Abstract
Aflatoxin is a toxic, carcinogenic mycotoxin primarily produced by Aspergillus parasiticus and Aspergillus flavus. Previous studies have predicted the existence of more than 20 genes in the gene cluster involved in aflatoxin biosynthesis. Among these genes, aflK encodes versicolorin B synthase, which converts versiconal to versicolorin B. Past research has investigated aflK in A. parasiticus, but few studies have characterized aflK in the animal, plant, and human pathogen A. flavus. To understand the potential role of aflK in A. flavus, its function was investigated here for the first time using gene replacement and gene complementation strategies. The aflK deletion‐mutant ΔaflK exhibited a significant decrease in sclerotial production and aflatoxin biosynthesis compared with wild‐type and the complementation strain ΔaflK::aflK. ΔaflK did not affect the ability of A. flavus to infect seeds, but downregulated aflatoxin production after seed infection. This is the first report of a relationship between aflK and sclerotial production in A. flavus, and our findings indicate that aflK regulates aflatoxin formation.
Keywords: aflatoxin, Aspergillus flavus, gene complementation strain, gene deletion strain, sclerotia
1. Introduction
The genus Aspergillus is a family of filamentous fungi with worldwide distribution, which is well‐studied for its important secondary metabolites, both beneficial and harmful. Within this genus, Aspergillus flavus infects a wide range of plants, animals, and humans as a disease‐causing pathogen (Amaike & Keller, 2011; Shieh et al., 1997). It can cause serious agricultural problems by contaminating important crops and producing extremely carcinogenic and highly toxic secondary metabolites known as aflatoxins (Amaike & Keller, 2011). Crops contaminated with aflatoxins pose a serious or even fatal threat to animals and humans. The gene cluster for aflatoxin biosynthesis in both A. flavus and Aspergillus parasiticus has been reported to encode at least 27 enzymes and regulatory factors within a 70 kb region (Yabe & Nakajima, 2004).
The functions of many genes within this cluster have been previously explored. For example, aflX is required for the conversion of versicolorin A, similar to the function of aflN, aflM, and aflY (Cary, Ehrlich, Bland, & Montalbano, 2006). The expression of aflR and aflQ of the aflatoxins biosynthesis cluster was analyzed Sweeney, Pamies, & Dobson (2000) using reverse transcription PCR, while Mayer, Bagnara, Farber, & Geisen (2003) examined the expression of aflD and the aflatoxin yield in wheat. Recently, Al‐Saad, Al‐Badran, Al‐Jumayli, Magan, & Rodriguez (2016) reported the impact of bacterial biocontrol agents on aflD and aflR expression and aflatoxin B1 production when A. flavus was grown under different environmental conditions and with various nutritional media. The expression of secondary metabolite biosynthesis cluster genes in A. flavus, A. parasiticus, and Aspergillus oryzae has also been compared (Ehrlich & Mack, 2014). Furthermore, aflatoxin biosynthesis gene expression was determined relative to water activity and temperature (Schmidt‐Heydt, Abdel‐Hadi, Magan, & Geisen, 2009), and we previously conducted a transcriptome analysis of A. flavus in response to water activity (Zhang et al., 2014).
Of the aflatoxin biosynthesis genes, vbs, also known as aflK, codes versicolorin B synthase (VBS) which is responsible for the conversion of versiconal to versicolorin B (Yu, Bhatnagar, & Ehrlich, 2002). vbs has been heterologous expressed, purified, isolated, and characterized in A. parasiticus (McGuire, Silva, Casillas, & Townsend, 1996; Silva, Minto, Barry, Holland, & Townsend, 1996; Silva & Townsend, 1997), and the distribution and subcellular localization of VBS was shown to change with respect to culture time in A. parasiticus (Chiou et al., 2004). Linz, Wee, & Roze (2014) hypothesized that VBS transport is tightly regulated by the timing and synthesis level of versicolorin A.
Almost all studies on aflK were performed in A. parasiticus, while few have investigated aflK in A. flavus. Therefore, to understand the potential role of aflK in A. flavus, we constructed the first known aflK deletion mutant and its complementation strain of A. flavus. We then analyzed the function of aflK, including its effect on growth, the formation of conidia and sclerotia, aflatoxin biosynthesis, and host colonization of A. flavus.
2. Materials and Methods
2.1. Phylogenetic tree and domain architecture
The protein sequence of AflK from A. flavus was identified and used in a BLAST search on the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/). All available homogenous sequences from different organisms were downloaded and used to construct a phylogenetic tree with MEGA6.0 software and the neighbor‐joining method (Tamura, Stecher, Peterson, Filipski, & Kumar, 2013). Bootstrap analysis was set as 1,000 replicates. To visualize the AflK domain, information from the SMART database (http://smart.embl-heidelberg.de/) was subjected to DOG2.0 (Yang et al., 2016).
2.2. Fungal strains and media
Aspergillus flavus CA14PTSΔpyrG, a uracil auxotrophic, was obtained from Prof. Chang P. K. (Chang, Scharfenstein, Wei, & Bhatnagar, 2010). Aspergillus fumigatus Af293 was a gift from Dr. Yang Liu (Institute of Agro‐Products Processing Science & Technology, Chinese Academy of Agricultural Sciences, Beijing, China). Aspergillus flavus was cultured on yeast extract sucrose (YES) agar plates for fungal growth, and YGTUU media was applied for the preparation of protoplasts. Yeast extract glucose (YGT) agar was used to screen gene deletion strains and to prepare protoplasts of deletion strains. Czapek's agar with pyrithiamine was used to screen complementation strains. Potato dextrose agar (PDA) was used for conidia production (Chang et al., 2011), and Wickerham media was used for sclerotial formation (Chang, Scharfenstein, Mack, & Ehrlich, 2012).
2.3. Protoplast preparation and fungal transformation
Protoplast preparation was performed as previously described (Chang et al., 2010; Gehret, Connelly, & Dumont, 2012; He, Price, OBrian, Georgianna, & Payne, 2007). Briefly, A. flavus CA14PTSΔpyrG conidia were activated with YGTUU media and collected. After enzymolysis by β‐glucuronidase (Sigma, USA), lysing enzyme (Sigma, USA), and driselase (Sigma, USA), protoplasts were resuspended in 1.0 ml of STC buffer (10.0 mmol/L Tris‐HCl, pH 7.5, 10.0 mmol/L CaCl2, 1.2 mol/L sorbitol), then subpackaged (about 105 protoplasts in 100 μl STC) and stored at −80°C. Deletion strain protoplasts were prepared using the same method.
