Abstract
The interactions of pathogens and phagocytes are complex. Our study demonstrated that Aeromonas hydrophila B11 can survive in the macrophagocytes of Tilapia mossambica. To explore the regulatory processes of A. hydrophila survival in the macrophagocytes, we used the mini‐Tn10 transposon mutagenesis system to build a mutant library by mixing Escherichia coli Sm10 (pLOFKm) and A. hydrophila B11. In total, 102 mutant colonies were detected, and 11 of them showed reduced survival in macrophagocytes. The mutant with the most severe phenotype, AM73, was chosen for further research. The ORF interrupted by mini‐Tn10 in AM73 was approximately 960 bp and was deposited in GenBank with the accession number SRP049226. The 319 amino acid protein encoded by the ORF showed a high degree of identity (89%) with proteins in the histone deacetylase/AcuC/AphA family of A. hydrophila subsp. hydrophila ATCC7966. A strain (AC73) in which the acuC mutation was complemented was constructed by generating the recombinant expression plasmid pACYC184‐acuC and introducing it into the AM73 mutant strain. Our experiments revealed that strain AM73 was deficient in biofilm formation, adhesion, survival in macrophagocytes, and virulence compared with A. hydrophila B11, and all of these biological properties were improved in strain AC73. The expression of 10 significant virulence genes was significantly inhibited in strain AM73. The results indicated that AcuC was an important regulatory protein contributing to the pathogenicity of A. hydrophila.
Keywords: Aeromonas hydrophila, histone deacetylase AcuC, intracellular survival, pathogenicity, phagocytes
1. Introduction
Aeromonas hydrophila, an opportunistic pathogen, is widespread in water, domestic animals, and various foods (Çiftci et al., 2016). A. hydrophila is reported to be the causative agent of several diseases (da Silva et al., 2012; Peyghan, 2010) in farmed and feral fish as well as gastrointestinal and extraintestinal diseases in humans (Janda & Abbott, 2010). A. hydrophila causes high mortality in aquaculture throughout the world and results in extensive economic losses (Reyes‐Becerril, López‐Medina, Ascencio‐Valle, & Esteban, 2011). Therefore, an increasing amount of research is being focused on the virulence of A. hydrophila.
The process of infection includes adhesion, colonization, invasion, proliferation in the host, and the secretion of toxins (Huang et al., 2016; Wang et al., 2015). Bacterial adhesion to host surfaces is one of the key steps in the initial infection process (Chen, Yan, Wang, Zhuang, & Wang, 2008; Luo et al., 2016), and many pathogens have been shown to develop the ability to adhere to their hosts (Huang et al., 2015; Lin et al., 2017; Qin, Lin, Chen, Xu, & Yan, 2016). During infection, pathogens are able to neutralize bactericidal and bacteriostatic mechanisms to survive and replicate in various host cells, including macrophagocytes (Beaz‐Hidalgo & Figueras, 2013). The interior of cells can protect pathogens from phagocytes and various antibiotics, which can lead to recrudescent infection (Garzoni & Kelley, 2009; Kaufmann, 2011). Therefore, the ability to survive in host phagocytes is an important virulence factor of pathogens (Qin et al., 2014). Biofilms can not only protect bacteria from host defense mechanisms, including phagocytosis, but also serve as a recalcitrant source of bacteria during antimicrobial therapy (Qin et al., 2014); therefore, biofilm formation is considered to be a virulence factor of pathogens. A. hydrophila has conventionally been considered an extracellular pathogen, but previous evidence has demonstrated that it can survive intracellularly (Qin et al., 2014). Other extracellular pathogens, including Staphylococcus aureus, have been shown to survive intracellularly (Foster, 2005; Thwaites & Gant, 2011). Currently, research on the genes that are conducive to the intracellular survival of nonobligate intracellular pathogens is limited.
The functions of the three‐gene operon acuABC are involved in acetoin catabolism (Grundy, Waters, Takova, & Henkin, 1993). Acetoin utilization A (AcuA) was deduced to be an acetyltransferase of the GNAT family (Sterner & Berger, 2000). The function of acetoin utilization B (AcuB) is unknown, but the acetoin utilization C (AcuC) protein and class I histone deacetylases (HDACs) were shown to be homologs (Thiagalingam et al., 2003). AcuA and AcuC are encoded by the acuABC operon, which comprises a protein acetylation/deacetylation posttranslational modification system to control the activity of acetyl‐coenzyme A (Ac‐CoA) synthetase in Bacillus subtilis (Leipe & Landsman, 1997). Acetyl‐CoA synthetase, a ubiquitous enzyme, is responsible for the reversible conversion of acetate to Ac‐CoA. The protein acetylation/deacetylation posttranslational modification is an efficient mechanism for controlling the activity of structural proteins, gene expression regulators, and enzymes in response to rapidly changing physiological conditions (Gardner, Grundy, Henkin, & Escalante‐Semerena, 2006). In Salmonella enterica, Acs activity is modulated by the protein acetylation/deacetylation system, which is critical to the synthesis of the acetyl‐AMP intermediate from acetate and ATP (Starai, Celic, Cole, Boeke, & Escalante‐Semerena, 2002). Studies of Escherichia coli demonstrated that the abundant lysine acetylation may modify or regulate the activities of many enzymes in pivotal metabolic processes and the synthesis of biological building blocks in response to changes in the environment (Gulick, Starai, Horswill, Homick, & Escalante‐Semerena, 2003). The conservation of AcuC suggested the possibility that the acuC gene might encode a protein acetylation/deacetylation posttranslational modification system in A. hydrophila. However, no studies of the acuC gene in A. hydrophila have been reported.
In this study, a mini‐Tn10 transposon mutagenesis system was used to build a mutant library, and a mutant defective in intracellular survival was chosen for further phenotypic analysis. The aim of this study is to explore the possible mechanisms by which A. hydrophila survives in macrophages.
2. Materials and Methods
2.1. Bacteria and culture conditions
The bacteria and plasmids used in this study are listed in Table 1. E. coli and A. hydrophila were cultured at 37°C and 28°C, respectively, in Luria‐Bertani medium. The bacteria were washed in phosphate‐buffered saline (PBS; pH 7.4) after 24 hr growth. The bacterial density was determined by OD550. Antibiotics were added to the medium at the following concentrations: 50 μg/ml ampicillin (Ap); 25 μg/ml chloromycetin (Cm); 100 μg/ml kanamycin (Km); and 50 μg/ml streptomycin (Sm).
Table 1.
Strains and plasmids
| Strains or plasmids | Description | Source |
|---|---|---|
| Strains | ||
| Aeromonas hydrophilaB11 | Wild‐type strain (SmR) | (Qin et al., 2014) |
| AM01~AM102 | Mini‐Tn10Km insertion mutant (SmRKmR) | Our study |
| AM73 (acuC−) | AcuC:: mini‐Tn10Km (SmRKmR) | Our study |
| AC73 (acuC+) | AM73 complemented with pACYC184‐AcuC (SmRKmRCmr) | Our study |
| E. coliSM10 | thithrleutonAlacYsupErecARP4 – 2 ‐ Tc:: Mu:: Km (λpir) | (Qin et al., 2013) |
| E. coliDH5α | F−, φ 80dlacZΔM15, ΔU169 (lacZYA‐argF), deoR, recA1endA1, hsdR17 (rK − ,mK + ), phoA, supE44, λ− , thi ‐1, gyrA96, relA1 | Takara (Qin et al., 2014) |
| Plasmids | ||
| pMD18 ‐ T | Cloning vector (ApR) | Takara |
| pLOF/Km | Tnl0‐connected transmit plasmid (KmRApR); | (Herrero et al., 1990) |
| pACYC184 | (CmRTcR) | afforded by Prof. Nie |
| pACYC184 ‐acuC | Recombination of pACYC184 and AcuC including the promoter and ORF (CmR) | Our study |
2.2. Preparation of Tilapia mossambica macrophage suspensions
Healthy Tilapia mossambica individuals were obtained from the market. The macrophages were prepared as previously described (Leung, Low, Lam, & Sin, 1995). In brief, the fish were incubated on ice to reduce activity, and head kidneys were removed, pooled under sterile conditions and then filtered with a 100 μm filter membrane. Leibovitz‐15 medium (Biological Industries, Israel) was used to suspend the samples, and heparin (10 IU/ml), streptomycin/penicillin (100 IU/ml), and 2% fetal calf serum were added. The cell suspension was layered over a 34%/51% discontinuous Percoll gradient. The cell suspension was centrifuged at 400g for 30 min at 4°C. The cells above the 34%/51% interface were collected, washed and resuspended in Leibovitz‐15 medium with 10% fetal calf serum, 100 IU/ml streptomycin/penicillin, and 10 IU/ml heparin. After the cell suspension was adjusted to 1 × 107 cells/ml, samples were transferred to six‐well plates (1 ml/well).
