ABSTRACT
Mycobacterial 6-kDa early secreted antigenic target (ESAT-6) system (ESX) exporters transport proteins across the cytoplasmic membrane. Many proteins transported by ESX systems are then translocated across the mycobacterial cell envelope and secreted from the cell. Although the mechanism underlying protein transport across the mycolate outer membrane remains elusive, the ESX systems are closely connected with and localize to the cell envelope. Links between ESX-associated proteins, cell wall synthesis, and the maintenance of cell envelope integrity have been reported. Genes encoding the ESX systems and those required for biosynthesis of the mycobacterial envelope are coregulated. Here, we review the interplay between ESX systems and the mycobacterial cell envelope.
KEYWORDS: ESX system, cell envelope, lipids, mycobacteria, protein secretion, transport
INTRODUCTION
Bacterial protein transporters are large molecular machines that assemble within the cytoplasm and bacterial cell envelope, which generally consists of the cytoplasmic membrane (CM) and extracytoplasmic compartments. All bacteria use protein secretion systems to actively transport protein substrates and/or nucleic acids from the bacterial cytoplasm to extracytoplasmic environments (1). In diderm-lipopolysaccharide (LPS) (Gram-negative) bacteria, which have an inner membrane (IM) and an outer membrane (OM) containing LPS, there are nine types of secretion systems (types I to IX) (1). Types I through VI, the most characterized systems, promote the secretion of proteins across the IM and the OM (recently reviewed in reference 2). In monoderm (Gram-positive) bacteria, which have only a CM, there are eight secretion systems that promote protein secretion across the CM (reviewed in reference 1). Based on microscopy studies and lipid analysis, mycobacteria are considered to be diderm bacteria (1, 3–5). However, the mycobacterial cell envelope contains a mycolate-OM (MOM), which differs in lipid content from other diderm OMs (Fig. 1) (reviewed in references 1 and 6 to 8). As such, mycobacteria are classified as diderm-mycolate bacteria (1).
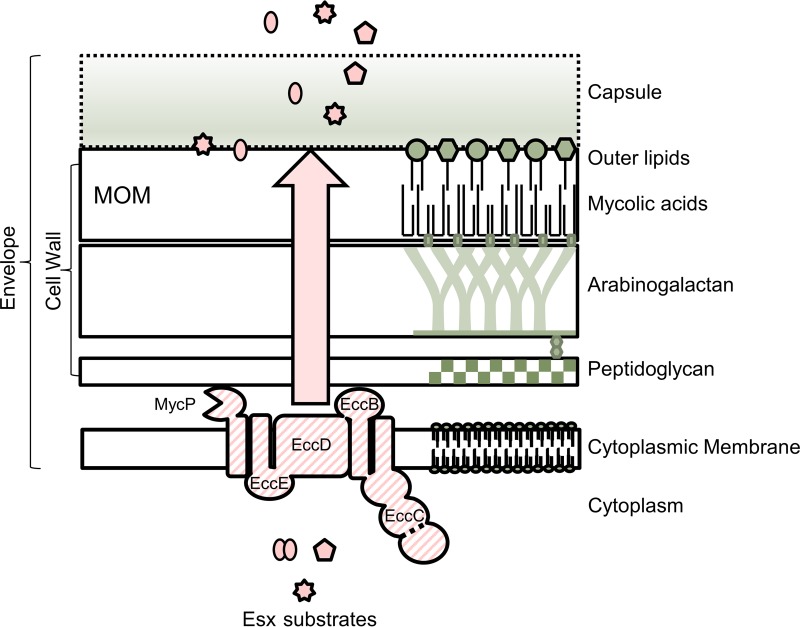
FIG 1.
ESX exporters in the mycobacterial cell envelope. The model shows a cartoon representation of the complex mycobacterial cell envelope, spanning from the cytoplasmic membrane through the mycolate outer membrane (MOM) and capsule. Proteins are excluded from the cell envelope, other than ESX proteins, for simplicity. Components of the envelope are not drawn to scale. For a recent review on the mycobacterial cell envelope, please see references 7, 80, and 81. The ESX membrane complex and MycP are indicated in the cytoplasmic membrane (striped). The dotted line in the Ecc protein refers to the fact that specifically in the ESX-1 system, the EccC protein is split into two proteins (EccCa1 and EccCb1). Additional ESX proteins required for transport are excluded for simplicity (Esp and additional Ecc proteins, PE and PPE). For recent reviews on the molecular mechanisms of ESX export, please see references 33 to 36. The cartoon was based on schematics in references 80 and 144. Ecc, ESX-conserved component. Large arrow indicates that proteins traverse the cell wall via an unknown mechanism.
Hundreds of mycobacterial proteins are routinely observed on the cell surface or in the culture medium during in vitro growth (9–12). As with other OMs, the MOM is frequently regarded as a permeability barrier (13–15). Yet, it is unknown how any mycobacterial proteins are secreted across the MOM (15). In contrast to secretion, export is the transport of substrates from the cytoplasm across the cytoplasmic membrane. The general secretory pathway (Sec) and the twin-arginine transporter (TAT) are well-characterized examples of protein exporters (1). In mycobacteria and several monoderm bacteria, there is a unique family of protein exporters referred to as 6-kDa early secreted antigenic target (ESAT-6) system/WXG-100 secretion system (ESX/WSS) or type VII secretion systems (16–21). There is no known ESX counterpart in diderm-LPS bacteria. However, proteins with WXG motifs, which are a hallmark of substrates of the ESX/WSS, have been described in diderm-LPS bacteria (22).
As of yet, there is no evidence that the ESX apparatus spans the envelope. Therefore, we refer to ESX-mediated protein transport as export. Yet, ESX substrates are secreted from the mycobacterial cell. Therefore, either the export machinery or the substrates themselves likely intimately interact with the MOM. Several recent studies have reported connections between ESX systems and the cell envelope in different mycobacterial species (23–25), prompting us to revisit the literature linking ESX systems with the mycobacterial cell envelope. Here, we focus on protein transport across the envelope, interaction between ESX proteins and the cell envelope, links between ESX and cell envelope integrity, and coregulation of the ESX systems and components of the cell envelope.
OVERVIEW OF MYCOBACTERIAL ESX SYSTEMS
Mycobacteria have several ESX systems encoded within the genome or on conjugative plasmids (26–29). Up to five ESX systems can be encoded in the genome (ESX-1 to ESX-5). Protein transport has been demonstrated for the ESX-1, ESX-5, and ESX-3 systems but not for the ESX-2 and ESX-4 systems (18, 30–32). ESX systems generally include ESX conserved components (Ecc proteins), a mycosin serine protease (MycP), and a pair of small secreted antigenic proteins with WXG-100 motifs (Esx proteins) (16, 17, 26). Interestingly, outside the conserved components, ESX systems vary in genetic composition and function across diverse mycobacterial species. In this section, we highlight the five ESX systems encoded in the mycobacterial genome and how these systems vary functionally in mycobacteria. The molecular details and functions of the ESX systems have been more comprehensively reviewed elsewhere (18, 33–36).
The genes encoding ESX systems are conserved across diverse mycobacterial species (27, 29). Mycobacterium tuberculosis is the causative agent of human tuberculosis (37). Because M. tuberculosis grows slowly and must be used in a biosafety level 3 facility, additional mycobacterial species have served as model systems to define mechanisms and functions of the ESX systems (18, 37–41). Importantly, studying ESX systems in distinct mycobacterial species has provided insight into the function of ESX exporters that would have been otherwise missed by studying a single mycobacterial species.
In M. tuberculosis and other mycobacterial pathogens (M. marinum and M. bovis), the ESX-1 and ESX-5 systems are required for virulence (30, 31, 42–45). The loss of ESX-1 or ESX-5 genes in pathogenic mycobacteria causes attenuation in cellular and animal models of infection (30, 31, 42–51). Pathogenic mycobacteria use the ESX-1 system to damage the phagosomal membrane while residing within phagocytes (52–56). The ESX-5 system, which is restricted to slow-growing mycobacterial species, likely promotes the uptake of nutrients essential for mycobacterial survival in the phagocyte (27, 50, 57, 58). Relative to the ESX-1 system, the ESX-5 system secretes a large number of proline-glutamate/proline-proline-glutamate (PE/PPE) proteins which promote virulence (51, 59–62). ESX-5 is also unique in that it is a modular system; accessory components outside the conserved locus promote the secretion of a specific subset of ESX-5 substrates (36, 63).
In Mycobacterium smegmatis, two ESX systems promote atypical conjugation (64–67). Conjugation is a process by which genetic material is transferred directionally from a donor cell to a recipient cell. M. smegmatis undergoes distributive conjugal transfer, in which donated DNA is integrated in an unpredictable genome-wide manner. Interestingly, in addition to regulating conjugation, genes encoding ESX-1 confer mating type designations (donor versus recipient) in M. smegmatis (66, 68). Recently, Gray et al. (67) found that the ESX-4 system was required for conjugation specifically in recipient strains. Moreover, expression of ESX-4 genes was induced in recipient cells in an ESX-1-dependent manner during donor-recipient coculture experiments. Therefore, in M. smegmatis, ESX-1 and ESX-4 systems coordinate communication between mycobacterial cells (67). In support of these findings, a recent study by Boritsch et al. indicated that a similar process of horizontal gene transfer was observed in Mycobacterium canettii, a close relative of M. tuberculosis. Although the dependence of the observed DNA transfer on the ESX-1 and ESX-4 systems was not tested, this study indicates that horizontal gene transfer is likely widespread in mycobacteria (69).
ESX-3 systems have two independent functional roles in mycobacteria. First, ESX-3 systems promote metal homeostasis (32, 70–74). Although ESX-3 promotes siderophore-mediated iron and zinc acquisition in M. tuberculosis (70, 71), in M. smegmatis, the ESX-3 system promotes iron homeostasis only (71, 72). Underscoring the importance of metal homeostasis, ESX-3 genes are essential in M. tuberculosis; survival in the absence of ESX-3 genes can be restored with metal supplementation (73). Second, the ESX-3 system promotes virulence of M. tuberculosis (73, 75). The ESX-3 substrate EsxH directly interacts with host endosomal sorting complexes required for transport (ESCRT) machinery, preventing phagosomal maturation and antigen presentation during macrophage infection with M. tuberculosis (75, 76).
Compared to the other ESX systems, little is known about ESX-2. Like ESX-5, ESX-2 systems are restricted to slow-growing mycobacterial species (27). ESX-2 genes are transcriptionally coregulated in M. tuberculosis with genes encoding additional ESX systems and were identified in a screen for genes necessary for survival in dendritic cells (77–79).
MYCOBACTERIAL CELL ENVELOPE
After crossing the CM, secreted proteins must transit several layers that form the mycobacterial envelope, including a covalently linked structure of peptidoglycan, arabinogalactan, and mycolic acids (Fig. 1) (7, 80). Because the mycobacterial peptidoglycan structure was recently reviewed by Alderwick et al., we will not discuss it in detail here (81). Covalently attached to the peptidoglycan is arabinogalactan, a macromolecule consisting of a chain of galactan that is modified with 2 to 3 branched arabinan chains (reviewed in references 6 and 80).
The MOM is linked to the arabinogalactan via covalent bonds between mycolic acids and the arabinan units of arabinogalactan (6). Mycolic acids and extractable lipids and glycolipids form the MOM. Other mycobacterial lipids may include, but are not limited to, trehalose-containing lipids (e.g., sulfolipids, polyacyltrehalose [PAT], diacyltrehalose [DAT], and lipooligosaccharide [LOS]), phthiocerol dimycocerosates (PDIM), and phenolic glycolipids (PGLs) (reviewed in reference 80). The mycobacterial cell envelope also includes a capsule layer (4).
