Abstract
Despite the existence of a commercial vaccine routinely used to protect salmonids against Yersinia ruckeri, outbreaks still occur, mainly caused by nonmotile and lipase‐negative strains (serotype O1 biotype 2). Moreover, epizootics caused by other uncommon serotypes have also been reported. At the moment, one of the main concerns for the aquaculture industry is the expanding range of hosts of this pathogen and the emergence of new biotypes and serotypes causing mortality in fish farms and against which the vaccine cannot protect. The comparative analysis of the genome sequences of five Y. ruckeri strains (150, CSF007‐82, ATCC29473, Big Creek 74, and SC09) isolated from different hosts and classified into different serotypes revealed important genetic differences between the genomes analyzed. Thus, a clear genetic differentiation was found between serotype O1 and O2 strains. The presence of 99 unique genes in Big Creek 74 and 261 in SC09 could explain the adaptation of these strains to salmon and catfish, respectively. Finally, the absence of 21 genes in ATCC29473 which are present in the other four virulent strains could underpin the attenuation described for this strain. The study reveals important genetic differences among the genomes analyzed. Further investigation of the genes highlighted in this study could provide insights into the understanding of the virulence and niche adaptive mechanisms of Y. ruckeri.
Keywords: adaptation, Genome, host, Serotype, Virulence, Yersinia ruckeri
1. Background
Yersinia ruckeri is a gram‐negative rod‐shaped bacterium able to infect different fish species such as rainbow trout, carp, catfish, sturgeon, burbot, and perch. In salmonids, it causes enteric red mouth disease (ERM), a serious septicemic fish disease which is a major problem for aquaculture industries all over the world. Since its isolation in the United States in the 1950s (Ross, Rucker, & Ewing, 1966), the number of host species and the geographic distribution of this pathogen have increased considerably (Bastardo, Ravelo, & Romalde, 2015). Despite the existence of a reasonably effective immersion vaccine, outbreaks still occur, produced mainly by nonmotile and lipase‐negative strains (Arias et al., 2007; Austin, Robertson, & Austin, 2003; Calvez, Gantelet, Blanc, Douet, & Daniel, 2014; Fouz, Zarza, & Amaro, 2006).
In 1993, Y. ruckeri strains were classified into four serotypes with different subgroups (Romalde, Magariños, Barja, & Toranzo, 1993). The vast majority of epizootics in salmonid fish farms are caused by motile serotype O1, although epizootics caused by other uncommon serotypes have also been reported (Romalde, Planas, Sotelo, & Toranzo, 2003). Recently, a significant positive correlation between genetic and geographical distances was observed by Bastardo et al., 2015. Their results revealed that Y. ruckeri has experienced population changes that were probably induced by biogeography forces in the past and, much more recently, by adaptive processes resulting from aquaculture expansion.
During the last few years, nine genome sequences of Y. ruckeri strains, isolated from different niches have been uploaded onto NCBI (MKFJ00000000, NZ_CP011078, NZ_CP009539, JPFO00000000, CQBN00000000, CPUZ00000000, JPPT00000000, CCYO00000000, and JRWX00000000). Here, we present for the first time in this species a comparative analysis of five of those genomes belonging to strains isolated from different hosts and classified into different serotypes. The study reveals data that are important for a better understanding of the mechanisms underlying the niche adaptation and virulence of Y. ruckeri.
2. Materials and Methods
2.1. Y. ruckeri strains used for genome comparison
Five previously sequenced Y. ruckeri strains were selected for comparative genome analysis based upon their characteristics and hosts (Table 1). Three strains were from serotype O1, isolated from rainbow trout (Oncorhynchus mykiss), of which two were virulent (Y. ruckeri 150 and Y. ruckeri CSF007‐82), while the other, ATCC29473 type strain, was described as nonvirulent (Furones, Gilpin, Alderman, & Munn, 1990). The other two strains included in the analysis were Y. ruckeri Big Creek 74, belonging to serotype O2 and isolated from salmon, and Y. ruckeri SC09 isolated from catfish and of unknown serotype.
Table 1.
List of Y. ruckeri strains used in comparative analysis
| Characteristic | 150 | ATCC29473 | CSF007‐82 | Big Creek 74 | SC09 |
|---|---|---|---|---|---|
| Host | Rainbow trout | Rainbow trout | Rainbow trout | Chinook salmon | Catfish |
| Genome size (Mb) | 3.82 | 3.77 | 3.83 | 3.69 | 3.92 |
| Scaffolds | 49 | 2 | 1 | 1 | 32 |
| Contigs | 169 | 15 | 1 | 1 | 32 |
| GC (%) | 46.8 | 47.4 | 47.5 | 47.6 | 47.45 |
| CDS | 3,538 | 3,377 | 3,530 | 3,136 | 3,651 |
| RNAs | 25 | 80 | 102 | 103 | 127 |
| Serotype | O1 | O1 | O1 | O2 | – |
| Accession Number | MKFJ00000000 | JPPT00000000 | CCYO00000000 | CP011078 | JRWX00000000 |
2.2. Comparative analysis of Y. ruckeri genomes
Identification of putative protein‐encoding genes and annotation of Y. ruckeri genomes were performed with Rapid Annotation using Subsystem Technology (RAST) (Brettin et al., 2015). Before comparative analysis, the set of proteins from the five genomes selected were compared using BLAST to UniRef90 to associate each translation product to a Uniref90 protein. It was considered that a protein from one genome was orthologous to another one when they were in the same cluster. Based on this clusterization process, Venn diagrams were constructed with shared proteins (orthologous proteins) using the Venn diagram package in R (Chen & Boutros, 2011). Pairwise genome alignments were performed with MAUVE (Darling, Mau, Blattner, & Perna, 2004).
3. Results and Discussion
3.1. Y. ruckeri whole‐genome comparisons
The pairwise full genome alignments revealed a mosaic pattern of homology organized in local collinear blocks (LCBs) between 150 and each of the other four strains (Figure 1). The 150 strain shares larger portions of genetic information with CSF007‐82 and ATCC24973, than it does with Big Creek74 and SC09. This result suggests that Y. ruckeri strains belonging to serotype O1 and having rainbow trout as a host (150, ATCC29473, CSF007‐82) are genetically more similar to each other than to other serotypes isolated from different animals, suggesting that differences in cell surface antigens and host specificity may have a markedly genetic base.
Figure 1.