2.3. Preparation of mucus
Healthy Anguilla japonica (297.5 ± 17.6 g) were obtained from the aquaculture farm. Briefly, the skin mucus was prepared by scraping the surface of A. japonica (Balebona et al., 1998). The gill mucus was obtained from the gill arches of A. japonica by scraping the surface (Lumsden, Ostland, Byrne, & Ferguson, 1993). The intestinal mucus was obtained by scraping the surface of the intestine from A. japonica (Yan, Chen, Ma, Zhuang, & Wang, 2007). Mucus was mixed with sterile PBS after incubation at 4°C for 3 or 4 hr. The mixtures were centrifuged for 30 min at 20,000g at 4°C. The supernatants were filtered through 0.45 μm and 0.22 μm filters. Then, the protein concentrations of the mucus mixtures were adjusted to 1 mg/ml with sterile PBS using the method of Bradford to measure protein concentration (Bradford, 1976).
2.4. Invasion and survival assays of the wild‐type strain in macrophages in vitro
The assays were performed as described previously (Leung, Lim, Lam, & Sin, 1996). In step I, the concentration of bacteria was normalized using colony‐forming units (CFUs) as described by Jin & Pancholi (2006). The exact number of CFUs was determined by plating the bacterial culture on Luria‐Bertani agar plates and counting the resulting colonies. The macrophage suspension was added to six‐well plates and incubated for 2 hr. In step II, the bacteria (100 bacteria/cell) were added, and the mixture was incubated at 28°C for 1 hr. The cells were collected and centrifuged at 100g for 5 min at 28°C, and the supernatant was carefully removed. After the macrophages were washed, they were suspended in ice‐cold PBS. The macrophage suspensions received 250 μg/ml gentamycin and were incubated for 20 min at 4°C to remove residual extracellular bacteria, then washed with PBS. The supernatant fluid was withdrawn and plated on solid Luria‐Bertani medium to detect the bacteria in the supernatant. The cells were resuspended in fresh Leibovitz‐15 medium with 10 IU/ml heparin, 10% fetal calf serum and 100 IU/ml streptomycin/penicillin. In step III, the cell suspension was incubated at 28°C in 5% CO2 and sampled at 0 hr, 1 hr, 2 hr, 4 hr, 12 hr, and 24 hr. Cells were centrifuged for 5 min at 100g and 28°C. After the supernatant was removed, sterile distilled water was used to lyse the cells but not the intracellular bacteria, and the lysate was incubated for 30 min. The cell lysate was diluted 10‐fold with PBS. The dilutions of 10−3, 10−4, 10−5, 10−6, 10−7 were plated on solid Luria‐Bertani medium and cultivated at 28°C for 24 hr. The resulting bacterial colonies were observed and counted.
2.5. Mutagenesis of A. hydrophila
E. coli Sm10 with pLOFKm (a type of suicide vector) was mated with A. hydrophila strain B11 to introduce the mini‐Tn10Km transposon into the A. hydrophila (Herrero, de Lorenzo, & Timmis, 1990). In brief, E. coli Sm10 (mini‐Tn10Km) and A. hydrophila were mixed at a ratio of 1:4 on 0.22 μm filters and cultivated on tryptone soya agar plates with 3 mmol/L isopropyl β‐D‐1‐thiogalactopyranoside for 4 hr at 28°C. Then, the filters were eluted with trypticase soy broth, and the tryptone soya agar plates were incubated with kanamycin and streptomycin at 28°C for 24 hr. Single colonies were selected for further study of intracellular survival. The procedure was the same as described earlier, except the incubation at the end was 1 hr.
2.6. Southern blots
Southern blots were performed as described by Qin et al. (2014). In brief, the genomic DNA of A. hydrophila B11 and the mutant strains were extracted with a DNA extraction kit (TaKaRa, Japan). The DNA was digested with SacI (TaKaRa) restriction endonuclease, electrophoresed on a 1% agarose gel, and transferred to a nylon membrane (Liu, Mitsukawa, Oosumi, & Whittier, 1995). Using a digoxigenin‐deoxyuridine‐triphosphate‐tagged probe (Roche, Switzerland), blotting was performed to detect the mini‐Tn10Km transposon. A region of the KmR gene was amplified to be the probe. The length of the probe was 176 bp, and the primers used were FKm3: 57‐CGG GGA TCG CAG TGG‐37 and FKm4: 5'‐TGG GAA GCC CGA TGC‐3' with the DIG‐PCR probe synthesis kit (Roche). Hybridization was performed at 42°C for 16 hr followed by washing and immunological detection with a digoxigenin detection kit (Roche). The results with single bands for the mutants were as expected.
2.7. TAIL‐PCR
TAIL‐PCR was performed as previously described by Qin et al. (2014). The random primer was provided by a genomic walking kit (TaKaRa, Japan), and the nested primers were designed on mini‐Tn10 using the primer Premier Version 5.0 Tool (PREMIER Biosoft International, Palo Alto, CA) (Table 2). The first PCR was performed with the primers L1/R1 and the random primer. The product of the first PCR and the primers L2/R2 were used for the second PCR. The third PCR was performed with the primers L3/R3 and the product of second PCR as template. The thermal cycling conditions are listed in Table 3. The DNA products were purified and cloned into pMD18‐T (TaKaRa, Japan) for sequencing and then analyzed by BLAST, ClustalW and MegAlign (DNAStar) to determine the site interrupted by mini‐Tn10.
Table 2.
Specific primers of TAIL‐PCR
| Primer | Sequence (5′→3′) | Application |
|---|---|---|
| L1 | ATGCTTGATGGTCGGAAGAGGC | upstream sequences |
| L2 | CATCGGGCTTCCCATACAATCG | |
| L3 | ATTATCGCGAGCCCATTTATACCC | |
| R1 | CCTGTTGAACAAGTCTGGAAAGAAATG | downstream sequences |
| R2 | GATCTTGCCATCCTATGGAACTG | |
| R3 | TTACGCTGACTTGACGGGACGG |
Table 3.
The thermal cycling conditions of TAIL‐PCR
| No. | Thermal cycling conditions |
|---|---|
| 1st | |
| 94°C 1 min | 1 cycle |
| 98°C 1 min | 1 cycle |
| 94°C 30 s; 60°C 1 min; 72°C 150 s | 5 cycles |
| 94°C 30 s; 25°C 3 min; 72°C 150 s | 1 cycle |
| 94°C 30 s; 60°C 1 min; 72°C 150 s | 15 cycles |
| 94°C 30 s; 60°C 1 min; 72°C 150 s | |
| 94°C 30 s; 44°C 1 min; 72°C 150 s | |
| 72°C 10 min | 1 cycle |
| 2st | |
| 94°C 30 s; 62°C 1 min; 72°C 150 s | 15 cycles |
| 94°C 30 s; 62°C 1 min; 72°C 150 s | |
| 94°C 30 s; 44°C 1 min; 72°C 150 s | |
| 72°C 10 min | 1 cycle |
| 3rd | |
| 94°C 30 s; 60°C 1 min; 72°C 150 s | 15 cycles |
| 94°C 30 s; 60°C 1 min; 72°C 150 s | |
| 94°C 30 s; 44°C 1 min; 72°C 150 s | |
| 72°C 10 min | 1 cycle |
2.8. Construction of the complemented strain AC73
The acuC gene of wild‐type B11 was amplified with the primers acuC‐BamHI‐f: GGA TCC AGC TGC AAA ACT GGT ACA AG and acuC‐2HA‐SalI‐r: GTC GAC TTA CTA GAG GCT AGC ATA ATC AGG AAC TAC GGA TAG CCG TAG CGT TTG TTC G (the enzyme restriction sites are italicized and the hemagglutinin‐tag is bold). The purified acuC gene was digested by BamHI and SalI (TaKaRa) and ligated into pACYC184 to obtain pACYC184‐acuC. To construct the complement of the acuC mutation, we introduced pACYC184‐acuC into the mutant strain AM73. We selected the complemented AC73 strain on plates with chloromycetin and detected the expression of the AcuC protein by western blot (Merino, Rubires, Aguilar, & Tomás, 1996). In brief, the complemented AC73 strain and A. hydrophila B11 were heated to 100°C for 5 min. The mixture was centrifuged at 12,000g for 15 min at 4°C, and the supernatant was carefully collected to detect the expression of the AcuC protein in cells. The extracellular proteins were obtained as described by Munro, Hastings, Ellis, & Liversidge (1980). Briefly, the cultures of the complemented AC73 strain and A. hydrophila B11 grown in Luria‐Bertani medium were spread on sterile cellophane films placed on the surface of solid Luria‐Bertani medium plates. After incubation at 28°C for 48 hr, the bacterial cells were washed off the cellophane sheet with PBS and removed by centrifugation at 12,000 rpm at 4°C for 15 min. Supernatants were sterilized by filtration through a 0.22 μm filter (Sartorius Stedim Biotech, Germany). The intracellular and extracellular proteins were freeze‐dried, dissolved in sterile water, and stained with Coomassie brilliant blue to measure the concentration. The protein solutions were adjusted to the same concentration and heated at 100°C for 5 min. Then, 20 μl samples were separated on a 12% gel and transferred to a polyvinylidenedifluoride membrane. The membrane was blocked with 1% bovine serum albumin and incubated with a 1:1,000 dilution of anti‐hemagglutinin antibody. Membranes were washed and incubated with a 1:3,000 dilution of goat anti‐rabbit horseradish peroxidase‐conjugated secondary antibody, and the signal was detected using a chemiluminescent substrate (PerkinElmer, Waltham). Lysates of A. hydrophila B11 were used as controls.
