ABSTRACT
The function and extracellular location of cell envelope proteins make them attractive candidates for developing vaccines against bacterial diseases, including challenging drug-resistant pathogens, such as Neisseria gonorrhoeae. A proteomics-driven reverse vaccinology approach has delivered multiple gonorrhea vaccine candidates; however, the biological functions of many of them remain to be elucidated. Herein, the functions of six gonorrhea vaccine candidates—NGO2121, NGO1985, NGO2054, NGO2111, NGO1205, and NGO1344—in cell envelope homeostasis were probed using phenotype microarrays under 1,056 conditions and a ΔbamE mutant (Δngo1780) as a reference of perturbed outer membrane integrity. Optimal growth conditions for an N. gonorrhoeae phenotype microarray assay in defined liquid medium were developed, which can be useful in other applications, including rapid and thorough antimicrobial susceptibility assessment. Our studies revealed 91 conditions having uniquely positive or negative effects on one of the examined mutants. A cluster analysis of 37 and 57 commonly beneficial and detrimental compounds, respectively, revealed three separate phenotype groups: NGO2121 and NGO1985; NGO1344 and BamE; and the trio of NGO1205, NGO2111, and NGO2054, with the last protein forming an independent branch of this cluster. Similar phenotypes were associated with loss of these vaccine candidates in the highly antibiotic-resistant WHO X strain. Based on their extensive sensitivity phenomes, NGO1985 and NGO2121 appear to be the most promising vaccine candidates. This study establishes the principle that phenotype microarrays can be successfully applied to a fastidious bacterial organism, such as N. gonorrhoeae.
IMPORTANCE Innovative approaches are required to develop vaccines against prevalent and neglected sexually transmitted infections, such as gonorrhea. Herein, we have utilized phenotype microarrays in the first such investigation into Neisseria gonorrhoeae to probe the function of proteome-derived vaccine candidates in cell envelope homeostasis. Information gained from this screening can feed the vaccine candidate decision tree by providing insights into the roles these proteins play in membrane permeability, integrity, and overall N. gonorrhoeae physiology. The optimized screening protocol can be applied in investigations into the function of other hypothetical proteins of N. gonorrhoeae discovered in the expanding number of whole-genome sequences, in addition to revealing phenotypic differences between clinical and laboratory strains.
KEYWORDS: Neisseria gonorrhoeae, vaccine antigens, cell envelope, phenotypic microarrays, antibiotics, Biolog
INTRODUCTION
In Gram-negative bacteria, the cell envelope is composed of the outer membrane, the cell wall, and the cytoplasmic or inner membrane, each of which can be further broken down into its constituents (1). The outer membrane is a bilayer composed of a lipopolysaccharide (LPS) or lipooligosaccharide (LOS) outer leaflet facing the extracellular milieu and an inner leaflet of phospholipids. Housed in the periplasm between the outer and inner membranes, repeating units of N-acetylglucosamine-N-acetylmuramic acid disaccharides cross-linked by pentapeptide side chains make up the peptidoglycan cell wall (2), which provides a rigid structure and allows cells to maintain their shape (1). The inner membrane is a phospholipid bilayer containing proteins involved in energy production, secretion, lipid synthesis, and transport (1).
Proteins in the cell envelope play integral roles in its function and in bacterial physiology. Most outer membrane proteins are either β-barrel proteins or lipoproteins (1), with lipoproteins secured to the membrane by lipid anchors covalently attached to an invariant N-terminal cysteine (3). β-Barrel proteins frequently function as porins or as transporters or gated channels for the transport of ions or large substrates (1). Little is known about the functions of most lipoproteins (1), but those for which information is available have been shown to be involved in nutrient acquisition, cell wall structure and metabolism, cell signaling, antibiotic resistance, attachment to host cells, and ligands for Toll-like receptor 2, which triggers an innate immune response and helps establish adaptive immunity (4). Because of this potential interaction between host and pathogen, we are currently pursuing 20 novel outer membrane-localized proteins, including lipoproteins, as vaccine candidates against Neisseria gonorrhoeae, the gonococcus (GC) (5, 6).
The sexually transmitted infection gonorrhea is caused by N. gonorrhoeae, a highly adaptable pathogen that has acquired resistance to nearly every antibiotic currently available. Treatment failures with the last viable class of antibiotics, third-generation cephalosporins, have been encountered in several countries (7, 8). The development of a protective vaccine(s) is of paramount importance (9–13). An exciting development in vaccine research has been the recent success of an experimental vaccine composed of membrane vesicles derived from N. gonorrhoeae FA1090 (14) coadministered with interleukin-12. Mice immunized with this treatment mount a type 1 T helper cell-driven immune response that accelerates the clearance of gonococcal infections and defends against subsequent infection for at least 6 months after immunization. Critically, vaccination with FA1090-derived membrane vesicles protects from challenges with the commonly used laboratory strains MS11 and FA19, as well as recent clinical isolates GC68 and GC69 (15). Using proteomics-based reverse vaccinology (16), we have investigated the composition of N. gonorrhoeae cell envelopes and naturally released membrane vesicles across common laboratory strains, including FA1090, MS11, and FA19 (6), as well as FA1090 cell envelopes under four different host-relevant growth conditions (5) to identify vaccine candidate targets. Among the ubiquitously expressed proteins in four examined isolates were novel vaccine candidates NGO2121, NGO1985, NGO2054, NGO2111, NGO1205, and NGO1344 (5, 6), with predicted functions associated with cell envelope homeostasis. Herein, we utilized Biolog Phenotype MicroArrays (PMs) to identify the phenotypes of gene knockout mutants in our six vaccine candidates. Elucidating the physiological roles of these proteins is important to our vaccine candidate decision tree, as antibodies against protein antigens can disable their function (17).
Assessing the functions of hypothetical proteins is difficult due to the lack of available information on the protein, and a prohibitive number of treatments may need to be examined in a mutant organism before a phenotype is associated with the loss of that protein. To increase the number of conditions available for testing, bacterial PMs have been designed to comprehensively screen bacterial strains for unique phenotypes (18). These arrays have been used to assess the fitness cost of antimicrobial resistance (19), to determine whether the increased pathogenicity of sequence type 131 extraintestinal pathogenic Escherichia coli is due to an altered metabolic profile (20), to examine the biological role of two-component systems in E. coli (21), and to identify the physiological functions of genes (22, 23). Here, we have optimized growth conditions (culture medium, inoculum size, and incubation time) for N. gonorrhoeae and used them for the first PM analysis of N. gonorrhoeae to assess the physiological functions of vaccine candidate proteins NGO1205, NGO2054, NGO2121, NGO1985, NGO2054, NGO2111, NGO1205, and NGO1344, in addition to our marker of compromised membrane integrity, NGO1780 (gonococcal homolog of BamE [BamEGC]).
RESULTS AND DISCUSSION
Selected vaccine candidate proteins.
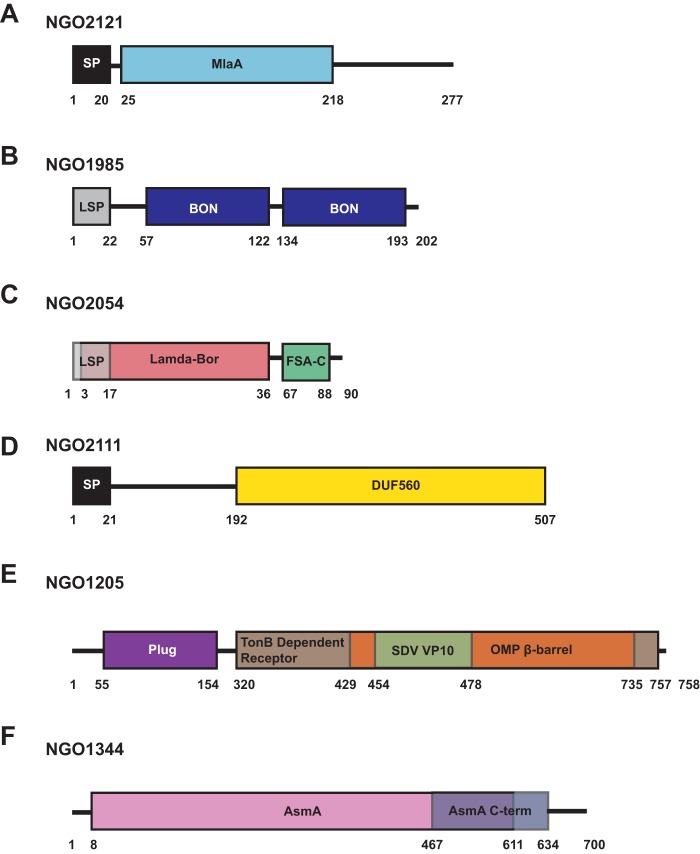
The open reading frames (ORFs) of our proteome-derived vaccine candidates chosen for further investigation, NGO2121, NGO1985, NGO2054, NGO2111, NGO1205 and NGO1344, encode hypothetical proteins. Homology, database, and literature searches were performed to gain information about their possible biological functions. All proteins contain predicted domains involved in cell envelope homeostasis (Fig. 1). NGO2121 is annotated as a predicted lipoprotein, maintenance of lipid asymmetry (MlaA; also known as VacJ [virulence-associated, chromosome locus J]); however, in N. gonorrhoeae, it does not contain an invariant cysteine residue, a hallmark feature of all lipoproteins (4). Residues 25 to 218 of NGO2121 in FA1090 form a conserved ABC transporter Mla domain (Fig. 1A). MlaA is involved in maintaining lipid asymmetry in the E. coli outer membrane (24), contributes to outer membrane vesicle formation (25), and is essential for Shigella flexneri to spread to adjacent cells during infection (26, 27).
FIG 1.
Protein domain schematics. ORFs were examined for the presence of putative domains using the Kyoto Encyclopedia of Genes and Genomes (KEGG) and for the presence of signal peptides using the SignalP 4.1 and LipoP 1.0 servers. (A) A predicted MlaA domain is present in NGO2121, in addition to a signal peptide. (B) NGO1985 is composed of two bacterial OsmY and nodulation (BON) domains and a lipoprotein signal peptide. (C) NGO2054 contains a Lambda-Bor domain that overlaps with a lipoprotein signal peptide, as well as a fragile site-associated protein C terminus (FSA-C). (D) NGO2111 is composed of a DUF560 domain and contains an N-terminal signal peptide. (E) NGO1205 comprises a plug domain, a Ton-B dependent receptor domain, an outer membrane protein (OMP) β-barrel domain, and a Seadornavirus (SDV) VP10 domain. (F) NGO1344 includes an AsmA domain and an AsmA C-terminal domain. Schematics are not to scale. SP, signal peptide; LSP, lipoprotein signal peptide; Mla, maintenance of lipid asymmetry; DUF, domain of unknown function.
Examination of NGO1985 showed that it is a predicted lipoprotein containing two bacterial OsmY and nodulation domains, BON domains, which are hypothesized to bind phospholipids (28) (Fig. 1B).
Among the selected candidates is NGO2054, a predicted lipoprotein containing a Lambda-Bor motif involved in resistance to serum in E. coli (29). Additionally, this protein contains a segment with homology to a fragile site-associated protein C terminus (FSA-C) involved in adipocyte differentiation in mammalian cells (30) (Fig. 1C). We have recently determined that NGO2054 is surface exposed, induces bactericidal antibodies, and is continuously expressed at the same level throughout all stages of gonococcal growth and under tested host-relevant conditions (5).
NGO2111 is annotated as a hypothetical protein containing a DUF560 domain of unknown function (Fig. 1D). In our proteomic profiling, we found that this protein was upregulated during growth under iron-limited conditions (5). The meningococcal homolog, NMB1971, has been described as a paralog of the surface lipoprotein assembly modulator Slam and was designated Slam2. In Neisseria meningitidis, this protein is responsible for specifically translocating the hemoglobin-haptoglobin utilization protein HpuA to the cell surface, and it has no effect on the localization of Slam1 substrate transferrin binding protein (TbpB), lactoferrin binding protein (LbpB), or factor H binding protein (fHbp). The presence of only the DUF560 domain was shown to be sufficient for translocation of Slam1 surface lipoprotein substrates, but the same experiment was not performed with the Slam2 DUF560 domain (31). It is likely that lipoprotein translocation is affected in the Δngo2111 mutant background.