Although many lipid biosynthesis genes are conserved among mycobacterial species, the lipid content of the envelope varies (82). PDIM and PGL biosynthesis genes are conserved in M. tuberculosis and in M. marinum but not in M. smegmatis (83). M. smegmatis has glycopeptidolipids (GPLs) on the cell surface (80). M. tuberculosis produces sulfolipids, which are not present in M. marinum (84, 85). M. marinum produces LOS, which are not found in M. tuberculosis (86–88). There are even differences in lipid content between isolates of the same species. For example, although the M. tuberculosis genome contains genes required for PDIM and PGL synthesis, some isolates of M. tuberculosis do not make PGLs (89). The differences in the mycobacterial cell envelope are important because a variety of ESX systems must interact with each unique cell envelope.
The mycobacterial cell envelope functions as a major virulence determinant. Pathogenic mycobacteria with mutations in PDIM biosynthesis genes are attenuated in cellular and animal infection models (90–92). Glycolipids can interact directly with host immune receptors (reviewed in reference 93). Many mycobacterial lipids are antigens that can activate CD1-restricted T cells (94). Intriguingly, it has been suggested that mycobacterial lipids may also insert directly into host cell membranes, altering membrane fluidity and affecting phagocytosis and trafficking (reviewed in references 94 and 95).
CELL WALL SYNTHESIS AND ESX SYSTEMS
ESX systems have been repeatedly linked to envelope biogenesis, primarily at the level of gene expression. Several transcription factors regulate the genes required for both cell wall processes and ESX transport. For example, the MprAB two-component system responds to cell envelope stress and regulates ESX-1 genes (96–99). Sodium chloride stress pathways influence the expression of genes required for cell wall remodeling and of ESX-1 genes (espACD) in M. tuberculosis CDC1551 (100). EspR is a transcriptional regulator of the ESX-1 system (77, 101). Loss of espR expression abrogates ESX-1 secretion and attenuates M. tuberculosis (101). In addition to the ESX-1 system, EspR regulates many cell wall genes, including those responsible for PDIM production, PE/PPE genes, and genes at the ESX-2 and ESX-5 loci (reference 77 and reviewed in reference 102). PhoP is part of a two-component system that regulates several virulence pathways in M. tuberculosis, including biosynthetic genes of the M. tuberculosis-specific lipids, sulfolipid, DAT, and PAT (103). In addition to regulating lipid biosynthesis genes, PhoP also regulates whiB6 and works directly with EspR, and both regulate ESX-1 gene expression (96, 104, 105). Therefore, under many of the conditions in which cell envelope remodeling may be occurring or where cell wall genes are regulated, ESX genes are also regulated.
Links between the regulation of genes encoding ESX systems and the cell envelope have been extended to cellular models. A 2015 study by Mendum et al. found that lipids (PDIM, TDM, sulfolipids, and PGL), ESX systems (ESX-1, ESX-2, and ESX-4), and ESX-related genes (PPE proteins) were prominent pathways needed for survival in dendritic cells (79). In a macrophage infection model, the expression of PDIM-related genes was downregulated during infection, while the expression of other lipid biosynthesis genes (including those encoding sulfolipids, DAT, and PAT), ESX-1-related genes, and espR was induced (106).
In addition to regulatory linkages, ESX-1 proteins have been linked directly to lipid composition and metabolism. In 2012, Joshi et al. (107) demonstrated that EccA1, an ESX-1-associated protein (44, 49), complexes with mycolic acid (Pks13, KasB, KasA, and MmaA4) and PDIM/PGL synthesis (Mas, Pks15/1, PpsD, and PpsE) proteins in M. marinum. Disruption of the eccA1 gene resulted in a 30 to 40% reduction in mycolic acid in M. marinum, suggesting that EccA1 promotes mycolic acid synthesis (107).
A recent study examined metabolic changes in the presence and absence of ESX-1 genes in M. smegmatis (23). ESX-1-deficient strains showed significantly elevated levels of 22 metabolites compared to the wild-type strains under growth on a variety of carbon sources (23). Several of the identified metabolites were linked to cell envelope biogenesis, including mycolic acid synthesis, peptidoglycan biosynthesis, and arabinogalactan and arabinomannan biogenesis (23). Metabolic changes were not addressed in a complemented strain, leaving some possibility that the metabolic changes were not due to a loss of ESX-1 export. Nevertheless, this study is consistent with the study linking EccA1 to mycolic acid biosynthesis (107).
Mycobacterial species can exhibit a smooth or rough colony morphology when grown on agar. Disruption of some individual genes required for lipid biosynthesis results in changes to colony morphology (from smooth to rough, or rough to smooth) when grown on agar (86, 88, 90, 108). Interestingly, the loss of individual ESX-1 genes in several mycobacterial species also results in changes in colony morphologies. For example, Mycobacterium bovis BCG and Mycobacterium microti bear natural deletions in ESX-1 genes. The introduction of ESX-1 genes from M. tuberculosis into the M. bovis BCG Pasteur and M. microti strains resulted in the conversion from a smooth to rough colony phenotype (48). In M. marinum, strains bearing deletions in ESX-1 or ESX-5 genes also display smooth/shiny colony morphologies (50, 51, 109, 110). M. tuberculosis H37Ra is an attenuated laboratory strain. H37Ra is attenuated primarily due to a mutation in the phoP gene. Restoration of a wide-type copy of the phoP gene was sufficient to restore ESX-1 secretion and virulence and to promote the conversion of the H37Ra strain from a smooth to a rough colony morphology (111). The smooth-colony phenotype observed in the absence of ESX systems may indicate that ESX systems impact the cell envelope composition, either directly or indirectly. However, the mechanism by which the loss of ESX systems impacts colony morphology remains unknown.
Bacterial secretion systems and other large machinery, such as flagella and conjugation systems, frequently require localized cell wall remodeling to insert into the cell envelope (reviewed in references 112 and 113). Mycobacterial cell growth occurs from both cell poles, with faster growth occurring at the old pole (reviewed in reference 114). Proteins involved in peptidoglycan, arabinogalactan, and mycolic acid synthesis and transport localize to polar regions (115, 116). Consistent with a link between ESX systems and cell envelope biogenesis, several protein components of the ESX-1 system have been localized to the mycobacterial cell pole in M. marinum and M. smegmatis (109, 117). Localization of components of other ESX systems has not been determined. However, the Rv1818c (PE_PGRS33) ESX-5 substrate has been localized to the mycobacterial pole, which may indicate that the ESX-5 system is also polar (61, 118). The polar localization of ESX-1 systems and cell wall biogenesis proteins may indicate that lipid or cell wall processes play a role in ESX-1 localization and assembly.
ESX SYSTEMS AND ENVELOPE PERMEABILITY
The lipid-rich MOM is thought to form a natural permeability barrier to hydrophilic molecules and nutrients, much like the OM of diderm-LPS bacteria (119, 120). However, the cell envelope from diverse mycobacterial species is reportedly 20- to 100-fold less permeable to hydrophilic solutes than the E. coli OM (120, 121). Both mycobacteria and diderm-LPS bacteria are intrinsically resistant to several antibiotics, including the large hydrophilic glycopeptide vancomycin (122, 123). Consistent with the idea that mycobacterial lipids provide a permeability barrier, mycobacterial strains lacking PDIM have increased susceptibility to vancomycin (124, 125).
In diderm-LPS bacteria, many outer membrane proteins (OMPs) have β-barrel structures and form channels or porins in the outer membrane that promote nutrient acquisition (126, 127). In mycobacteria, relatively few proteins that function as channels through the MOM have been characterized (128–131). The MspA protein in M. smegmatis is the best characterized mycobacterial porin but has no orthologue in M. tuberculosis (129). More recently, CpnT (Rv3903c) was identified as a mycobacterial toxin and water-filled protein channel that promotes the uptake of glycerol and hydrophilic and hydrophobic antibiotics (131, 132).
There are conflicting data linking the ESX-1 system and membrane permeability. Garces et al. reported that M. tuberculosis ESX-1-deficient strains (specifically lacking the EspA substrate) are more sensitive to SDS treatment and other envelope stresses, indicating increased permeability (133). In contrast, Chen et al. found that loss of the espA gene or deletion of other ESX-1 genes did not impact cell wall integrity. They found no differences between the wild-type and espA mutant strains in Nile red or ethidium bromide (EtBr) uptake or in sensitivity to SDS, which are common measures of permeability (134). We observed no differences in EtBr uptake between ESX-1-deficient and wild-type M. marinum strains (135). Differences in strain background could account for the divergent conclusions.
In contrast to ESX-1, the ESX-5 system is clearly involved in OM permeability. The ESX-5 system is essential for in vitro growth of M. tuberculosis, M. bovis, and M. marinum (58, 136). The essential nature of a subset of ESX-5 genes, primarily encoding components, has been reported by several groups (31, 51, 136–138). Interestingly, essentiality could be bypassed under certain conditions where the MOM was permeabilized. Individual ESX-5 genes could be deleted from the M. marinum genome when PDIM/PGL biosynthesis genes were mutated or by expressing the MspA porin from M. smegmatis to permeabilize the envelope. From these findings, the ESX-5 system may include the uptake of essential nutrients (58). While the essential nutrient capable of passing through the MspA porin was unidentified, Ates et al. demonstrated that ESX-5 promotes the uptake of fatty acids (58). The ESX-5 system was recently shown to be induced by phosphate limitation (139, 140). However, it has not been tested whether ESX-5 or ESX-5 substrates can promote the uptake of phosphate.
Ates et al. proposed that the activity of ESX-5 substrates, rather than the ESX-5 system itself, is essential (58). Although the precise substrates have yet to be identified, overexpression of the PE19 substrate in M. tuberculosis leads to increased membrane permeability (139, 141).
The fact that deletions of essential ESX-5 genes could be generated when the envelope was permeabilized raises the possibility that spontaneous mutations in lipid biogenesis genes are present in strains with defective ESX-5 systems. In support of this idea, strains with transposon insertions in the eccC5 and eccD5 genes, which are essential in M. tuberculosis (136), were isolated in the clinical M. tuberculosis strain CDC1551 (59). Although both strains exhibited increased membrane permeability, the permeability phenotype was genetically unlinked from the ESX-5 genes (58, 59).
A new study has linked ESX-5 to a drug resistance phenotype. Resistance to ofloxacin and other fluoroquinolones is generally established through mutation of the genes encoding DNA gyrase (142). Interestingly, ofloxacin-monoresistant M. tuberculosis clinical isolates lacking DNA gyrase mutations were reported to have mutations in ESX-5 genes (143). The eccC5 V762G mutation found in the clinical strains was sufficient to promote ofloxacin resistance when recapitulated in the M. tuberculosis H37Rv laboratory strain (143). While the mechanism linking ESX-5 to ofloxacin resistance is unknown (143), based on the studies presented above (58, 141), mutation of an ESX-5 component could cause increased ofloxacin resistance by decreasing OM permeability and reducing the uptake of ofloxacin.
Not all ESX-5 genes are essential. Transposon insertions in the eccA5 and espG5 genes have been previously described (31, 51, 86). Disruption of espG5 and the substrate-containing gene ppe10 in M. marinum or the espG5 gene in M. tuberculosis impacted the appearance and composition of the capsule, indicating a role for ESX-5 in maintaining capsule integrity (50).