Transformation was performed according to previously published techniques with minor modifications (Cary et al., 2006; He et al., 2007; Szewczyk et al., 2006). Protoplasts, the gel extraction product of fusion PCR or complementation vectors, and polyethylene glycol (PEG) buffer (50% PEG 4,000, 10.0 mmol/L NaH2PO4, pH 5.8, 10.0 mmol/L Tris‐HCl, pH 7.5, 50.0 mmol/L CaCl2, and 0.6 mmol/L KCl) were mixed with 10 mL of upper regeneration media (50.0 g/L of Czapek Solution Agar (BD Bioscience, NJ, USA), 1.0 mol/L sucrose, and 5.0 g/L agar), then plated on 10 ml of lower regeneration media (50.0 g/L of Czapek Solution Agar (BD Bioscience), 1.0 mol/L sucrose, and 15.0 g/L agar). The plates were cultured at 37°C for about 3 days.
2.4. Construction of deletion and complementation mutants
A previous publication was used as a reference to construct the aflK deletion strain ΔaflK and complementation strain ΔaflK::aflK (Ren et al., 2016). All gene sequences, including the open reading frame of aflK, the upstream and downstream sequence of aflK, and the pyrG sequence of A. fumigatus Af293, were searched for and downloaded from the NCBI website and Aspergillus Comparative Database (http://www.broadinstitute.org/annotation/genome/aspergillus_group/MultiHome.html). The PCR fusion program was performed as described by Szewczyk et al. (2006), and the PCR‐amplified complementary fragment was inserted into chromosomal integrating shuttle vector pPTR I (containing a pyrithiamine resistance gene as the selection marker, TAKARA, Japan) using Kpn I and Hind III (Thermo, USA) digestion and the ClonExpress II One Step Cloning Kit (C112‐01, Vazyme Biotech Co., Ltd, Nanjing, China). After bacterial transformation and PCR verification, the vectors were transformed into protoplasts of ΔaflK to construct the complementary strain ΔaflK::aflK. pyrG was amplified from A. fumigatus Af293 and transformed into A. flavus CA14PTSΔpyrG protoplasts, which were then used as wild‐type (WT).
2.5. RNA extraction and reverse transcription (RT)‐PCR
The mycelia of WT, ΔaflK, and ΔaflK::aflK were harvested after cultured on YES agar for 48 hr. After grinding in liquid nitrogen, the result powder was suspended with TRIzol reagent (Biomarker Technologies, Beijing, China) for RNA extract. RNA molecules were purified with the DNA‐free kit (TransGen Biotech, Beijing, China) to remove genomic DNA. First‐strand cDNA was synthesized with the TransScript® All‐in‐One First‐Strand cDNA Synthesis SuperMix.
2.6. Analysis of colony growth, conidial production, and sclerotial formation
To record the colony growth, 2 μl of 106 spores/ml suspension of WT, ΔaflK, and ΔaflK::aflK were inoculated onto YES media. The diameters were measured until colonies filled the plate. For the enumeration of conidia, 2 μl spores suspension was inoculated onto PDA media, and cultured for 6 days at 37°C. Three pieces of 1‐cm diameter were harvested from the edge to the centre of each colony, and homogenized in 3 ml of distilled water. Then, spore number was counted manually with a hemocytometer. Conidiophores were observed according to a previous report (Yang et al., 2016). To analyze sclerotial production, 2 μl spores suspension of the three strains was cultured at 37°C for 10 days, then 75% ethanol was used to wash away conidia for the enumeration of sclerotia (Yang et al., 2016). The sclerotia were also observed microscopically. Each experiment was performed three times using four replicates. One‐way analysis of variance (ANOVA) was used for statistical testing.
2.7. Aflatoxin analysis
For aflatoxin biosynthesis analysis, 106 spores of the three strains were incubated into YES liquid media, and cultured at 28°C for 6 days in the dark with shaking at 180 rpm. Aflatoxin was extracted with chloroform, then 10 ml of chloroform was transferred to a new tube and evaporated to dryness. Thin‐layer chromatography (TLC) was used to analyze aflatoxin production, and the outcome observed under ultraviolet light at 365 nm. The JD‐801 Computer‐aided Image Analysis System (JEDA Co., Nanjing, China) was used for the quantitative analysis of aflatoxin biosynthesis (Yang et al., 2016). Each experiment was performed three times using four replicates and analyzed by one‐way ANOVA. High‐performance liquid chromatography (HPLC) was used to confirm the presence of aflatoxin. The samples were filtered with 0.22 mm organic filtration, then analyzed by HPLC (Breeze™ HPLC, Waters, USA) on a MYCOTOX™ C18 column (NO. 1612124, 250 × 4.6 mm, Pickering Laboratories, USA) at 42°C. The running solvent was water:methanol:acetonitrile (56:22:22). A total of 20 μl of all samples was injected and run isocratically for 15 min at a flow rate of 1.0 ml/min. Aflatoxin was detected by a fluorescent detector with an emission wavelength of 455 nm and an excitation wavelength of 365 nm (Yang et al., 2016).
2.8. Seed infection
The ability of the three strains to infect peanut and maize was determined as described previously (Tsitsigiannis & Keller, 2006). After asepsis with 0.05% sodium hypochlorite and 75% ethanol, peanut cotyledons and maize kernels were inoculated with 105 spores/ml suspension for 30 min, with shaking at 80 rpm. Then 20 cotyledons and 10 maize kernels were placed in culture dishes lined with three pieces of moist sterile filter paper to maintain humidity. A blank control was included in which the cotyledons within sterile water. Peanuts and maize were incubated at 28°C for 4 days in the dark, and humidity was maintained. Peanut seeds and maize kernels were then harvested in 50 ml tubes, and vortexed to release the spores into 15 ml of 0.05% Tween 20 (v/v in water); conidia were counted hemocytometrically. An equal amount of chloroform was used to extract aflatoxins. Each experiment was performed three times using four replicates and analyzed by one‐way ANOVA.
2.9. Formula of media
Yeast extract sucrose (YES) plate: 20.0 g/L yeast extract, 1.0 g/L MgSO4•7H2O, 150.0 g/L sucrose, and 15.0 g/L agar (Abdel‐Hadi, Schmidt‐Heydt, Parra, Geisen, & Magan, 2012; Calvo, Bok, Brooks, & Keller, 2004). Yeast extract glucose (YGT) agar: 5.0 g/L yeast extract, 20.0 g/L glucose, 1 ml of trace element solution per liter of media, and 15.0 g/L agar (Yang et al., 2016). YGTUU plate: 5.0 g/L yeast extract, 20.0 g/L glucose, 1 ml of trace element solution per liter of media, 1.0 mg/ml uracil, 1.0 mg/ml uridine, and 15.0 g/L agar (Yang et al., 2016). Czapek's agar: 50.0 g/L of Czapek solution agar (BD Bioscience, NJ, USA). Potato dextrose agar (PDA): 39.0 g/L of potato dextrose agar (BD Bioscience, NJ, USA). Wickerham media: 30.0 g/L sucrose, 3.0 g/L peptone, 5.0 g/L corn steep liquid, 2.0 g/L yeast extract, 2.0 g/L NaNO3, 0.1 g/L FeSO4 •7H2O, 1.0 g/L K2HPO4•H2O, 2.0 g/L dextrose, 0.2 g/L KCl, 0.5 g/L MgSO4•7H2O, pH 5.5 (Chang et al., 2012).