Our searches showed that NGO1205, also known as TdfJ, is a member of the ferric uptake regulator regulon (32, 33) and has homology to the meningococcal zinc uptake protein ZnuD (34). Supporting this role, NGO1205 includes a predicted TonB-dependent receptor domain, as well as a plug domain. An outer membrane protein β-barrel domain within the TonB-dependent receptor domain further suggests that this protein is a receptor channel. NGO1205 also comprises a domain with homology to the Seadornavirus structural protein VP10 of unknown function (Fig. 1E). Structural studies of N. meningitidis ZnuD indicated that zinc binding induces multiple conformational changes of surface loops, potentially allowing this protein to avoid detection by the immune system despite high sequence conservation among meningococcal strains (35). Further, a meningococcal ZnuD mutant is less virulent in a mouse model of systemic infection (35) and is more sensitive to killing by neutrophil extracellular traps (36), which contain calprotectin to impose nutritional immunity by sequestering zinc ions (37). In N. gonorrhoeae, NGO1205 was not required for survival within cervical epithelial cells (38) but was upregulated in biofilms, where anaerobic respiration is primarily employed for survival (39). Corroborating these findings, high-throughput proteomics of cell envelopes showed increased levels of NGO1205 during the growth of N. gonorrhoeae under anaerobic conditions (5).
Finally, NGO1344 has homology to the assembly suppressor mutation protein AsmA of E. coli (40), where removal of this protein resulted in reduced LPS biosynthesis (41). However, when it was knocked out in N. meningitidis, no effect on LPS was observed (42). Examination of NGO1344 indicated the presence of an AsmA domain from residues 8 to 611, as well as an AsmA C-terminal domain from amino acids 467 to 634 (Fig. 1F).
Rationale of experimental design.
To assess the functions of our six vaccine candidate proteins in the N. gonorrhoeae cell envelope, we chose to use the Phenotype MicroArray (PM) technology developed by Biolog (22). PMs are 96-well microtiter plates formulated with various concentrations of compounds selected to evaluate a wide range of phenotypes, ranging from nutrient requirements and antibiotic sensitivities to responses to osmolytes and chemicals (18). For this study, we have selected PM9, which contains various osmolytes, and PM11 to PM20, which are formulated with a series of compounds to probe cellular membrane integrity, including metals, antimicrobial peptides, small hydrophobic molecules, and dyes. We did not employ the PMs designed to test nutrient utilization, because N. gonorrhoeae is a fastidious organism that can catabolize only glucose, pyruvate, and lactate (43).
We have chosen N. gonorrhoeae strain FA1090 as our type strain for vaccine candidate research. This strain was isolated in 1983 from a woman with disseminated gonococcal infection (14). Its infection characteristics have been extensively characterized, both in human male volunteer trials (44) and in the murine model for female infection (45–48). FA1090 is naturally streptomycin resistant (46), which allows for selection from normal human or murine flora without additional genetic manipulation. Our criteria for suitable vaccine candidates include that antigens are universally present and do not display phase or antigenic variation. The FA1090 genome is one of the smallest gonococcal genomes, with only 2,157 annotated coding sequences (RefSeq accession no. NC_002946.2). We hypothesize that proteins encoded in this compact genome are more likely to be conserved across multiple strains and are important for bacterial physiology. Therefore, we predict that vaccine candidates selected from the FA1090 proteome will provide protection against heterologous strains. In support of our strategy, mice treated with interleukin-12 and immunized with membrane vesicles derived from FA1090 that contain proteins investigated in the current study (6) are protected against infection by both laboratory strains and clinical isolates (15).
Screening was performed in WT FA1090 and isogenic strains deficient in NGO2121, NGO1985, NGO2054, NGO2111, NGO1205, and NGO1344. To have a reference of phenotypes associated with compromised cell envelope integrity, we included a mutant deficient in NGO1780, the gonococcal homolog of BamE (BamEGC). BamE is a member of the β-barrel assembly machinery (BAM) complex together with BamA to BamD. Neisseria spp., in contrast to E. coli, do not possess BamB (49). In E. coli, BamE is thought to influence the structure of the integral member of the complex, BamA, through its interaction with BamD (50). The ΔbamEGC knockout mutant in N. gonorrhoeae FA1090 was obtained easily but displayed characteristics of perturbed outer membrane permeability, including increased sensitivity to antibiotics and detergents, alterations in cell envelope composition, and increased vesiculation (A. E. Sikora, I. H. Wierzbicki, N. Noinaj, R. A. Zielke, R. F. Ryner, and K. V. Korotkov, unpublished data).
For the screening, we used Graver-Wade (GW) liquid medium (51), as it is clear, colorless, and free of protein, unlike gonococcal base liquid (GCBL) medium. Additionally, it has been successfully utilized to establish a time course of bacterial killing with antibiotics of different mechanisms of action (52).
Protein profiling.
Our initial investigations into the effects of knockout mutations of NGO1985, NGO2121, NGO2111, or NGO1344 when probed with bile salts, polymyxin B, Tween 20, sodium dodecyl sulfate, urea, and chloramphenicol indicated that the cell envelope was disrupted to various extents, depending on the missing vaccine candidate. An N. gonorrhoeae FA1090 mutant lacking NGO1985 was the most severely affected, exhibiting increased sensitivity to all of the compounds tested, while the Δngo2121 mutant was sensitive to bile salts and polymyxin B. In contrast, gonococci lacking NGO1344 or NGO2111 were not affected by any of the compounds tested (6).
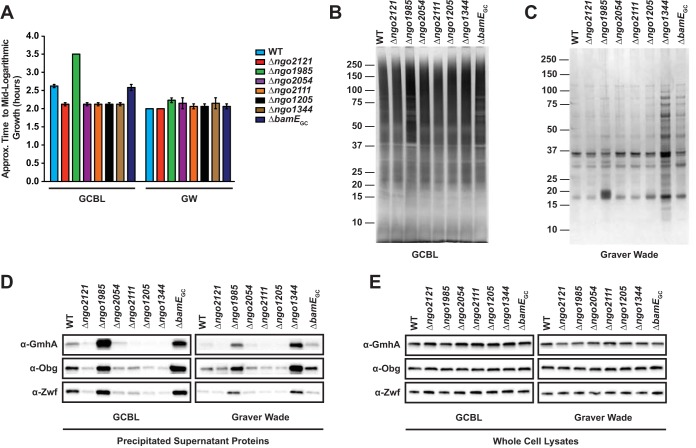
First, we aimed to examine the possible differences between the phenotypes of gonococci cultured in GCBL and GW media by monitoring bacterial growth and assessing the protein profiles of supernatants as well as whole-cell lysates. A comparison of the time the strains required to reach mid-logarithmic growth (optical density at 600 nm [OD600], 0.6 to 0.8) indicated that the overall growth rates in GW medium were more consistent than those in GCBL medium. While the eight strains required 2.0 to 2.3 h to reach mid-log phase in GW medium, the same stage of growth was achieved within 2.08 to 3.5 h in GCBL (Fig. 2A).
FIG 2.
Growth comparison and protein profiling of Neisseria gonorrhoeae cultured in GCBL and GW media. (A) FA1090 (WT) and all mutant strains were cultured in GCBL or GW liquid medium until mid-logarithmic growth (OD600, 0.6 to 0.8), and the time required to achieve the appropriate turbidity was recorded. n = 2; bars represent mean ± the standard error of the mean (SEM). Culture supernatants were precipitated with a pyrogallol red-molybdate-methanol procedure and concentrated 15-fold. (B and C) Equivalent material, based on the OD600 of the original culture, was separated with a 4 to 12% Bis-Tris NuPAGE gel and silver stained. Migration of molecular weight markers and their masses (in kilodaltons) are indicated on the left. Supernatants from cultures grown in GCBL (B) or GW (C) medium are presented. (D and E) Immunoblotting analysis of supernatants (D) or whole-cell lysates (E) from cultures grown in GCBL or GW medium, as indicated, were normalized based on OD600 values of the original cultures and resolved on either Bio-Rad Any kD or 4 to 20% TGX precast protein gels. Proteins were transferred and probed with polyclonal rabbit antiserum, as indicated. WT and mutant strains are indicated at the top of the immunoblot. Supernatant collection was performed on two separate occasions, and representative gels and immunoblots are presented. GCBL, gonococcal base liquid medium; GW, Graver-Wade liquid medium; OD600, optical density at 600 nm.
Supernatant samples were obtained from all strains grown in both GCBL and GW media to mid-logarithmic phase, and the proteins were precipitated with a modified pyrogallol red-molybdate-methanol procedure, as described previously (53). Visualization of their protein contents by silver staining (Fig. 2B and C) revealed dramatic differences. While GW medium is protein free, GCBL medium is predominantly proteolytically digested peptone, resulting in high background staining and protein bands that were less distinct than those seen in GW medium, making it difficult to assess differences in protein abundance and in the overall number of protein species (Fig. 2B). Silver staining of supernatants derived from the WT and the mutants cultured in GW medium revealed that the Δngo1344 mutant released the most protein species into the extracellular milieu, followed by the ΔbamEGC and Δngo1985 mutants (Fig. 2C). To assess whether this elevated protein content was due to cell lysis, the supernatants were also probed with antibodies against cytoplasmic proteins GmhA, Obg, and Zwf (54–57). During growth in GCBL medium, the Δngo1985 and ΔbamEGC mutants had elevated amounts of all cytoplasmic protein markers compared to the WT (Fig. 2D, left), indicating increased cell lysis. However, when the mutants were grown in GW medium (Fig. 2D, right), the release of cytoplasmic proteins appeared to be greatly decreased, suggesting that a component(s) of the GW medium partially compensated the phenotype. In contrast, the Δngo1344 mutant did not exhibit enhanced cell lysis during growth in GCBL medium. However, when cultured in GW medium, there was increased cell lysis, based on the elevated levels of GmhA, Obg, and Zwf. These striking differences were not observed when the abundance of the proteins was compared to the whole-cell lysates (Fig. 2E), which suggests that the increase in protein abundance observed in the culture supernatants was not due to alterations in expression.
Our protein profiling indicated that the culture medium played an important role in the growth characteristics of the mutants, which should be considered when investigating protein biological function. It also further validated our choice of GW medium for the screening, as the growth rates for all mutants were highly consistent and reproducible.
Screening optimization.
Initially, our screening workflow was based on the time-kill experiments performed by Foerster et al. (52), who suspended N. gonorrhoeae in phosphate-buffered saline to a turbidity equivalent to a 0.5 McFarland standard and then inoculated 15 ml of GW medium with 30 μl of suspension. The inoculum was dispensed into microtiter plates and grown for 4 h, at which point antibiotics were added, and cultures were grown up to 6 h longer (52). However, we were unable to achieve consistent, reliable growth with this method using N. gonorrhoeae strain FA1090, likely due to the various degrees of cell lysis observed between different gonococci (58–62) as well as of our knockout strains. Additionally, there were no means of ensuring that bacteria were actively dividing before dispensing the culture into the screening plates. In a subsequent method optimization attempt, N. gonorrhoeae strains were collected from solid medium, suspended in GW medium to an OD600 of 0.1, cultured for 3 h, back diluted to an OD600 of 0.1, and then dispensed into each well of the screening plate, which was subsequently cultured for 6 h. This approach resulted in consistent, predictable growth and provided a way to ensure growth before screening (as the turbidity of the cultures at OD600 was monitored before dispensing into plates), but the bacteria reached stationary phase between 4 and 6 h. As a result, some of the gonococci likely underwent autolysis, yielding incomparable growth curves. Finally, minimized autolysis was achieved by diluting the culture 10-fold more, to an OD600 of 0.01, before dispensing it into the screening plates. With our optimized protocol, gonococci were actively dividing before initiating the screening and showed consistent, reproducible growth kinetics without reaching stationary phase before the end of the experiment. Typically, a tetrazolium dye is added to the suspension used to inoculate the PM plates as a measure of bacterial viability (18); however, we found that this dye prevented gonococcal growth in GW medium, so it was omitted.