CROSSING THE ENVELOPE
The mechanism of protein secretion across the envelope and the MOM is perhaps one of the most prominent unanswered questions in the field. Because ESX substrates are secreted from the mycobacterial cell, there are several options to consider for transport across the mycobacterial envelope. If the mycobacterial envelope acts as an impermeability barrier, as discussed above, there must be at least one apparatus that spans the envelope. Each ESX system could have independent means to cross the MOM, making the ESX systems true secretion systems. Alternatively, there could be a shared mechanism used by several exporters (ESX, Sec, or Tat) by which proteins cross the envelope. If the mycobacterial envelope is permeable, no apparatus may be needed to promote substrate transit across the envelope. Instead, properties intrinsic to secreted proteins could promote transit across the envelope. Unlike other secretion systems, the structure of an assembled ESX apparatus remains unknown (1). However, consideration of the localization of ESX-associated proteins in the envelope may inform potential mechanisms of transport.
CM.
Four ESX component proteins (EccB, EccC, EccD, and EccE) form a complex in the CM (59, 144). The ESX-5 membrane complex from Mycobacterium xenopi was recently resolved to 13 Å by electron microscopy and revealed a novel oligomeric complex with 6-fold symmetry. The complex resides exclusively within the cytoplasmic membrane and hints at a novel mechanism of protein translocation (145). Higher-resolution structures have been solved for individual components in the membrane complex (146–150). Importantly, the membrane complex is restricted to the CM and does not span the envelope. The mycosin proteases may function to stabilize the membrane complex (144). In addition to having a structural role, the mycosin proteases cleave ESX substrates. The ESX-1-secreted protein EspB is cleaved by MycP1 upon secretion (151). The mature processed form of EspB binds phospholipids (phosphatidic acid and phosphatidyl serine), suggesting that EspB may interact with either the cytoplasmic membrane or with phospholipids in the host cell (152).
Cell wall.
For bacterial protein secretion systems and other molecular machinery, the apparatus is inserted into the bacterial cell envelope, where it interacts directly with the peptidoglycan. Peptidoglycan-binding proteins promote pilus assembly and anchoring in the cell envelope. In the Neisseria gonorrhoeae type IVa pilus system, a peptidoglycan-binding protein anchors the apparatus in the membrane (153). In the Pseudomonas aeruginosa type IV pilus system FimV, an inner membrane protein that binds peptidoglycan is required for multimerization of PilQ (outer membrane component of the type VI pilus [154]).
The interaction of any ESX component with peptidoglycan/arabinogalactan has yet to be demonstrated in mycobacteria. Based upon the structure of the EccB1 component (subscripted “1” indicates that this protein is part of ESX-1 [17]), part of EccB1 may extend into the periplasm and interact with peptidoglycan (148). The peptidoglycan-binding properties of EccB1 have not been directly tested.
The characterization of MOM proteins may provide insight into their secretory mechanism. It is possible that known proteins integral to the MOM promote the transit of proteins across the envelope. For example, in silico analyses have suggested that Mce1 family proteins, which have a role in lipid homeostasis and transport, may promote ESX-1-mediated translocation across the MOM (130, 155, 156). The proposed link between the Mce1 proteins and ESX-1 is based on phylogenetic profiles, predicted protein-protein relationships, and analyses suggesting the Mce proteins may be MOM pore-forming proteins (155). A direct role for Mce1 family proteins in protein transport has not been demonstrated.
Alternatively, the ESX substrates may direct their own transit across the MOM, by either forming the apparatus spanning the MOM or by transiting the MOM directly (48, 104, 157–159). Notably, the EspC substrate of the ESX-1 system in M. tuberculosis (160) was recently reported by Lou et al. to self-assemble into filaments in vitro. EspC filaments were localized to the membrane fraction and were visualized on the surface/capsule of M. tuberculosis (157). The assembly of EspC into filaments led to several intriguing ideas, including one that EspC is a component of the MOM channel or a “needle” for the ESX-1 system (157, 161). EspC is encoded with the EspA and EspD substrates by the espACD operon (160, 162, 163). There are no recognized espACD counterparts for the ESX-2 through ESX-5 systems. The EspACD proteins may fulfill a function that is specific to the ESX-1 system, for example by promoting interaction with the host cell. Alternatively, additional systems may have genes which are functionally redundant with the espACD genes.
ESX substrates have been localized to the mycobacterial cell surface, extrinsically associated with the MOM (4, 31, 58, 60, 109, 164–167). Surface-localized ESX-1 substrates include EsxA and EspE. The EsxA substrate has been visualized and quantified on the mycobacterial cell surface (164, 166, 167). Several PE_PGRS proteins, a subfamily of PE proteins with polymorphic GC-rich sequences, are surface-localized ESX-5 substrates (51). The mechanism determining whether ESX substrates are targeted to the cell surface or the extracellular environment and the relationship between the surface-localized and secreted populations is unknown. However, a link between PE_PGRS secretion and LOS biosynthesis genes was reported in M. marinum (86). Interestingly, in strains lacking LOS, PE_PGRS proteins and the ESX-1 substrate EspE were more strongly attached to the cell surface, possibly linking LOS to the localization of ESX substrates on the cell surface (86).
Extracellular locations.
In vitro mycobacterial growth conditions preclude retention of the mycobacterial capsule. Specifically, mycobacterial strains are grown in vitro in the presence of detergent, usually either Tween 80 or tyloxapol, to reduce bacterial clumping during growth. Growth in detergent promotes the release of the mycobacterial capsule into the growth medium (4, 168). When mycobacteria are grown without detergent, to promote the retention of the capsule, several ESX-1 and ESX-5 substrates are localized to the capsule layer (4, 165, 169). Mycobacterial protein secretion is largely studied by the presence or absence of proteins in spent medium during in vitro growth. Indeed, a long list of proteins found in spent medium are dependent on ESX systems (30–32, 42–44, 51, 57, 59, 65, 73, 74, 101, 110, 162, 163, 165, 169–172).
ESX-1, A MEMBRANOLYTIC SYSTEM
The ESX-1 system has long been known to promote membrane lysis. In mycobacterial pathogens, the ESX-1 system damages the phagosomal membrane, promoting interaction between the bacteria and cytoplasm of host macrophages (46, 52, 55, 56, 173–175). Cytosolic signaling is required for mycobacterial virulence (173, 174, 175). Therefore, mycobacterial strains lacking ESX-1 genes are attenuated likely because they are retained in the phagosome and cannot trigger cytosolic signaling. Indeed, the expression of a secreted lysin from Listeria monocytogenes (LLO) in the absence of a functional ESX-1 exporter was sufficient to bypass the need for ESX-1 export in macrophage infections (173).
ESX-1 substrates have been considered prime candidates for mycobacterial membrane lysins. In 2003, it was proposed that EsxA, a major ESX-1 substrate, was the major membrane lysin secreted by the ESX-1 system. Genetic analysis of the function of EsxA is complicated. EsxA and its binding partner, EsxB, are encoded from the esxBA operon (176). Deletion of the esxBA operon, or of the esxA gene, abrogates ESX-1 export in pathogenic mycobacteria (30, 43). Because the loss of EsxA and EsxB secretion from pathogenic mycobacteria prevents the secretion of all of the other known ESX-1 substrates (165), it is difficult to ascribe a function to individual ESX-1 substrates using genetics alone. As such, the proposed membranolytic activity of EsxA has been supported by biochemical and biophysical analyses conducted by several research groups.
EsxA was first proposed to be a membrane lysin by Hsu et al. in a landmark paper which linked the loss of the region of deletion 1 (RD1) to attenuation of the BCG vaccine strain (42). The authors conducted a screen designed to identify genes required for cytolysis of lung epithelial cells, and one of the strains they identified was an M. tuberculosis strain bearing a transposon insertion in the esxB gene, which was polar on esxA expression. The authors then demonstrated that purified EsxA, in the presence or absence of EsxB, was sufficient to cause membrane destruction, as determined by measuring changes in conductance across an artificial membrane bilayer. These initial biophysical data were extended by several independent reports, including those demonstrating EsxA-dependent lysis of physiologically relevant liposome membranes (177), characterizing pore formation in sheep red blood cell (sRBC) membranes by EsxA (53), and the lysis of type 1 and type 2 pneumocytes by either purified EsxA or EsxA applied to the surface of ESX-1-deficient mycobacterial strains (167). Several more recent studies have focused on the mechanism of EsxA membrane lysis by investigating changes to membrane lysis as a function of pH (177), by comparing the activities of EsxA proteins from pathogenic and nonpathogenic mycobacterial species (178, 179), and by generating specific point mutations which disrupt the membranolytic activity of EsxA (180). For a recent comprehensive review of the evidence of EsxA pore-forming activity, please see the study by Peng and Sun (181).
Despite the aforementioned studies, the ability of EsxA to lyse membranes independently has been recently challenged. Conrad et al. found undeniably that EsxA is not sufficient to promote membrane lysis (182). Recombinant EsxA is widely acquired directly from the BEI repository or produced using a nondenaturing protocol, which includes the addition of detergent (183). In a series of well-controlled and clearly interpretable experiments, Conrad et al. demonstrated that the detergent present in the EsxA preparations, and not the EsxA protein itself, was responsible for the observed membranolytic activity of EsxA. Treatment of recombinant preparations of EsxA or Staphylococcus aureus α-hemolysin or Streptococcus pneumoniae pneumolysin (Hla and PLY, respectively, established bacterial lysins) with proteinase K abrogated Hla and PLY activity but did not alter the lytic activity of the EsxA preparation. Although the authors observed pH-dependent lytic activity of recombinant EsxA prepared without detergent, they show that changes in pH are not required to mediate ESX-1-mediated phagosomal lysis in a cellular model of infection. Thus, either EsxA is not required for phagosomal lysis, or additional ESX-1-associated proteins are required (182).
Although EsxA appears to be insufficient to promote ESX-1-mediated membrane lysis, the study by Conrad et al. definitively showed that the M. marinum ESX-1 system functions to lyse membranes in a contact-dependent manner (182). Contact-dependent membrane lysis by mycobacteria has been suggested but not directly demonstrated in several earlier studies (44, 167, 184). Coupled with evidence that ESX-1 substrates are present in the capsule and on the cell surface, discovered by our group and several others, direct translocation of ESX-1 substrates into the host macrophage may not occur. Rather, the presence of ESX-1 substrates on the cell surface may be sufficient for promoting phagosomal lysis (134, 166–185). Alternatively, contact with membranes may induce ESX-1 secretion through an unknown signal transduction mechanism.
Although strains bearing mutations in genes required for PDIM production are still competent to secrete ESX-1 substrates in vitro, recent studies have suggested that PDIM works synergistically with ESX-1 to promote phagosomal damage (24, 25). A recent study by Quigley et al. linked the levels of PDIM production with the ability of M. tuberculosis to promote phagosomal damage and downstream events in the macrophage (25). MmpL7 transports PDIM from the cytoplasm, where it is generated, to the MOM. In the absence of Mmpl7, PDIM accumulates in the mycobacterial cytoplasm (90). Quigley et al. demonstrated that M. tuberculosis strains bearing a transposon insertion in the mmpL7 gene damage the phagosomal membrane of THP-1 cells significantly less than the wild type (WT) or the complemented strain. As such, the mmpL7::Tn strain promoted less autophagy and host cell necrosis, events that are downstream of phagosomal damage. Importantly, a clean deletion of the mmpL7 gene did not impact the secretion of EsxA by M. tuberculosis in vitro (25). In another recent study, Augenstreich et al. demonstrated that M. tuberculosis strains lacking genes required for PDIM production led to reduced phagosomal damage in human monocyte-derived macrophages. The same study investigated if the mycobacterial lipid PDIM enhances phagosomal lysis by the ESX-1 system by incubating EsxA with vesicles with and without PDIM. Membrane lysis was enhanced in the PDIM-containing vesicles (24). In light of the paper by Conrad et al. (182), the mechanism linking PDIM to ESX-1 lysis is unlikely directly through EsxA. Yet, both studies concluded that PDIM aids in ESX-1-mediated membrane damage by likely inserting into and changing the biophysical properties of the phagosomal membrane, making it more susceptible to lysis by the ESX-1 system (24, 25).