3. Results
3.1. Phylogenetic analysis and domain prediction of AflK in A. flavus
Protein sequences of AflK (versicolorin B synthase) from A. flavus and 21 homologous proteins were used to construct a phylogenetic tree. The protein sequences chosen as analogous to A. flavus AflK (XP_002379930.1) for the analysis were: versicolorin B synthase in A. oryzae RIB40 (XP_001821530.1), choline dehydrogenase in A. oryzae 3.042 (EIT81342.1), versicolorin B synthase in Aspergillus nomius (AAS90066.1), conserved hypothetical protein in Aspergillus nidulans FGSC A4 (CBF80156.1), hypothetical protein ARAM_004338 in Aspergillus rambellii (KKK21812.1), hypothetical protein AOCH_000141 in Aspergillus ochraceoroseus (KKK22281.1), versicolorin B synthase in Dothistroma septosporum (ABO72541.2), versicolorin B synthase in Passalora arachidicola (ADN06239.1), hypothetical protein in Podospora anserina S mat+ (XP_001911514.1), hypothetical protein HK57_00552 in Aspergillus ustus (KIA75620.1), versicolorin B synthase in Aspergillus sojae (BAJ83509.1), versicolorin B synthase in A. parasiticus (Q12062.1), a protein similar to glucose‐methanol‐choline (GMC) oxidoreductase in Leptosphaeria maculans JN3 (XP_003844946.1), putative versicolorin B synthase in Rosellinia necatrix (GAP92978.1), GMC oxidoreductase in Penicillium expansum (AIG62134.1), putative GMC oxidoreductase in Diplodia seriata (KKY14472.1), putative GMC oxidoreductase protein in Neofusicoccum parvum UCRNP2 (XP_007589463.1), hypothetical protein V499_02164 in Pseudogymnoascus sp. VKM F‐103 (KFY78724.1), GMC oxidoreductase in Aspergillus clavatus NRRL 1 (XP_001273087.1), GMC oxidoreductase‐like protein in Aspergillus ruber CBS 135680 (EYE92427.1), and versicolorin B synthase in Aspergillus pseudotamarii (BAJ83519.1). Among these protein sequences, the consistency between versicolorin B synthase in A. oryzae RIB40 (XP_001821530.1) and AflK in A. flavus (XP_002379930.1) was 99.69%, between choline dehydrogenase in A. oryzae 3.042 (EIT81342.1) and XP_002379930.1 was 99.53%, between versicolorin B synthase in A. parasiticus (Q12062.1) and XP_002379930.1 was 98.76%, and between conserved hypothetical protein in A. nidulans FGSC A4 (CBF80156.1) and XP_002379930.1 was 75.70%. As shown in Figure 1a, the phylogenetic tree demonstrated that AflK from A. flavus and VBS from other Aspergillus species belonged to the same clade. Within this clade, A. flavus AflK and A. oryzae VBS belonged to the same grade, VBS from A. parasiticus and A. sojae were in the same grade, but A. flavus AflK and A. parasiticus VBS were not. Based on the results of SMART, three domains were identified in AflK of A. flavus, including the transmembrane domain (from 13th to 35th amino acid), the C‐terminal of GMC oxidoreductase (GMC_oxred_C, from 496th to 635th amino acid) and D‐amino acid oxidase (DAO) domain (from 76th to 382nd amino acid) or the N‐terminal of GMC oxidoreductase (GMC_oxred_N, from 75th to 394th amino acid) (Figure 1b). These results suggested that AflK was conserved but relatively unique.
Figure 1.
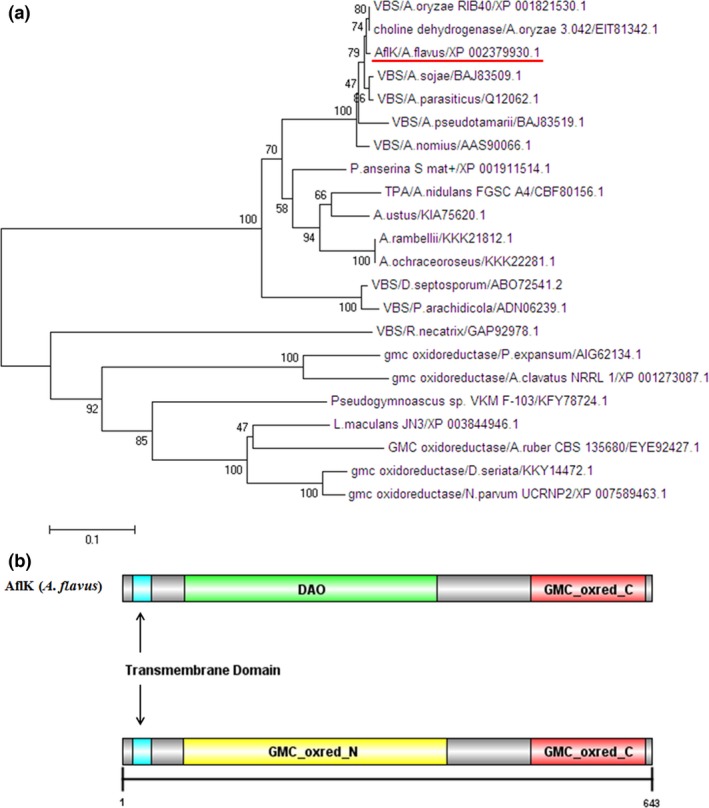
Phylogenetic tree and domain analysis of AflK in A. flavus. (a) Phylogenetic tree of AflK (versicolorin B synthase). AflK of A. flavus is marked with a red line. (b) Domain analysis of AflK
3.2. Growth and conidia production of WT, ΔaflK, and ΔaflK::aflK
aflK is one element of the aflatoxin biosynthesis cluster. Although it has been fully studied in A. parasiticus, our phylogenetic analysis showed that AflK of A. flavus and VBS of A. parasiticus were in different grades of one clade suggesting a possible difference or specialty of AflK in A. flavus. To study the potential function of A. flavus aflK, we constructed an aflK deletion mutant by replacing its open reading frame with a selection marker, pyrG. All primers used in this study are listed in Table 1, and a schematic of this method is given in Figure 2a. Deletion and complementation mutants of aflK were successfully obtained as verified by PCR of selected transformants (data not shown); the failure or restoration of gene expression in the ΔaflK mutant and ΔaflK::aflK strain was confirmed by RT‐PCR (Figure 2b). Next, we studied the role of AflK in A. flavus development. As shown in Figures 2c and d, no significant differences were observed in the colonial phenotypes or diameters when WT, ΔaflK, and ΔaflK::aflK were cultured on YES plates. The effect of aflK on conidiation was also studied by culturing WT, ΔaflK, and ΔaflK::aflK onto PDA plates and YES agar at 37°C. No significant difference was found in the conidia production of the three strains (Figure 3a and Figure 3b). Figure 3c shows that the conidiophores of these strains cultured on YES agar were identical. These results indicated that aflK does not affect the growth and conidia production of A. flavus.