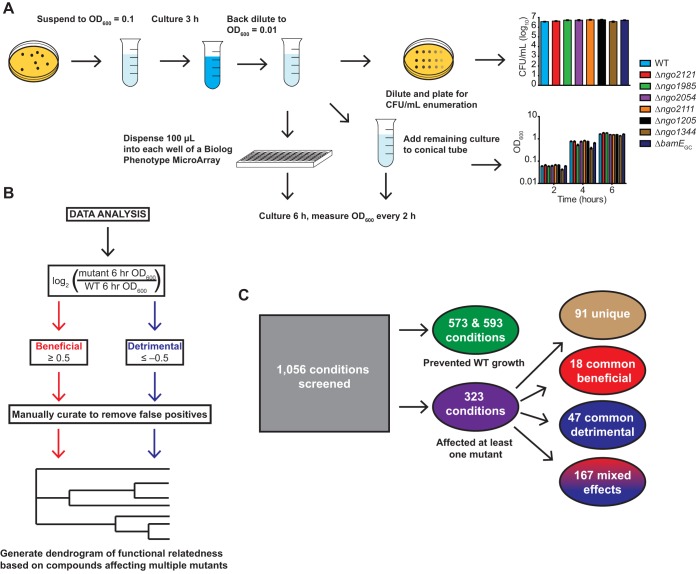
Our final experimental workflow is outlined in Fig. 3A. Bacteria were suspended to an OD600 of 0.1 in prewarmed GW medium and cultured in a 5% CO2 atmosphere at 37°C with shaking for 3 h. Cultures were then diluted to an OD600 of 0.01. We enumerated the CFU to quantify the starting material for each strain. Between 3.8 × 106 and 5.8 × 106 CFU/ml (Fig. 3A) were dispensed into individual wells of the PM plate, and the remaining suspension was cultured separately in conical tubes to monitor growth without compounds. Cultures inoculated into 96-well plates and tubes were both treated for 6 h in the same manner as described above, and the turbidity of the cultures maintained on the PM plates was measured every 2 h. The raw data from all plates at all time points can be found in File S1 in the supplemental material, and the growth monitoring data are shown in Fig. 3A. Each compound at each concentration tested was designated one condition. Two sets of screening experiments were performed. In the first, the WT, Δngo2121 mutant, and Δngo1985 mutant were tested, and the WT and Δngo2054, Δngo2111, Δngo1205, Δngo1344, and ΔbamEGC mutants were examined in the second set.
FIG 3.
Phenotype screening procedure. (A) Flowchart outlining the optimized phenotype screening procedure. Nonpiliated colonies of FA1090 (WT) and mutant bacteria were collected from GCB plates, suspended in GW medium to an OD600 of 0.1, cultured for 3 h in a 5% CO2 environment at 37°C, and back diluted to an OD600 of 0.01. Cultures were split for CFU enumeration, overall growth monitoring, and phenotype screening. Inoculum for screening plates was serially diluted 10-fold from −1 to −4, and 5 μl was spotted on GCB plates for enumeration. OD600 values of screening plates and cultures without compounds were measured every 2 h for each strain. Bars represent means ± SEM of cultures without compounds. WT, n = 17; Δngo2121 and Δngo1985 mutants, n = 6; Δngo2054, Δngo2111, Δngo1205, Δngo1344, and ΔbamEGC mutants, n = 11. (B) Flowchart outlining data analysis. Endpoint OD600 values were used to calculate a log2-transformed ratio of mutant to WT growth. Beneficial conditions were designated those that resulted in a ratio of ≥0.5. Detrimental conditions were those with a ratio of −0.5 or lower. Data were manually curated to remove false positives. Hits were used to construct a functional relatedness dendrogram of vaccine candidates. (C) Out of 1,056 conditions screened, 573 and 593 conditions prevented growth of WT in the first and second screenings, respectively. Three hundred twenty-three conditions had some effect on at least one mutant. Ninety-one conditions uniquely affected one mutant. Of the conditions with effects on multiple mutants, 18 conditions were commonly beneficial, 47 were commonly detrimental, and 167 had mixed effects. GCB, gonococcal base medium; GW, Graver-Wade liquid medium; OD600, optical density at 600 nm.
Phenomes of N. gonorrhoeae FA1090 and isogenic vaccine candidate mutants.
Overall, we screened 1,056 conditions for each of eight N. gonorrhoeae strains: the WT and the Δngo2121, Δngo1985, Δngo2054, Δngo2111, Δngo1205, Δngo1344, and ΔbamEGC mutants. For each condition, mutant growth was compared to that of the WT by calculating the log2 ratio of the mutant to WT OD600 value at the experimental endpoint. If the log2 ratio was 0.5 or above, the condition was considered beneficial to the mutant, while a ratio of −0.5 or below was considered detrimental. Next, the hits were manually curated to remove any false positives where (i) the mutant and WT OD600 values decreased over the course of the experiment, (ii) the mutant OD600 decreased or the increase was not consistent for a beneficial hit, or (iii) the WT OD600 decreased or the increase was not consistent for a detrimental hit. These criteria were applied to each condition independently for each mutant. Mutant-to-WT ratios were calculated based on the WT data collected in the respective experiment (Fig. 3B).
In the first and second sets of experiments, 573 and 593 conditions, respectively, prevented growth of the wild type, determined by counting the conditions under which the OD600 of the well increased by less than 0.01 over the course of the experiment (Fig. 3C and File S1). Three hundred twenty-three conditions influenced at least one of the tested mutants, with 60 conditions being beneficial to only one of the seven mutants screened and 31 conditions that were uniquely detrimental (Fig. 3C). No compound was universally beneficial or detrimental. The structures or molecular formulas, with molecular weights and mechanism of action, of the compounds that affected at least one of the mutants are provided in Fig. S1. In both the previous study (6) and the current one, the Δngo1985 mutant exhibited the most striking phenotypes with three and 46 detrimental and beneficial conditions, respectively (Fig. 4). As might be expected from our preliminary phenotypic screen (6), the Δngo2121 mutant was the second most affected mutant, with six unique beneficial conditions and 15 detrimental conditions (Fig. 5). None of the other mutants tested was affected by as many unique conditions as the Δngo2121 or Δngo1985 mutant. Both the Δngo2054 and Δngo1205 mutants were benefitted by two conditions (red and orange bars, Fig. 6), while two and one conditions were detrimental to the growth of these mutants, respectively (blue and light blue bars, Fig. 6). One condition enhanced the growth of the Δngo1344 mutant, while four conditions attenuated the mutant's growth (bright pink and dark blue bars, Fig. 6). Finally, three conditions were beneficial to the ΔbamEGC mutant, and six conditions were detrimental (light pink and teal bars, Fig. 6). Notably, no conditions had a unique effect on the Δngo2111 mutant.
FIG 4.
Phenome of Δngo1985 mutant. Forty-nine conditions affected the Δngo1985 mutant without having an effect on any of the other mutants tested. Forty-six conditions resulted in a log2 ratio of ≥0.5 for the Δngo1985 mutant (red bars, beneficial). The ratio was −0.5 or lower for 3 conditions (blue bars, detrimental). mut, mutant.
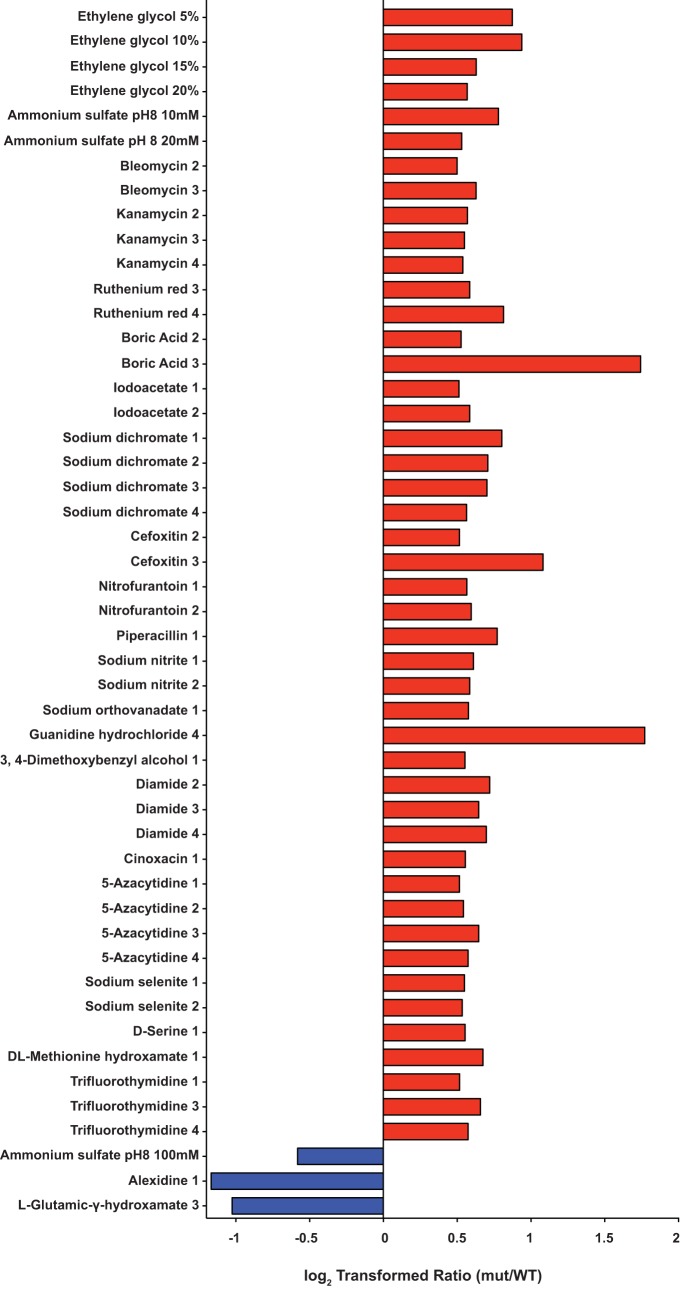
FIG 5.
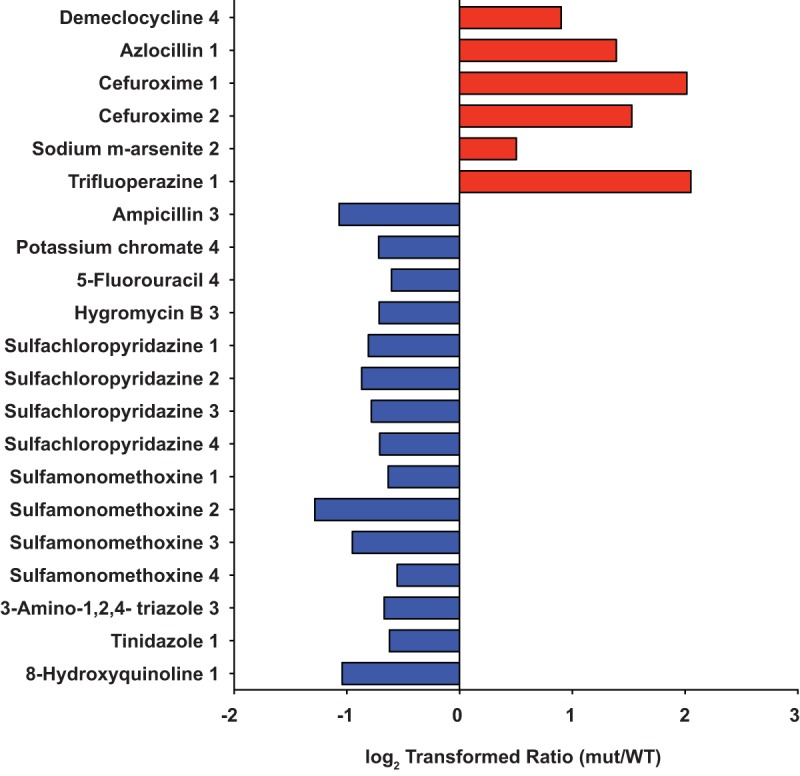
Phenome of Δngo2121 mutant. Twenty-one conditions affected the Δngo2121 mutant without having an effect on any of the other mutants tested. Six conditions resulted in a log2 ratio of ≥0.5 for the Δngo2121 mutant (red bars, beneficial). The ratio was −0.5 or lower for 15 conditions (blue bars, detrimental). mut, mutant.