We also recently observed a connection between PDIM biosynthesis and ESX-1 function (135). We found a spontaneous ochre mutation in the eccCb1 gene, which encodes a component of the ESX-1 system (30, 43, 44). The ochre mutation in eccCb1 resulted in a loss of ESX-1 function in M. marinum. Interestingly, we identified several suppressor strains which allowed the production of EccCb1 and restoration of ESX-1 function. In our efforts to map the suppressor of the ochre mutation, we found that the suppressor strains all had the same 13-bp insertion in the ppsC gene. ppsC is part of the pps operon, which is required for the production of PDIM (90, 186). While we do not yet understand the mechanism of suppression, our findings genetically link the ESX-1 system to PDIM production.
Additional links between the ESX-1 system and mycobacterial lipids have been reported. For example, pathogenic mycobacteria prevent the maturation of the phagosome within the host macrophage. In a screen designed to elucidate genes required for phagosome maturation arrest (PMA), lipid biosynthesis genes and genes encoding ESX-1 components were both identified (187). The mechanisms connecting ESX-1 genes and lipid biosynthetic genes in PMA are unknown.
CONCLUSIONS
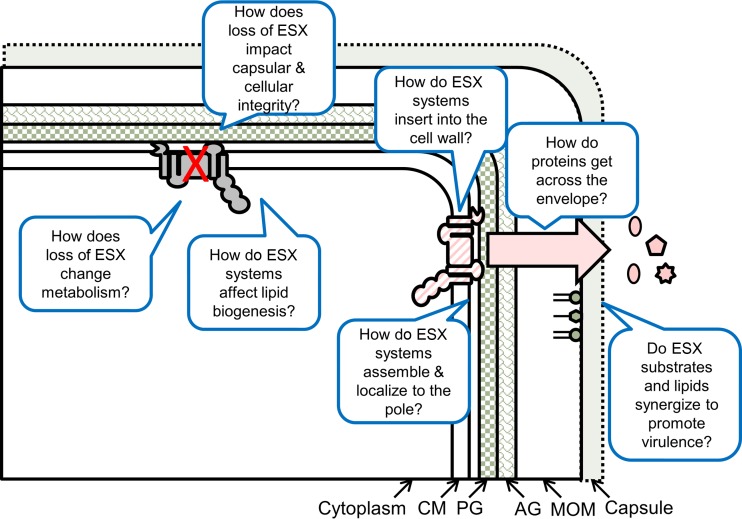
In summary, the relationship between ESX systems with the mycobacterial envelope is a complex and evolving story. Many details and unanswered questions remain (Fig. 2). How do secreted proteins cross the mycolate outer membrane? Do ESX substrates directly interact with the envelope, or are the ESX systems true secretion machines with components that span the envelope? How do ESX systems impact lipid biogenesis, and what are the downstream consequences? Do ESX systems provide additional functions for the mycobacterial cell, for example, by maintaining lipid biogenesis or envelope integrity in addition to roles in protein secretion? And finally, do these two complex systems, the mycobacterial cell envelope and the ESX exporters, work together to promote virulence within the host? It is clear that the answers to these questions will provide many exciting and revealing findings in the future.
FIG 2.
Unanswered questions regarding the interactions between ESX systems and the mycobacterial cell envelope. CM, cytoplasmic membrane; PG, peptidoglycan; AG, arabinogalactan; MOM, mycolate outer membrane.
ACKNOWLEDGMENTS
The Champion laboratory is supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01 AI106872 to P.A.C. R.E.B. is supported by the National Science Foundation Graduate Research Fellowship Program under grant DGE-1313583.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation or the National Institutes of Health.
We thank Micah Ferrell, Kevin Sanchez, Cristal Reyna Thompson, and Alexandra Chirakos for the critical reading of the manuscript.
REFERENCES
- 1.Chagnot C, Zorgani MA, Astruc T, Desvaux M. 2013. Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective. Front Microbiol 4:303. doi: 10.3389/fmicb.2013.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green ER, Mecsas J. 2016. Bacterial secretion systems: an overview. Microbiol Spectr 4:215–239. doi: 10.1128/microbiolspec.VMBF-0012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. 2008. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci U S A 105:3963–3967. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sani M, Houben EN, Geurtsen J, Pierson J, de Punder K, van Zon M, Wever B, Piersma SR, Jimenez CR, Daffe M, Appelmelk BJ, Bitter W, van der Wel N, Peters PJ. 2010. Direct visualization by cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog 6:e1000794. doi: 10.1371/journal.ppat.1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bansal-Mutalik R, Nikaido H. 2014. Mycobacterial outer membrane is a lipid bilayer and the inner membrane is unusually rich in diacyl phosphatidylinositol dimannosides. Proc Natl Acad Sci U S A 111:4958–4963. doi: 10.1073/pnas.1403078111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jankute M, Cox JA, Harrison J, Besra GS. 2015. Assembly of the mycobacterial cell wall. Annu Rev Microbiol 69:405–423. doi: 10.1146/annurev-micro-091014-104121. [DOI] [PubMed] [Google Scholar]
- 7.Daffé M. 2015. The cell envelope of tubercle bacilli. Tuberculosis (Edinb) 95(Suppl 1):S155–S158. doi: 10.1016/j.tube.2015.02.024. [DOI] [PubMed] [Google Scholar]
- 8.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abou-Zeid C, Smith I, Grange JM, Ratliff TL, Steele J, Rook GA. 1988. The secreted antigens of Mycobacterium tuberculosis and their relationship to those recognized by the available antibodies. J Gen Microbiol 134:531–538. [DOI] [PubMed] [Google Scholar]
- 10.Andersen P, Askgaard D, Ljungqvist L, Bennedsen J, Heron I. 1991. Proteins released from Mycobacterium tuberculosis during growth. Infect Immun 59:1905–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonnenberg MG, Belisle JT. 1997. Definition of Mycobacterium tuberculosis culture filtrate proteins by two-dimensional polyacrylamide gel electrophoresis, N-terminal amino acid sequencing, and electrospray mass spectrometry. Infect Immun 65:4515–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge Y, El-Naggar M, Sze SK, Oh HB, Begley TP, McLafferty FW, Boshoff H, Barry CE III. 2003. Top down characterization of secreted proteins from Mycobacterium tuberculosis by electron capture dissociation mass spectrometry. J Am Soc Mass Spectrom 14:253–261. doi: 10.1016/S1044-0305(02)00913-3. [DOI] [PubMed] [Google Scholar]
- 13.Delcour AH. 2009. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta 1794:808–816. doi: 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambert PA. 2002. Cellular impermeability and uptake of biocides and antibiotics in Gram-positive bacteria and mycobacteria. J Appl Microbiol 92(Suppl):46S–54S. [PubMed] [Google Scholar]
- 15.Niederweis M, Danilchanka O, Huff J, Hoffmann C, Engelhardt H. 2010. Mycobacterial outer membranes: in search of proteins. Trends Microbiol 18:109–116. doi: 10.1016/j.tim.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CM, Appelmelk BJ, Bitter W. 2007. Type VII secretion–mycobacteria show the way. Nat Rev Microbiol 5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- 17.Bitter W, Houben EN, Bottai D, Brodin P, Brown EJ, Cox JS, Derbyshire K, Fortune SM, Gao LY, Liu J, Gey van Pittius NC, Pym AS, Rubin EJ, Sherman DR, Cole ST, Brosch R. 2009. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog 5:e1000507. doi: 10.1371/journal.ppat.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houben EN, Korotkov KV, Bitter W. 2014. Take five–type VII secretion systems of mycobacteria. Biochim Biophys Acta 1843:1707–1716. doi: 10.1016/j.bbamcr.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Pallen MJ. 2002. The ESAT-6/WXG100 superfamily–and a new Gram-positive secretion system? Trends Microbiol 10:209–212. doi: 10.1016/S0966-842X(02)02345-4. [DOI] [PubMed] [Google Scholar]
- 20.Burts ML, Williams WA, DeBord K, Missiakas DM. 2005. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc Natl Acad Sci U S A 102:1169–1174. doi: 10.1073/pnas.0405620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garufi G, Butler E, Missiakas D. 2008. ESAT-6-like protein secretion in Bacillus anthracis. J Bacteriol 190:7004–7011. doi: 10.1128/JB.00458-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Unnikrishnan M, Constantinidou C, Palmer T, Pallen MJ. 2017. The enigmatic Esx proteins: looking beyond mycobacteria. Trends Microbiol 25:192–204. doi: 10.1016/j.tim.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Loots DT, Swanepoel CC, Newton-Foot M, Gey van Pittius NC. 2016. A metabolomics investigation of the function of the ESX-1 gene cluster in mycobacteria. Microb Pathog 100:268–275. doi: 10.1016/j.micpath.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 24.Augenstreich J, Arbues A, Simeone R, Haanappel E, Wegener A, Sayes F, Chevalier FL, Chalut C, Malaga W, Guilhot C, Brosch R, Astarie-Dequeker C. 2017. ESX-1 and phthiocerol dimycocerosates of Mycobacterium tuberculosis act in concert to cause phagosomal rupture and host cell apoptosis. Cell Microbiol 19:e12726. doi: 10.1111/cmi.12726. [DOI] [PubMed] [Google Scholar]
- 25.Quigley J, Hughitt VK, Velikovsky CA, Mariuzza RA, El-Sayed NM, Briken V. 2017. The cell wall lipid PDIM contributes to phagosomal escape and host cell exit of Mycobacterium tuberculosis. mBio 8(2):e00148-17. doi: 10.1128/mBio.00148-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gey Van Pittius NC, Gamieldien J, Hide W, Brown GD, Siezen RJ, Beyers AD. 2001. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol 2:RESEARCH0044. doi: 10.1186/gb-2001-2-10-research0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newton-Foot M, Warren RM, Sampson SL, van Helden PD, Gey van Pittius NC. 2016. The plasmid-mediated evolution of the mycobacterial ESX (type VII) secretion systems. BMC Evol Biol 16:62. doi: 10.1186/s12862-016-0631-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ummels R, Abdallah AM, Kuiper V, Aajoud A, Sparrius M, Naeem R, Spaink HP, van Soolingen D, Pain A, Bitter W. 2014. Identification of a novel conjugative plasmid in mycobacteria that requires both type IV and type VII secretion. mBio 5(5):e01744-14. doi: 10.1128/mBio.01744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dumas E, Boritsch EC, Vandenbogaert M, Rodriguez de la Vega RC, Thiberge JM, Caro V, Gaillard JL, Heym B, Girard-Misguich F, Brosch R, Sapriel G. 2016. Mycobacterial pan-genome analysis suggests important role of plasmids in the radiation of type VII secretion systems. Genome Biol Evol 8:387–402. doi: 10.1093/gbe/evw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanley SA, Raghavan S, Hwang WW, Cox JS. 2003. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc Natl Acad Sci U S A 100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abdallah AM, Verboom T, Hannes F, Safi M, Strong M, Eisenberg D, Musters RJ, Vandenbroucke-Grauls CM, Appelmelk BJ, Luirink J, Bitter W. 2006. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol Microbiol 62:667–679. doi: 10.1111/j.1365-2958.2006.05409.x. [DOI] [PubMed] [Google Scholar]
- 32.Siegrist MS, Unnikrishnan M, McConnell MJ, Borowsky M, Cheng TY, Siddiqi N, Fortune SM, Moody DB, Rubin EJ. 2009. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc Natl Acad Sci U S A 106:18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ates LS, Houben EN, Bitter W. 2016. Type VII secretion: a highly versatile secretion system. Microbiol Spectr 4:357–384. doi: 10.1128/microbiolspec.VMBF-0011-2015. [DOI] [PubMed] [Google Scholar]