Table 1.
Primers used in this study
| Primers | Sequence(5′‐3′) | Application |
|---|---|---|
| aflK‐1 | CGGAGCCTCGTTCCAAAT | To amplify upstream fragment of aflK |
| aflK‐3 | ACTGACAATTTGAAGCTGGAATCCCTCAACGGGCTTTC | |
| aflK‐4 | TCAGGACCACATACTCTACCATACCGTTCTCGAACTCGC | To amplify downstream fragment of aflK |
| aflK‐6 | GTTGGTGGACGTTGATGG | |
| aflK‐2 | CTCGTCCTTCCATCCTCT | To amplify overlap fragment to replace aflK |
| aflK‐5 | CTGTTCGTCAAAGCCCTA | |
| pyrG‐F | GCCTCAAACAATGCTCTTCACCC | To amplify pyrG |
| pyrG‐R | GTCTGAGAGGAGGCACTGATGC | |
| P801‐R | CAGGAGTTCTCGGGTTGTCG | To verify overlap fragment of aflK |
| P1020‐F | ATCGGCAATACCGTCCAGAAGC | |
| aflK‐OF | CTCAGGGCTTCTCCAATG | To verify ORF of aflK |
| aflK‐OR | CTCGGGTCCAGCAACG | |
| aflK‐com‐F | CTATGACCATGATTACGCCAAGCTTAGGATCAGATCTGGGATG | To amplify complementation vector of aflK |
| aflK‐com‐R | CCAGTGAATTCGAGCTCGGTACCTGATAGTTTCATCGCC | |
| actin‐F | CAGCCGCTAAGAGTTCCAG | To amplify actin |
| actin‐R | CACCGATCCAAACCGAGTAC |
Figure 2.
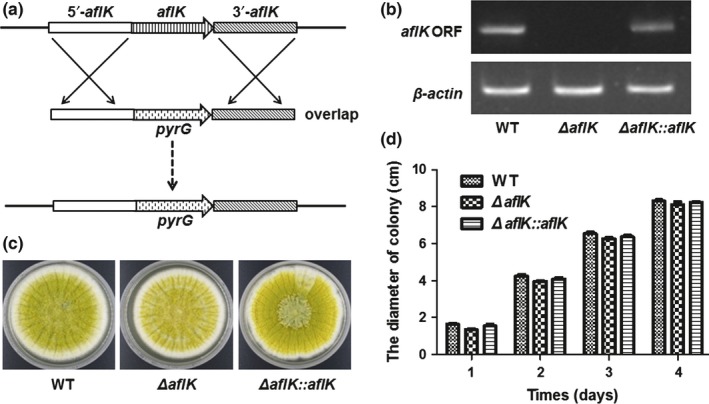
Construction of ∆aflK and growth analysis of WT, ∆aflK, and ∆aflK::aflK. (a) Schematic of the replacement of aflK (versicolorin B synthase gene) with pyrG. (b) cDNA verification of ∆aflK and ∆aflK::aflK. The actin gene was used as a reference. ORF, open reading frame of aflK. (c) Morphological phenotypes of the three strains cultivated with YES. (d) Quantification analysis of colony diameter
Figure 3.
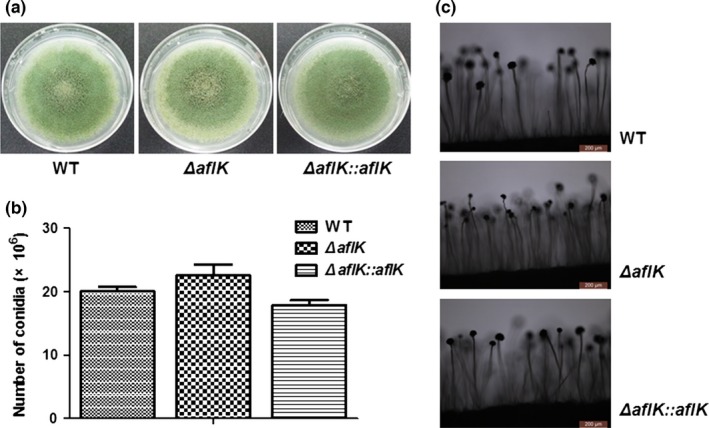
Conidiation phenotype of WT, ∆aflK, and ∆aflK::aflK. (a) Morphological phenotypes of WT, ∆aflK, and ∆aflK::aflK cultivated with PDA media. (b) Quantification analysis of conidia. (c) Microphotograph of conidiophore cultivated with YES media
3.3. Reduction of aflatoxin biosynthesis in ΔaflK
aflK is located in the aflatoxin synthesis gene cluster of A. flavus, so we next investigated aflatoxin biosynthesis after the deletion of aflK. To examine the role played by aflK in aflatoxin synthesis, we tested aflatoxin production in WT, ∆aflK, and ∆aflK::aflK by TLC at 6th day. The aflatoxin B1 (AFB1) produced by ΔaflK was reduced compared with that of WT and ΔaflK::aflK (Figure 4a), and Figure 4b shows the quantitative analysis of AFB1 production. HPLC was also used to confirm these results, which indicated that AFB1 production of ΔaflK was lower than that of WT and ΔaflK::aflK strains (Figure 4c). The original maps of TLC and HPLC are shown as the representative result. These findings suggested that aflK plays a vital role in aflatoxin biosynthesis in A. flavus.
Figure 4.