2.9. Enzyme activity of the AcuC protein
The AcuC protein is a homologue of the histone deacetylase family of proteins. The activity of AcuC in the intracellular proteins of the wild‐type B11 and the mutant strain AM73 was measured. In this assay, the bacteria were washed twice with ice‐cold PBS and resuspended in sterile distilled water containing protease inhibitors. After being swollen for 60 min, the bacteria were subjected to a freeze‐thaw cycle. The homogenate was clarified by centrifugation at 8,000g for 30 min at 4°C; the supernatant was then lyophilized, and the proteins were dissolved in water at a concentration of 1 mg/ml. The Epigenase HDAC Activity Assay Kit (Epigentek, USA) was used to detect the enzymatic activity following the manufacturer's instructions. The proteins were incubated for 90 min with the assay buffer and with the substrate stably coated on the microplate wells. The wells were washed, and the capture antibody was added. Then, the wells were washed again, and the detection antibody and the solution for color development were added. Finally, the enzyme activity was measured colorimetrically by reading the absorbance at 450 nm using a microplate spectrophotometer. The activity of AcuC was proportional to the A450.
2.10. Bacterial biofilm formation assays of wild‐type strain B11, AM73, and AC73
Biofilm formation was assayed as previously described by Chan & Chua (2005) with some modification. Briefly, 50 μl of an overnight bacterial culture (OD590 of 0.2) and 150 μl Luria‐Bertani medium were added to a 96‐well microtiter plate. The plate was incubated at 28°C for 20 hr and washed with PBS and then fixed at 60°C. Two hundred microliters of 1% (wt/vol) crystal violet (Sigma, China) was added, and after 10 min at room temperature, samples were carefully washed with PBS. We added 200 μl of 33% (vol/vol) glacial acetic acid to solubilize the stain and then tested the extent of biofilm formation by reading the absorbance of the solution at 590 nm. The biofilm‐forming ability was calculated using the wild‐type strain as the control.
2.11. Bacterial adhesion assays
The assays were carried out as described by Ofek, Courtney, Schifferli, & Beachey (1986) with some modification. Briefly, 100 μl gill mucus, intestinal mucus or skin mucus was added to a microtiter plate and fixed by incubation overnight at 4°C; the unbound mucus was removed by washing the wells twice with 200 μl of sterile physiological saline (0.85% NaCl). Bacterial suspensions were adjusted to OD550=0.20 (≈108 CFU/ml), and equal volumes were added to the wells and incubated at 28°C for 2.5 hr. After washing the wells twice to remove the nonadhering bacteria, the microtiter plate was air‐dried at 60°C for 0.5 hr. Then, 300 μl calf serum was added to each well and incubated at 37°C for 1 hr. After being washed with PBS containing Tween‐20, the microtiter plate was incubated with a 1:100 dilution of anti‐Aeromonas hydrophila antiserum (prepared from rabbits in our laboratory) at 37°C for 1 hr. The microtiter plate was washed and incubated at 37°C for 1 hr with 200 μl of a 1:500 dilution of goat anti‐rabbit IgG‐horseradish peroxidase antibody. After the wells were washed, 100 μl fresh o‐phenylenediamine‐H2O2 was added to the wells, and the samples were allowed to react at room temperature for 0.5 hr in the dark. The reaction was terminated by adding 50 μl 2 mol/l H2SO4 to each well. The OD492 was measured using a microtiter plate reader (Thermo Scientific Varioskan Flash).
2.12. Bacterial survival assays of AM73, AC73, and the wild‐type strain
The assays were performed according to the procedure described for the invasion and survival assays in macrophages described above except that the cell suspensions were incubated at 28°C in 5% CO2 only for 0 hr and 1 hr. The results at 0 hr and 1 hr indicate the ability of the bacteria to invade and survive, respectively, in macrophages.
2.13. Microscopic analyses
To construct the fluorescence labeled wild‐type B11, the EX‐EGFP‐B01 (pReceiver‐B01 with enhanced green fluorescent protein) was introduced into the bacteria and measured by direct visualization with an inverted microscope using UV for excitation (Leica DM‐4000B). The invasion and survival assays were performed as described above. The cell suspension was incubated at 28°C in 5% CO2 for 0 hr and 1 hr and observed with a confocal microscope to evaluate the status of A. hydrophila B11 in cells.
2.14. Bacterial infection assays
Infection of Danio rerio was performed as described by (Neely, Pfeifer, & Caparon (2002). The infection was carried out using wild‐type B11, AM73, and AC73, and PBS was used as a control. Briefly, the bacterial concentration was adjusted to OD550=0.20 (108 CFU/ml). The Danio rerio were incubated on ice to reduce their activity. Twenty fishes per group were injected intraperitoneally using an ultrafine insulin syringe to inject 30 μl of PBS or suspensions of B11, AM73, or AC73 into each fish. The head kidneys of infected fishes were homogenized in PBS and spread on Luria‐Bertani agar plates. The identity of individual colonies was verified by 16S rRNA gene sequence analysis. The log‐rank test was used to evaluate mortality.
2.15. Quantitative real‐time PCR
To test the effect of the acuC mutation on other significant virulence genes, quantitative real‐time reverse transcription‐PCR was performed to quantify the expression of the outer membrane protein genes (ompA and ompTS), the type Ш secretion system gene (ascV), pilin and flagellar family protein genes (traA, flgE, flgL, and pilB), the serine peptidase gene (degQ), the type VI secretion system‐related protein gene (vash), the lipopolysaccharide gene (rfaF), the adhesin gene (cblA), and the aerolysin and hemolysin genes (hlyA and aerA) in A. hydrophila B11 and the mutant, AM73, as described by Lü et al. (2015) with some modifications. Total RNA was isolated from A. hydrophila B11 and AM73 with Trizol. The first‐strand cDNA was synthesized from the total RNA using the PrimeScript® RT kit. The acuC gene in the wild‐type strain was detected by PCR with the cDNA as the template. The sequences of 13 virulence genes were obtained from the NCBI. The primers designed using Premier 5.0 software are listed in Table 4. The analysis was performed on the Step One Plus Real‐Time PCR system (ABI, USA) using SYBR green I fluorescent dye. The reactions were performed in a 10 μl volume containing 0.2 μl SYBR Green I, 5 pmol/l primers and approximately 50 ng cDNA. The cycling parameters were 95°C for 10 min, followed by 45 cycles of 95°C for 20 s, 55°C for 20 s, and 72°C for 20 s. Threshold cycles and dissociation curves were determined with Rotor Gene 6,000 software, and the gene expression levels were normalized to those of 16S rRNA (Kong et al., 2015).
Table 4.
Primers designed for qRT‐PCR
| Gene | Primer | sequence (5′ → 3′) |
|---|---|---|
| acuC | Ac‐F | AGGACGATGCCTACCTCACC |
| Ac‐R | GCGTTTGTTCGCCTCTTCA | |
| cblA | Ad‐F | CCGAGGCGTTCTATGTGCA |
| Ad‐R | TTGGTCAGGTAGCCGGTGAT | |
| aerA | Ae‐F | GGTCTGTGGCGACAAGTATCG |
| Ae‐R | AGAGCAGACAGAGTCGGTATTTCTC | |
| ascV | As‐F | GGGTATTCACCTGCGTTTCA |
| As‐R | GATGTTCATTAGCGACCCACA | |
| hlyA | Hl‐F | CCGCCCAGTCCTTCATCTAT |
| Hl‐R | AGGGTCCGTAGGCTCACATT | |
| ompA | Om‐F | CTCACGATCTGGGTGACTTTG |
| Om‐R | CGCCGTTGATGGACTTGA | |
| ompTS | Pr‐F | AATGGCTCCTTCCCTGATCG |
| Pr‐R | TGGCACCCTGGTTCTCGTAA | |
| vash | Va‐F | AAACTGGCACGGGGAAAGAG |
| Va‐R | GCTTGTAAGGTGAGCGGCATAT | |
| traA | Tr‐F | GTGATGGTCGTCGCCTTTCT |
| Tr‐R | GATAACCTTCTCCGCATTTTCC | |
| pilB | Pi‐F | CTAATGCGAATGCAGCACGTA |
| Pi‐R | CGCTTCAACAGTTCCAACCA | |
| rfaf | Rf‐F | TACCTGGCACTGGCCTATCC |
| Rf‐R | CTCGTCGAGGTGCTTTTGTG | |
| degQ | De‐F | CACCGAGCTTACCTCCGAAAT |
| De‐R | CCGCCTTCTTCAGCGTGAC | |
| flgE | Fe‐F | CCCGCTCAGACATTGGAGAT |
| Fe‐R | GTCGCATTGCTGTAGGTCGC | |
| flgL | Fl‐F | GCCCCAGAACAACAACATCC |
| Fl‐R | GCCGCATCCTCTTTTGACA | |
| 16s | 16s‐F | GGGGAGTACGGTCGCAAGAT |
| 16s‐R | CGCTGGCAAACAAGGATAAGG |
2.16. Statistical analysis
The results were statistically analyzed by ANOVA using SPSS18.0 and are reported as the means ± SD. A value of p < .05 indicated a significant difference between samples.