- 34.Gröschel MI, Sayes F, Simeone R, Majlessi L, Brosch R. 2016. ESX secretion systems: mycobacterial evolution to counter host immunity. Nat Rev Microbiol 14:677–691. doi: 10.1038/nrmicro.2016.131. [DOI] [PubMed] [Google Scholar]
- 35.Majlessi L, Prados-Rosales R, Casadevall A, Brosch R. 2015. Release of mycobacterial antigens. Immunol Rev 264:25–45. doi: 10.1111/imr.12251. [DOI] [PubMed] [Google Scholar]
- 36.Shah S, Briken V. 2016. Modular organization of the ESX-5 secretion system in Mycobacterium tuberculosis. Front Cell Infect Microbiol 6:49. doi: 10.3389/fmicb.2016.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koch R. 1882. Die Atiologie der Tuberkulose. Berliner Klin Wochenschr 15:221–230. https://www.asm.org/ccLibraryFiles/FILENAME/0000000228/1882p109.pdf. [Google Scholar]
- 38.Shiloh MU, Champion PA. 2010. To catch a killer. What can mycobacterial models teach us about Mycobacterium tuberculosis pathogenesis? Curr Opin Microbiol 13:86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamm LM, Brown EJ. 2004. Mycobacterium marinum: the generalization and specialization of a pathogenic mycobacterium. Microbes Infect 6:1418–1428. doi: 10.1016/j.micinf.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 40.Pozos TC, Ramakrishnan L. 2004. New models for the study of Mycobacterium-host interactions. Curr Opin Immunol 16:499–505. doi: 10.1016/j.coi.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 41.Tobin DM, Ramakrishnan L. 2008. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell Microbiol 10:1027–1039. doi: 10.1111/j.1462-5822.2008.01133.x. [DOI] [PubMed] [Google Scholar]
- 42.Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, Marks CB, Padiyar J, Goulding C, Gingery M, Eisenberg D, Russell RG, Derrick SC, Collins FM, Morris SL, King CH, Jacobs WR Jr. 2003. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci U S A 100:12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guinn KM, Hickey MJ, Mathur SK, Zakel KL, Grotzke JE, Lewinsohn DM, Smith S, Sherman DR. 2004. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol Microbiol 51:359–370. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao LY, Guo S, McLaughlin B, Morisaki H, Engel JN, Brown EJ. 2004. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol Microbiol 53:1677–1693. doi: 10.1111/j.1365-2958.2004.04261.x. [DOI] [PubMed] [Google Scholar]
- 45.Abdallah AM, Savage ND, van Zon M, Wilson L, Vandenbroucke-Grauls CM, van der Wel NN, Ottenhoff TH, Bitter W. 2008. The ESX-5 secretion system of Mycobacterium marinum modulates the macrophage response. J Immunol 181:7166–7175. doi: 10.4049/jimmunol.181.10.7166. [DOI] [PubMed] [Google Scholar]
- 46.Kennedy GM, Morisaki JH, Champion PA. 2012. Conserved mechanisms of Mycobacterium marinum pathogenesis within the environmental amoeba, Acanthamoeba castellanii. Appl Environ Microbiol 8:2049–2052. doi: 10.1128/AEM.06965-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lewis KN, Liao R, Guinn KM, Hickey MJ, Smith S, Behr MA, Sherman DR. 2003. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guérin attenuation. J Infect Dis 187:117–123. doi: 10.1086/345862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pym AS, Brodin P, Brosch R, Huerre M, Cole ST. 2002. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol Microbiol 46:709–717. doi: 10.1046/j.1365-2958.2002.03237.x. [DOI] [PubMed] [Google Scholar]
- 49.Brodin P, Majlessi L, Marsollier L, de Jonge MI, Bottai D, Demangel C, Hinds J, Neyrolles O, Butcher PD, Leclerc C, Cole ST, Brosch R. 2006. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect Immun 74:88–98. doi: 10.1128/IAI.74.1.88-98.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ates LS, van der Woude AD, Bestebroer J, van Stempvoort G, Musters RJ, Garcia-Vallejo JJ, Picavet DI, Weerd R, Maletta M, Kuijl CP, van der Wel NN, Bitter W. 2016. The ESX-5 system of pathogenic mycobacteria is involved in capsule integrity and virulence through its substrate PPE10. PLoS Pathog 12:e1005696. doi: 10.1371/journal.ppat.1005696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abdallah AM, Verboom T, Weerdenburg EM, Gey van Pittius NC, Mahasha PW, Jimenez C, Parra M, Cadieux N, Brennan MJ, Appelmelk BJ, Bitter W. 2009. PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol Microbiol 73:329–340. doi: 10.1111/j.1365-2958.2009.06783.x. [DOI] [PubMed] [Google Scholar]
- 52.Simeone R, Bobard A, Lippmann J, Bitter W, Majlessi L, Brosch R, Enninga J. 2012. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog 8:e1002507. doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith J, Manoranjan J, Pan M, Bohsali A, Xu J, Liu J, McDonald KL, Szyk A, LaRonde-LeBlanc N, Gao LY. 2008. Evidence for pore formation in host cell membranes by ESX-1-secreted ESAT-6 and its role in Mycobacterium marinum escape from the vacuole. Infect Immun 76:5478–5487. doi: 10.1128/IAI.00614-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stamm LM, Morisaki JH, Gao LY, Jeng RL, McDonald KL, Roth R, Takeshita S, Heuser J, Welch MD, Brown EJ. 2003. Mycobacterium marinum escapes from phagosomes and is propelled by actin-based motility. J Exp Med 198:1361–1368. doi: 10.1084/jem.20031072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Houben D, Demangel C, van Ingen J, Perez J, Baldeon L, Abdallah AM, Caleechurn L, Bottai D, van Zon M, de Punder K, van der Laan T, Kant A, Bossers-de Vries R, Willemsen P, Bitter W, van Soolingen D, Brosch R, van der Wel N, Peters PJ. 2012. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol 14:1287–1298. doi: 10.1111/j.1462-5822.2012.01799.x. [DOI] [PubMed] [Google Scholar]
- 56.van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, Brenner M, Peters PJ. 2007. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell 129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- 57.Bottai D, Di Luca M, Majlessi L, Frigui W, Simeone R, Sayes F, Bitter W, Brennan MJ, Leclerc C, Batoni G, Campa M, Brosch R, Esin S. 2012. Disruption of the ESX-5 system of Mycobacterium tuberculosis causes loss of PPE protein secretion, reduction of cell wall integrity and strong attenuation. Mol Microbiol 83:1195–1209. doi: 10.1111/j.1365-2958.2012.08001.x. [DOI] [PubMed] [Google Scholar]
- 58.Ates LS, Ummels R, Commandeur S, van de Weerd R, Sparrius M, Weerdenburg E, Alber M, Kalscheuer R, Piersma SR, Abdallah AM, Abd El Ghany M, Abdel-Haleem AM, Pain A, Jimenez CR, Bitter W, Houben EN. 2015. Essential role of the ESX-5 secretion system in outer membrane permeability of pathogenic mycobacteria. PLoS Genet 11:e1005190. doi: 10.1371/journal.pgen.1005190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Houben EN, Bestebroer J, Ummels R, Wilson L, Piersma SR, Jimenez CR, Ottenhoff TH, Luirink J, Bitter W. 2012. Composition of the type VII secretion system membrane complex. Mol Microbiol 85:472–484. doi: 10.1111/j.1365-2958.2012.08206.x. [DOI] [PubMed] [Google Scholar]
- 60.Daleke MH, Cascioferro A, de Punder K, Ummels R, Abdallah AM, van der Wel N, Peters PJ, Luirink J, Manganelli R, Bitter W. 2011. Conserved Pro-Glu (PE) and Pro-Pro-Glu (PPE) protein domains target LipY lipases of pathogenic mycobacteria to the cell surface via the ESX-5 pathway. J Biol Chem 286:19024–19034. doi: 10.1074/jbc.M110.204966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cascioferro A, Daleke MH, Ventura M, Dona V, Delogu G, Palu G, Bitter W, Manganelli R. 2011. Functional dissection of the PE domain responsible for translocation of PE_PGRS33 across the mycobacterial cell wall. PLoS One 6:e27713. doi: 10.1371/journal.pone.0027713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daleke MH, van der Woude AD, Parret AH, Ummels R, de Groot AM, Watson D, Piersma SR, Jimenez CR, Luirink J, Bitter W, Houben EN. 2012. Specific chaperones for the type VII protein secretion pathway. J Biol Chem 287:31939–31947. doi: 10.1074/jbc.M112.397596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shah S, Cannon JR, Fenselau C, Briken V. 2015. A duplicated ESAT-6 region of ESX-5 is involved in protein export and virulence of mycobacteria. Infect Immun 83:4349–4361. doi: 10.1128/IAI.00827-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Flint JL, Kowalski JC, Karnati PK, Derbyshire KM. 2004. The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proc Natl Acad Sci U S A 101:12598–12603. doi: 10.1073/pnas.0404892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coros A, Callahan B, Battaglioli E, Derbyshire KM. 2008. The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol Microbiol 69:794–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gray TA, Krywy JA, Harold J, Palumbo MJ, Derbyshire KM. 2013. Distributive conjugal transfer in mycobacteria generates progeny with meiotic-like genome-wide mosaicism, allowing mapping of a mating identity locus. PLoS Biol 11:e1001602. doi: 10.1371/journal.pbio.1001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gray TA, Clark RR, Boucher N, Lapierre P, Smith C, Derbyshire KM. 2016. Intercellular communication and conjugation are mediated by ESX secretion systems in mycobacteria. Science 354:347–350. doi: 10.1126/science.aag0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Derbyshire KM, Gray TA. 2014. Distributive conjugal transfer: new insights into horizontal gene transfer and genetic exchange in mycobacteria. Microbiol Spectr 2:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boritsch EC, Khanna V, Pawlik A, Honore N, Navas VH, Ma L, Bouchier C, Seemann T, Supply P, Stinear TP, Brosch R. 2016. Key experimental evidence of chromosomal DNA transfer among selected tuberculosis-causing mycobacteria. Proc Natl Acad Sci U S A 113:9876–9881. doi: 10.1073/pnas.1604921113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Serafini A, Boldrin F, Palu G, Manganelli R. 2009. Characterization of a Mycobacterium tuberculosis ESX-3 conditional mutant: essentiality and rescue by iron and zinc. J Bacteriol 191:6340–6344. doi: 10.1128/JB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Serafini A, Pisu D, Palu G, Rodriguez GM, Manganelli R. 2013. The ESX-3 secretion system is necessary for iron and zinc homeostasis in Mycobacterium tuberculosis. PLoS One 8:e78351. doi: 10.1371/journal.pone.0078351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tinaztepe E, Wei JR, Raynowska J, Portal-Celhay C, Thompson V, Philips JA. 2016. Role of metal-dependent regulation of ESX-3 secretion in intracellular survival of Mycobacterium tuberculosis. Infect Immun 84:2255–2263. doi: 10.1128/IAI.00197-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tufariello JM, Chapman JR, Kerantzas CA, Wong KW, Vilcheze C, Jones CM, Cole LE, Tinaztepe E, Thompson V, Fenyo D, Niederweis M, Ueberheide B, Philips JA, Jacobs WR Jr. 2016. Separable roles for Mycobacterium tuberculosis ESX-3 effectors in iron acquisition and virulence. Proc Natl Acad Sci U S A 113:E348–E357. doi: 10.1073/pnas.1523321113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Siegrist MS, Steigedal M, Ahmad R, Mehra A, Dragset MS, Schuster BM, Philips JA, Carr SA, Rubin EJ. 2014. Mycobacterial Esx-3 requires multiple components for iron acquisition. mBio 5(3):e01073-14. doi: 10.1128/mBio.01073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Portal-Celhay C, Tufariello JM, Srivastava S, Zahra A, Klevorn T, Grace PS, Mehra A, Park HS, Ernst JD, Jacobs WR Jr, Philips JA. 2016. Mycobacterium tuberculosis EsxH inhibits ESCRT-dependent CD4+ T-cell activation. Nat Microbiol 2:16232. doi: 10.1038/nmicrobiol.2016.