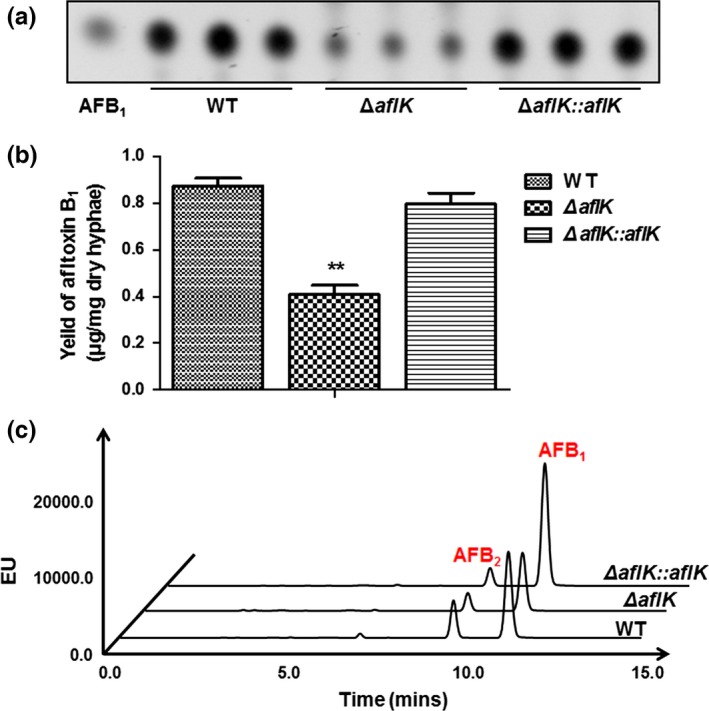
Aflatoxin production of WT, ∆aflK, and ∆aflK::aflK. (a) TLC analysis of aflatoxin B1 (AFB 1) production. (b) Quantification analysis of AFB 1 as in (a). Asterisks ** represent a significant difference (p‐value < .01). (c) HPLC analysis of aflatoxin production. The peak of AFB 1 is almost at 10.9 min and AFB 2 is almost at 9.1 min
3.4. Negative role of aflK in sclerotial production
Previous studies have shown that sclerotial production may be associated with aflatoxin biosynthesis (Calvo et al., 2004; Duran, Cary, & Calvo, 2007). Therefore, we determined the effect of aflK on sclerotial production by culturing the strains on Wickerham agar at 37°C for 7 days in the dark. The conidia were washed away with 75% ethanol to visualize the sclerotial phenotypes, as shown in Figure 5a and b. Surprisingly, the sclerotial production of ΔaflK was significantly decreased compared with that of WT and ΔaflK::aflK strains (Figure 5c), indicating that aflK plays an important role in the sclerotial production of A. flavus.
Figure 5.
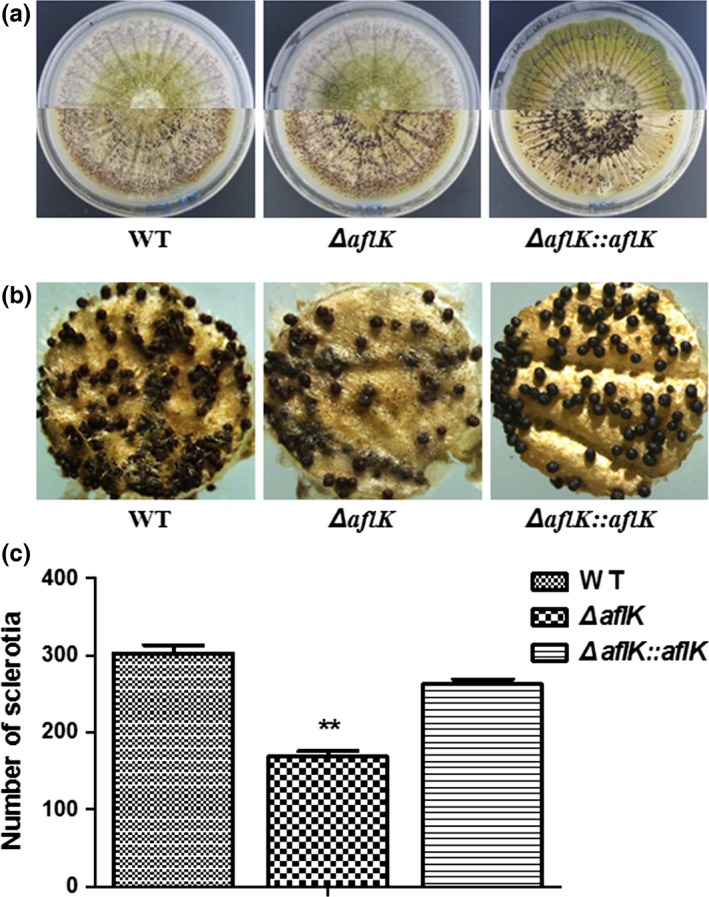
Sclerotial phenotype of WT, ∆aflK, and ∆aflK::aflK. (a) Phenotypic characterization of WT, ∆aflK, and ∆aflK::aflK before (upper section) and after (lower section) ethanol treatment. (b) Microphotograph of sclerotia. (c) Quantification analysis of sclerotia. Asterisks ** represent a significant difference (p‐value < .01)
3.5. Seed infections of WT, ΔaflK, and ΔaflK::aflK
A. flavus has a high number of diverse virulence factors and is a biological hazard to crops (Amaike & Keller, 2011). Based on the observed changes in aflatoxin production and sclerotial production caused by aflK deletion, we determined whether the aflK deletion also affected the ability of the strain to infect seeds. Peanut and maize were treated with conidia from WT, ΔaflK, and ΔaflK::aflK strains, but no obvious infection ability of A. flavus was observed (Figure 6a), and there was no significant difference in the number of conidia after infection of seeds by WT, ΔaflK, and ΔaflK::aflK (Figure 6b). Aflatoxin was extracted with chloroform from infected peanut cotyledons and maize kernels, showing that the deletion of aflK reduced AFB1 production after seed infection (Figure 6c). Quantification analysis of AFB1 was shown in Figure 6d, based on the results of Figure 6c. Together, these results illustrated that the deletion of aflK did not influence the infection ability of A. flavus, but reduced the level of aflatoxin biosynthesis after seed infection.
Figure 6.