3. Results
3.1. Invasion and survival assays of the wild‐type bacteria in macrophages in vitro
To validate the idea that A. hydrophila could invade and survive in macrophages, we analyzed the invasion and survival of A. hydrophila B11 in macrophages. The data indicated that 2.0 × 105 CFU/ml A. hydrophila B11 invaded the cells, and 1.5 × 105 CFU/ml survived in the macrophages after 1 hr. The survival at 12 hr and 24 hr was 28% and 2.1%, respectively, of the invading bacteria (Figure 1). Thus, we determined that A. hydrophila B11 had the ability to invade macrophages, and some bacteria survived more than 24 hr.
Figure 1.
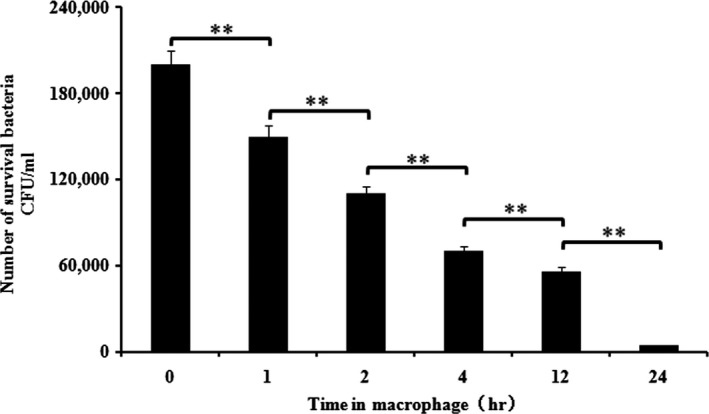
The survival of A. hydrophila B11 in macrophages. The X‐axis represents the time in the macrophages, and the Y‐axis represents the number of surviving bacteria per ml of cells. The number of bacteria that survived in the macrophages gradually decreased with time. However, 2.1% of the invasive bacteria still survived after 24 hr. The data are expressed as the mean ± standard deviation and were statistically analyzed using SPSS18.0. The differences between the mean values were determined by an analysis of variance (ANOVA). Values denoted by different numbers of asterisks were significantly different when compared by ANOVA (“**” p < .01; “*” p < .05)
3.2. The isolation of mutant strains
To screen for crucial virulence genes of A. hydrophila, a mutagenesis library was constructed by introducing the mini‐Tn10Km transposon on the suicide plasmid pLOFKm into A. hydrophila B11. The mutants were selected on tryptone soya agar containing 100 μg/ml kanamycin and 50 μg/ml streptomycin. Eleven colonies that exhibited significant declines in intracellular survival were selected from 102 mutants. The survival rates of the 11 mutant strains were lower than that of the wild‐type B11 (p < .01) (Figure 2a). The survival rates of AM78, AM73, AM71, AM16, AM10, and AM9 were lower than those of the other mutants. These mutants have been shown to carry mutations of flgE, acuC, asmA, glpC, ahHr, and merR, respectively. The AM73 mutant, which had the lowest survival among the mutants, was chosen for further examination, and the remaining mutants were studied in our other research.
Figure 2.
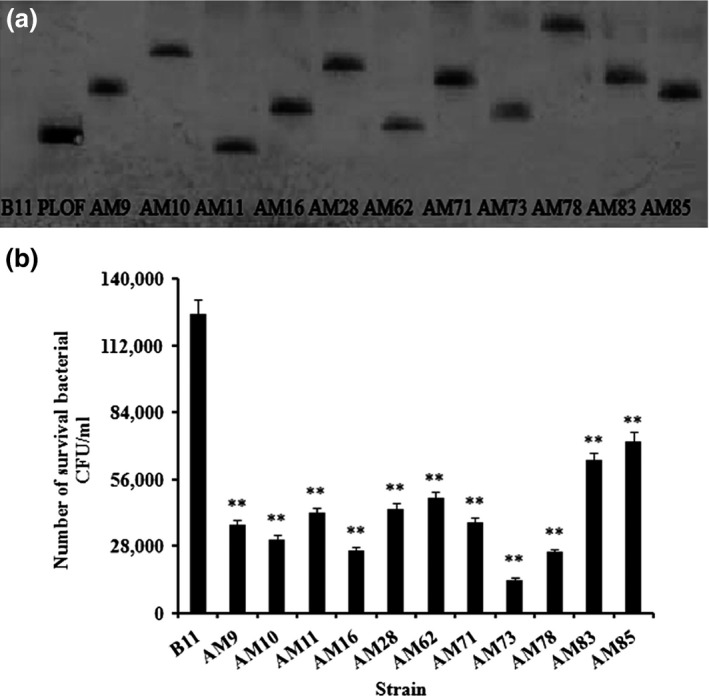
(a) Southern blotting of DNA from wild‐type B11 and mutant strains after digestion with restriction enzymes. The DNA extracted from wild‐type B11 and the mutant strains was digested with PstI. The restricted DNAs were electrophoresed, transferred to nitrocellulose, and hybridized to mixed DIG‐High Prime. The single band indicated that only a single mini‐Tn10 transposon was inserted in the mutants. (b) The number of surviving mutant bacteria in 1 ml of cells was analyzed 1 hr after invasion of the macrophages. Wild‐type B11 was the control. Values denoted by different numbers of asterisks were significantly different when compared by ANOVA (“**” p < .01; “*” p < .05)
3.3. Southern blot analysis
Southern blots were performed to detect the number of mini‐Tn10Km transposons inserted into the bacterial genome. The mutants and the positive control pLOFKm showed only one copy of the transposon, whereas the wild‐type strain B11 showed no transposon signal (Figure 2b). Thus, we determined that the insertion of a single copy of the transposon mini‐Tn10Km could lead to a mutation.
3.4. Identification of the acuC gene
To identify the gene that was mutated in AM73, a series of experiments were performed. The sequence 1,500 bp upstream of the inserted transposon and the sequence 2,000 bp downstream of the inserted transposon were analyzed. The results showed that the acuC gene, including the 960 bp ORF, was interrupted by mini‐Tn10 (GenBank accession numbers: SRP049226) (Figure 3). The protein encoded by the ORF was deduced to be 319 amino acids, and it showed the highest identity (89%) with the histone deacetylase/AcuC/AphA family proteins of A. hydrophila subsp. hydrophila, ATCC 7966.
Figure 3.

The nucleotide sequence alignment of the ORFs of A. hydrophila B11 to Aeromonas hydrophila subsp. hydrophila ATCC 7966. The initiation codon is shown in bold, and the insertion site is indicated in red arrow
3.5. Complementation
To validate the effects caused by the mutation in AM73, acuC with a hemagglutinin‐tag and pACYC184 were modified to generate the pACYC184‐acuC recombinant expression plasmid, which was introduced into the mutant strain AM73 to restore the expression and function of acuC. We assessed the AcuC protein in the extracellular and intracellular proteins of AC73 by western blot with the intracellular proteins of B11 as the control. The size of AcuC is approximately 33.02 kDa. The corresponding band was absent in the extracellular proteins of B11, but it was present in the intracellular proteins of AC73. The band probed with anti‐hemagglutinin antibody only appeared in the intracellular proteins of AC73 because the hemagglutinin‐tag is located on the pACYC184‐acuC recombinant plasmid (Figure 4). These results confirmed the integrity of acuC in AC73 and the AcuC protein as an intracellular protein in A. hydrophila.
Figure 4.
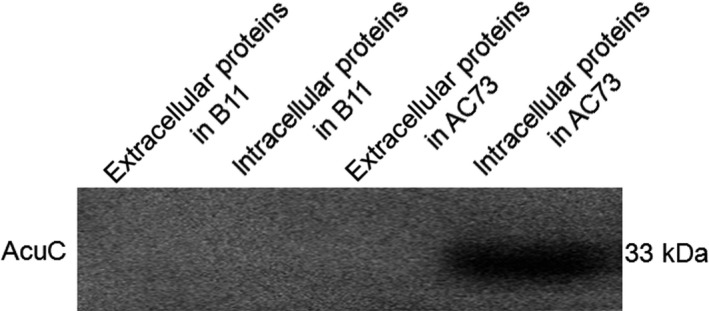
Western blot analysis showing a band of approximately 33 kb exclusively in the intracellular protein of AC73. The intracellular and extracellular protein of the wild‐type B11 was used as the control. Each lane was loaded with 20 μg protein
3.6. Enzyme activity
To assess the function of acuC in A. hydrophila, the corresponding enzyme activity was measured. The activity of the intracellular protein in the B11 strain was threefold higher than the enzyme activity in AM73 (Figure 5). This result demonstrated that the AcuC protein had HDAC enzyme activity.