FIG 6.
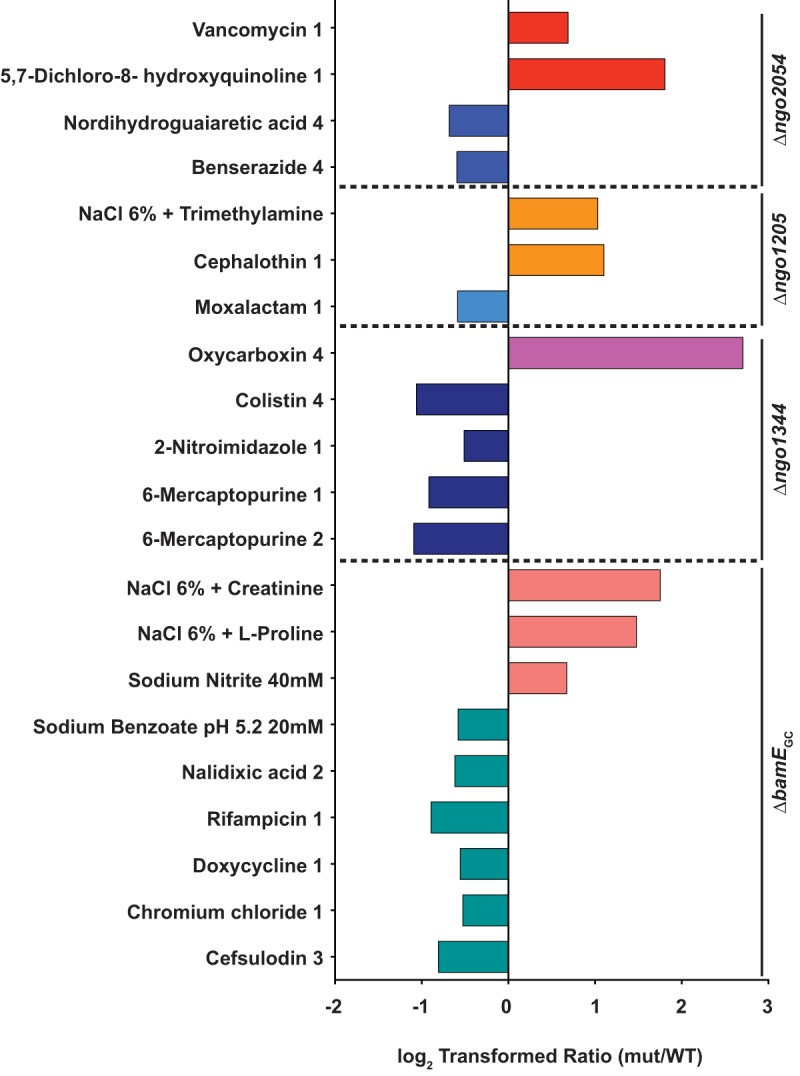
Conditions with unique effects on Δngo2054, Δngo1205, Δngo1344, and ΔbamEGC mutants. The Δngo2054 mutant was uniquely affected by two conditions with a log2 ratio of ≥0.5 (red bars, beneficial) and two conditions with a ratio of −0.5 or lower (blue bars, detrimental). The Δngo1205 mutant was uniquely affected by two conditions with a log2 ratio of ≥0.5 (orange bars, beneficial) and one condition with a ratio of −0.5 or lower (light blue bar, detrimental). The Δngo1344 mutant was uniquely affected by one condition with a ratio of ≥0.5 (bright pink bar, beneficial) and four conditions with a ratio of −0.5 or lower (dark blue bars, detrimental). The ΔbamEGC mutant was uniquely affected by three conditions with a ratio ≥ 0.5 (light pink bars, beneficial) and six conditions with a ratio of −0.5 or lower (teal bars, detrimental). mut, mutant.
Phenome of Δngo1985 mutant.
Our experiments revealed 46 unique conditions under which the Δngo1985 mutant grew better than the WT and three unique conditions where mutant growth was attenuated compared to the WT (Fig. 4). Among the unique beneficial compounds was ethylene glycol at all concentrations tested in the osmolyte screening plate. A common component of antifreeze, ethylene glycol can act as an osmoprotectant, blocking membrane pores caused by antimicrobial peptides (63). In our screening, ethylene glycol may have partially compensated for the cell envelope defects caused by the loss of NGO1985, allowing the mutant to grow to an OD600 up to 0.442, while the WT grew to an OD600 of 0.298 at an ethylene glycol concentration of 20% (Fig. 4 and File S1). Ammonium sulfate was beneficial to the Δngo1985 mutant at 10 to 20 mM but was detrimental to the mutant at 100 mM (Fig. 4 and File S1). This chemical has been used to induce bacterial aggregation, which is dependent on cell surface hydrophobicity and can be quantified by measuring the decrease in turbidity after ammonium sulfate application (64). It is also commonly used to precipitate proteins (65). Our results suggest that the loss of NGO1985 affects the surface hydrophobicity of the mutant; therefore, the presence of ammonium sulfate might result in cell aggregation lower than that of the WT, rather than enhancing the growth of the mutant. Higher concentrations may impose high ionic or osmotic stress that the mutant cannot withstand due to compromised membrane integrity.
The two most beneficial conditions for the Δngo1985 mutant were boric acid at concentration 3 and guanidine hydrochloride at concentration 4, with log2 ratios of 1.74 and 1.77, respectively. Boric acid is a weak Lewis acid that exhibits antimicrobial activity against another sexually transmitted disease-causing agent, Trichomonas vaginalis, although the mechanism of action is unknown (66). Guanidine hydrochloride is a chaotropic denaturant commonly used in the purification of otherwise insoluble proteins (67). Finally, not surprisingly, kanamycin was designated a beneficial compound in comparison to the WT. When knocking out the ngo1985 locus, we replaced it with the kanamycin resistance cassette under the control of its own promoter (6). The other mutants tested in this study were constructed in the same manner, and although they did grow in the presence of kanamycin throughout the experiment, the mutant-to-WT ratios did not meet our cutoff criteria. Compound concentrations are not provided for the PM chemical sensitivity plates. Instead, the concentrations are listed from 1 to 4, from lowest to highest. It is therefore not possible to compare the concentration of kanamycin in the screening plate to the concentration we used to select for mutant bacteria.
The most detrimental compounds were alexidine and l-glutamic-γ-hydroxamate, resulting in ratios of −1.17 and −1.02, respectively. Alexidine is a bisbiguanide disinfectant that binds to LPS and increases the permeability of the cell membrane, potentially by forming lipid domains within the membrane (68, 69). The already-perturbed Δngo1985 mutant cell envelope likely cannot compensate for the additional disruption, leading to inhibition of growth. While the WT OD600 increased from 0.039 to 0.11 over the time course, the Δngo1985 mutant increased from 0.046 to 0.051 within 2 h and then decreased to 0.049 by the end of the experiment. l-Glutamic-γ-hydroxamate is a glutamic acid hydroxamate derivative that irreversibly inhibits E. coli γ-glutamyl-cysteine synthase in an ATP-dependent manner (70). The inhibition of this biosynthetic enzyme depletes cellular glutathione stores, reducing the cell's ability to defend against toxic molecules, including reactive oxygen species (71). Glutathione is also involved in protecting the cell against the effects of low pH, chlorine compounds, and osmotic stress (72). In N. gonorrhoeae, glutathione is responsible for defense against the reactive nitrogen species nitric oxide (73). Thus, the reduction in available glutathione likely exacerbates the cellular stress inherent to the Δngo1985 mutant, preventing its growth.
Phenome of Δngo2121 mutant.
The screening indicated a total of 56 compounds with a positive or negative effect on the Δngo2121 mutant. Six conditions improved the growth of only the Δngo2121 mutant versus the wild type, and 15 conditions were detrimental to the mutant uniquely (Fig. 5). Azlocillin and cefuroxime, a ureidopenicillin and a second-generation cephalosporin, respectively, resulted in better growth of the mutant. While both compounds interfere with cell wall synthesis by binding to penicillin-binding proteins (74), another penicillin derivative with a similar mode of action, ampicillin, was detrimental to mutant viability. Additionally, sulfachloropyridazine and sulfamonomethoxine both attenuated the growth of the Δngo2121 mutant at all concentrations, while sulfamethoxazole and sulfanilamide had no effect at any concentration (Fig. 6 and File S1). All are sulfonamide antibiotics, which act to disrupt folate production by inhibiting dihydropteroate synthase (75). Analysis of computed partition coefficient (XlogP3) values indicated that the lipophilicities of these compounds increased in the following order: sulfanilamide (−0.6), sulfamonomethoxine (0.8), sulfamethoxazole (0.9), and sulfachloropyridazine (1.0). Based on their biological half-lives, sulfanilamide and sulfachloropyridazine are short-acting sulfa drugs, sulfamethoxazole is intermediate acting, and sulfamonomethoxine is long acting (https://pubchem.ncbi.nlm.nih.gov) (76). These lines of evidence indicate that neither solubility nor duration of action are adequate predictors of activity against the Δngo2121 mutant. The Δngo2121 mutant appeared more sensitive to oxidative stress, as potassium chromate and 3-amino-1,2,4-triazole were both detrimental to the growth of the mutant. This is consistent with observations in other bacteria, as a Campylobacter jejuni mutant deficient in the VacJ homolog, Cj1371, was more sensitive to oxidative stress induced by exposure to paraquat (77). As a catalase inhibitor, 3-amino-1,2,4-triazole increases cellular sensitivity to hydrogen peroxide (78). In Shewanella oneidensis, exposure to potassium chromate induced increased expression of numerous proteins with roles in protection against oxidative stress (79). Additionally, potassium chromate induced the SOS response in both S. oneidensis and E. coli (79, 80). Notably, another compound detrimental to the Δngo2121 mutant was 5-fluorouracil, an antitumor agent which induces the E. coli SOS response (81) and synergistically increases the sensitivity of Gram-negative bacteria to β-lactam antimicrobials (82). While N. gonorrhoeae strain MS11-A does not have a classical SOS system (83), the FA1090 genome does encode SOS system homologs that are activated by DNA damage and oxidative stress (84). Combined, the N. gonorrhoeae FA1090 mutant lacking NGO2121 displays an oxidative stress phenotype and a specific membrane permeability defect.
Other unique phenotypes.
None of the remaining five mutants showed as many phenotypes as the Δngo1985 or Δngo2121 mutant, and the Δngo2111 mutant was not uniquely affected by any conditions (Fig. 6). A mutant lacking NGO2054 was positively affected by vancomycin and 5,7-dichloro-8-hydroxyquinoline (Fig. 6, red bars). Vancomycin is a glycopeptide antibiotic that blocks cell wall synthesis by binding to peptidoglycan subunits. Typically, Gram-negative bacteria are resistant to vancomycin, because vancomycin is too large to easily diffuse through the outer membrane (74). The loss of NGO2054 may further fortify the cell envelope against larger molecules, as the mutant grew to an OD600 of 0.079, while the WT did not grow at all. The copper chelator 5,7-dichloro-8-hydroxyquinoline is has been shown to inhibit DNA polymerase activity and inhibit Rous sarcoma virus and herpes simplex virus 1 (85). Nordihydroguaiaretic acid and benserazide were both detrimental to the growth of the Δngo2054 mutant (blue bars). The primary active component in extracts of the creosote plant, nordihydroguaiaretic acid is a redox-active compound that appears to target the cellular membrane and can disrupt biofilm structure (86). Benserazide is an aromatic l-amino acid decarboxylase inhibitor that is coadministered with levodopa to treat Parkinson's disease and has been shown to increase the sensitivity of multidrug-resistant Pseudomonas aeruginosa to minocycline (87).