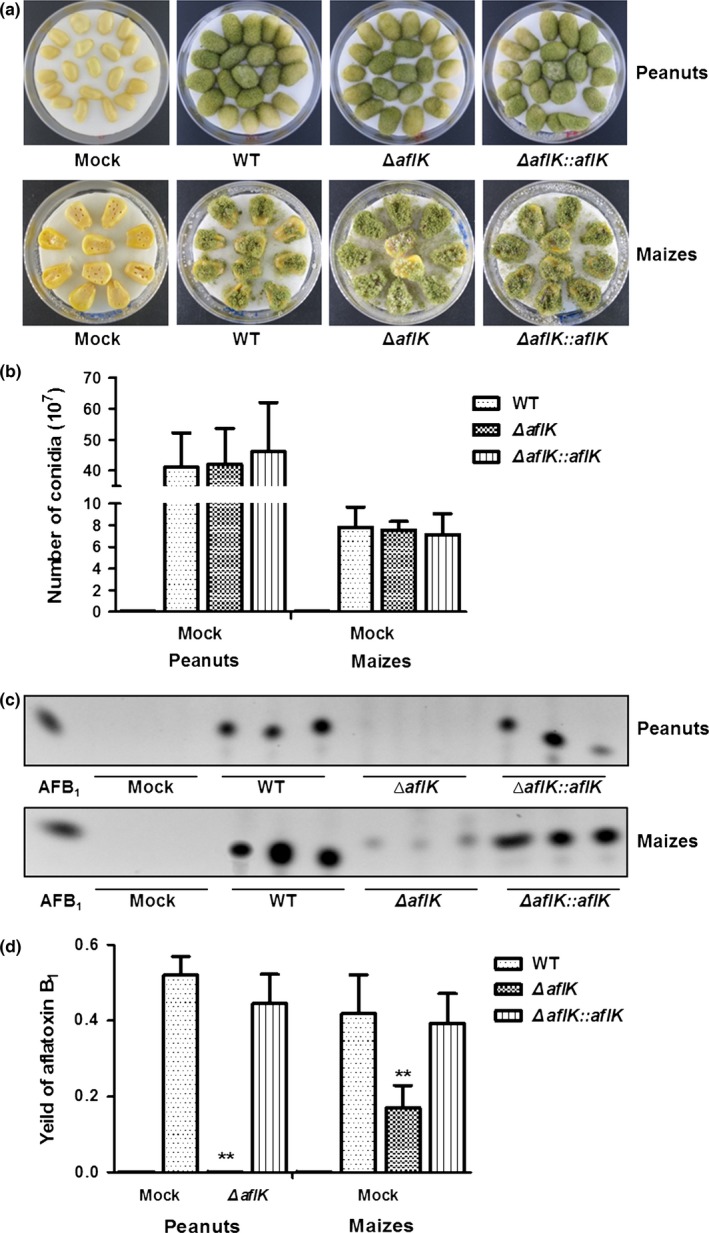
Seed infection ability and aflatoxin production (after infection) of WT, ∆aflK, and ∆aflK::aflK. (a) Peanut cotyledons and maize kernels were infected by the three strains. (b) Quantification analysis of conidia production after infection. (c) Thin‐layer chromatography analysis of AFB 1 production. (d) Quantification analysis of AFB 1. Asterisks ** represent a significant difference (p‐value < .01)
4. Discussion
In this study, we found that VBS was relative conserved in Aspergillus sp., and that it also shares a high level of identity with GMC oxidoreductase in other pathogenic microorganisms. The domain prediction analysis further confirmed this finding, revealing a probable DAO domain or GMC domain in the AflK protein. The DAO domain is the characteristic structure of D‐amino acid oxidase, which catalyses the oxidation of neutral and basic D‐amino acids (Miyano et al., 1991). D‐amino acid oxidase is located in peroxisomes (Pollegioni, Piubelli, Sacchi, Pilone, & Molla, 2007), in which the early steps of aflatoxin biosynthesis are likely to occur, and were shown by Chanda et al. (2009) to supply acetyl‐CoA for aflatoxin synthesis. This suggests that the A. flavus AflK protein plays a pivotal role in aflatoxin biosynthesis and is determined by its domain. However, the overall level of homology between A. flavus AflK and common Aspergillus sp. (A. oryzae, A. parasiticus, and A. nidulans) did not always exceed 99%. Phylogenetic tree analysis showed further differences between A. flavus AflK and its homologous proteins.
Previous investigations of the aflatoxin biosynthesis pathway fully researched its enzymes, substrates, and products, and largely focused on the detection and control of aflatoxin production (Bennett & Christensen, 1983; Bhatnagar, Ehrlich, & Cleveland, 2003; Biollaz, Buchi, & Milne, 1970; Cary, Szerszen, & Calvo, 2009; Ellis, Smith, Simpson, & Oldham, 1991; Gupta, Prasanna, Viswanathan, & Venkitasubramanian, 1975; Maggon, Gupta, & Venkitasubramanian, 1977). Thus, such studies have included the examination of aflK, encoding versicolorin B synthase (Yabe & Nakajima, 2004; Yu et al., 2002, 2004), revealing its heterologous expression, purification, isolation, characterization, distribution, and subcellular localization in A. parasiticus (Chiou et al., 2004; McGuire et al., 1996; Silva & Townsend, 1997; Silva et al., 1996). Because few aflK studies have been carried out in A. flavus, we performed homologous recombination to obtain an aflK deletion mutant and used a commercial chromosomal integrating shuttle vector to produce an aflK complementation strain. It is the first time that a gene knockout method is conducted for A. flavus ∆aflK. Our research focused not only on the role of aflK in aflatoxin biosynthesis, but also in development and infection of A. flavus.
Our results showed that the deletion of aflK had no effect on growth and conidiation of A. flavus. However, aflK appeared to play an important role in aflatoxin production, which is consistent with previous studies about aflatoxin biosynthesis (Yabe & Nakajima, 2004; Yu et al., 2002, 2004). Moreover, the aflK deletion did not completely inhibit the production of aflatoxin, as previously seen following the deletion of A. parasiticus aflE (Trail, Chang, Cary, & Linz, 1994). This may be because an alternative protein compensated for the role of AflK. A BLAST search of A. flavus AflK identified other proteins with different accession numbers in A. flavus homologous to VBS. Interestingly, we found that the inactivation of aflK decreased sclerotial production in A. flavus. The synchronous variation between sclerotia and aflatoxin was comparable with previous studies of veA and laeA (Amaike & Keller, 2009; Calvo et al., 2004; Duran et al., 2007; Kale et al., 2008). Aflatoxin biosynthesis in A. flavus and A. parasiticus was previously shown to be regulated by veA and laeA, which are also necessary for sclerotial formation (Calvo et al., 2004; Kale et al., 2008). We therefore speculate that sclerotial formation was decreased in response to the deletion of aflK, under the effects of these regulatory genes. But this is the first time to reveal the effect of aflatoxin biosynthesis gene aflK on sclerotial formation. Our results therefore indicate that aflK has an important role in aflatoxin biosynthesis and sclerotial production in A. flavus.