Figure 5.
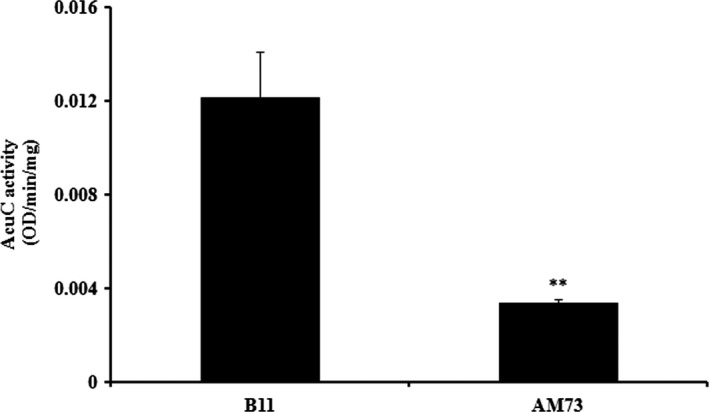
The HDAC enzyme activity in the intracellular proteins of the wild‐type B11 and the mutant AM73 was measured. The protein concentration was 1 mg/ml. Values denoted by different numbers of asterisks were significantly different when compared by ANOVA (“**” p < .01; “*” p < .05)
3.7. Biofilm formation
To ascertain whether acuC could affect other virulence factors as well as survival, biofilm formation by wild‐type B11, AM73, and AC73 was quantified by absorbance readings (A590). We found that the biofilm formation by AM73 was only 18.75% of that of the wild‐type B11 and that the biofilm formation of the complemented strain was 56.25% of that of the wild type (Figure 6a). The extent of biofilm formation can be determined based on the depth of color generated by adding glacial acetic acid to the slides. The color of the B11 strain and the complemented strain AC73 was darker than that of AM73 (Figure 6b). This result demonstrated that acuC affects the biofilm formation of A. hydrophila and that acuC might be involved in the regulation of multiple virulence factors.
Figure 6.
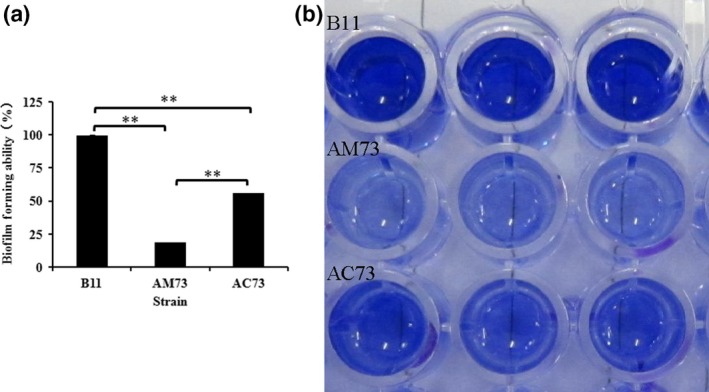
(a) The biofilm‐forming ability of wild‐type B11, AM73, and AC73. Values denoted by different numbers of asterisks were significantly different when compared by ANOVA (“**” p < .01; “*” p < .05). (b) The picture shows the color of crystal violet dissolved in 33% acetic acid
3.8. Bacterial adhesion
To investigate the ability of acuC to affect other virulence factors in addition to survival, the adhesive capacity of wild‐type B11, AM73, and AC73 was also tested and quantified. We employed gill mucus, intestinal mucus, and skin mucus for this test. The adhesion of wild‐type B11, AM73, and AC73 was 0.85, 0.43 and 0.64, respectively, in the gill mucus, 0.68, 0.28, and 0.49 in the intestinal mucus, and 1.61, 0.64, and 1.15 in the skin mucus. The results showed that the largest number of A. hydrophila adhered to the skin mucus and that the adhesive capacity of the AM73 mutant was significantly less than that of the wild type (Figure 7). These results indicated that the acuC gene plays an important role in bacterial adhesion and that acuC might be involved in the regulation of multiple virulence factors.
Figure 7.
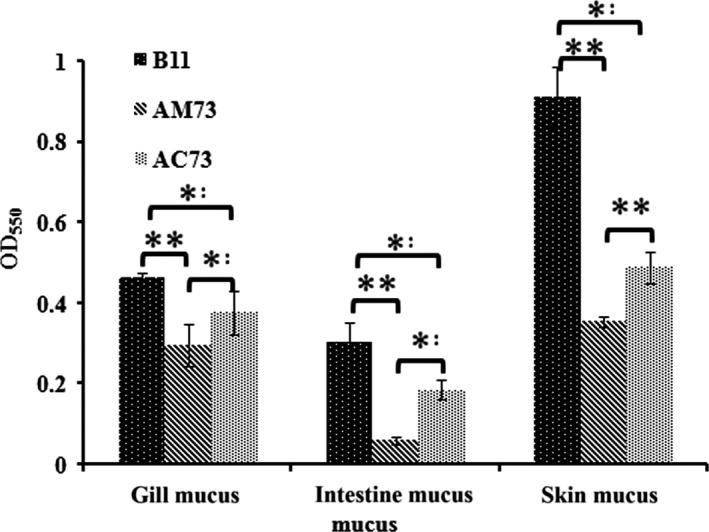
The adhesion of B11, AM73, and AC73 to gill mucus, intestinal mucus, and skin mucus. Values denoted by different numbers of asterisks were significantly different when compared by ANOVA (“**” p < .01; “*” p < .05)
3.9. Invasion and survival assays of AM73, AC73, and the wild‐type strain
The ability of B11, AM73, and AC73 to invade and survive in macrophages was compared to assess the role of acuC in invasion and survival. The data showed that the numbers of wild‐type B11, AM73, and AC73 that invaded macrophages were 2.1 × 105 CFU/ml, 2.3 × 105 CFU/ml, and 2.0 × 105 CFU/ml, respectively, at 0 hr and that 1.4 × 105 CFU/ml, 1.4 × 104 CFU/ml, and 1.0 × 105 CFU/ml, respectively, survived for 1 hr in macrophages. The survival rates of the wild‐type B11 and AC73 were 64.5% and 48.6%, respectively, whereas the survival of AM73 was only 6.1% at 1 hr (Figure 8). The results at 0 hr and 1 hr indicated the invasion and intracellular survival ability of the strains. The results showed that the number of intracellular AM73 was slightly, but not significantly, larger than the number of B11 at 0 hr. However, the data at 1 hr suggested a significantly impaired survival of AM73 in host macrophages. These results showed that acuC plays a vital role in the survival of A. hydrophila in host macrophages.
Figure 8.
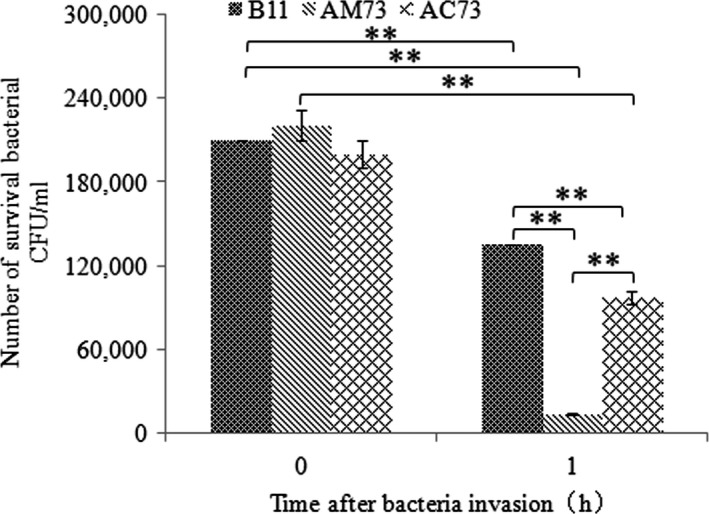
The invasion and survival of B11, AM73, and AC73 in macrophages. The X‐axis represents time after bacterial invasion; the Y‐axis represents the number of surviving bacteria per ml of macrophages. Values denoted by different numbers of asterisks were significantly different when compared by ANOVA (“**” p < .01; “*” p < .05)
3.10. Fluorescence microscopy analyses
Fluorescence microscopy and labeled bacteria were employed to evaluate the status of A. hydrophila B11 in macrophages. The pictures taken at 0 hr (Figure 9a) and 1 hr (Figure 9b) show the invasion and intracellular survival, respectively. Numerous bacterial cells were found in the macrophages at 0 hr, and most of the bacterial cells remained alive at 1 hr post invasion. These results suggested that the bacteria could invade and survive in the macrophages.