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mehra A, Zahra A, Thompson V, Sirisaengtaksin N, Wells A, Porto M, Koster S, Penberthy K, Kubota Y, Dricot A, Rogan D, Vidal M, Hill DE, Bean AJ, Philips JA. 2013. Mycobacterium tuberculosis type VII secreted effector EsxH targets host ESCRT to impair trafficking. PLoS Pathog 9:e1003734. doi: 10.1371/journal.ppat.1003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blasco B, Chen JM, Hartkoorn R, Sala C, Uplekar S, Rougemont J, Pojer F, Cole ST. 2012. Virulence regulator EspR of Mycobacterium tuberculosis is a nucleoid-associated protein. PLoS Pathog 8:e1002621. doi: 10.1371/journal.ppat.1002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Casonato S, Cervantes Sanchez A, Haruki H, Rengifo Gonzalez M, Provvedi R, Dainese E, Jaouen T, Gola S, Bini E, Vicente M, Johnsson K, Ghisotti D, Palu G, Hernandez-Pando R, Manganelli R. 2012. WhiB5, a transcriptional regulator that contributes to Mycobacterium tuberculosis virulence and reactivation. Infect Immun 80:3132–3144. doi: 10.1128/IAI.06328-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mendum TA, Wu H, Kierzek AM, Stewart GR. 2015. Lipid metabolism and type VII secretion systems dominate the genome scale virulence profile of Mycobacterium tuberculosis in human dendritic cells. BMC Genomics 16:372. doi: 10.1186/s12864-015-1569-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Daffé M, Crick DC, Jackson M. 2014. Genetics of capsular polysaccharides and cell envelope (glyco)lipids. Microbiol Spectr 2:MGM2–0021-2013. doi: 10.1128/microbiolspec.MGM2-0021-2013. [DOI] [PubMed] [Google Scholar]
- 81.Alderwick LJ, Harrison J, Lloyd GS, Birch HL. 2015. The mycobacterial cell wall–peptidoglycan and arabinogalactan. Cold Spring Harb Perspect Med 5:a021113. doi: 10.1101/cshperspect.a021113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Onwueme KC, Vos CJ, Zurita J, Ferreras JA, Quadri LE. 2005. The dimycocerosate ester polyketide virulence factors of mycobacteria. Prog Lipid Res 44:259–302. doi: 10.1016/j.plipres.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 83.Kapopoulou A, Lew JM, Cole ST. 2011. The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb) 91:8–13. doi: 10.1016/j.tube.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 84.Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PD, Abdellah Z, Arrowsmith C, Chillingworth T, Churcher C, Clarke K, Cronin A, Davis P, Goodhead I, Holroyd N, Jagels K, Lord A, Moule S, Mungall K, Norbertczak H, Quail MA, Rabbinowitsch E, Walker D, White B, Whitehead S, Small PL, Brosch R, Ramakrishnan L, Fischbach MA, Parkhill J, Cole ST. 2008. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res 18:729–741. doi: 10.1101/gr.075069.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brennan PJ, Nikaido H. 1995. The envelope of mycobacteria. Annu Rev Biochem 64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 86.van der Woude AD, Sarkar D, Bhatt A, Sparrius M, Raadsen SA, Boon L, Geurtsen J, van der Sar AM, Luirink J, Houben EN, Besra GS, Bitter W. 2012. Unexpected link between lipooligosaccharide biosynthesis and surface protein release in Mycobacterium marinum. J Biol Chem 287:20417–20429. doi: 10.1074/jbc.M111.336461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Etienne G, Malaga W, Laval F, Lemassu A, Guilhot C, Daffe M. 2009. Identification of the polyketide synthase involved in the biosynthesis of the surface-exposed lipooligosaccharides in mycobacteria. J Bacteriol 191:2613–2621. doi: 10.1128/JB.01235-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Boritsch EC, Frigui W, Cascioferro A, Malaga W, Etienne G, Laval F, Pawlik A, Le Chevalier F, Orgeur M, Ma L, Bouchier C, Stinear TP, Supply P, Majlessi L, Daffe M, Guilhot C, Brosch R. 2016. pks5-recombination-mediated surface remodelling in Mycobacterium tuberculosis emergence. Nat Microbiol 1:15019. doi: 10.1038/nmicrobiol.2015.19. [DOI] [PubMed] [Google Scholar]
- 89.Constant P, Perez E, Malaga W, Laneelle MA, Saurel O, Daffe M, Guilhot C. 2002. Role of the pks15/1 gene in the biosynthesis of phenolglycolipids in the Mycobacterium tuberculosis complex. Evidence that all strains synthesize glycosylated p-hydroxybenzoic methyl esters and that strains devoid of phenolglycolipids harbor a frameshift mutation in the pks15/1 gene. J Biol Chem 277:38148–38158. [DOI] [PubMed] [Google Scholar]
- 90.Cox JS, Chen B, McNeil M, Jacobs WR Jr. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 91.Yu J, Tran V, Li M, Huang X, Niu C, Wang D, Zhu J, Wang J, Gao Q, Liu J. 2012. Both phthiocerol dimycocerosates and phenolic glycolipids are required for virulence of Mycobacterium marinum. Infect Immun 80:1381–1389. doi: 10.1128/IAI.06370-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Camacho LR, Ensergueix D, Perez E, Gicquel B, Guilhot C. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol Microbiol 34:257–267. doi: 10.1046/j.1365-2958.1999.01593.x. [DOI] [PubMed] [Google Scholar]
- 93.Ishikawa E, Mori D, Yamasaki S. 2017. Recognition of mycobacterial lipids by immune receptors. Trends Immunol 38:66–76. doi: 10.1016/j.it.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 94.Stanley SA, Cox JS. 2013. Host-pathogen interactions during Mycobacterium tuberculosis infections. Curr Top Microbiol Immunol 374:211–241. doi: 10.1007/82_2013_332. [DOI] [PubMed] [Google Scholar]
- 95.Rhoades ER, Ullrich HJ. 2000. How to establish a lasting relationship with your host: lessons learned from Mycobacterium spp. Immunol Cell Biol 78:301–310. doi: 10.1046/j.1440-1711.2000.00938.x. [DOI] [PubMed] [Google Scholar]
- 96.Cao G, Howard ST, Zhang P, Wang X, Chen XL, Samten B, Pang X. 2015. EspR, a regulator of the ESX-1 secretion system in Mycobacterium tuberculosis, is directly regulated by the two-component systems MprAB and PhoPR. Microbiology 161:477–489. doi: 10.1099/mic.0.000023. [DOI] [PubMed] [Google Scholar]
- 97.Datta P, Ravi J, Guerrini V, Chauhan R, Neiditch MB, Shell SS, Fortune SM, Hancioglu B, Igoshin OA, Gennaro ML. 2015. The Psp system of Mycobacterium tuberculosis integrates envelope stress-sensing and envelope-preserving functions. Mol Microbiol 97:408–422. doi: 10.1111/mmi.13037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martin C, Williams A, Hernandez-Pando R, Cardona PJ, Gormley E, Bordat Y, Soto CY, Clark SO, Hatch GJ, Aguilar D, Ausina V, Gicquel B. 2006. The live Mycobacterium tuberculosis phoP mutant strain is more attenuated than BCG and confers protective immunity against tuberculosis in mice and guinea pigs. Vaccine 24:3408–3419. doi: 10.1016/j.vaccine.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 99.Pang X, Samten B, Cao G, Wang X, Tvinnereim AR, Chen XL, Howard ST. 2013. MprAB regulates the espA operon in Mycobacterium tuberculosis and modulates ESX-1 function and host cytokine response. J Bacteriol 195:66–75. doi: 10.1128/JB.01067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hatzios SK, Baer CE, Rustad TR, Siegrist MS, Pang JM, Ortega C, Alber T, Grundner C, Sherman DR, Bertozzi CR. 2013. Osmosensory signaling in Mycobacterium tuberculosis mediated by a eukaryotic-like Ser/Thr protein kinase. Proc Natl Acad Sci U S A 110:E5069–E5077. doi: 10.1073/pnas.1321205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Raghavan S, Manzanillo P, Chan K, Dovey C, Cox JS. 2008. Secreted transcription factor controls Mycobacterium tuberculosis virulence. Nature 454:717–721. doi: 10.1038/nature07219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fishbein S, van Wyk N, Warren RM, Sampson SL. 2015. Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Mol Microbiol 96:901–916. doi: 10.1111/mmi.12981. [DOI] [PubMed] [Google Scholar]
- 103.Walters SB, Dubnau E, Kolesnikova I, Laval F, Daffe M, Smith I. 2006. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol Microbiol 60:312–330. doi: 10.1111/j.1365-2958.2006.05102.x. [DOI] [PubMed] [Google Scholar]
- 104.Anil Kumar V, Goyal R, Bansal R, Singh N, Sevalkar RR, Kumar A, Sarkar D. 2016. EspR-dependent ESAT-6 protein secretion of Mycobacterium tuberculosis requires the presence of virulence regulator PhoP. J Biol Chem 291:19018–19030. doi: 10.1074/jbc.M116.746289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Solans L, Aguilo N, Samper S, Pawlik A, Frigui W, Martin C, Brosch R, Gonzalo-Asensio J. 2014. A specific polymorphism in Mycobacterium tuberculosis H37Rv causes differential ESAT-6 expression and identifies WhiB6 as a novel ESX-1 component. Infect Immun 82:3446–3456. doi: 10.1128/IAI.01824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rohde KH, Veiga DF, Caldwell S, Balazsi G, Russell DG. 2012. Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection. PLoS Pathog 8:e1002769. doi: 10.1371/journal.ppat.1002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Joshi SA, Ball DA, Sun MG, Carlsson F, Watkins BY, Aggarwal N, McCracken JM, Huynh KK, Brown EJ. 2012. EccA1, a component of the Mycobacterium marinum ESX-1 protein virulence factor secretion pathway, regulates mycolic acid lipid synthesis. Chem Biol 19:372–380. doi: 10.1016/j.chembiol.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 108.Fujiwara N, Ohara N, Ogawa M, Maeda S, Naka T, Taniguchi H, Yamamoto S, Ayata M. 2015. Glycopeptidolipid of Mycobacterium smegmatis J15cs affects morphology and survival in host cells. PLoS One 10:e0126813. doi: 10.1371/journal.pone.0126813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Carlsson F, Joshi SA, Rangell L, Brown EJ. 2009. Polar localization of virulence-related Esx-1 secretion in mycobacteria. PLoS Pathog 5:e1000285. doi: 10.1371/journal.ppat.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McLaughlin B, Chon JS, MacGurn JA, Carlsson F, Cheng TL, Cox JS, Brown EJ. 2007. A Mycobacterium ESX-1-secreted virulence factor with unique requirements for export. PLoS Pathog 3:e105. doi: 10.1371/journal.ppat.0030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Frigui W, Bottai D, Majlessi L, Monot M, Josselin E, Brodin P, Garnier T, Gicquel B, Martin C, Leclerc C, Cole ST, Brosch R. 2008. Control of M. tuberculosis ESAT-6 secretion and specific T cell recognition by PhoP. PLoS Pathog 4:e33. doi: 10.1371/journal.ppat.0040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Scheurwater EM, Burrows LL. 2011. Maintaining network security: how macromolecular structures cross the peptidoglycan layer. FEMS Microbiol Lett 318:1–9. doi: 10.1111/j.1574-6968.2011.02228.x. [DOI] [PubMed] [Google Scholar]