We found that an aflK deletion did not affect the infection of A. flavus, but caused a notable decrease in aflatoxin production after infection of peanut and maize. The aflK deletion also had no effect on conidia production after infection, consistent with our findings that aflK did not affect growth or conidia production, but regulated aflatoxin biosynthesis of A. flavus. This could reflect the fact that A. flavus virulence is a multifactorial process closely connected with secondary metabolism, developmental linkage of sporulation, and other aspects of A. flavus (Amaike & Keller, 2011). Hence, because aflK is only one part of the aflatoxin biosynthesis gene cluster, it did not influence the infection of A. flavus, but decreased aflatoxin production after infection.
In conclusion, AflK appears to be a conservative and unique oxidoreductase of Aspergillus sp. This is the first report of the influence of aflK on both sclerotial formation and aflatoxin biosynthesis in A. flavus. A more comprehensive investigation of the role of aflK in the development and pathogenicity of A. flavus will be conducted in future studies.
Conflict of Interest
None declared.
Acknowledgments
This work was funded by National 973 Program of the Ministry of Science and Technology of China (No. 2013CB127802), and the grants of the National Natural Science Foundation of China (No. 31172297, No.31400100). We are grateful to all other teachers and students of Key Laboratory of Pathogenic Fungi and Mycotoxins of Fujian Province for assistance provided.
Ren S, Yue Y, Li Y, Guo X, Wang S. Functional analyses of the versicolorin B synthase gene in Aspergillus flavus . MicrobiologyOpen. 2017;6:e471 https://doi.org/10.1002/mbo3.471
References
- Abdel‐Hadi, A. , Schmidt‐Heydt, M. , Parra, R. , Geisen, R. , & Magan, N. (2012). A systems approach to model the relationship between aflatoxin gene cluster expression, environmental factors, growth and toxin production by Aspergillus flavus . Journal of the Royal Society Interface, 9, 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Saad, L. A. , Al‐Badran, A. I. , Al‐Jumayli, S. A. , Magan, N. , & Rodriguez, A. (2016). Impact of bacterial biocontrol agents on aflatoxin biosynthetic genes, aflD and aflR expression, and phenotypic aflatoxin B1 production by Aspergillus flavus under different environmental and nutritional regimes. International Journal of Food Microbiology, 217, 123–129. [DOI] [PubMed] [Google Scholar]
- Amaike, S. , & Keller, N. P. (2009). Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus . Eukaryotic Cell, 8, 1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaike, S. , & Keller, N. P. (2011). Aspergillus flavus . Annual Review of Phytopathology, 49, 107–133. [DOI] [PubMed] [Google Scholar]
- Bennett, J. W. , & Christensen, S. B. (1983). New perspectives on aflatoxin biosynthesis. Advances in Applied Microbiology, 29, 53–92. [DOI] [PubMed] [Google Scholar]
- Bhatnagar, D. , Ehrlich, K. C. , & Cleveland, T. E. (2003). Molecular genetic analysis and regulation of aflatoxin biosynthesis. Applied Microbiology and Biotechnology, 61, 83–93. [DOI] [PubMed] [Google Scholar]
- Biollaz, M. , Buchi, G. , & Milne, G. (1970). The biosynthesis of the aflatoxins. Journal of the American Chemical Society, 92, 1035–1043. [DOI] [PubMed] [Google Scholar]
- Calvo, A. M. , Bok, J. , Brooks, W. , & Keller, N. P. (2004). veA is required for toxin and sclerotial production in Aspergillus parasiticus . Applied and Environmental Microbiology, 70, 4733–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary, J. W. , Ehrlich, K. C. , Bland, J. M. , & Montalbano, B. G. (2006). The aflatoxin biosynthesis cluster gene, aflX, encodes an oxidoreductase involved in conversion of versicolorin A to demethylsterigmatocystin. Applied and Environmental Microbiology, 72, 1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary, J. W. , Szerszen, L. , & Calvo, A. M. (2009). Regulation of Aspergillus flavus aflatoxin biosynthesis and development. Mycotoxin Prevention and Control in Agriculture, 1031, 183–203. [Google Scholar]
- Chanda, A. , Roze, L. V. , Kang, S. , Artymovich, K. A. , Hicks, G. R. , Raikhel, N. V. , & Linz, J. E. (2009). A key role for vesicles in fungal secondary metabolism. Proceedings of the National Academy of Science of the United States of America, 106, 19533–19538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, P. K. , Scharfenstein, L. L. , Luo, M. , Mahoney, N. , Molyneux, R. J. , Yu, J. , & Campbell, B. C. (2011). Loss of msnA, a putative stress regulatory gene, in Aspergillus parasiticus and Aspergillus flavus increased production of conidia, aflatoxins and kojic acid. Toxins, 3, 82–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, P. K. , Scharfenstein, L. L. , Mack, B. , & Ehrlich, K. C. (2012). Deletion of the Aspergillus flavus orthologue of A. nidulans fluG reduces conidiation and promotes production of sclerotia but does not abolish aflatoxin biosynthesis. Applied and Environmental Microbiology, 78, 7557–7563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, P. K. , Scharfenstein, L. L. , Wei, Q. , & Bhatnagar, D. (2010). Development and refinement of a high‐efficiency gene‐targeting system for Aspergillus flavus . Journal of Microbiological Methods, 81, 240–246. [DOI] [PubMed] [Google Scholar]
- Chiou, C. H. , Lee, L. W. , Owens, S. A. , Whallon, J. H. , Klomparens, K. L. , Townsend, C. A. , & Linz, J. E. (2004). Distribution and sub‐cellular localization of the aflatoxin enzyme versicolorin B synthase in time‐fractionated colonies of Aspergillus parasiticus . Archives of Microbiology, 182, 67–79. [DOI] [PubMed] [Google Scholar]
- Duran, R. M. , Cary, J. W. , & Calvo, A. M. (2007). Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Applied Microbiology and Biotechnology, 73, 1158–1168. [DOI] [PubMed] [Google Scholar]
- Ehrlich, K. C. , & Mack, B. M. (2014). Comparison of expression of secondary metabolite biosynthesis cluster genes in Aspergillus flavus, A. parasiticus, and A. oryzae . Toxins, 6, 1916–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, W. O. , Smith, J. P. , Simpson, B. K. , & Oldham, J. H. (1991). Aflatoxins in food: Occurrence, biosynthesis, effects on organisms, detection, and methods of control. Critical Reviews in Food Science and Nutrition, 30, 403–439. [DOI] [PubMed] [Google Scholar]