Figure 9.
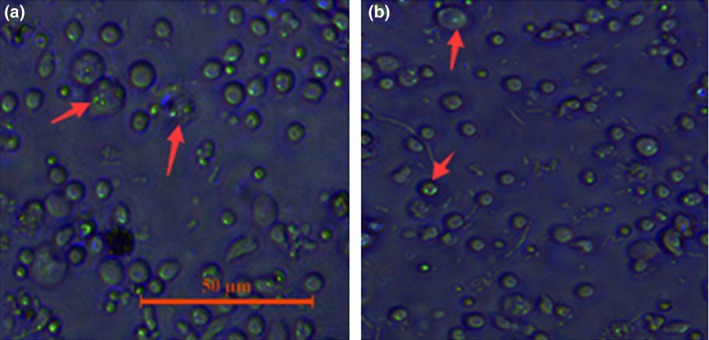
The status of wild‐type B11 labeled with EGFP in macrophages. Red arrows point to cells. (a) invasion; (b) intracellular survival after 1 hr
3.11. Infection of Danio rerio
The defects in biofilm formation, adherence and intracellular survival of the mutant indicated that acuC plays an important role in the pathogenesis of A. hydrophila. To test this conclusion, we infected the Danio rerio with A. hydrophila. Danio rerio infected intraperitoneally with wild‐type B11 or AC73 showed 40% and 30% mortality in 3–4 days, whereas the virulence of the mutant AM73 was significantly lower (p = .019). After 7 days of observation, only 10% mortality was observed in the Danio rerio infected with AM73 (Figure 10). The bacteria isolated from the infected fish were identified by PCR as the wild‐type B11 and AM73. This result demonstrated that the mutation of acuC in AM73 significantly impaired the lethality of A. hydrophila.
Figure 10.
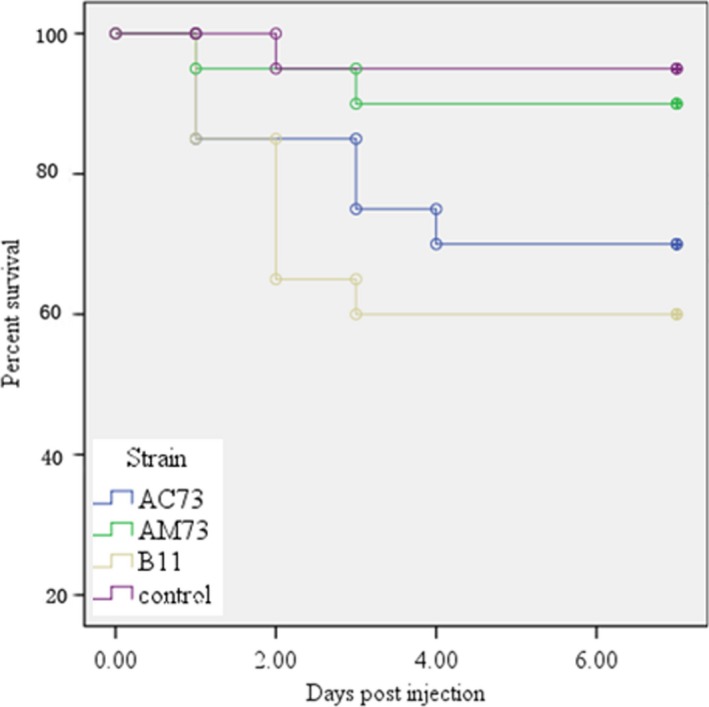
Infection of Danio rerio (n = 20) with wild‐type B11, the mutant AM73, and the complemented AC73 bacteria. Survival/mortality was monitored for 7 days. The X‐axis represents the days, and the Y‐axis represents the percent survival. The data were statistically evaluated and analyzed by the log‐rank test using GraphPad Prism 4 software (p < .05)
3.12. Expression of various virulence genes
Quantitative real‐time reverse transcription‐PCR showed that a lipopolysaccharide gene (rfaF) and one of the type VI secretion system‐related protein genes (vash) were significantly up‐regulated in AM73, while the other type VI secretion system‐related protein gene (traA) showed no obvious change. The expression of two outer membrane protein genes (ompA and ompTS), the type Ш secretion system gene (ascV), pilin and flagellar family protein genes (flgE, flgL, and pilB), the serine peptidase gene (degQ), the hemolysin gene (hlyA), the adhesin subunit gene (cblA), and the aerolysin gene (aerA) were significantly down‐regulated in AM73 (Figure 11).
Figure 11.
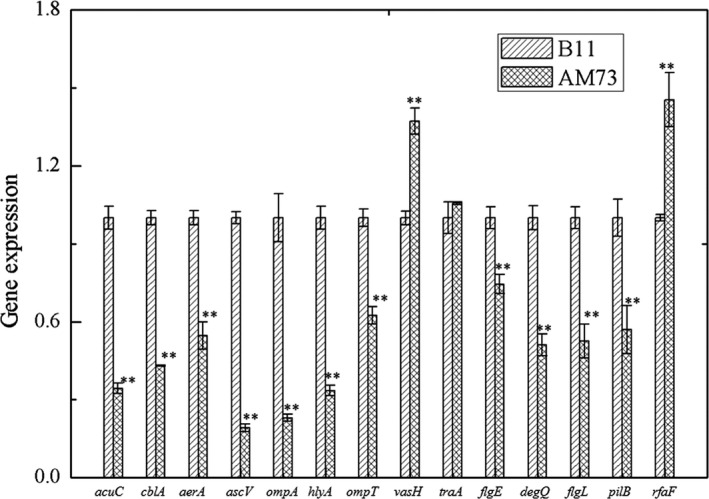
Expression profiles of A. hydrophila virulence‐related genes assayed by qRT‐PCR. The expression levels of acuC and 13 virulence genes in the wild‐type strain and AM73. The 16S rRNA was used as the internal control. Each bar represents the mean value of quintuplicates ± SD. Values denoted by different numbers of asterisks were significantly different when compared by ANOVA (“**” p < .01; “*” p < .05)
4. Discussion
In this study, a pathogenic strain of A. hydrophila B11 was found to invade the macrophages of Tilapia mossambica in vitro, and many of the bacteria survived in the macrophages for at least 24 hr. Once the bacteria entered the phagocytes, they must resist disruption by various enzymes present in the cells. The ability to survive in cells, even briefly, might open a window of opportunity for A. hydrophila and other pathogens to survive and trigger disease.
In this study, a mutant library of A. hydrophila was constructed by introducing the mini‐Tn10 transposon into the bacteria. The mutant strain AM73 exhibited the poorest survival in macrophagocytes. The mutant gene was found to have the highest homology with a histone deacetylase/AcuC/AphA family protein gene of A. hydrophila subsp. hydrophila ATCC 7966. AcuC was found to be an intracellular protein by western blotting assay. The results of further assays showed that the enzymatic activity of the intracellular protein in AM73 was significantly reduced. These results demonstrated that the acuC gene, which encodes the histone deacetylase enzymatic activity, was mutated in AM73.
The protein acetylation/deacetylation posttranslational modification is an efficient mechanism for controlling the activity of structural proteins, gene expression regulators, and enzymes in response to rapidly changing physiological conditions (Gardner et al., 2006; Hu, Lima, & Wolfe, 2010). In Bacillus subtilis, the activity of acetyl‐coenzyme A (Ac‐CoA) synthetase is regulated by the protein acetylation/deacetylation posttranslational modification system comprised of AcuA and AcuC (Leipe & Landsman, 1997). OatA, a peptidoglycan O‐acetyltransferase, has been shown to be involved in Listeria monocytogenes immune escape and is critical for virulence (Aubry et al., 2011). In Legionella pneumophila, histone acetylation was demonstrated to be important for virulence (Schmeck et al., 2008). In this study, the deletion of acuC was found to reduce the intracellular survival ability of A. hydrophila. This result is consistent with the previous findings of Aubry et al. and Schmeck et al.
Quantitative real‐time reverse transcription‐PCR showed that deletion of acuC resulted in the up‐regulation of vash and rfaF and the down‐regulation of 10 genes, including ompA, ompTS, traA, flgE, flgL, pilB, degQ, cblA, hlyA, and aerA. These genes are closely related to the adhesion and virulence of A. hydrophila (Ho, Sohel, & Schoolnik, 1992; Qin et al., 2014; Quinn et al., 1994). Flagellar motility has been demonstrated to be necessary for A. hydrophila adhesion (Qin et al., 2016). Outer membrane proteins are important adhesion factors and protective antigens closely related to the virulence of A. hydrophila (Kawai, Liu, Ohnishi, & Oshima, 2004). OmpA has also been reported to be correlated with the adhesion of A. hydrophila (Quinn et al., 1994). FlgE, the structural gene encoding the flagellar hook protein, has also been reported to be correlated with the adhesion of A. hydrophila (Qin et al., 2014). It has previously been reported that the pilin gene, the gene coding for the flexible pilus subunit, was correlated with adhesion in A. hydrophila (Ho et al., 1992). In this study, the adhesion ability of A. hydrophila was significantly inhibited after deletion of acuC; this result is consistent with the gene expression results.