- 113.Koraimann G. 2003. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell Mol Life Sci 60:2371–2388. doi: 10.1007/s00018-003-3056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kieser KJ, Rubin EJ. 2014. How sisters grow apart: mycobacterial growth and division. Nat Rev Microbiol 12:550–562. doi: 10.1038/nrmicro3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Carel C, Nukdee K, Cantaloube S, Bonne M, Diagne CT, Laval F, Daffe M, Zerbib D. 2014. Mycobacterium tuberculosis proteins involved in mycolic acid synthesis and transport localize dynamically to the old growing pole and septum. PLoS One 9:e97148. doi: 10.1371/journal.pone.0097148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Meniche X, Otten R, Siegrist MS, Baer CE, Murphy KC, Bertozzi CR, Sassetti CM. 2014. Subpolar addition of new cell wall is directed by DivIVA in mycobacteria. Proc Natl Acad Sci U S A 111:E3243–E3251. doi: 10.1073/pnas.1402158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wirth SE, Krywy JA, Aldridge BB, Fortune SM, Fernandez-Suarez M, Gray TA, Derbyshire KM. 2012. Polar assembly and scaffolding proteins of the virulence-associated ESX-1 secretory apparatus in mycobacteria. Mol Microbiol 83:654–664. doi: 10.1111/j.1365-2958.2011.07958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Delogu G, Pusceddu C, Bua A, Fadda G, Brennan MJ, Zanetti S. 2004. Rv1818c-encoded PE_PGRS protein of Mycobacterium tuberculosis is surface exposed and influences bacterial cell structure. Mol Microbiol 52:725–733. doi: 10.1111/j.1365-2958.2004.04007.x. [DOI] [PubMed] [Google Scholar]
- 119.Trias J, Benz R. 1994. Permeability of the cell wall of Mycobacterium smegmatis. Mol Microbiol 14:283–290. doi: 10.1111/j.1365-2958.1994.tb01289.x. [DOI] [PubMed] [Google Scholar]
- 120.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hett EC, Rubin EJ. 2008. Bacterial growth and cell division: a mycobacterial perspective. Microbiol Mol Biol Rev 72:126–156, table of contents. doi: 10.1128/MMBR.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vaara M, Nurminen M. 1999. Outer membrane permeability barrier in Escherichia coli mutants that are defective in the late acyltransferases of lipid A biosynthesis. Antimicrob Agents Chemother 43:1459–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brown-Elliott BA, Nash KA, Wallace RJ Jr. 2012. Antimicrobial susceptibility testing, drug resistance mechanisms, and therapy of infections with nontuberculous mycobacteria. Clin Microbiol Rev 25:545–582. doi: 10.1128/CMR.05030-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Soetaert K, Rens C, Wang XM, De Bruyn J, Laneelle MA, Laval F, Lemassu A, Daffe M, Bifani P, Fontaine V, Lefevre P. 2015. Increased vancomycin susceptibility in mycobacteria: a new approach to identify synergistic activity against multidrug-resistant mycobacteria. Antimicrob Agents Chemother 59:5057–5060. doi: 10.1128/AAC.04856-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rens C, Laval F, Daffe M, Denis O, Frita R, Baulard A, Wattiez R, Lefevre P, Fontaine V. 2016. Effects of lipid-lowering drugs on vancomycin susceptibility of mycobacteria. Antimicrob Agents Chemother 60:6193–6199. doi: 10.1128/AAC.00872-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fairman JW, Noinaj N, Buchanan SK. 2011. The structural biology of β-barrel membrane proteins: a summary of recent reports. Curr Opin Struct Biol 21:523–531. doi: 10.1016/j.sbi.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Selkrig J, Leyton DL, Webb CT, Lithgow T. 2014. Assembly of β-barrel proteins into bacterial outer membranes. Biochim Biophys Acta 1843:1542–1550. doi: 10.1016/j.bbamcr.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 128.van der Woude AD, Mahendran KR, Ummels R, Piersma SR, Pham TV, Jimenez CR, de Punder K, van der Wel NN, Winterhalter M, Luirink J, Bitter W, Houben EN. 2013. Differential detergent extraction of mycobacterium marinum cell envelope proteins identifies an extensively modified threonine-rich outer membrane protein with channel activity. J Bacteriol 195:2050–2059. doi: 10.1128/JB.02236-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Niederweis M, Ehrt S, Heinz C, Klocker U, Karosi S, Swiderek KM, Riley LW, Benz R. 1999. Cloning of the mspA gene encoding a porin from Mycobacterium smegmatis. Mol Microbiol 33:933–945. doi: 10.1046/j.1365-2958.1999.01472.x. [DOI] [PubMed] [Google Scholar]
- 130.Forrellad MA, McNeil M, Santangelo Mde L, Blanco FC, Garcia E, Klepp LI, Huff J, Niederweis M, Jackson M, Bigi F. 2014. Role of the Mce1 transporter in the lipid homeostasis of Mycobacterium tuberculosis. Tuberculosis (Edinb) 94:170–177. doi: 10.1016/j.tube.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Danilchanka O, Pires D, Anes E, Niederweis M. 2015. The Mycobacterium tuberculosis outer membrane channel protein CpnT confers susceptibility to toxic molecules. Antimicrob Agents Chemother 59:2328–2336. doi: 10.1128/AAC.04222-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Danilchanka O, Sun J, Pavlenok M, Maueroder C, Speer A, Siroy A, Marrero J, Trujillo C, Mayhew DL, Doornbos KS, Munoz LE, Herrmann M, Ehrt S, Berens C, Niederweis M. 2014. An outer membrane channel protein of Mycobacterium tuberculosis with exotoxin activity. Proc Natl Acad Sci U S A 111:6750–6755. doi: 10.1073/pnas.1400136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Garces A, Atmakuri K, Chase MR, Woodworth JS, Krastins B, Rothchild AC, Ramsdell TL, Lopez MF, Behar SM, Sarracino DA, Fortune SM. 2010. EspA acts as a critical mediator of ESX1-dependent virulence in Mycobacterium tuberculosis by affecting bacterial cell wall integrity. PLoS Pathog 6:e1000957. doi: 10.1371/journal.ppat.1000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen JM, Zhang M, Rybniker J, Basterra L, Dhar N, Tischler AD, Pojer F, Cole ST. 2013. Phenotypic profiling of Mycobacterium tuberculosis EspA point-mutants reveals blockage of ESAT-6 and CFP-10 secretion in vitro does not always correlate with attenuation of virulence. J Bacteriol 195:5421–5430. doi: 10.1128/JB.00967-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Williams EA, Mba Medie F, Bosserman RE, Johnson BK, Reyna C, Ferrell MJ, Champion MM, Abramovitch RB, Champion PA. 2017. A nonsense mutation in Mycobacterium marinum that is suppressible by a novel mechanism. Infect Immun 85:e00653-16. doi: 10.1128/IAI.00653-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Di Luca M, Bottai D, Batoni G, Orgeur M, Aulicino A, Counoupas C, Campa M, Brosch R, Esin S. 2012. The ESX-5 associated eccB-eccC locus is essential for Mycobacterium tuberculosis viability. PLoS One 7:e52059. doi: 10.1371/journal.pone.0052059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sassetti CM, Rubin EJ. 2003. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A 100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tischler AD, Leistikow RL, Kirksey MA, Voskuil MI, McKinney JD. 2013. Mycobacterium tuberculosis requires phosphate-responsive gene regulation to resist host immunity. Infect Immun 81:317–328. doi: 10.1128/IAI.01136-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Elliott SR, Tischler AD. 2016. Phosphate responsive regulation provides insights for ESX-5 function in Mycobacterium tuberculosis. Curr Genet 62:759–763. doi: 10.1007/s00294-016-0604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ramakrishnan P, Aagesen AM, McKinney JD, Tischler AD. 2015. Mycobacterium tuberculosis resists stress by regulating PE19 expression. Infect Immun 84:735–746. doi: 10.1128/IAI.00942-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Almeida Da Silva PE, Palomino JC. 2011. Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J Antimicrob Chemother 66:1417–1430. doi: 10.1093/jac/dkr173. [DOI] [PubMed] [Google Scholar]
- 143.Eilertson B, Maruri F, Blackman A, Guo Y, Herrera M, van der Heijden Y, Shyr Y, Sterling TR. 2016. A novel resistance mutation in eccC5 of the ESX-5 secretion system confers ofloxacin resistance in Mycobacterium tuberculosis. J Antimicrob Chemother 71:2419–2427. doi: 10.1093/jac/dkw168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.van Winden VJC, Ummels R, Piersma SR, Jimenez CR, Korotkov KV, Bitter W, Houben ENG. 2016. Mycosins are required for the stabilization of the ESX-1 and ESX-5 type VII secretion membrane complexes. mBio 7(5):e01471-16. doi: 10.1128/mBio.01471-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Beckham KS, Ciccarelli L, Bunduc CM, Mertens HD, Ummels R, Lugmayr W, Mayr J, Rettel M, Savitski MM, Svergun DI, Bitter W, Wilmanns M, Marlovits TC, Parret AH, Houben EN. 2017. Structure of the mycobacterial ESX-5 type VII secretion system membrane complex by single-particle analysis. Nat Microbiol 2:17047. doi: 10.1038/nmicrobiol.2017.47. [DOI] [PubMed] [Google Scholar]