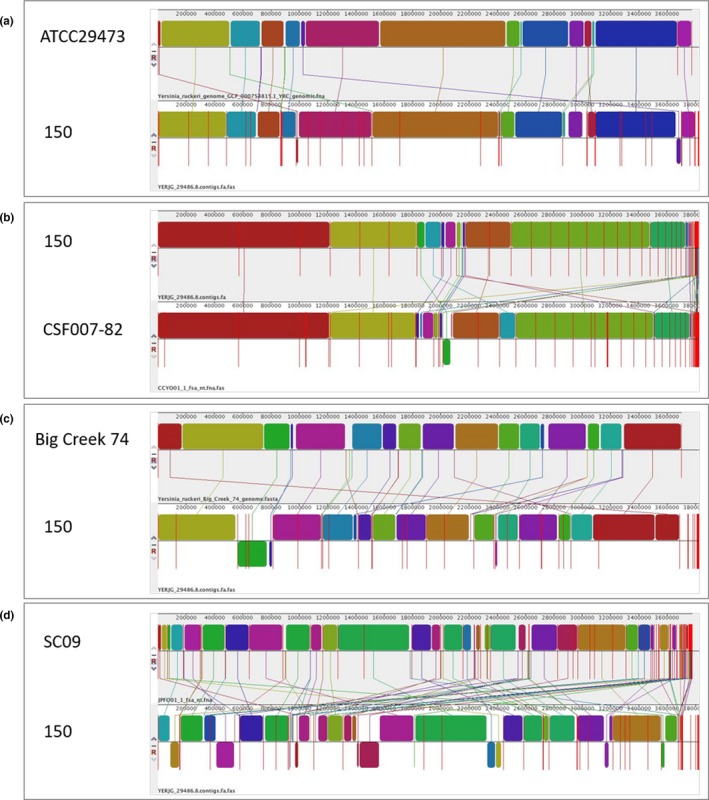
Chromosome alignments of Y. ruckeri 150 and (a) ATCC29473, (b) CSF007‐82, (c) Big Creek 74, and (d) SC09 using progressive Mauve. Local collinear blocks (LCBs) of conserved sequences among the strains are represented by rectangles of the same color. When these are above the chromosome (black line) they indicate the forward orientation and when they are positioned under the chromosome, the reverse orientation. Red lines represent contig limits. Connecting lines can be used to visualize genetic rearrangements
To identify orthologs shared by Y. ruckeri strains, a five‐way Venn diagram was made (Figure 2). The pangenome consists of 4,117 protein‐coding genes with a core of 3,090 genes (75.05%). A total of 370 genes were found to be strain‐specific, two genes corresponding to CSF007‐82, eight to ATCC29473, 99 to Big Creek 74, and 261 to SC09 genomes. Approximately, half of these unique genes (57%) were annotated as coding for hypothetical proteins. Interestingly, while serotype O1 strains isolated from rainbow trout have few unique genes (150 has none), the other two strains, Big Creek74 and SC09, have a great number, 99 and 261, respectively. Most of these genes could be related to host adaptation processes, in particular to survival in salmon in the first case and in catfish in the second.
Figure 2.
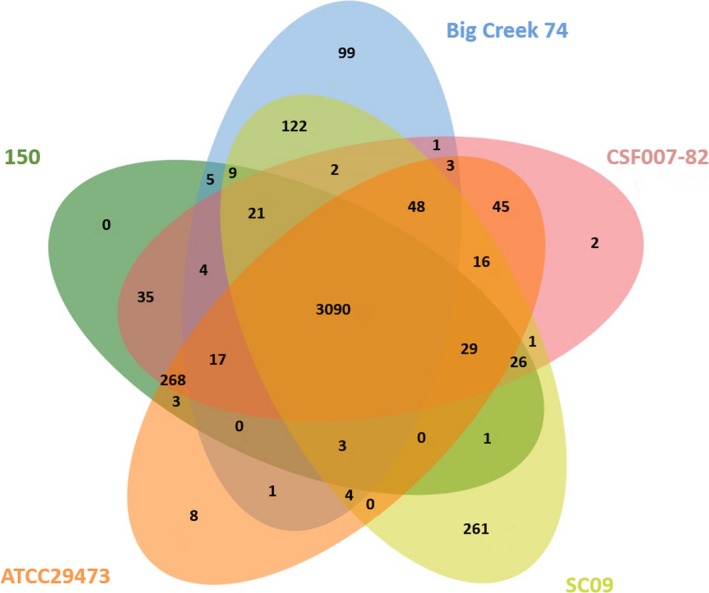
Venn diagram of Y. ruckeri 150, ATCC29473, CSF007‐82, Big Creek 74, and SC09. The number in the center of the diagram represents the genes shared by all species, whereas the digit on each branch indicates the number of unique or shared genes of the different strains
As can be seen from the Venn diagram, the 150, ATCC29473, and CSF007‐82 strains share between them, and not with the other two strains, a total of 268 genes (6.5%). Similarly, the Big Creek 74 and SC09 strains share 122 (3%) genes which are absent in the genomes of serotype O1 strains. These data suggest a clear separation between serotype O1 strains and the Big Creek 74 and SC09 strains; so, it is tempting to speculate that SC09, whose serotype has not been described yet, could belong to serotype O2, as does Big Creek 74. This differentiation may constitute the genetic basis for the variation in serotype‐associated features among the strains.
3.2. Genes exclusively shared by serotype O1 strains
As mentioned above, a total of 268 genes were shared by 150, ATCC29473, and CSF007‐82 strains, all of them belonging to serotype O1 and isolated from rainbow trout. These genes include 113 which encode for hypothetical proteins, 33 are mobile genetic elements, 24 encode for phage‐related proteins, and 98 for proteins with different functions (Table 2). Thus, some of them are associated with restriction‐modification and toxin‐antitoxin systems. Both these systems have in common the death of cells that have lost one of the components (the antitoxin or the modification enzyme) and also their effect on global gene expression, which results in altered adaptive phenotypes. Thus, the antitoxin of the Escherichia coli MqsR–MqsA toxin‐antitoxin system directly represses the transcription of the gene encoding the master stress regulator RpoS, while the degradation of the antitoxin during stress leads to a switch from the high‐motility state to biofilm formation (Wang et al., 2011). In the same way, methylation events produced by restriction‐modification systems may affect nearby gene expression. Thus, methylation by Type III RM systems controls the expression of certain genes leading to two distinct cell types with two distinct phenotypes (“phasevarion”) (Srikhanta, Fox, & Jennings, 2010).
Table 2.