The growth of a mutant deficient in NGO1205 was enhanced compared to that of the wild type by a 6% solution of NaCl supplemented with trimethylamine, as well as by cephalothin. The presence of trimethylamine increases the pH of the culture medium (88), suggesting that the mutant may better tolerate a more basic environment. Cephalothin, a first-generation cephalosporin that is 325-fold less effective than ceftriaxone against N. gonorrhoeae (89), permitted the growth of the Δngo1205 mutant, resulting in a final mutant OD600 of 0.101, while the WT did not grow at all (File S1). However, the presence of moxalactam, a third-generation cephalosporin that is over 20-fold more effective than cephalothin (89), allowed the WT to grow to an OD600 of 0.195, while the mutant achieved an OD600 of 0.13 (File S1).
The growth of a Δngo1344 mutant was enhanced by oxycarboxin (Fig. 6, bright pink bar), an antifungal that interferes with fungal mitochondrial electron transport and inhibits succinate oxidation in the Gram-negative bacterium Micrococcus denitrificans (90). Although the turbidity of both the WT and mutant started out high, at OD600 values of 0.209 and 0.212, respectively, over the course of the experiment, the WT OD600 dropped to 0.051, while that of the Δngo1344 mutant increased to 0.333 (File S1). The mutant-to-WT ratio was therefore inflated; however, oxycarboxin still met our curation criteria. Growth of the Δngo1344 mutant was attenuated by 6-mercaptopurine at concentrations 1 and 2. This compound is typically used as an immunosuppressant in patients with Crohn's disease but has also been shown to suppress the growth of Mycobacterium paratuberculosis, a possible cause or trigger of Crohn's disease (91). The Δngo1344 mutant was also negatively affected by both colistin and 2-nitroimidazole. Colistin is a cationic antimicrobial peptide, also known as polymyxin E. Like polymyxin B, it targets the cell envelope, increasing permeability and causing cell death (92). 2-Nitroimidazole exhibits antitubercular activity, even to Mycobacterium tuberculosis within macrophages (93). These data suggest that the loss of NGO1344 causes a membrane defect that is more specific than that caused by a lack of NGO1985.
Three osmolytes were beneficial to the growth of the ΔbamEGC mutant exclusively (Fig. 6, light pink bars). A 6% NaCl solution supplemented with either creatinine or l-proline improved the growth of the mutant to a log2 ratio of 1.75 or 1.48, respectively. Sodium nitrite is commonly used as a packaged meat preservative and is bacteriostatic toward Listeria monocytogenes (94). It also kills P. aeruginosa, Staphylococcus aureus, and Burkholderia cepacia during both planktonic and biofilm growth (95). In contrast, a 20 mM solution of sodium benzoate at pH 5.2 attenuated the growth of the ΔbamEGC mutant, resulting in a log2 ratio of −0.58. The mutant was also negatively affected by five other compounds (Fig. 6, teal bars), including four antibiotics: nalidixic acid, rifampin, doxycycline, and cefsulodin. All four antibiotics employ different mechanisms to kill bacteria. Nalidixic acid hinders DNA synthesis, rifampin inhibits RNA synthesis, doxycycline interferes with protein synthesis, and cefsulodin prevents cell wall synthesis (74). Additionally, the ΔbamEGC mutant was more sensitive than the WT to chromium chloride, which inhibits DNA synthesis (96). The diversity of the activities of compounds that affect the BamEGC knockout indicated that the lack of BamEGC caused a more general defect in membrane permeability. A lack of BamE affects membrane permeability in both N. meningitidis and E. coli, as measured by an increase in susceptibility to antibiotics, including vancomycin (49, 97), bacitracin and erythromycin (50), and the antimicrobial peptide polymyxin B (98). The effects of BamE deletion have also been studied in Caulobacter crescentus, where mutant bacteria exhibited increased sensitivity to nalidixic acid, rifampin, and carbenicillin (99). No previous study utilizing PMs has examined bacteria deficient in BamE, nor have sensitivity phenotypes to sodium benzoate or chromium chloride been reported.
Cluster analysis of phenotypes.
To gain further insight into the physiological functions of the vaccine candidate proteins under investigation, we grouped them together based on which compounds were beneficial or detrimental to two or more mutants. For this analysis, as long as the mutant-to-WT log2 ratio meet our cutoff criteria, the magnitude of the effect was ignored. Additionally, if a compound affected two mutants at two different concentrations, the mutants were clustered together, regardless of concentration. Based on the compounds that affected the mutants in common, we generated a dendrogram of functional relatedness (Fig. 7). This analysis indicated that NGO2121 and NGO1985 formed a phenotype cluster distinct from the other five mutants tested, suggesting that they perform tasks unique from the other proteins. NGO1344 and BamEGC clustered together, and NGO2054, NGO1205, and NGO2111 formed a third cluster, with NGO1205 and NGO2111 being the most closely related of the three. This phenotype clustering, combined with the fact that no conditions had a unique effect on the Δngo2111 mutant, suggests that NGO2111 may have a redundant role in the cell envelope; however, additional studies are needed to further elucidate its function.
FIG 7.
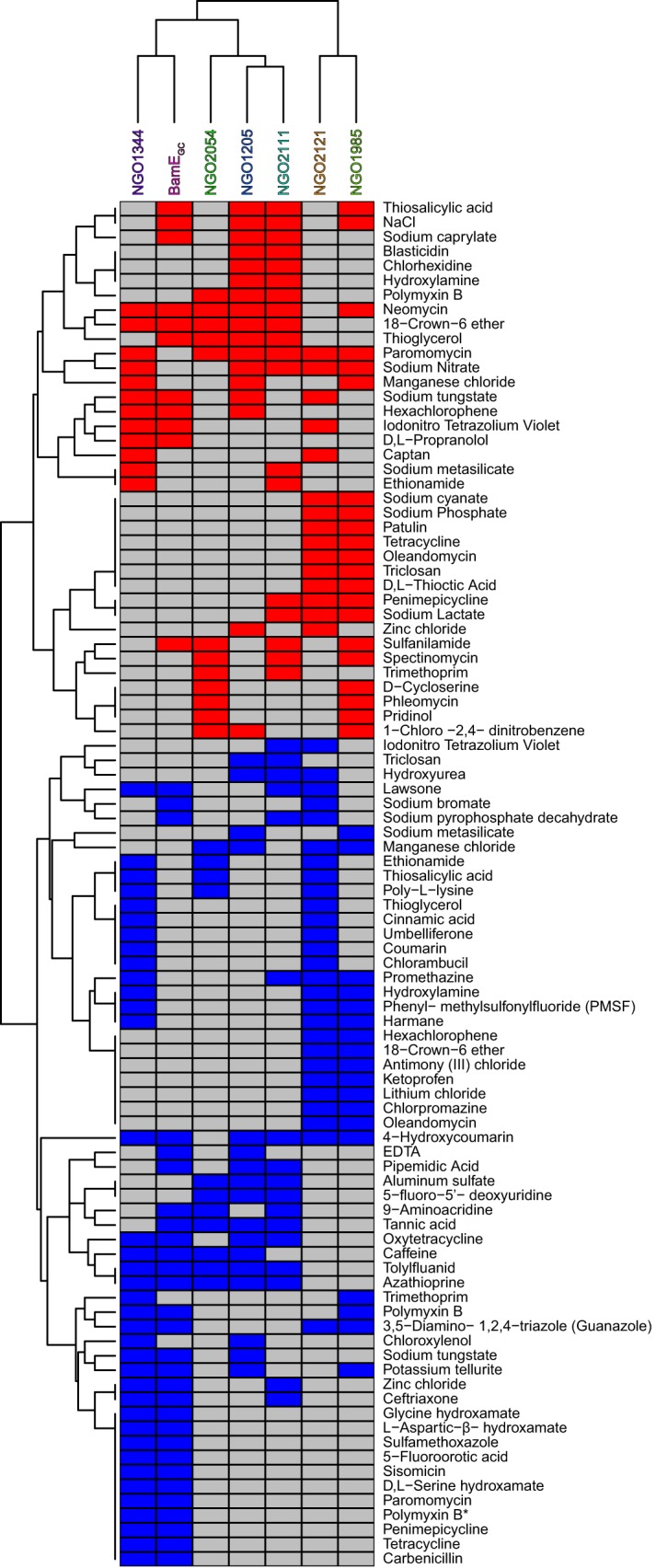
Dendrogram of vaccine candidates' functional relatedness. A clustering analysis was performed based on the mutants' common responses to 37 beneficial compounds (red boxes) and 57 detrimental compounds (blue boxes) that affected two or more mutants at any concentration. *, polymyxin B, present on two screening plates, was detrimental to the Δngo1985 mutant on one plate and had no effect on the Δngo1985 mutant on the other plate.
We expected that kanamycin and derivative compounds, such as neomycin and paromomycin (Fig. S1), would have less of an effect on the mutant strains than the WT because of the kanamycin resistance cassette used to replace the gene during generation of the knockout strains (5, 6). Aminoglycoside-modifying enzymes confer resistance to kanamycin, neomycin, and paromomycin (100), so these antibiotics can serve as a control for a beneficial compound. The log2 ratio of the Δngo2121 mutant did not reach our 0.5 cutoff for neomycin, nor did the ΔbamEGC mutant ratio for paromomycin. At lower concentrations, paromomycin was detrimental to the Δngo1344 and ΔbamEGC mutants (Fig. 7 and File S1). However, the presence of paromomycin allowed the two mutants to reach turbidities with OD600 values of 0.124 and 0.12, respectively, at the highest concentration, while WT growth was limited to an OD600 of 0.086.
Among the tested compounds that differentially affected more than two mutants was 18-crown-6 ether, which inhibited growth of the Δngo2121 and Δngo1985 mutants, while it enhanced the growth of the other five mutants to log2 ratios of 0.82 to 4.30 (File S1). The toxicity and antimicrobial activity (101) of this planar lipophilic compound (Fig. S1) involve binding and solubilizing potassium ions within the cell membrane (102). It is possible that the alterations in the membrane caused by the loss of NGO2121 or NGO1985 increase the susceptibility of the mutants to this compound by allowing easier penetration into the cell envelope. The other mutants may have cell envelope modifications that limit the diffusion of 18-crown-6 ether into the cell, or the compound may prevent the loss of potassium ions through channels in the membrane (103).
Also, thioglycerol had mixed effects on six mutants. It was beneficial to the growth of the ΔbamEGC, Δngo2054, Δngo1205, and Δngo2111 mutants and detrimental to the growth of the Δngo1344 and Δngo2121 mutants. This compound is able to inhibit the growth of E. coli by depressing RNA synthesis, inhibiting aerobic respiration, and reducing S-adenosylmethionine concentrations by acting as a methyl acceptor (104–106). As noted previously, the Δngo2121 mutant appeared to be more sensitive to oxidative stress, which likely contributed to its increased susceptibility to thioglycerol.
The sulfonamide antibacterial sulfanilamide was beneficial to the growth of the Δngo1985, Δngo2111, Δngo2054, and ΔbamEGC mutants. This compound is the active component of the prodrug prontosil, and it interferes with folate synthesis by acting as a competitive inhibitor of dihydropteroate synthase (107). In contrast, the antibiotic oxytetracycline, which inhibits protein synthesis (74), was detrimental to the growth of the Δngo2111, Δngo1205, ΔbamEGC, and Δngo1344 mutants.
Caffeine has previously been shown to exhibit antibacterial activity against E. coli through interfering with DNA synthesis (108, 109). It prevented the removal of thymidine dimers induced by UV radiation, contributing to increased mutational frequency (110). In our screening, the Δngo1205, Δngo2054, ΔbamEGC, and Δngo1344 mutants were negatively affected by the presence of caffeine. The transport of membrane proteins has also been shown to be inhibited by caffeine (111). These lines of evidence suggest that the loss of NGO1205, NGO2054, BamEGC, or NGO1344 allowed increased influx of caffeine into the cell, impeding appropriate protein transport while simultaneously impairing DNA synthesis.
Chemical sensitivity complementation testing.