- Gehret, A. U. , Connelly, S. M. , & Dumont, M. E. (2012). Functional and physical interactions among Saccharomyces cerevisiae alpha‐factor receptors. Eukaryotic Cell, 11, 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S. R. , Prasanna, H. R. , Viswanathan, L. , & Venkitasubramanian, T. A. (1975). Factors influencing the biosynthesis of aflatoxins. Indian Journal of Biochemistry and Biophysics, 12, 179–181. [PubMed] [Google Scholar]
- He, Z. M. , Price, M. S. , OBrian, G. R. , Georgianna, D. R. , & Payne, G. A . (2007). Improved protocols for functional analysis in the pathogenic fungus Aspergillus flavus . BMC Microbiology, 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale, S. P. , Milde, L. , Trapp, M. K. , Frisvad, J. C. , Keller, N. P. , & Bok, J. W. (2008). Requirement of LaeA for secondary metabolism and sclerotial production in Aspergillus flavus . Fungal Genetics and Biology, 45, 1422–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linz, J. E. , Wee, J. M. , & Roze, L. V . (2014). Aflatoxin biosynthesis: Regulation and subcellular localization. In: Martín J. F., García‐Estrada C., Zeilinger S. (Eds.), Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites, Fungal Biology (89–110). NewYork: Springer. [Google Scholar]
- Maggon, K. K. , Gupta, S. K. , & Venkitasubramanian, T. A. (1977). Biosynthesis of Aflatoxins. Bacteriological Reviews, 41, 822–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, Z. , Bagnara, A. , Farber, P. , & Geisen, R. (2003). Quantification of the copy number of nor‐1, a gene of the aflatoxin biosynthetic pathway by real‐time PCR, and its correlation to the cfu of Aspergillus flavus in foods. International Journal of Food Microbiology, 82, 143–151. [DOI] [PubMed] [Google Scholar]
- McGuire, S. M. , Silva, J. C. , Casillas, E. G. , & Townsend, C. A. (1996). Purification and characterization of versicolorin B synthase from Aspergillus parasiticus. Catalysis of the stereodifferentiating cyclization in aflatoxin biosynthesis essential to DNA interaction. Biochemistry, 35, 11470–11486. [DOI] [PubMed] [Google Scholar]
- Miyano, M. , Fukui, K. , Watanabe, F. , Takahashi, S. , Tada, M. , Kanashiro, M. , & Miyake, Y. (1991). Studies on Phe‐228 and Leu‐307 recombinant mutants of porcine kidney D‐amino acid oxidase: Expression, purification, and characterization. Journal of Biochemistry, 109, 171–177. [DOI] [PubMed] [Google Scholar]
- Pollegioni, L. , Piubelli, L. , Sacchi, S. , Pilone, M. S. , & Molla, G. (2007). Physiological functions of D‐amino acid oxidases: From yeast to humans. Cellular and Molecular Life Sciences, 64, 1373–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, S. , Yang, M. , Li, Y. , Zhang, F. , Chen, Z. , Zhang, J. , & Wang, S. (2016). Global phosphoproteomic analysis reveals the involvement of phosphorylation in aflatoxins biosynthesis in the pathogenic fungus Aspergillus flavus . Scientific Reports, 6, 34078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt‐Heydt, M. , Abdel‐Hadi, A. , Magan, N. , & Geisen, R. (2009). Complex regulation of the aflatoxin biosynthesis gene cluster of Aspergillus flavus in relation to various combinations of water activity and temperature. International Journal of Food Microbiology, 135, 231–237. [DOI] [PubMed] [Google Scholar]
- Shieh, M. T. , Brown, R. L. , Whitehead, M. P. , Cary, J. W. , Cotty, P. J. , Cleveland, T. E. , & Dean, R. A. (1997). Molecular genetic evidence for the involvement of a specific polygalacturonase, P2c, in the invasion and spread of Aspergillus flavus in cotton bolls. Applied and Environmental Microbiology, 63, 3548–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, J. C. , Minto, R. E. , Barry, C. E. 3rd , Holland, K. A. , & Townsend, C. A. (1996). Isolation and characterization of the versicolorin B synthase gene from Aspergillus parasiticus. Expansion of the aflatoxin B1 biosynthetic gene cluster. Journal of Biological Chemistry, 271, 13600–13608. [DOI] [PubMed] [Google Scholar]
- Silva, J. C. , & Townsend, C. A. (1997). Heterologous expression, isolation, and characterization of versicolorin B synthase from Aspergillus parasiticus. A key enzyme in the aflatoxin B1 biosynthetic pathway. Journal of Biological Chemistry, 272, 804–813. [DOI] [PubMed] [Google Scholar]
- Sweeney, M. J. , Pamies, P. , & Dobson, A. D. (2000). The use of reverse transcription‐polymerase chain reaction (RT‐PCR) for monitoring aflatoxin production in Aspergillus parasiticus 439. International Journal of Food Microbiology, 56, 97–103. [DOI] [PubMed] [Google Scholar]
- Szewczyk, E. , Nayak, T. , Oakley, C. E. , Edgerton, H. , Xiong, Y. , Taheri‐Talesh, N. , & Oakley, B. R. (2006). Fusion PCR and gene targeting in Aspergillus nidulans . Nature Protocols, 1, 3111–3120. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. , & Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trail, F. , Chang, P. K. , Cary, J. , & Linz, J. E. (1994). Structural and functional analysis of the nor‐1 gene involved in the biosynthesis of aflatoxins by Aspergillus parasiticus . Applied and Environmental Microbiology, 60, 4078–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsigiannis, D. I. , & Keller, N. P. (2006). Oxylipins act as determinants of natural product biosynthesis and seed colonization in Aspergillus nidulans . Molecular Microbiology, 59, 882–892. [DOI] [PubMed] [Google Scholar]
- Yabe, K. , & Nakajima, H. (2004). Enzyme reactions and genes in aflatoxin biosynthesis. Applied Microbiology and Biotechnology, 64, 745–755. [DOI] [PubMed] [Google Scholar]
- Yang, K. L. , Liang, L. L. , Ran, F. L. , Liu, Y. H. , Li, Z. G. , Lan, H. H. , & Wang, S. H. (2016). The DmtA methyltransferase contributes to Aspergillus flavus conidiation, sclerotial production, aflatoxin biosynthesis and virulence. Scientific Reports, 6, 23259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. , Bhatnagar, D. , & Ehrlich, K. C. (2002). Aflatoxin biosynthesis. Revista Iberoamericana de Micologia, 19, 191–200. [PubMed] [Google Scholar]
- Yu, J. , Chang, P. K. , Ehrlich, K. C. , Cary, J. W. , Bhatnagar, D. , Cleveland, T. E. , & Bennett, J. W. (2004). Clustered pathway genes in aflatoxin biosynthesis. Applied and Environmental Microbiology, 70, 1253–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F. , Guo, Z. , Zhong, H. , Wang, S. , Yang, W. , Liu, Y. , & Wang, S. (2014). RNA‐Seq‐based transcriptome analysis of aflatoxigenic Aspergillus flavus in response to water activity. Toxins, 6, 3187–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]