Proteins exclusively shared by serotype O1 strains
| Protein | 150 | ATCC2947 | CSF007 |
|---|---|---|---|
| Restriction‐modification systems | |||
| Type I restriction‐modification system, specificity subunit S | BI323_00005 | DJ39_RS07815 | CSF007_RS15880 |
| Eco57I restriction‐modification methylase family protein | BI323_06175 | DJ39_RS04570 | CSF007_RS05790 |
| Type I restriction‐modification system, restriction subunit R | BI323_00080 | DJ39_RS07890 | CSF007_RS15805 |
| Type I restriction‐modification system, DNA‐methyltransferase subunit M | BI323_00010 | DJ39_RS07820 | CSF007_RS15875 |
| Restriction methylase | BI323_06275 | DJ39_RS04465 | CSF007_RS05895 |
| Restriction methylase | BI323_07120 | DJ39_RS07795 | CSF007_RS06135 |
| Antirestriction family protein | BI323_06250 | DJ39_RS04490 | CSF007_RS05870 |
| Antirestriction family protein | BI323_000601 | DJ39_RS07870 | CSF007_RS15825 |
| Toxin‐antitoxin systems | |||
| YfjZ protein (Antitoxin to YpjF) | BI323_00070 | DJ39_RS07880 | CSF007_RS15815 |
| Toxin YkfI | BI323_00075 | DJ39_RS07885 | CSF007_RS15810 |
| Toxin_HigB‐2_ | BI323_15350 | DJ39_RS16625 | CSF007_RS00015 |
| Antitoxin ParD | BI323_16600 | DJ39_RS11355 | CSF007_RS09305 |
| Toxin YkfI | BI323_06265 | DJ39_RS04475 | CSF007_RS05885 |
| Legionaminic acid biosynthesis | |||
| Dehydratase/C‐5‐epimerase | BI323_14300 | DJ39_RS00115 | CSF007_RS08290 |
| Aminotransferase | BI323_14305 | DJ39_RS00110 | CSF007_RS08285 |
| UDP‐N‐acetylglucosamine 2‐epimerase | BI323_14310 | DJ39_RS00105 | CSF007_RS08280 |
| N‐acetylneuraminate synthase | BI323_14315 | DJ39_RS00100 | CSF007_RS08275 |
| 4‐amino‐6‐deoxy‐N‐Acetyl‐D‐hexosaminyl‐(Lipid carrier) acetyltrasferase | BI323_14320 | DJ39_RS00095 | CSF007_RS08270 |
| Mannose‐1‐phosphate guanyltransferase | BI323_14325 | DJ39_RS00090 | CSF007_RS08265 |
| Oxidoreductase, NAD‐binding Rossmann fold family protein | BI323_14330 | DJ39_RS00085 | CSF007_RS08260 |
| Acylneuraminate cytidylyltransferase | BI323_14335 | DJ39_RS00080 | CSF007_RS08255 |
| Dehydrogenase | BI323_14340 | DJ39_RS00075 | CSF007_RS08250 |
| Polysaccharide biosynthesis family protein | BI323_14345 | DJ39_RS00070 | CSF007_RS08245 |
| Aminotransferase | BI323_14360 | DJ39_RS00055 | CSF007_RS08230 |
| Imidazole glycerol phosphate synthase subunit HisH | BI323_14365 | DJ39_RS00050 | CSF007_RS08225 |
| Imidazole glycerol phosphate synthase | BI323_14370 | DJ39_RS00045 | CSF007_RS08220 |
| Epimerase/dehydratase | BI323_14375 | DJ39_RS00040 | CSF007_RS08215 |
| UDP‐2‐acetamido‐2,6‐dideoxy‐beta‐L‐talose‐4‐dehy drogenase | BI323_14380 | DJ39_RS00035 | CSF007_RS08210 |
| UDP‐N‐acetylglucosamine 2‐epimerase | BI323_14385 | DJ39_RS00030 | CSF007_RS08205 |
| Glycosyl transferases group 1 family protein | BI323_14390 | DJ39_RS00025 | CSF007_RS08200 |
| Tryptophan synthase beta chain like | BI323_14395 | DJ39_RS00020 | CSF007_RS08195 |
| DNA repair | |||
| DNA repair RadC family protein | BI323_00065 | DJ39_RS07875 | CSF007_RS15820 |
| ATPase involved in DNA repair | BI323_15390 | DJ39_RS16660 | CSF007_RS00050 |
| DNA_repair_ATPase_ | BI323_06180 | DJ39_RS04565 | CSF007_RS05795 |
| ATPase involved in DNA repair | BI323_06215 | DJ39_RS04530 | CSF007_RS05830 |
| RadC family DNA repair protein | BI323_06255 | DJ39_RS04485 | CSF007_RS05875 |
| Transcriptional regulators | |||
| Regulator | BI323_01350 | DJ39_RS09205 | CSF007_RS01385 |
| XRE_family_transcriptional_regulator | BI323_15355 | DJ39_RS16630 | CSF007_RS00020 |
| Transcription_factor | BI323_16050 | DJ39_RS16900 | CSF007_RS16935 |
| XRE_family_transcriptional_regulator | BI323_05810 | DJ39_RS16260 | CSF007_RS05430 |
| Type IV secretion system | |||
| Type IV secretion‐system coupling DNA‐binding domain protein | BI323_15400 | DJ39_RS16670 | CSF007_RS00060 |
| TrbA | BI323_15410 | DJ39_RS16680 | CSF007_RS00070 |
| RelB/StbD replicon stabilization protein (Antitoxin to RelE/StbE) | BI323_15420 | DJ39_RS16690 | CSF007_RS00080 |
| Conjugal transfer/type IV secretion DotA/TraY family protein | BI323_15425 | DJ39_RS16680 | CSF007_RS00085 |
| IncI1 plasmid conjugative transfer protein TraW | BI323_15435 | DJ39_RS16705 | CSF007_RS00095 |
| IncI1 plasmid conjugative transfer protein TraU | BI323_15440 | DJ39_RS16710 | CSF007_RS00100 |
| IncI1 plasmid conjugative transfer protein | BI323_15455 | DJ39_RS16725 | CSF007_RS00115 |
| IncI1 plasmid conjugative transfer protein TraO | BI323_15465 | DJ39_RS16735 | CSF007_RS00125 |
| TraN | BI323_15470 | DJ39_RS16740 | CSF007_RS00130 |
| TraM | BI323_15475 | DJ39_RS16745 | CSF007_RS00135 |
| TraC | BI323_15485 | DJ39_RS16755 | CSF007_RS00145 |
| TraK | BI323_15490 | DJ39_RS16760 | CSF007_RS00150 |
| Plasmid transfer ATPase TraJ | BI323_15495 | DJ39_RS16765 | CSF007_RS00155 |
| TraI | BI323_15500 | DJ39_RS16770 | CSF007_RS00160 |
| IncI1 plasmid conjugative transfer protein TraH | BI323_15505 | DJ39_RS16775 | CSF007_RS00165 |
| Prepilin | BI323_15525 | DJ39_RS16795 | CSF007_RS00185 |
| General_secretion_pathway_protein_GspF | BI323_15530 | DJ39_RS16800 | CSF007_RS00190 |
| PilO | BI323_15545 | DJ39_RS16815 | CSF007_RS00205 |
| IncI1 plasmid conjugative transfer lipoprotein PilN | BI323_15550 | DJ39_RS16820 | CSF007_RS00210 |
| Type IVB pilus formation outer membrane protein, R64 PilN family | BI323_15555 | DJ39_RS16825 | CSF007_RS00215 |
| Transferases | |||
| Methyltransferase domain protein | BI323_16780 | DJ39_RS04020 | CSF007_RS16255 |