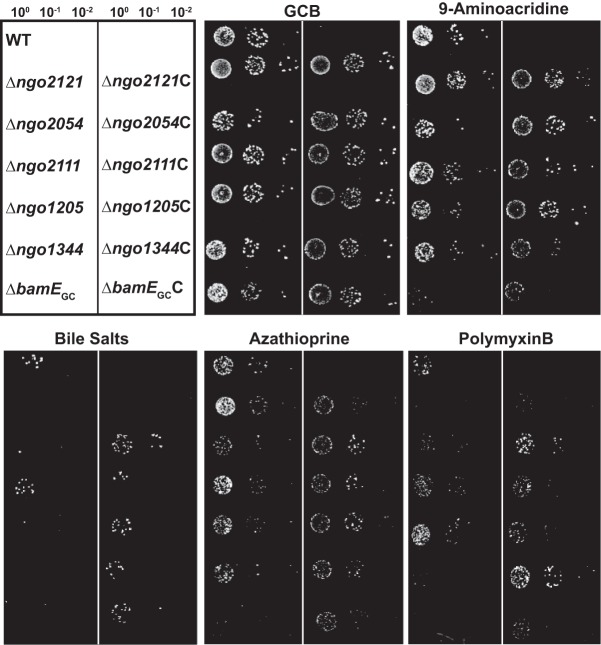
To ensure that the phenotypes observed in the screening were due to the loss of the proteins under investigation, we generated complemented strains of all null mutations by placing the individual genes under the control of the lactose promoter in an unlinked chromosomal locus. We have previously demonstrated full complementation of phenotypes associated with deletion of NGO1985 (6) (I. H. Wierzbicki, R. A. Zielke, J. Li, A. A. Begum, L. E. Peterson, R. W. Reed, M. Unemo, K. V. Korotkov, A. E. Jerse, and A. E. Sikora, unpublished data). WT, knockout mutants, and all complemented strains were spotted on gonococcal base (GCB) solid medium plates supplemented with isopropyl-β-d-1-thiogalactopyranoside (IPTG) and 9-aminoacridine, bile salts, azathioprine, or polymyxin B. Exposure to 9-aminoacridine, a DNA binding dye that inhibits the activity of E. coli RNA polymerase (112), affected the Δngo2054, Δngo2111, Δngo1205, and ΔbamEGC mutants. Resistance to this compound was restored to the WT level upon expression of the genes (Fig. 8). Bile salts nearly abolished gonococcal viability in mutants lacking NGO2054, NGO1205, NGO1344, and BamEGC, while they grew similarly to the WT upon complementation (Fig. 8). We previously examined the effect of bile salts on the Δngo2121, Δngo1985, Δngo2111, and Δngo1344 knockout mutants, which resulted in strongly attenuated phenotypes for the Δngo1985 and Δngo2121 mutants. The growth of the Δngo2111 and Δngo1344 mutants was not noticeably affected in our earlier investigation (6); however, using a 2.6-fold higher concentration of bile salts in the current study revealed a growth defect for the Δngo1344 mutant. The Δngo2111 mutant was not affected by exposure to bile salts in either study. The phenotype observed for the Δngo2121 mutant when exposed to bile salts was not complemented (Fig. 8). This may be due to insufficient induction of protein expression, reflecting a requirement for increased expression of this protein during exposure to amphipathic membrane-perturbing agents. Complementation reversed the sensitivity of the Δngo2054, Δngo2111, Δngo1205, Δngo1344, and ΔbamEGC mutants to azathioprine, a prodrug used as an immunosuppressant (Fig. 8). Azathioprine is metabolized to 6-mercaptopurine, which, as a purine analog, acts by inhibiting DNA synthesis (113). Exposure to polymyxin B was detrimental to the Δngo2121, Δngo2054, Δngo1344, and ΔbamEGC mutants (Fig. 8), and viability was completely restored upon expression of all proteins from an unlinked chromosomal locus. Notably, the complementation of NGO2054 resulted in larger colonies than those of the WT, suggesting that an increased level of this protein could confer a fitness benefit when cells are exposed to antimicrobial peptides. Together, these complementation studies showed that the lack of vaccine candidate proteins was responsible for the compound sensitivities observed in the screening.
FIG 8.
Phenotypic complementation assessment. FA1090 (WT), isogenic knockout strains, and complemented strains were suspended to 5 × 105 CFU/ml. Suspensions were serially diluted and spotted onto a gonococcal base solid medium (GCB) plate, as well as GCB supplemented with 0.1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) and 9-aminoacridine (12 μM), bile salts (0.13%), azathioprine (80 μg/ml), or polymyxin B (600 U/ml). Plates were incubated at 37°C in a 5% CO2 atmosphere and inspected after 22 h. Experiments were performed in triplicate, and representative images are presented. C, complemented strain.
Phenotype testing in WHO X background.
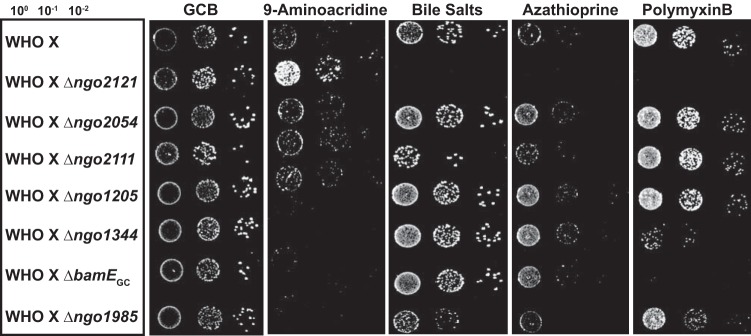
Finally, we examined the role of the vaccine candidates in a contemporary clinical isolate. Knockout mutations of each gene were constructed in one of the 2016 World Health Organization (WHO) reference strains, WHO X (11). This strain, formerly known as H041, was isolated in Kyoto, Japan, in 2009 and was the first strain identified with high-level resistance to ceftriaxone (114). Further examination revealed resistance to all β-lactams, fluoroquinolones, macrolides, tetracycline, trimethoprim-sulfamethoxazole, chloramphenicol, and nitrofurantoin, as well as elevated MICs for extended spectrum cephalosporins. However, it was still sensitive to spectinomycin and rifampin (115). WHO X and isogenic knockouts were spotted onto GCB solid medium containing polymyxin B, bile salts, azathioprine, or 9-aminoacridine (Fig. 9). As expected, when vaccine candidate proteins were knocked out in the WHO X background, the chemical sensitivity profiles were similar to those of the deletion mutants in the FA1090 background. However, the WHO X strain exhibited increased resistance to all compounds tested; therefore, higher concentrations were required to observe similar sensitivity phenotypes. Exposure to 9-aminoacrine was detrimental to the Δngo1344, ΔbamEGC, and Δngo1985 mutants. In stark contrast, however, the viability of the Δngo2121 mutant was far greater than that of the wild type when subjected to this compound. Bile salts completely abolished Δngo2121 mutant growth and diminished growth of the Δngo2111 and Δngo1985 mutants. Azathioprine had an effect only on the Δngo2121 mutant, while polymyxin B was detrimental to mutants lacking NGO2121, NGO1344, BamEGC, and NGO1985. In the WHO X background, the loss of NGO2054 or NGO1205 did not result in a phenotype with the compounds selected. However, the Δngo2111 mutant was sensitive to bile salts in the WHO X background but was the only mutant resistant to this compound in the FA1090 background. The altered sensitivity profile of the Δngo2111 mutant between the two strains tested suggested that alternative lipoproteins could depend on translocation by NGO2111 in the WHO X strain if this protein acts as Slam2. The results of our phenotypic assessment in WHO X illustrated the difference between strains and likely reflected the adaptations this highly antibiotic-resistant strain has undergone. Importantly, these studies indicated that vaccine candidate proteins NGO2121 and NGO1985 serve similar functions in a drug-resistant contemporary clinical isolate, as their sensitivity phenomes were comparable in the two strains. The evidence gathered from comparing these mutants in the two strains further illustrated their potential utility in a protective vaccine(s). Finally, the similarity in phenotypic profiles between mutants in the WHO X background compared to the same null mutations in FA1090 provides additional validation of our choice of N. gonorrhoeae FA1090 as a type strain for vaccine development.
FIG 9.
Phenotypic analysis in a contemporary clinical isolate. WHO X (WT) and isogenic knockouts, as indicated, were suspended to an optical density at 600 nm (OD600) of 0.1, cultured for 3 h at 37°C, and diluted to 5 × 105 CFU/ml. Cultures were serially diluted and spotted onto a gonococcal base solid medium (GCB) plate, in addition to GCB plates supplemented with 9-aminoacridine (20 μM), bile salts (0.2%), azathioprine (118 μg/ml), or polymyxin B (1,600 U/ml). Plates were incubated at 37°C in a 5% CO2 atmosphere and inspected after 22 h. Experiments were performed in triplicate, and representative images are presented.
Conclusions.
We present here the first phenotype microarray screening of N. gonorrhoeae to investigate the function of gonococcal vaccine candidate proteins NGO2121, NGO1985, NGO2054, NGO2111, NGO1205, and NGO1344, in addition to a marker for increased membrane permeability, BamEGC. Our optimized screening method can be employed to search for inhibitors of gonococcal growth in a 96-well plate format or for the assessment of novel antigonococcal therapeutics. The insights gained into the functions of vaccine candidates will allow us to better understand the biology of the gonococcus and the effects of targeting these proteins with a vaccine. Using our screening results, we constructed a dendrogram illustrating the functional relatedness of the proteins under investigation, revealing that NGO2121 and NGO1985 have the most distinct functions compared to the other five proteins. We also confirmed our previous observations about the extent of the membrane defects caused by the loss of NGO1985 (6). It was the most highly affected mutant, with 64 compounds positively or negatively influencing its growth. Additionally, for the first time, we identified conditions under which the growth of the Δngo2111 or Δngo1344 mutant was affected, as our previous investigation did not reveal any phenotypes for either mutant (6). The lack of any conditions that uniquely affected the Δngo2111 mutant also suggested that this protein may have a redundant function and could compensate for the loss of another protein. For the Δngo2121 mutant, our results revealed a sensitivity to oxidative stress, as well as a specific outer membrane defect. This study can act as a springboard for further characterization of these proteins in the cell envelope. Furthermore, by revealing specific mutant sensitivities, the results of this study can allow us to focus our attention on vaccine candidates that are likely to have a more substantial physiological effect if their function is disrupted by the antibodies generated against them. For example, the increased susceptibility to oxidative stress observed for the Δngo2121 mutant and the sensitivity of the Δngo1985 mutant to a multitude of compounds with widely varying modes of action illustrate the strong potential of these proteins as vaccine candidates. Additionally, the similarity in phenotypic profiles in mutants lacking NGO2121 or NGO1985 in both a laboratory strain and a highly antibiotic-resistant contemporary clinical isolate provide further support for their inclusion in a future vaccine. With the increasing abundance of genomic and proteomic data available for N. gonorrhoeae, screenings such as the one presented here have the potential to dramatically reduce the amount of time required to evaluate novel vaccine candidates.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
For this study, we used Neisseria gonorrhoeae FA1090 as the wild type (WT) (14). Isogenic knockout mutations of NGO2121, NGO1985, NGO1344, and NGO2111 were constructed previously (6), as were knockout mutations of NGO2054 (5) and NGO1780 (BamEGC) (Sikora et al., unpublished). The Δngo1205 mutant and the complemented strains were generated for this study as described below or in a previous study (5, 6). All mutations were introduced into the FA1090 background and WHO X (H041 [11]). Isolates of N. gonorrhoeae were streaked from frozen stocks stored in glycerol at −80°C onto gonococcal base (GCB) agar solid medium (Difco) with Kellogg's supplements I and II, diluted 1:100 and 1:1,000, respectively (116). Strains were cultured for 18 to 20 h at 37°C in a 5% CO2 atmosphere. Piliated or nonpiliated transparent colonies were subcultured onto fresh GCB agar plates and cultured as described above. Piliated colonies were used for transformation experiments, and nonpiliated colonies were used in all other experiments. To initiate growth in liquid medium, nonpiliated colonies were collected from plates incubated for ∼18 h and suspended to an OD600 of 0.1 in prewarmed GCB liquid (GCBL) medium supplemented as described above with the addition of 0.042% sodium bicarbonate, or in Graver-Wade (GW) medium (51). Suspensions in GCBL medium were incubated at 37°C with shaking at 220 rpm for 3 h. GW medium suspensions were cultured in the same manner in a 5% CO2 environment.
