: 10.1111/hel.12297. [DOI] [PubMed] [Google Scholar]
- 73.Human Microbiome Project Consortium . Structure function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. Understanding the role of gut microbiome-host metabolic signal disruption in health and disease. Trends Microbiol. 2011;19(7):349–359. doi: 10.1016/j.tim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 75.Nguyen KV. Epigenetic regulation in amyloid precursor protein and the Lesch-Nyhan syndrome. Biochem Biophys Res Commun. 2014;446(4):1091–1095. doi: 10.1016/j.bbrc.2014.03.062. [DOI] [PubMed] [Google Scholar]
- 76.Anand R, Gill KD, Mahdi AA. Therapeutics of Alzheimer’s disease: Past, present and future. Neuropharmacology. 2014;76(Pt A):27–50. doi: 10.1016/j.neuropharm.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 77.Le Y, et al. Amyloid (beta)42 activates a G-protein-coupled chemoattractant receptor, FPR-like-1. J Neurosci. 2001;21(2):RC123. doi: 10.1523/JNEUROSCI.21-02-j0003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fulgione A, et al. Lactoferrin Adsorbed onto Biomimetic Hydroxyapatite Nanocrystals Controlling - In Vivo - the Helicobacter pylori Infection. PLoS One. 2016;11(7):e0158646. doi: 10.1371/journal.pone.0158646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26(6):509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 80.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 84.Rau A, Gallopin M, Celeux G, Jaffrézic F. Data-based filtering for replicated high-throughput transcriptome sequencing experiments. Bioinformatics. 2013;29(17):2146–52. doi: 10.1093/bioinformatics/btt350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tarazona S, et al. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015;43(21):e140. doi: 10.1093/nar/gkv711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