| Glycosyl transferase, family 2 | BI323_06290 | DJ39_RS04450 | CSF007_RS05910 |
| Other proteins | |||
| ATP/GTP‐binding_protein | BI323_00035 | DJ39_RS07845 | CSF007_RS15850 |
| Bipolar DNA helicase HerA | BI323_10945 | DJ39_RS01005 | CSF007_RS11255 |
| Phosphopantetheine attachment site family protein | BI323_13885 | DJ39_RS06465 | CSF007_RS15645 |
| AMP‐dependent synthetase | BI323_13890 | DJ39_RS06470 | CSF007_RS15640 |
| 3‐oxoacyl‐[acyl‐carrier protein] reductase | BI323_13895 | DJ39_RS06475 | CSF007_RS15635 |
| Polysaccharide deacetylase | BI323_13905 | DJ39_RS06485 | CSF007_RS15625 |
| Endonuclease | BI323_15395 | DJ39_RS16665 | CSF007_RS00055 |
| Putative ATP‐binding protein involved in virulence | BI323_16055 | DJ39_RS16905 | CSF007_RS16930 |
| ATP‐dependent DNA helicase RecG | BI323_16060 | DJ39_RS16910 | CSF007_RS16925 |
| Chromosome segregation ATPase | BI323_16110 | DJ39_RS16960 | CSF007_RS16875 |
| Initiator Replication family protein | BI323_16595 | DJ39_RS11350 | CSF007_RS09310 |
| Plasmid_stabilization_protein_ | BI323_16605 | DJ39_RS11360 | CSF007_RS09300 |
| Cobyrinic acid a,c‐diamide synthase | BI323_16640 | DJ39_RS11395 | CSF007_RS09265 |
| Glycosaminoglycan attachment site | BI323_16685 | DJ39_RS14755 | CSF007_RS03940 |
| Retron‐type RNA‐directed DNA polymerase | BI323_16690 | DJ39_RS14760 | CSF007_RS03935 |
| Sulfur transport family protein | BI323_16730 | DJ39_RS00135 | CSF007_RS17245 |
| Cytotoxic family protein | BI323_16760 | DJ39_RS17105 | CSF007_RS17365 |
| Low calcium response locus protein T | BI323_16800 | DJ39_RS04040 | CSF007_RS16275 |
| ParB/RepB/Spo0J family partition domain protein | BI323_16820 | DJ39_RS16600 | CSF007_RS16685 |
| Plasmid partition protein A | BI323_16825 | DJ39_RS16605 | CSF007_RS16690 |
| ATP‐dependent Lon protease | BI323_06155 | DJ39_RS04590 | CSF007_RS05770 |
| alkaline_phosphatase | BI323_06160 | DJ39_RS04585 | CSF007_RS05775 |
| ABC_transporter_ATP‐binding_protein_ | BI323_06170 | DJ39_RS04575 | CSF007_RS05785 |
| DNA‐binding_protein_ | BI323_06200 | DJ39_RS04545 | CSF007_RS05815 |
| AlpA family protein | BI323_06210 | DJ39_RS04535 | CSF007_RS05825 |
| GTPase | BI323_06225 | DJ39_RS04520 | CSF007_RS05840 |
| YagBYeeUYfjZ family protein | BI323_06270 | DJ39_RS04470 | CSF007_RS05890 |
| DNA‐binding protein | BI323_06285 | DJ39_RS04455 | CSF007_RS05905 |
| 37‐kD nucleoid‐associated bacterial protein | BI323_06340 | DJ39_RS00940 | CSF007_RS06920 |
| (p)ppGpp_synthetase_ | BI323_06345 | DJ39_RS00935 | CSF007_RS06915 |
| Colicin‐Ib | BI323_06365 | DJ39_RS17065 | CSF007_RS17470 |
| AAA ATPase | BI323_06375 | DJ39_RS00905 | CSF007_RS06885 |
| AAA ATPase | BI323_06400 | DJ39_RS00875 | CSF007_RS06855 |
| Invasin | BI323_06460 | DJ39_RS17045 | CSF007_RS17450 |
| DNA‐directed RNA polymerase subunit sigma70 | BI323_09490 | DJ39_RS13885 | CSF007_RS05930 |
| ATP‐binding_protein_ | BI323_09540 | DJ39_RS13835 | CSF007_RS05980 |
A relevant finding shared by all serotype O1 strains was related to a cluster of genes which are involved in the biosynthesis of the legionaminic acid, a nine‐carbon diamino monosaccharide that is found coating the surface of various bacterial human pathogens, being the major component of the LPS. Interestingly, these genes which are grouped in a cluster of at least 18 genes are absent in other Yersinia species but present in other aquatic bacteria such as Vibrio vulnificus, Aeromonas salmonicida, Vibrio fischeri, or Photobacterium profundum. It is possible that this cluster provides an adaptive advantage for surviving in the aquatic environment or, as happens in some organisms such as Campylobacter jejuni (Zebian et al., 2016), it is related to virulence. This is because legionaminic acid is essential for flagella assembly in several species (Morrison & Imperiali, 2014) and for this reason, the genes involved in its biosynthesis are novel targets for the development of antivirulence agents (Table 2).
Other genes which are exclusive to O1 serotype strains code for a bacteriocin similar to colicin‐Ib of Escherichia coli (WP_062877260) and virulence factors such as a type IV secretion system previously analyzed by Méndez et al., (2009) and an invasin present in other enterobacteriaceae such as Yersinia pestis (EIR59646), Y. pseudotuberculosis (WP_050128752), and Edwarsiella tarda (WP_047059316) (Table 2).
3.3. Genes exclusively shared by Big Creek 74 and SC09 strains
As indicated in the Venn diagram (Figure 2), Big Creek 74 and SC09 share a total of 122 genes which include 37 ORFs encoding for proteins of unknown function, 50 for phage‐related proteins, and 35 encode for proteins with similarity to proteins involved in a variety of functions such as restriction‐modification systems, toxin‐antitoxin systems or proteins involved in fimbriae synthesis (Table 3). One such case is that of a cluster involved in fimbriae biosynthesis, similar to the Stf cluster of Salmonella typhimurium which has been associated with differences in virulence and host range between the different serotypes (Emmerth, Goebel, Miller, & Hueck, 1999). Although one Stf cluster copy is present in the five genomes analyzed (Figure 3a), an additional complete copy of this cluster was only found in the genome of SC09 and, with the exception of the gene encoding the minor fimbriae subunit (stfE), also in Big Creek 74 (Figure 3b). The last copy seems to be the result of several genetic rearrangements so it is probably not functional in those strains (Figure 3b).