Phenotype screening.
After an initial 3 h of incubation in GW medium, cultures were back diluted to a 20-ml volume at an OD600 of 0.01 in prewarmed GW medium, and 100 μl was dispensed into each well of the Biolog Phenotype MicroArray. Plates were incubated at 37°C in a 5% CO2 atmosphere with agitation (220 rpm) for 6 h, and the OD600 values were measured every 2 h using a Synergy HT BioTek plate reader. In parallel, the remaining volume of culture was incubated in a conical tube under the same conditions to monitor overall bacterial growth. OD600 values were measured every 2 h. Additionally, samples of each culture were taken immediately after dilution and serially diluted in GW medium, and 5 μl of each dilution was spotted onto a GCB plate for CFU enumeration. The GCB plate was cultured as described above for 18 to 20 h, and colonies were counted. The WT, Δngo2121 mutant, and Δngo1985 mutant were screened in one set of experiments, and the WT and Δngo2054, Δngo2111, Δngo1205, Δngo1344, and ΔbamEGC mutants were screened in a second set of experiments.
Supernatant protein profiling.
After initial incubation, cultures were diluted to an OD600 of 0.1 in prewarmed GCBL or GW medium. Strains were cultured as described above until they reached approximately mid-logarithmic growth (OD600 between 0.6 and 0.8). Supernatants were separated by low-speed centrifugation (6,000 × g, 10 min, 25°C) and filtered through a 0.22-μm polyethersulfone (PES) filter (VWR). Proteins in the supernatant were precipitated through a pyrogallol red-molybdate-methanol procedure, as described previously (53), except that supernatants were treated with 10 U/ml DNase I (Thermo Scientific), and membrane vesicles were not removed prior to precipitation. Supernatant samples from cultures grown in both GCBL and GW media were subsequently analyzed by SDS-PAGE and immunoblotting.
SDS-PAGE and immunoblotting.
Precipitated supernatant protein samples were standardized by the OD600 of the source culture, separated in either an Any kD Criterion TGX stain-free protein gel (Bio-Rad) or a 4 to 20% Criterion TGX precast protein gel (Bio-Rad), and transferred to a 0.2-μm polyvinylidene difluoride (PVDF) or nitrocellulose membrane (Bio-Rad) with a Trans-Blot Turbo transfer system (Bio-Rad). Membranes were blocked in blocking buffer (5% skim milk in phosphate-buffered saline [PBS {pH 7.4}; Molecular Biologicals International] with 0.1% Tween 20 [PBST]) for 1 h at room temperature. Polyclonal rabbit antisera were diluted in blocking buffer as follows: anti-ObgGC, 1:2,500 (54); anti-Zwf, 1:2,500 (55); and anti-GmhA, 1:2,500 (55). Horseradish peroxidase-conjugated goat anti-rabbit IgG antiserum (Bio-Rad) was used as secondary antibody and diluted 1:10,000 in blocking buffer. Membranes were incubated in primary antiserum solutions overnight at 4°C and in secondary antiserum solution for 1 h at room temperature. Membranes were washed in PBST after antiserum applications. Proteins were visualized by development with Clarity Western ECL substrate (Bio-Rad) and imaging with a ChemiDoc MP system (Bio-Rad).
Whole-cell lysate samples were analyzed as described above, except that primary antisera were diluted an additional 4-fold, and membranes were blocked overnight at 4°C and incubated in primary antisera for 1 h at room temperature.
For visualization of the overall supernatant protein profile, samples were normalized as described above, separated on a NuPAGE Novex 4 to 12% Bis-Tris protein gel (Thermo Fisher Scientific), and visualized by staining with a SilverQuest silver staining kit (Thermo Fisher Scientific).
DNA manipulations.
The N. gonorrhoeae FA1090 genome sequence (accession no. NC_002946) was used to design oligonucleotides with the SnapGene software version 2.8 (GSL Biotech LLC). Primers were synthesized by Integrated DNA Technologies (IDT), and genomic DNA was purified with the Wizard genomic DNA purification kit (Promega). The primers used in this study are listed in Table S1 in the supplemental material. A QIAprep Spin miniprep kit (Qiagen) was used to purify plasmid DNA and PCR products. Chromosomal or plasmid DNA was amplified by PCR using the appropriate primers and Q5 high-fidelity polymerase (New England BioLabs). Escherichia coli strain MC1061 was used to perform genetic manipulations. Cloned fragment sequences were verified at the Oregon State University Center for Genome Research and Biocomputing. Transformation into N. gonorrhoeae was performed as described previously (6).
The deletion of ngo1205 was obtained by the following steps. The 522-bp region of chromosomal DNA upstream of NGO1205 was amplified with primers NGO1205-Up-F and NGO1205-Up-R, cleaved with EcoRI and KpnI, and inserted into similarly digested pUC18K, resulting in pUC18K-NGO1205Up. The region 523 bp downstream from NGO1205 was amplified with NGO1205-Down-F and NGO1205-Down-R, treated with BamHI and HindIII, and cloned into pUC18K-NGO1205Up cleaved with the same enzymes. The resulting plasmid, pUC18K-NGO1205, was used to transform FA1090. Replacement of ngo1205 with the nonpolar kanamycin resistance cassette was verified by PCR using primers NGO1205-Ver-F and NGO1205-Ver-R, with FA1090 genomic DNA as a control.
For complementations, individual genes (ngo2121, ngo2111, ngo1205, and ngo1344) were amplified by PCR using genomic DNA of FA1090 as the template and primers listed in Table S1. PCR products were subsequently cloned into the pGCC4 vector and placed at an unlinked chromosomal locus between the lctP and aspC genes using the neisserial insertional complementation system (117).
Null mutations in ngo1205, ngo1344, ngo1985, ngo2111, ngo2121, ngo1205, ngo2054, and bamEGC were also created in one of the 2016 World Health Organization (WHO) gonococcal reference strains, WHO X (H041) (11), using genetic constructs described above and previously (5, 6) (Sikora et al., unpublished). Due to the elevated kanamycin resistance exhibited by WHO X, selection for transformants was performed on GCB plates containing 50 μg/ml kanamycin. Genetic locus replacement with a kanamycin resistance cassette was confirmed with primers listed in Table S1 and previously (5, 6) (Sikora et al., unpublished).
Phenotypic microarray data analysis.
The ratio of mutant to wild-type (WT) growth was calculated by dividing the endpoint (6 h) OD600 of the mutant by the endpoint OD600 of the WT. Ratios were then log2 transformed. Hits were deemed significantly beneficial if the compound resulted in a ratio of least 0.5 or significantly detrimental if the compound resulted in a ratio of −0.5 or lower. Hits were manually curated and removed from analysis if (i) the OD600 values for both mutant and WT bacteria decreased over the course of the experiment, (ii) mutant bacteria exhibited no or inconsistent growth in “beneficial” hits, or (iii) WT bacteria exhibited no or inconsistent growth in “detrimental” hits. Each compound at each concentration was termed a “condition,” and each condition for each mutant was evaluated independently.
Susceptibility studies.
Sensitivity to polymyxin B sulfate, bile salts, azathioprine, and 9-aminoacridine was assessed as described by Zielke et al. (6) using 5 × 105 CFU/ml of nonpiliated N. gonorrhoeae FA1090 WT, isogenic mutants, and complemented strains. Azathioprine was dissolved in dimethyl sulfoxide (DMSO). 9-Aminoacridine was dissolved in ethanol. Bacteria were serially diluted in GCBL, supplemented as described above, with the addition of 0.1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG). Five microliters of each dilution was spotted onto GCB plates, with the addition of 0.1 mM IPTG and polymyxin B (600 U/ml), bile salts (0.13%), azathioprine (80 μg/ml), or 9-aminoacridine (12 μM). WHO X WT and knockout strains were suspended to an OD600 of 0.1 in GCBL medium, supplemented as described above, and cultured at 37°C with shaking until the OD600 had approximately doubled. Cultures were diluted to 5 × 105 CFU/ml and serially diluted in GCBL medium, and 5 μl was spotted on plates supplemented with polymyxin B (1,600 U/ml), bile salts (0.2%), azathioprine (118 μg/ml), or 9-aminoacridine (20 μM). Titrations of each compound with all mutants in both strain backgrounds were performed, and experiments with concentrations resulting in phenotypes were repeated three times. Bacterial growth was examined after incubation for 22 h in a 5% CO2 atmosphere at 37°C.
Clustering analysis.
The seven strains were clustered using hierarchical clustering, based on the binary matrices that indicated which strains had beneficial (and, respectively, detrimental) hits for each compound. Hamming distance, which counts how many elements differ between two vectors, and is equivalent to Manhattan distance on binary data, was used to determine the distance between strains. Average linkage was used to determine distances between clusters; other linkage methods (single and complete) were also explored and produced very similar results.
Compound structural analysis.
The structures or molecular formulas and the molecular weights of the compounds screened in the phenotypic microarray that affected at least one of the mutants are provided in Fig. S1. The structures were analyzed to determine if there was a correlation between observed phenotype and chemical structure.
Supplementary Material
ACKNOWLEDGMENTS
We thank Igor H. Wierzbicki for creating the Δngo1205 knockout strain as well as complementation constructs.
Funding for this work was provided to A.E.S. by grant R01-AI117235 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00037-17.