- 146.Solomonson M, Huesgen PF, Wasney GA, Watanabe N, Gruninger RJ, Prehna G, Overall CM, Strynadka NC. 2013. Structure of the mycosin-1 protease from the mycobacterial ESX-1 protein type VII secretion system. J Biol Chem 288:17782–17790. doi: 10.1074/jbc.M113.462036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wagner JM, Evans TJ, Chen J, Zhu H, Houben EN, Bitter W, Korotkov KV. 2013. Understanding specificity of the mycosin proteases in ESX/type VII secretion by structural and functional analysis. J Struct Biol 184:115–128. doi: 10.1016/j.jsb.2013.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wagner JM, Chan S, Evans TJ, Kahng S, Kim J, Arbing MA, Eisenberg D, Korotkov KV. 2016. Structures of EccB1 and EccD1 from the core complex of the mycobacterial ESX-1 type VII secretion system. BMC Struct Biol 16:5. doi: 10.1186/s12900-016-0056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wagner JM, Evans TJ, Korotkov KV. 2014. Crystal structure of the N-terminal domain of EccA(1) ATPase from the ESX-1 secretion system of Mycobacterium tuberculosis. Proteins 82:159–163. doi: 10.1002/prot.24351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rosenberg OS, Dovala D, Li X, Connolly L, Bendebury A, Finer-Moore J, Holton J, Cheng Y, Stroud RM, Cox JS. 2015. Substrates control multimerization and activation of the multi-domain ATPase motor of type VII secretion. Cell 161:501–512. doi: 10.1016/j.cell.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ohol YM, Goetz DH, Chan K, Shiloh MU, Craik CS, Cox JS. 2010. Mycobacterium tuberculosis MycP1 protease plays a dual role in regulation of ESX-1 secretion and virulence. Cell Host Microbe 7:210–220. doi: 10.1016/j.chom.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen JM, Zhang M, Rybniker J, Boy-Rottger S, Dhar N, Pojer F, Cole ST. 2013. Mycobacterium tuberculosis EspB binds phospholipids and mediates EsxA-independent virulence. Mol Microbiol 89:1154–1166. doi: 10.1111/mmi.12336. [DOI] [PubMed] [Google Scholar]
- 153.Siewering K, Jain S, Friedrich C, Webber-Birungi MT, Semchonok DA, Binzen I, Wagner A, Huntley S, Kahnt J, Klingl A, Boekema EJ, Sogaard-Andersen L, van der Does C. 2014. Peptidoglycan-binding protein TsaP functions in surface assembly of type IV pili. Proc Natl Acad Sci U S A 111:E953–E961. doi: 10.1073/pnas.1322889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wehbi H, Portillo E, Harvey H, Shimkoff AE, Scheurwater EM, Howell PL, Burrows LL. 2011. The peptidoglycan-binding protein FimV promotes assembly of the Pseudomonas aeruginosa type IV pilus secretin. J Bacteriol 193:540–550. doi: 10.1128/JB.01048-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Das C, Ghosh TS, Mande SS. 2011. Computational analysis of the ESX-1 region of Mycobacterium tuberculosis: insights into the mechanism of type VII secretion system. PLoS One 6:e27980. doi: 10.1371/journal.pone.0027980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ekiert DC, Bhabha G, Isom GL, Greenan G, Ovchinnikov S, Henderson IR, Cox JS, Vale RD. 2017. Architectures of lipid transport systems for the bacterial outer membrane. Cell 169:273–285 e217. doi: 10.1016/j.cell.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lou Y, Rybniker J, Sala C, Cole ST. 2017. EspC forms a filamentous structure in the cell envelope of Mycobacterium tuberculosis and impacts ESX-1 secretion. Mol Microbiol 103:26–38. doi: 10.1111/mmi.13575. [DOI] [PubMed] [Google Scholar]
- 158.Okkels LM, Brock I, Follmann F, Agger EM, Arend SM, Ottenhoff TH, Oftung F, Rosenkrands I, Andersen P. 2003. PPE protein (Rv3873) from DNA segment RD1 of Mycobacterium tuberculosis: strong recognition of both specific T-cell epitopes and epitopes conserved within the PPE family. Infect Immun 71:6116–6123. doi: 10.1128/IAI.71.11.6116-6123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Demangel C, Brodin P, Cockle PJ, Brosch R, Majlessi L, Leclerc C, Cole ST. 2004. Cell envelope protein PPE68 contributes to Mycobacterium tuberculosis RD1 immunogenicity independently of a 10-kilodalton culture filtrate protein and ESAT-6. Infect Immun 72:2170–2176. doi: 10.1128/IAI.72.4.2170-2176.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.MacGurn JA, Raghavan S, Stanley SA, Cox JS. 2005. A non-RD1 gene cluster is required for Snm secretion in Mycobacterium tuberculosis. Mol Microbiol 57:1653–1663. doi: 10.1111/j.1365-2958.2005.04800.x. [DOI] [PubMed] [Google Scholar]
- 161.Ates LS, Brosch R. 2017. Discovery of the type VII ESX-1 secretion needle? Mol Microbiol 103:7–12. doi: 10.1111/mmi.13579. [DOI] [PubMed] [Google Scholar]
- 162.Fortune SM, Jaeger A, Sarracino DA, Chase MR, Sassetti CM, Sherman DR, Bloom BR, Rubin EJ. 2005. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc Natl Acad Sci U S A 102:10676–10681. doi: 10.1073/pnas.0504922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chen JM, Boy-Rottger S, Dhar N, Sweeney N, Buxton RS, Pojer F, Rosenkrands I, Cole ST. 2012. EspD is critical for the virulence-mediating ESX-1 secretion system in Mycobacterium tuberculosis. J Bacteriol 194:884–893. doi: 10.1128/JB.06417-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Champion MM, Williams EA, Kennedy GM, Champion PA. 2012. Direct detection of bacterial protein secretion using whole colony proteomics. Mol Cell Proteomics 11:596–604. doi: 10.1074/mcp.M112.017533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Champion MM, Williams EA, Pinapati RS, Champion PA. 2014. Correlation of phenotypic profiles using targeted proteomics identifies mycobacterial Esx-1 substrates. J Proteome Res 13:5151–5164. doi: 10.1021/pr500484w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kennedy GM, Hooley GC, Champion MM, Mba Medie F, Champion PA. 2014. A novel ESX-1 locus reveals that surface-associated ESX-1 substrates mediate virulence in Mycobacterium marinum. J Bacteriol 196:1877–1888. doi: 10.1128/JB.01502-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kinhikar AG, Verma I, Chandra D, Singh KK, Weldingh K, Andersen P, Hsu T, Jacobs WR Jr, Laal S. 2010. Potential role for ESAT6 in dissemination of M. tuberculosis via human lung epithelial cells. Mol Microbiol 75:92–106. doi: 10.1111/j.1365-2958.2009.06959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Prados-Rosales R, Carreno LJ, Weinrick B, Batista-Gonzalez A, Glatman-Freedman A, Xu J, Chan J, Jacobs WR Jr, Porcelli SA, Casadevall A. 2016. The type of growth medium affects the presence of a mycobacterial capsule and is associated with differences in protective efficacy of BCG vaccination against Mycobacterium tuberculosis. J Infect Dis 214:426–437. doi: 10.1093/infdis/jiw153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Champion PA, Champion MM, Manzanillo P, Cox JS. 2009. ESX-1 secreted virulence factors are recognized by multiple cytosolic AAA ATPases in pathogenic mycobacteria. Mol Microbiol 73:950–962. doi: 10.1111/j.1365-2958.2009.06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Converse SE, Cox JS. 2005. A protein secretion pathway critical for Mycobacterium tuberculosis virulence is conserved and functional in Mycobacterium smegmatis. J Bacteriol 187:1238–1245. doi: 10.1128/JB.187.4.1238-1245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Daleke MH, Ummels R, Bawono P, Heringa J, Vandenbroucke-Grauls CM, Luirink J, Bitter W. 2012. General secretion signal for the mycobacterial type VII secretion pathway. Proc Natl Acad Sci U S A 109:11342–11347. doi: 10.1073/pnas.1119453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Andersen AB, Andersen P, Ljungqvist L. 1992. Structure and function of a 40,000-molecular-weight protein antigen of Mycobacterium tuberculosis. Infect Immun 60:2317–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. 2012. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe 11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Stanley SA, Johndrow JE, Manzanillo P, Cox JS. 2007. The type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J Immunol 178:3143–3152. doi: 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- 175.Watson RO, Manzanillo PS, Cox JS. 2012. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Berthet FX, Rasmussen PB, Rosenkrands I, Andersen P, Gicquel B. 1998. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10). Microbiology 144:3195–3203. [DOI] [PubMed] [Google Scholar]
- 177.de Jonge MI, Pehau-Arnaudet G, Fretz MM, Romain F, Bottai D, Brodin P, Honore N, Marchal G, Jiskoot W, England P, Cole ST, Brosch R. 2007. ESAT-6 from Mycobacterium tuberculosis dissociates from its putative chaperone CFP-10 under acidic conditions and exhibits membrane-lysing activity. J Bacteriol 189:6028–6034. doi: 10.1128/JB.00469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.De Leon J, Jiang G, Ma Y, Rubin E, Fortune S, Sun J. 2012. Mycobacterium tuberculosis ESAT-6 exhibits a unique membrane-interacting activity that is not found in its ortholog from non-pathogenic Mycobacterium smegmatis. J Biol Chem 287:44184–44191. doi: 10.1074/jbc.M112.420869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Peng X, Jiang G, Liu W, Zhang Q, Qian W, Sun J. 2016. Characterization of differential pore-forming activities of ESAT-6 proteins from Mycobacterium tuberculosis and Mycobacterium smegmatis. FEBS Lett 590:509–519. doi: 10.1002/1873-3468.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ma Y, Keil V, Sun J. 2015. Characterization of Mycobacterium tuberculosis EsxA membrane insertion: roles of N- and C-terminal flexible arms and central helix-turn-helix motif. J Biol Chem 290:7314–7322. doi: 10.1074/jbc.M114.622076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Peng X, Sun J. 2016. Mechanism of ESAT-6 membrane interaction and its roles in pathogenesis of Mycobacterium tuberculosis. Toxicon 116:29–34. doi: 10.1016/j.toxicon.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Conrad WH, Osman MM, Shanahan JK, Chu F, Takaki KK, Cameron J, Hopkinson-Woolley D, Brosch R, Ramakrishnan L. 2017. Mycobacterial ESX-1 secretion system mediates host cell lysis through bacterium contact-dependent gross membrane disruptions. Proc Natl Acad Sci U S A 114:1371–1376. doi: 10.1073/pnas.1620133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.College of Veterinary Medicine and Biomedical Sciences. 2016. Production of recombinant ESat-6 under non-denaturing conditions SOP. College of Veterinary Medicine and Biomedical Sciences, Colorado State University, Fort Collins, CO: http://csu-cvmbs.colostate.edu/Documents/dobos-rp004.pdf. [Google Scholar]
- 184.King CH, Mundayoor S, Crawford JT, Shinnick TM. 1993. Expression of contact-dependent cytolytic activity by Mycobacterium tuberculosis and isolation of the genomic locus that encodes the activity. Infect Immun 61:2708–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Champion PA. 2013. Disconnecting in vitro ESX-1 secretion from mycobacterial virulence. J Bacteriol 195:5418–5420. doi: 10.1128/JB.01145-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Azad AK, Sirakova TD, Fernandes ND, Kolattukudy PE. 1997. Gene knockout reveals a novel gene cluster for the synthesis of a class of cell wall lipids unique to pathogenic mycobacteria. J Biol Chem 272:16741–16745. doi: 10.1074/jbc.272.27.16741. [DOI] [PubMed] [Google Scholar]
- 187.MacGurn JA, Cox JS. 2007. A genetic screen for Mycobacterium tuberculosis mutants defective for phagosome maturation arrest identifies components of the ESX-1 secretion system. Infect Immun 75:2668–2678. doi: 10.1128/IAI.01872-06. [DOI] [PMC free article] [PubMed] [Google Scholar]