Table 3.
Proteins shared by Big Creek 74 and SC09 strains
| Protein | SC09 | Big creek 74 |
|---|---|---|
| Restriction‐modification system | ||
| TypeI R‐M System, specificity subunitS | NJ56_RS02590 | UGYR_RS06280 |
| Toxin‐antitoxin system | ||
| Antitoxin to RelE/StbE | NJ56_RS02580 | UGYR_RS12460 |
| Replicon stabilization toxin RelE | NJ56_RS02585 | UGYR_RS12465 |
| Fimbria | ||
| Fimbrial protein StfD | NJ56_RS14520 | UGYR_RS0378 |
| Exotoxin/MrfF | NJ56_RS14525 | UGYR_RS03790 |
| Putative fimbrial membrane protein | NJ56_RS14530 | UGYR_RS03795 |
| Exotoxin | NJ56_RS14540 | UGYR_RS03805 |
| Fimbrial anchoring protein FimD | NJ56_RS10070 | UGYR_RS06285 |
| Fimbrial subunit | NJ56_RS17965 | UGYR_RS16650 |
| Exotoxin/minnor fimbrial subunit | NJ56_RS14535 | UGYR_RS03800 |
| Fimbria‐like adhesine SfmA | NJ56_RS16255 | UGYR_RS05635 |
| Fimbrial periplasmic chaperone SfmC | NJ56_RS10065 | UGYR_RS06280 |
| Fimbrial like adhesine prot | NJ56_RS17890 | UGYR_RS16665 |
| PilN family type IV pilus biogenesis protein | NJ56_RS10080 | UGYR_RS06295 |
| GCN5‐related N‐acetyltransferase | NJ56_RS10095 | UGYR_RS06310 |
| Insecticidal virulence protein | ||
| Probable insecticidal protein | NJ56_RS10100 | UGYR_RS06315 |
| Putative toxin subunit | NJ56_RS10110 | UGYR_RS06325 |
| Putative insecticidal toxin complex | NJ56_RS10115 | UGYR_RS06330 |
| Sorbitol sorbose utilization | ||
| Arabinose 5‐phosphate isomerase | NJ56_RS11170 | UGYR_RS07380 |
| Transcriptional regulator/glucitol operon activator prot | NJ56_RS11160 | UGYR_RS07370 |
| Transcriptional repressor of fructose operon DeoR family | NJ56_RS11165 | UGYR_RS07375 |
| Sorbitol‐6‐phosphate 2‐dehydrogenase | NJ56_RS11155 | UGYR_RS07365 |
| PTS glucitol/sorbitol transporter subunit IIB | NJ56_RS11145 | UGYR_RS07355 |
| PTS glucitol/sorbitol transporter subunit IIA | NJ56_RS11150 | UGYR_RS07360 |
| PTS glucitol/sorbitol transporter subunit IIC | NJ56_RS11140 | UGYR_RS07350 |
| Other proteins | ||
| Transcriptional regulator LysR family | NJ56_RS10085 | UGYR_RS06300 |
| S‐adenosylhomocysteine hydrolase | NJ56_RS02625 | UGYR_RS12500 |
| rimosomal protein | NJ56_RS14055 | UGYR_RS02835 |
| Outer memb component of tripartite multidrug resistance system | NJ56_RS10260 | UGYR_RS06475 |
| Chromosome partitioning protein ParA | NJ56_RS11555 | UGYR_RS07765 |
| Tfp pilus assembly protein%2C major pilin PilA | NJ56_RS09235 | UGYR_RS02515 |
| yclopropane‐fatty‐acyl‐phospholipid synthase | NJ56_RS11560 | UGYR_RS07770 |
| FAD‐dependent oxidoreductase | NJ56_RS11550 | UGYR_RS07760 |
| Inorganic pyrophosphatase/exopoliphosphatase | NJ56_RS02630 | UGYR_RS12505 |
| Spermidine/putrescine ABC trasporter permease | NJ56_RS06425 | UGYR_RS16290 |
Figure 3.
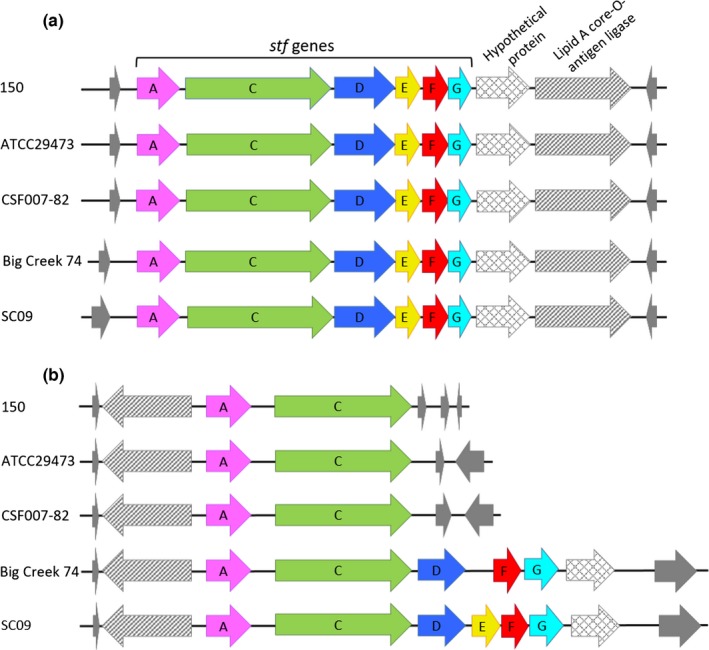
Analysis of the stf genes in Y. ruckeri genomes. Two copies of the stf cluster were found in Y. ruckeri strains. One copy of the cluster stfACDEFG is complete in the five strains (a), while a second copy is only complete in SC09, and with the exception of stfE gene, in Big Creek 74. The second copy of the stf cluster in 150, ATCC29473, and CSF007‐82 strains is only constituted by stfA and stfC genes (b). Note that the gene represented by a striped arrow, which encodes a lipid A core‐O‐antigen ligase, was affected by a translocation and an inversion event, resulting in a different localization in the two clusters
Three insecticidal toxin complexes (tc)‐like proteins were also identified as unique in these strains. They are similar to the TcdA, TcdB, and TcdC proteins of Vibrio parahaemolyticus, which are involved in the production of acute hepatopancreatic necrosis disease in penaeid shrimp (Tang & Lightner, 2014).