Biofilm formation has previously been reported to occur in three main stages: (1) attachment, specific protein‐based binding to abiotic surfaces; (2) proliferation and formation of mature biofilm structures; and (3) detachment, also called dispersal (O'Toole, Kaplan, & Kolter, 2000). Biofilm assays showed that the mutant strain AM73 was significantly defective in forming biofilms. This result indicated that acuC is involved in biofilm formation by A. hydrophila.
Unlike the well‐studied bacterial pathogenic processes, information on the intracellular survival of A. hydrophila is still very limited. Qin et al. (2014) found that a ΔflgE mutant of A. hydrophila showed significantly reduced intracellular survival in phagocytes. In this study, the intracellular survival of AM73 was significantly lower than the survival of the wild type. In addition, the expression of motility‐related genes, including flgE, traA, flgL, and pilB was significantly down‐regulated in AM73. Previous studies have shown that bacterial motility affects bacterial survival in phagocytes (Qin et al., 2014). In this study, the expression of rfaF was significantly down‐regulated in AM73. The rfaF gene is associated with the synthesis of lipopolysaccharide heptosyltransferase II. Guo et al. (1997) found that the proteins of lipopolysaccharide modification contribute to bacterial survival in macrophages by conferring resistance to antimicrobial peptides and nutrient scavenging. The expression of ascV, one of the type III secretion system genes, was also significantly down‐regulated in AM73. Some previous reports have revealed that the secreted molecules of the type III secretion system affect bacterial internalization (Cirillo, Valdivia, Monack, & Falkow, 1998).
In this study, the deletion of acuC resulted in the reduction in mortality of infected Danio rerio. The deletion of acuC also resulted in the down‐regulation of several genes, including hlyA, in the mutant strains. A hemolytic toxin, the product of hlyA, has also been reported to induce bacterial infection (Qian, Chen, Shen, & Shen, 1995). The results indicated that acuC is involved in the pathogenicity of A. hydrophila by affecting the expression of other virulence genes.
The significant reduction in the number of bacteria surviving in the host suggests a close relationship between acuC and survival, although the mechanisms of A. hydrophila survival in host phagocytes remain unclear. Further study is needed to reveal the form of A. hydrophila B11 residing in the macrophages and the activities of A. hydrophila B11 used to facilitate survival.
In conclusion, this study revealed that A. hydrophila B11 is able to invade the macrophages of Tilapia mossambica in vitro and that the bacteria can also survive more than 24 hr. The acuC gene can influence the expression of crucial virulence genes and biofilm formation, adhesion, and mortality in A. hydrophila, which indicates that AcuC is an important regulatory protein that contributes to the survival and pathogenicity of A. hydrophila.
Conflict of Interest
None declared.
Acknowledgments
This work was supported by grants from The Science and Technology Program of Xiamen Southern Oceanographic Center under contract No. 14CZP032HJ06, The Regional Demonstration of Marine Economy Innovative Development Project under contracts No. 14PYY050SF03 and 12PYY001SF08, The National Natural Science Foundation of China under contracts No. 31272699 and 31502194, Fujian Provincial Department of Science & Technology under contracts No. JA15289, 2015I1002 and 2015R1036‐4, and The Natural Science Foundation of Fujian Province under contract No. 2016J05080. We gratefully thank Prof. Nie and Dr. Huang for providing some of the strains and plasmids used in this study.
Jiang Q, Chen W, Qin Y, et al. AcuC, a histone deacetylase, contributes to the pathogenicity of Aeromonas hydrophila . MicrobiologyOpen. 2017;6:e468 https://doi.org/10.1002/mbo3.468
References
- Aubry, C. , Goulard, C. , Nahori, M. A. , Cayet, N. , Decalf, J. , Sachse, M. , … Dussurget, O . (2011). OatA, a peptidoglycan O‐acetyltransferase involved in Listeria monocytogenes immune escape, is critical for virulence. Journal of Infectious Diseases, 204(5), 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balebona, M. , Krovacek, K. , Morinigo, M. , Mansson, I. , Faris, A. , & Borrego, J. (1998). Neurotoxic effect on two fish species and a PC12 cell line of the supernate of Vibrio alginolyticus and Vibrio anguillarum. Veterinary Microbiology, 63, 61–69. [DOI] [PubMed] [Google Scholar]
- Beaz‐Hidalgo, R. , & Figueras, M. J. (2013). Aeromonas spp. whole genomes and virulence factors implicated in fish disease. Journal of Fish Diseases, 36(4), 371–388. [DOI] [PubMed] [Google Scholar]
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Analytical Biochemistry, 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Chan, Y. Y. , & Chua, K. L. (2005). The Burkholderia pseudomallei BpeAB‐OprB efflux pump: Expression and impact on quorum sensing and virulence. Journal of Bacteriology, 187, 4707–4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Q. , Yan, Q. , Wang, K. , Zhuang, Z. , & Wang, X. (2008). Portal of entry for pathogenic Vibrio alginolyticus into large yellow croaker Pseudosciaena crocea, and characteristics of bacterial adhesion to mucus. Diseases of Aquatic Organisms, 80(3), 181–188. [DOI] [PubMed] [Google Scholar]
- Çiftci, A. , Onuk, E. E. , Çiftci, G. , Fındık, A. , Söğüt, M. Ü. , Didinen, B. I. , … Altun, S . (2016). Development and validation of glycoprotein‐based native‐subunit vaccine for fish against Aeromonas hydrophila. Journal of Fish Diseases, 39(8), 981–992. [DOI] [PubMed] [Google Scholar]
- Cirillo, D. M. , Valdivia, R. H. , Monack, D. M. , & Falkow, S. (1998). Macrophage‐dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Molecular Microbiology, 30, 175–188. [DOI] [PubMed] [Google Scholar]
- da Silva, B. C. , Mouriño, J. L. P. , Vieira, F. N. , Jatobá, A. , Seiffert, W. Q. , & Martins, M. L. (2012). Haemorrhagic septicaemia in the hybrid surubim (Pseudoplatystoma corruscans×Pseudoplatystoma fasciatum) caused by Aeromonas hydrophila. Aquaculture Research, 43, 908–916. [Google Scholar]
- Foster, T. J. (2005). Immune evasion by staphylococci. Nature Reviews Microbiology, 3(12), 948–958. [DOI] [PubMed] [Google Scholar]
- Gardner, J. G. , Grundy, F. J. , Henkin, T. M. , & Escalante‐Semerena, J. C. (2006). Control of acetyl‐coenzyme A synthetase (AcsA) activity by acetylation/deacetylation without NAD+ involvement in Bacillus subtilis . Journal of Bacteriology, 188, 5460–5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzoni, C. , & Kelley, W. L. (2009). Staphylococcus aureus: New evidence for intracellular persistence. Trends in Microbiology, 17(2), 59–65. [DOI] [PubMed] [Google Scholar]
- Grundy, F. J. , Waters, D. A. , Takova, T. Y. , & Henkin, T. M. (1993). Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis . Molecular Microbiology, 10, 259–271. [DOI] [PubMed] [Google Scholar]
- Gulick, A. M. , Starai, V. J. , Horswill, A. R. , Homick, K. M. , & Escalante‐Semerena, J. C. (2003). The 1.75 Å crystal structure of acetyl‐CoA synthetase bound to adenosine‐5′‐propylphosphate and coenzyme A. Biochemistry, 42, 2866–2873. [DOI] [PubMed] [Google Scholar]
- Guo, L. , Lim, K. B. , Gunn, J. S. , Bainbridge, B. , Darveau, R. P. , Hackett, M. , & Miller, S. I. (1997). Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP‐phoQ. Science, 276, 250–253. [DOI] [PubMed] [Google Scholar]
- Herrero, M. , de Lorenzo, V. , & Timmis, K. N. (1990). Transposon vectors containing non‐antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram‐negative bacteria. Journal of Bacteriology, 172, 6557–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, A. S. , Sohel, I. , & Schoolnik, G. K. (1992). Cloning and characterization of fxp, the flexible pilin gene of Aeromonas hydrophila. Molecular Microbiology, 6, 2725–2732. [DOI] [PubMed] [Google Scholar]