One of the most interesting findings was that Big Creek 74 and SC09 strains share a cluster of seven genes involved in the utilization of sorbitol (Figure 4), a previously described characteristic associated with Y. ruckeri serotype O2 strains (Davies & Frerichs, 1989), which supports the hypothesis that SC09 belongs to this serotype. The genes are similar to gutAEBD of E. coli (Yamada & Saier, 1987). The first three genes encode the three subunits of the sorbitol transporter of the phosphoenolpyruvate‐dependent phosphotransferase system (PTS), involved in the uptake and phosphorylation of sorbitol, while gutD encodes a sorbitol‐6‐phosphate 2‐dehydrogenase that synthesizes D‐fructose 6‐phosphate from D‐sorbitol 6‐phosphate. In Y. ruckeri strains, as occurs in E. coli, downstream of gutAEBD genes, there are two transcriptional regulators, an activator and a repressor of the sorbitol operon, similar to gutM and gutR of E. coli, respectively. Downstream of gutR, is located gutQ, which encodes an arabinose 5‐phosphate isomerase involved in LPS biosynthesis. Although the role of this protein in the sorbitol metabolism is unclear, it could be a regulatory molecule involved in expression of the gut operon (Meredith & Woodard, 2005). In the plant pathogen Erwinia amylovora, the presence of this operon has been linked to virulence and suggested to contribute to host specificity (Aldridge, Metzger, & Geider, 1997).
Figure 4.
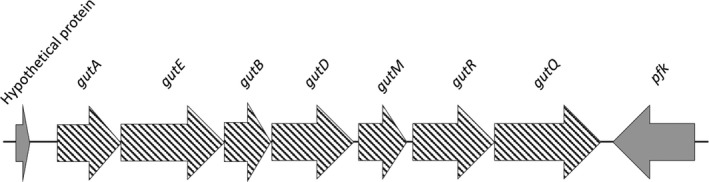
Genetic organization of the gut operon in Y. ruckeri Big Creek 74 and SC09 strains. gutA, gutE, and gutB: subunits of a glucitol/sorbitol‐specific transporter, gutD: sorbitol‐6‐phosphate 2‐dehydrogenase, gutM: glucitol operon activator protein, gutR: glucitol operon repressor protein, gutQ: an arabinose 5‐phosphate isomerase. pfk encodes for a 6‐phosphofructokinase class II
3.4. Unique genes of Big Creek 74
As was seen in the Venn diagram (Figure 2), Big Creek 74 strain has a total of 99 unique genes, which include 53 encoding hypothetical proteins, eight phage genes, four mobile genetic elements, and 34 genes which encode proteins with known function. As was mentioned above, the presence of some of these genes may underpin its adaptation to salmon, since the host of the other four strains is rainbow trout or catfish.
Among the proteins with known function (Table S1), we can find restriction‐modification systems, transcriptional regulators, transferases, or proteins involved in polysaccharide biosynthesis. Especially interesting is the gene encoding an ATP‐dependent Clp protease proteolytic subunit, a relevant regulatory enzyme in different bacteria, related also to virulence, environmental adaptation, and antibiotic resistance in microorganisms such as Staphylococcus aureus (Frees, Gerth, & Ingmer, 2014) or the fish pathogen Pseudomonas fluorescens (Liu, Chi, & Sun, 2015).
3.5. Unique genes of SC09
SC09 has a total of 261 genes that are not present in the other strains, 148 of them encode hypothetical proteins, 17 are phage‐related genes, nine mobile genetic elements, and the rest encode proteins with different functions. As was suggested for Big Creek 74, some of these genes may underpin the adaptation of this strain to survive inside the host (catfish) or under certain environmental conditions. Among these unique proteins are transcriptional regulators, proteins related to type IV secretion systems, restriction‐modification, and toxin‐antitoxin components and proteins associated with cellular energy homeostasis (Table S2). One of the most interesting proteins is a thymidylate synthase, an enzyme linked to virulence in several microorganisms such as Staphylococcus aureus (Kriegeskorte et al., 2014) or Salmonella typhimurium, in which it was necessary for intracellular growth, both in macrophage‐like and Hep‐2 human epithelial cell lines (1) and also for complete virulence in a BALB/c mice model (Kok, Bühlmann, & Pechère, 2001).
A finding which is worthy of further investigation was the presence, only in this strain, of a cluster of 12 genes related to cell wall polysaccharide biosynthesis, in particular the O‐antigen.
3.6. Genes solely absent in the avirulent strain ATCC29473
Among the five strains included in the study, ATCC29473 was defined as avirulent. In this sense, it was intriguing to analyze which genes are absent in this strain and present in the others, in order to elucidate the genetic basis of its attenuation. A total of 21 genes were found (Table 4), all of them encoding proteins with an assigned function, which were probably lost during the evolution of this strain. It is significant that 17 out of 21 genes are adjacent in the other four genomes from virulent strains (Figure 5). This region of 19,566 bp contains genes encoding for a Crp‐Fnr family transcriptional regulator, a hypothetical protein, an enzyme related to an enterobactin‐like siderophore and three different gene clusters: one formed by three genes involved in iron transport, a group of three genes related to hexose phosphate uptake; and a region containing nine genes involved in the uptake and metabolism of citrate. Since most of these genes are related to virulence (Gray, Freitag, & Boor, 2006; Moisi et al., 2013; Urbany & Neuhaus, 2008), it is possible that the absence of this region could explain, in some way, the attenuation of Y. ruckeri ATCC29473. This is important for future studies and may help to shed light on the virulence of the species.
Table 4.
Proteins absent in the avirulent strain ATCC29473 and present in the other strains
| Protein | 150 | BIG CREEK 74 | CSF007‐82 | SC09 |
|---|---|---|---|---|
| Aromatic amino acid decarboxylase | BI323_03940 | UGYR_RS01810 | CSF007_RS12055 | NJ56_RS08540 |
| Serine hydrolase family protein | BI323_03955 | UGYR_RS01805 | CSF007_RS12040 | NJ56_RS08535 |
| Virulence factor | BI323_03960 | UGYR_RS01800 | CSF007_RS12035 | NJ56_RS08530 |
| Crp‐like helix‐turn‐helix domain protein | BI323_16505 | UGYR_RS04050 | CSF007_RS14555 | NJ56_RS14785 |
| Fe(3 + ) ions import ATP‐binding protein FbpC | BI323_16510 | UGYR_RS04045 | CSF007_RS14560 | NJ56_RS14780 |
| Putative binding protein‐dependent transport system%2C inner‐membrane component | BI323_16515 | UGYR_RS04040 | CSF007_RS14565 | NJ56_RS14775 |
| Bacterial extracellular solute‐binding family protein | BI323_16520 | UGYR_RS04035 | CSF007_RS14570 | NJ56_RS14770 |
| Major Facilitator Superfamily protein | BI323_16525 | UGYR_RS04030 | CSF007_RS14575 | NJ56_RS14765 |
| Sensor histidine protein kinase UhpB, glucose‐6‐phosphate specific | BI323_16530 | UGYR_RS04025 | CSF007_RS14580 | NJ56_RS14760 |
| Bacterial regulatory s, luxR family protein | BI323_16535 | UGYR_RS04020 | CSF007_RS14585 | NJ56_RS14755 |
| Transcriptional regulatory protein | BI323_16540 | UGYR_RS04015 | CSF007_RS14590 | NJ56_RS14750 |
| Sensor histidine kinase DpiB | BI323_16545 | UGYR_RS04010 | CSF007_RS14595 | NJ56_RS14745 |
| [citrate (Pro‐3S)‐lyase] ligase | BI323_16550 | UGYR_RS04005 | CSF007_RS14600 | NJ56_RS14740 |
| Citrate lyase acyl carrier protein | BI323_16555 | UGYR_RS04000 | CSF007_RS14605 | NJ56_RS14735 |
| Citrate lyase subunit beta | BI323_16560 | UGYR_RS03995 | CSF007_RS14610 | NJ56_RS14730 |
| Citrate lyase alpha chain | BI323_16565 | UGYR_RS03990 | CSF007_RS14615 | NJ56_RS14725 |
| Holo‐ACP synthase CitX | BI323_16570 | UGYR_RS03985 | CSF007_RS14620 | NJ56_RS14720 |
| Probable 2‐(5’’‐triphosphoribosyl)‐3′‐dephosphocoenzyme‐A synthase | BI323_16575 | UGYR_RS03980 | CSF007_RS14625 | NJ56_RS14715 |
| Citrate carrier | BI323_16580 | UGYR_RS03975 | CSF007_RS14630 | NJ56_RS14710 |
| 4′‐phosphopantetheinyl transferase siderophore | BI323_16585 | UGYR_RS03970 | CSF007_RS14635 | NJ56_RS14705 |
| Holin | BI323_09445 | UGYR_RS04445 | CSF007_RS17025 | NJ56_RS17590 |
Figure 5.
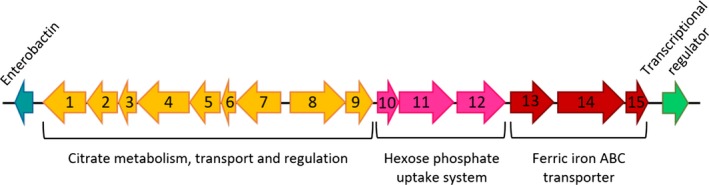
Genetic organization of the DNA region absent in ATCC29473 and present in the other strains. The region contains genes encoding for an enterobactin‐like siderophore (blue), nine genes involved in the uptake and metabolism of citrate (yellow), a group of three genes related to hexose phosphate uptake (pink), three genes involved in iron transport (red), and a Crp‐Fnr family transcriptional regulator (green). 1: Citrate succinate antiporter, 2: 2‐(5′’‐triphosphoribosyl)‐3′‐dephosphocoenzyme‐A synthase, 3: Apo‐citrate lyase phosphoribosyl‐dephospho‐CoA transferase, 4: Citrate lyase alpha chain, 5: Citrate lyase beta chain, 6: Citrate lyase gamma chain acyl carrier protein, 7: [Citrate [pro‐3S]‐lyase] ligase, 8: Sensor kinase, 9: Transcriptional regulatory protein, 10: Transcriptional regulatory protein, 11: Sensor histidine protein kinase glucose‐6‐phosphate specific, 12: Hexose phosphate uptake regulatory protein, 13: Ferric iron ABC transporter iron‐binding protein, 14: Ferric iron ABC transporter permease protein, 15: Ferric iron ABC transporter binding subunit
4. Conclusion
In this study, is presented for the first time, the comparative analysis of five genome sequences of Y. ruckeri. Although the five strains shared approximately 75% of their genes, our study has revealed important genetic differences between the five genomes. Aside from the genetic differentiation found between serotype O1 and O2 strains, especially relevant are the high number of unique genes found in Big Creek 74 and SC09 in relation to serotype O1 strains and the 21 genes absent in the avirulent strain ATCC29473. These findings could explain the host specificity of the first two strains or the virulence attenuation of ATCC29473. Further investigation of those genes will provide insights into understanding the pathogenesis and the adaptive mechanisms to different environments of Y. ruckeri.
Conflict of Interests
The authors declare no competing interests.
Ethics Statement
This research did not involve any human or animal subjects, materials, or data and therefore did not require any ethics oversight or approval in these respects.
Supporting information
Acknowledgments
We thank Pilar Reimundo for her valuable suggestions at the beginning of this project. This work was financially supported by the projects AGL2012‐35808 and AGL2015‐66018‐R, from the Ministry of Economy and Competitiveness (MINECO) of Spain and the FEDER fund. Desirée Cascales and Ana I García‐Torrico were the recipient of grants from the Spanish Ministry of Education, Culture and Sport (MEC) and from the Ministry of Economy and Competitiveness (MINECO), respectively.