- Hu, L. I. , Lima, B. P. , & Wolfe, A. J. (2010). Bacterial protein acetylation: The dawning of a new age. Molecular Microbiology, 77, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L. , Hu, J. , Su, Y. , Qin, Y. , Kong, W. , Ma, Y. , … Yan, Q . (2015). Identification and characterization of three Vibrio alginolyticus non‐coding RNAs involved in adhesion, chemotaxis, and motility processes. Frontiers in Cellular and Infection Microbiology, 5, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L. , Huang, L. , Yan, Q. , Qin, Y. , Ma, Y. , Lin, M. , … Zheng, J . (2016). The TCA pathway is an important player in the regulatory network governing Vibrio alginolyticus adhesion under adversity. Frontiers in Microbiology, 7, 40, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda, J. M. , & Abbott, S. L. (2010). The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clinical Microbiology Reviews, 23, 35–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H. , & Pancholi, V. (2006). Identification and biochemical characterization of a eukaryotic‐type serine/threonine kinase and its cognate phosphatase in Streptococcus pyogenes: Their biological functions and substrate identification. Journal of Molecular Biology, 357, 1351–1372. [DOI] [PubMed] [Google Scholar]
- Kaufmann, S. H. E. (2011). Intracellular pathogens: Living in an extreme environment. Immunological Reviews, 240, 5–10. [DOI] [PubMed] [Google Scholar]
- Kawai, K. , Liu, Y. , Ohnishi, K. , & Oshima, S.‐I. (2004). A conserved 37 kDa outer membrane protein of Edwardsiella tarda is an effective vaccine candidate. Vaccine, 22, 3411–3418. [DOI] [PubMed] [Google Scholar]
- Kong, W. , Huang, L. , Su, Y. , Qin, Y. , Ma, Y. , Xu, X. , … Yan, Q. (2015). Investigation of possible molecular mechanisms underlying the regulation of adhesion in Vibrio alginolyticus with comparative transcriptome analysis. Antonie van Leeuwenhoek, 107, 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe, D. D. , & Landsman, D. (1997). Histone deacetylases, acetoin utilization proteins and acetylpolyamine amidohydrolases are members of an ancient protein superfamily. Nucleic Acids Research, 25, 3693–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, K. , Lim, T. , Lam, T. , & Sin, Y. (1996). Morphological changes in carp epithelial cells infected with Aeromonas hydrophila. Journal of Fish Diseases, 19, 167–174. [Google Scholar]
- Leung, K. , Low, K. , Lam, T. , & Sin, Y. (1995). Interaction of the fish pathogen Aeromonas hydrophila with tilapia, Oreochromis aureus (Steindachner), phagocytes. Journal of Fish Diseases, 18, 435–447. [Google Scholar]
- Lin, G. , Chen, W. , Su, Y. , Qin, Y. , Huang, L. , & Yan, Q. (2017). Ribose operon repressor (RbsR) contributes to the adhesion of Aeromonas hydrophila to Anguilla japonica mucus. MicrobiologyOpen., 00, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.‐G. , Mitsukawa, N. , Oosumi, T. , & Whittier, R. F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T‐DNA insert junctions by thermal asymmetric interlaced PCR. The Plant Journal, 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Lü, A. J. , Hu, X. C. , Wang, Y. , Zhu, A. H. , Shen, L. L. , Tian, J. , … Feng, Z. J. (2015). Skin immune response in the zebrafish, Danio rerio (Hamilton), to Aeromonas hydrophila infection: A transcriptional profiling approach. Journal of Fish Diseases, 38, 137–150. [DOI] [PubMed] [Google Scholar]
- Lumsden, J. , Ostland, V. , Byrne, P. , & Ferguson, H. (1993). Detection of a distinct gill‐surface antibody response following horizontal infection and bath challenge of brook trout Salvelinus fontinalis with Flavobacterium branchiophilum, the causative agent of bacterial gill disease. Diseases of Aquatic Organisms, 16, 21–27. [Google Scholar]
- Luo, G. , Huang, L. , Su, Y. , Qin, Y. , Xu, X. , Zhao, L. , & Yan, Q. (2016). flrA, flrB and flrC regulate adhesion by controlling the expression of critical virulence genes in Vibrio alginolyticus. Emerging Microbes & Infections, 5(8), e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino, S. , Rubires, X. , Aguilar, A. , & Tomás, J. M. (1996). The O:34‐antigen lipopolysaccharide as an adhesin in Aeromonas hydrophila. FEMS Microbiology Letters, 139, 97–101. [DOI] [PubMed] [Google Scholar]
- Munro, A. L. S. , Hastings, T. S. , Ellis, A. E. , & Liversidge, J . (1980). Studies on an ichthyotoxic material produced extracellularly by the furunculosis bacterium Aeromonas salmonicida [salmon and trout] In Cooperative Programme of Research on Aquaculture (Fish Diseases)‐3. Session, Munich (Germany, FR): Springer‐Verlag. [Google Scholar]
- Neely, M. N. , Pfeifer, J. D. , & Caparon, M. (2002). Streptococcus‐zebrafish model of bacterial pathogenesis. Infection and Immunity, 70, 3904–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek, I. , Courtney, H. , Schifferli, D. , & Beachey, E. (1986). Enzyme‐linked immunosorbent assay for adherence of bacteria to animal cells. Journal of Clinical Microbiology, 24, 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole, G. , Kaplan, H. B. , & Kolter, R. (2000). Biofilm formation as microbial development. Annual Reviews in Microbiology, 54, 49–79. [DOI] [PubMed] [Google Scholar]
- Peyghan, R. (2010). Effect of intraperitoneal and intramuscular injection of killed aeromonas hydrophila on lymphocytes and serum proteins of common carp, cyprinus carpio. Advances in Bioscience and Biotechnology, 01, 26–29. [Google Scholar]
- Qian, D. , Chen, Y. , Shen, J. , & Shen, Z . (1995). Serogroups, virulence and hemolytic activity of Aeromonas hydrophila which caused fish bacterial septicaemia. Acta Microbiologica Sinica, 35, 460–464. [PubMed] [Google Scholar]
- Qin, Y. , Yan, Q. , Su, Y. , Li, H. , & Zou, W. (2013). Disruption of chemotaxis‐related genes affects multiple cellular processes and the virulence of pathogenic Vibrio harveyi. Acta Oceanologica Sinica, 32, 55–60. [Google Scholar]
- Qin, Y. , Lin, G. , Chen, W. , Huang, B. , Huang, W. , & Yan, Q. (2014). Flagellar motility contributes to the invasion and survival of Aeromonas hydrophila in Anguilla japonica macrophages. Fish & Shellfish Immunology, 39, 273–279. [DOI] [PubMed] [Google Scholar]
- Qin, Y. , Lin, G. , Chen, W. , Xu, X. , & Yan, Q. (2016). Flagellar motility is necessary for Aeromonas hydrophila adhesion. Microbial Pathogenesis, 98, 160–166. [DOI] [PubMed] [Google Scholar]
- Quinn, D. M. , Atkinson, H. M. , Bretag, A. H. , Tester, M. , Wong, C. , & Flower, R. (1994). Carbohydrate‐reactive, pore‐forming outer membrane proteins of Aeromonas hydrophila. Infection and Immunity, 62, 4054–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes‐Becerril, M. , López‐Medina, T. , Ascencio‐Valle, F. , & Esteban, M. Á. (2011). Immune response of gilthead seabream (Sparus aurata) following experimental infection with Aeromonas hydrophila. Fish & Shellfish Immunology, 31, 564–570. [DOI] [PubMed] [Google Scholar]
- Schmeck, B. , Lorenz, J. , Opitz, B. , vanLaak, V. , Zahlten, J. , Slevogt, H. , … Hippenstiel, S . (2008). Histone acetylation and flagellin are essential for Legionella pneumophila‐induced cytokine expression. The Journal of Immunology, 181(2), 940–947. [DOI] [PubMed] [Google Scholar]
- Starai, V. , Celic, I. , Cole, R. , Boeke, J. , & Escalante‐Semerena, J. (2002). Sir2‐dependent activation of acetyl‐CoA synthetase by deacetylation of active lysine. Science, 298, 2390–2392. [DOI] [PubMed] [Google Scholar]
- Sterner, D. E. , & Berger, S. L. (2000). Acetylation of histones and transcription‐related factors. Microbiology and Molecular Biology Reviews, 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiagalingam, S. , Cheng, K. H. , Lee, H. J. , Mineva, N. , Thiagalingam, A. , & Ponte, J. F . (2003). Histone deacetylases: Unique players in shaping the epigenetic histone code. Annals of the New York Academy of Sciences, 983, 84–100. [DOI] [PubMed] [Google Scholar]
- Thwaites, G. E. , & Gant, V. (2011). Are bloodstream leukocytes trojan horses for the metastasis of staphylococcus aureus? Nature Reviews Microbiology, 9(3), 215–222. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Huang, L. , Su, Y. , Qin, Y. , Kong, W. , Ma, Y. , … Yan, Q . (2015). Involvement of the flagellar assembly pathway in Vibrio alginolyticus adhesion under environmental stresses. Frontiers in Cellular and Infection Microbiology, 5, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Q. , Chen, Q. , Ma, S. , Zhuang, Z. , & Wang, X. (2007). Characteristics of adherence of pathogenic Vibrio alginolyticus to the intestinal mucus of large yellow croaker (Pseudosciaena crocea). Aquaculture, 269, 21–30. [Google Scholar]