Cascales D, Guijarro JA, García‐Torrico AI, Méndez J. Comparative genome analysis reveals important genetic differences among serotype O1 and serotype O2 strains of Y. ruckeri and provides insights into host adaptation and virulence. MicrobiologyOpen. 2017;6:e460 https://doi.org/10.1002/mbo3.460
References
- Aldridge, P. , Metzger, M. , & Geider, K. (1997). Genetics of sorbitol metabolism in Erwinia amylovora and its influence on bacterial virulence. Molecular and General Genetics, 256, 611–619. [DOI] [PubMed] [Google Scholar]
- Arias, C. R. , Olivares‐Fuster, O. , Hayden, K. , Shoemaker, C. A. , Grizzle, J. M. , & Klesius, P. H. (2007). First report of Yersinia ruckeri biotype 2 in the USA. Journal of Aquatic Animal Health, 19, 35–40. [DOI] [PubMed] [Google Scholar]
- Austin, D. A. , Robertson, P. A. , & Austin, B. (2003). Recovery of a new biogroup of Yersinia ruckeri from diseases rainbow trout (Oncorhynchus mykiss, Walbaum). Systematic and Applied Microbiology, 26, 127–131. [DOI] [PubMed] [Google Scholar]
- Bastardo, A. , Ravelo, C. , & Romalde, J. L. (2015). Phylogeography of Yersinia ruckeri reveals effects of past evolutionary events on the current strain distribution and explains variations in the global transmission of enteric redmouth (ERM) disease. Frontiers in Microbiology, 6, 1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettin, T. , Davis, J. J. , Disz, T. , Edwards, R. A. , Gerdes, S. , Olsen, G. J. , … Xia, F. (2015). RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Scientific Reports, 5, 8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvez, S. , Gantelet, H. , Blanc, G. , Douet, D. , & Daniel, P. (2014). Yersinia ruckeri biotypes 1 and 2 in France: Presence and antibiotic susceptibility. Diseases of Aquatic Organisms, 109, 117–126. [DOI] [PubMed] [Google Scholar]
- Chen, H. , & Boutros, P. C. (2011). VennDiagram: A package for the generation of highly‐customizable Venn and Euler diagrams in R. BMC Bioinformatics, 12, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling, A. C. E. , Mau, B. , Blattner, F. R. , & Perna, N. T. (2004). Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Research, 14, 1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, R. L. , & Frerichs, G. N. (1989). Morphological and biochemical differences among isolates of Yersinia ruckeri obtained from wide geographical areas. Journal of Fish Diseases, 12, 357–365. [Google Scholar]
- Emmerth, M. , Goebel, W. , Miller, S. I. , & Hueck, C. J. (1999). Genomic subtraction identifies Salmonella typhimurium prophages, F‐related plasmid sequences, and a novel fimbrial operon, stf, which are absent in Salmonella typhi . Journal of Bacteriology, 181, 5652–5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouz, B. , Zarza, C. , & Amaro, C. (2006). First description of non‐motile Yersinia ruckeri serovar I strains causing disease in rainbow trout, Oncorhynchus mykiss (Walbaum), cultured in Spain. Journal of Fish Diseases, 29, 339–346. [DOI] [PubMed] [Google Scholar]
- Frees, D. , Gerth, U. , & Ingmer, H. (2014). Clp chaperones and proteases are central in stress survival, virulence and antibiotic resistance of Staphylococcus aureus . International Journal of Medical Microbiology, 304, 142–149. [DOI] [PubMed] [Google Scholar]
- Furones, M. D. , Gilpin, M. J. , Alderman, D. J. , & Munn, C. B. (1990). Virulence of Yersinia ruckeri serotype I strains is associated with a heat sensitive factor (HSF) in cell extracts. FEMS Microbiology Letters, 54, 339–343. [DOI] [PubMed] [Google Scholar]
- Gray, M. J. , Freitag, N. E. , & Boor, K. J. (2006). How the bacterial pathogen Listeria monocytogenes mediates the switch from environmental Dr. Jekyll to pathogenic Mr Hyde. Infection and Immunity, 74, 2505–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok, M. , Bühlmann, E. , & Pechère, J.C . (2001). Salmonella typhimurium thyA mutants fail to grow intracellularly in vitro and are attenuated in mice. Microbiology (Reading, England), 147, 727–733. [DOI] [PubMed] [Google Scholar]
- Kriegeskorte, A. , Block, D. , Drescher, M. , Windmüller, N. , Mellmann, A. , Baum, C. , … Kahl, B. C. (2014). Inactivation of thyA in Staphylococcus aureus attenuates virulence and has a strong impact on metabolism and virulence gene expression. MBio, 5, e01447–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L. , Chi, H. , & Sun, L. (2015). Pseudomonas fluorescens: Identification of Fur‐regulated proteins and evaluation of their contribution to pathogenesis. Diseases of Aquatic Organisms, 115, 67–80. [DOI] [PubMed] [Google Scholar]
- Méndez, J. , Fernández, L. , Menéndez, A. , Reimundo, P. , Pérez‐Pascual, D. , Navais, R. , & Guijarro, J. A. (2009). A chromosomally located traHIJKCLMN operon encoding a putative type IV secretion system is involved in the virulence of Yersinia ruckeri . Applied and Environmental Microbiology, 75, 937–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith, T. C. , & Woodard, R. W. (2005). Identification of GutQ from Escherichia coli as a D‐arabinose 5‐phosphate isomerase. Journal of Bacteriology, 187, 6936–6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisi, M. , Lichtenegger, S. , Tutz, S. , Seper, A. , Schild, S. , & Reidl, J. (2013). Characterizing the hexose‐6‐phosphate transport system of Vibrio cholerae, a utilization system for carbon and phosphate sources. Journal of Bacteriology, 195, 1800–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, M. J. , & Imperiali, B. (2014). The renaissance of bacillosamine and its derivatives: Pathway characterization and implications in pathogenicity. Biochemistry, 53, 624–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romalde, J. L. , Magariños, B. , Barja, J. L. , & Toranzo, A. E. (1993). Antigenic and molecular characterization of Yersinia ruckeri proposal for a new intraspecies classification. Systematic and Applied Microbiology, 16, 411–419. [Google Scholar]
- Romalde, J. L. , Planas, E. , Sotelo, J. M. , & Toranzo, A. E. (2003). First description of Yersinia ruckeri serotype O2 in Spain. Bulletin European Association of Fish Pathologists, 23, 135–138. [Google Scholar]
- Ross, A. J. , Rucker, R. R. , & Ewing, W. H. (1966). Description of a bacterium associated with redmouth disease of rainbow trout (Salmo Gairdneri). Canadian Journal of Microbiology, 12, 763–770. [DOI] [PubMed] [Google Scholar]
- Srikhanta, Y. N. , Fox, K. L. , & Jennings, M. P. (2010). The phasevarion: Phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nature Reviews Microbiology, 8, 196–206. [DOI] [PubMed] [Google Scholar]
- Tang, K. F. J. , & Lightner, D. V. (2014). Homologues of insecticidal toxin complex genes within a genomic island in the marine bacterium Vibrio parahaemolyticus . FEMS Microbiology Letters, 361, 34–42. [DOI] [PubMed] [Google Scholar]
- Urbany, C. , & Neuhaus, H. E. (2008). Citrate uptake into Pectobacterium atrosepticum is critical for bacterial virulence. Molecular Plant‐Microbe Interactions, 21, 547–554. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Kim, Y. , Hong, S. H. , Ma, Q. , Brown, B. L. , Pu, M. , … Wood, T. K. (2011). Antitoxin MqsA helps mediate the bacterial general stress response. Nature Chemical Biology, 7, 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, M. , & Saier, M. H. (1987). Physical and genetic characterization of the glucitol operon in Escherichia coli . Journal of Bacteriology, 169, 2990–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebian, N. , Merkx‐Jacques, A. , Pittock, P. P. , Houle, S. , Dozois, C. M. , Lajoie, G. A. , & Creuzenet, C. (2016). Comprehensive analysis of flagellin glycosylation in Campylobacter jejuni NCTC 11168 reveals incorporation of legionaminic acid and its importance for host colonization. Glycobiology, 26, 386–397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