REFERENCES
- 1.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vollmer W, Blanot D, de Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 3.Buddelmeijer N. 2015. The molecular mechanism of bacterial lipoprotein modification–how, when and why? FEMS Microbiol Rev 39:246–261. doi: 10.1093/femsre/fuu006. [DOI] [PubMed] [Google Scholar]
- 4.Nakayama H, Kurokawa K, Lee BL. 2012. Lipoproteins in bacteria: structures and biosynthetic pathways. FEBS J 279:4247–4268. doi: 10.1111/febs.12041. [DOI] [PubMed] [Google Scholar]
- 5.Zielke RA, Wierzbicki IH, Baarda BI, Gafken PR, Soge OO, Holmes KK, Jerse AE, Unemo M, Sikora AE. 2016. Proteomics-driven antigen discovery for development of vaccines against gonorrhea. Mol Cell Proteomics 15:2338–2355. doi: 10.1074/mcp.M116.058800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zielke RA, Wierzbicki IH, Weber JV, Gafken PR, Sikora AE. 2014. Quantitative proteomics of the Neisseria gonorrhoeae cell envelope and membrane vesicles for the discovery of potential therapeutic targets. Mol Cell Proteomics 13:1299–1317. doi: 10.1074/mcp.M113.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Unemo M, Del Rio C, Shafer WM. 2016. Antimicrobial resistance expressed by Neisseria gonorrhoeae: a major global public health problem in the 21st century Microbiol Spectr 4:213–237. doi: 10.1128/microbiolspec.EI10-0009-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Unemo M, Shafer WM. 2014. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev 27:587–613. doi: 10.1128/CMR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jerse AE, Deal CD. 2013. Vaccine research for gonococcal infections: where are we? Sex Transm Infect 89(Suppl 4):iv63–iv68. doi: 10.1136/sextrans-2013-051225. [DOI] [PubMed] [Google Scholar]
- 10.Zhu W, Chen CJ, Thomas CE, Anderson JE, Jerse AE, Sparling PF. 2011. Vaccines for gonorrhea: can we rise to the challenge? Front Microbiol 2:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unemo M, Golparian D, Sanchez-Buso L, Grad Y, Jacobsson S, Ohnishi M, Lahra MM, Limnios A, Sikora AE, Wi T, Harris SR. 2016. The novel 2016 WHO Neisseria gonorrhoeae reference strains for global quality assurance of laboratory investigations: phenotypic, genetic and reference genome characterization. J Antimicrob Chemother 71:3096–3108. doi: 10.1093/jac/dkw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization. 2012. Global action plan to control the spread and impact of antimicrobial resistance in Neisseria gonorrhoeae. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 13.Centers for Disease Control and Prevention. 2013. Gonorrhea treatment guidelines. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/nchhstp/newsroom/docs/factsheets/archive/gonorrhea-treatment-guidelines-factsheet.pdf. [Google Scholar]
- 14.Cohen MS, Cannon JG, Jerse AE, Charniga LM, Isbey SF, Whicker LG. 1994. Human experimentation with Neisseria gonorrhoeae: rationale, methods, and implications for the biology of infection and vaccine development. J Infect Dis 169:532–537. doi: 10.1093/infdis/169.3.532. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Hammer LA, Liu W, Hobbs MM, Zielke RA, Sikora AE, Jerse AE, Egilmez NK, Russell MW. 8 March 2017. Experimental vaccine induces Th1-driven immune responses and resistance to Neisseria gonorrhoeae infection in a murine model. Mucosal Immunol. doi: 10.1038/mi.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baarda BI, Sikora AE. 2015. Proteomics of Neisseria gonorrhoeae: the treasure hunt for countermeasures against an old disease. Front Microbiol 6:1190. doi: 10.3389/fmicb.2015.01190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shewell LK, Ku SC, Schulz BL, Jen FE, Mubaiwa TD, Ketterer MR, Apicella MA, Jennings MP. 2013. Recombinant truncated AniA of pathogenic Neisseria elicits a non-native immune response and functional blocking antibodies. Biochem Biophys Res Commun 431:215–220. doi: 10.1016/j.bbrc.2012.12.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shea A, Wolcott M, Daefler S, Rozak DA. 2012. Biolog phenotype microarrays. Methods Mol Biol 881:331–373. doi: 10.1007/978-1-61779-827-6_12. [DOI] [PubMed] [Google Scholar]
- 19.Reales-Calderon JA, Blanco P, Alcalde-Rico M, Corona F, Lira F, Hernando-Amado S, Bernardini A, Sanchez MB, Martinez JL. 2016. Use of phenotype microarrays to study the effect of acquisition of resistance to antimicrobials in bacterial physiology. Res Microbiol 167:723–730. doi: 10.1016/j.resmic.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Alqasim A, Emes R, Clark G, Newcombe J, La Ragione R, McNally A. 2014. Phenotypic microarrays suggest Escherichia coli ST131 is not a metabolically distinct lineage of extra-intestinal pathogenic E. coli. PLoS One 9:e88374. doi: 10.1371/journal.pone.0088374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou L, Lei XH, Bochner BR, Wanner BL. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J Bacteriol 185:4956–4972. doi: 10.1128/JB.185.16.4956-4972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bochner BR, Gadzinski P, Panomitros E. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res 11:1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pletzer D, Braun Y, Dubiley S, Lafon C, Kohler T, Page MG, Mourez M, Severinov K, Weingart H. 2015. The Pseudomonas aeruginosa PA14 ABC transporter NppA1A2BCD is required for uptake of peptidyl nucleoside antibiotics. J Bacteriol 197:2217–2228. doi: 10.1128/JB.00234-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malinverni JC, Silhavy TJ. 2009. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc Natl Acad Sci U S A 106:8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roier S, Zingl FG, Cakar F, Durakovic S, Kohl P, Eichmann TO, Klug L, Gadermaier B, Weinzerl K, Prassl R, Lass A, Daum G, Reidl J, Feldman MF, Schild S. 2016. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat Commun 7:10515. doi: 10.1038/ncomms10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carpenter CD, Cooley BJ, Needham BD, Fisher CR, Trent MS, Gordon V, Payne SM. 2014. The Vps/VacJ ABC transporter is required for intercellular spread of Shigella flexneri. Infect Immun 82:660–669. doi: 10.1128/IAI.01057-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki T, Murai T, Fukuda I, Tobe T, Yoshikawa M, Sasakawa C. 1994. Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol Microbiol 11:31–41. doi: 10.1111/j.1365-2958.1994.tb00287.x. [DOI] [PubMed] [Google Scholar]
- 28.Yeats C, Bateman A. 2003. The BON domain: a putative membrane-binding domain. Trends Biochem Sci 28:352–355. doi: 10.1016/S0968-0004(03)00115-4. [DOI] [PubMed] [Google Scholar]
- 29.Barondess JJ, Beckwith J. 1990. A bacterial virulence determinant encoded by lysogenic coliphage lambda. Nature 346:871–874. doi: 10.1038/346871a0. [DOI] [PubMed] [Google Scholar]
- 30.Wei Y, Lin-Lee YC, Yang X, Dai W, Zhao S, Rassool FV, Elgart GW, Feun L, Savaraj N, Kuo MT. 2006. Molecular cloning of Chinese hamster 1q31 chromosomal fragile site DNA that is important to mdr1 gene amplification reveals a novel gene whose expression is associated with spermatocyte and adipocyte differentiation. Gene 372:44–52. doi: 10.1016/j.gene.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 31.Hooda Y, Lai CC, Judd A, Buckwalter CM, Shin HE, Gray-Owen SD, Moraes TF. 2016. Slam is an outer membrane protein that is required for the surface display of lipidated virulence factors in Neisseria. Nat Microbiol 1:16009. doi: 10.1038/nmicrobiol.2016.9. [DOI] [PubMed] [Google Scholar]
- 32.Ducey TF, Carson MB, Orvis J, Stintzi AP, Dyer DW. 2005. Identification of the iron-responsive genes of Neisseria gonorrhoeae by microarray analysis in defined medium. J Bacteriol 187:4865–4874. doi: 10.1128/JB.187.14.4865-4874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu C, Genco CA. 2012. Fur-mediated activation of gene transcription in the human pathogen Neisseria gonorrhoeae. J Bacteriol 194:1730–1742. doi: 10.1128/JB.06176-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stork M, Bos MP, Jongerius I, de Kok N, Schilders I, Weynants VE, Poolman JT, Tommassen J. 2010. An outer membrane receptor of Neisseria meningitidis involved in zinc acquisition with vaccine potential. PLoS Pathog 6:e1000969. doi: 10.1371/journal.ppat.1000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calmettes C, Ing C, Buckwalter CM, El Bakkouri M, Chieh-Lin Lai C, Pogoutse A, Gray-Owen SD, Pomes R, Moraes TF. 2015. The molecular mechanism of zinc acquisition by the neisserial outer-membrane transporter ZnuD. Nat Commun 6:7996. doi: 10.1038/ncomms8996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lappann M, Danhof S, Guenther F, Olivares-Florez S, Mordhorst IL, Vogel U. 2013. In vitro resistance mechanisms of Neisseria meningitidis against neutrophil extracellular traps. Mol Microbiol 89:433–449. doi: 10.1111/mmi.12288. [DOI] [PubMed] [Google Scholar]
- 37.Urban CF, Ermert D, Schmid M, Abu-Abed U, Goosmann C, Nacken W, Brinkmann V, Jungblut PR, Zychlinsky A. 2009. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog 5:e1000639. doi: 10.1371/journal.ppat.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hagen TA, Cornelissen CN. 2006. Neisseria gonorrhoeae requires expression of TonB and the putative transporter TdfF to replicate within cervical epithelial cells. Mol Microbiol 62:1144–1157. doi: 10.1111/j.1365-2958.2006.05429.x. [DOI] [PubMed] [Google Scholar]
- 39.Phillips NJ, Steichen CT, Schilling B, Post DM, Niles RK, Bair TB, Falsetta ML, Apicella MA, Gibson BW. 2012. Proteomic analysis of Neisseria gonorrhoeae biofilms shows shift to anaerobic respiration and changes in nutrient transport and outermembrane proteins. PLoS One 7:e38303. doi: 10.1371/journal.pone.0038303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Misra R, Miao Y. 1995. Molecular analysis of asmA, a locus identified as the suppressor of OmpF assembly mutants of Escherichia coli K-12. Mol Microbiol 16:779–788. doi: 10.1111/j.1365-2958.1995.tb02439.x. [DOI] [PubMed] [Google Scholar]
- 41.Deng M, Misra R. 1996. Examination of AsmA and its effect on the assembly of Escherichia coli outer membrane proteins. Mol Microbiol 21:605–612. doi: 10.1111/j.1365-2958.1996.tb02568.x. [DOI] [PubMed] [Google Scholar]
- 42.Putker F, Grutsch A, Tommassen J, Bos MP. 2014. Ght protein of Neisseria meningitidis is involved in the regulation of lipopolysaccharide biosynthesis. J Bacteriol 196:780–789. doi: 10.1128/JB.00943-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morse SA. 1978. The biology of the gonococcus. CRC Crit Rev Microbiol 7:93–189. [DOI] [PubMed] [Google Scholar]
- 44.Hobbs MM, Sparling PF, Cohen MS, Shafer WM, Deal CD, Jerse AE. 2011. Experimental gonococcal infection in male volunteers: cumulative experience with Neisseria gonorrhoeae strains FA1090 and MS11mkC. Front Microbiol 2:123. doi: 10.3389/fmicb.2011.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cole JG, Fulcher NB, Jerse AE. 2010. Opacity proteins increase Neisseria gonorrhoeae fitness in the female genital tract due to a factor under ovarian control. Infect Immun 78:1629–1641. doi: 10.1128/IAI.00996-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feinen B, Jerse AE, Gaffen SL, Russell MW. 2010. Critical role of Th17 responses in a murine model of Neisseria gonorrhoeae genital infection. Mucosal Immunol 3:312–321. doi: 10.1038/mi.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jerse AE. 1999. Experimental gonococcal genital tract infection and opacity protein expression in estradiol-treated mice. Infect Immun 67:5699–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jerse AE, Wu H, Packiam M, Vonck RA, Begum AA, Garvin LE. 2011. Estradiol-treated female mice as a surrogate for Neisseria gonorrhoeae genital tract infections. Front Microbiol 2:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Volokhina EB, Beckers F, Tommassen J, Bos MP. 2009. The β-barrel outer membrane protein assembly complex of Neisseria meningitidis. J Bacteriol 191:7074–7085. doi: 10.1128/JB.00737-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rigel NW, Schwalm J, Ricci DP, Silhavy TJ. 2012. BamE modulates the Escherichia coli beta-barrel assembly machine component BamA. J Bacteriol 194:1002–1008. doi: 10.1128/JB.06426-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wade JJ, Graver MA. 2007. A fully defined, clear and protein-free liquid medium permitting dense growth of Neisseria gonorrhoeae from very low inocula. FEMS Microbiol Lett 273:35–37. doi: 10.1111/j.1574-6968.2007.00776.x. [DOI] [PubMed] [Google Scholar]
- 52.Foerster S, Golparian D, Jacobsson S, Hathaway LJ, Low N, Shafer WM, Althaus CL, Unemo M. 2015. Genetic resistance determinants, in vitro time-kill curve analysis and pharmacodynamic functions for the novel topoisomerase II inhibitor ETX0914 (AZD0914) in Neisseria gonorrhoeae. Front Microbiol 6:1377. doi: 10.3389/fmicb.2015.01377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sikora AE, Zielke RA, Lawrence DA, Andrews PC, Sandkvist M. 2011. Proteomic analysis of the Vibrio cholerae type II secretome reveals new proteins, including three related serine proteases. J Biol Chem 286:16555–16566. doi: 10.1074/jbc.M110.211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zielke RA, Wierzbicki IH, Baarda BI, Sikora AE. 2015. The Neisseria gonorrhoeae Obg protein is an essential ribosome-associated GTPase and a potential drug target. BMC Microbiol 15:129. doi: 10.1186/s12866-015-0453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wierzbicki IH, Zielke RA, Korotkov KV, Sikora AE. 2017. Functional and structural studies on the Neisseria gonorrhoeae GmhA, the first enzyme in the glycero-manno-heptose biosynthesis pathways, demonstrate a critical role in lipooligosaccharide synthesis and gonococcal viability. Microbiologyopen 6:e00432. doi: 10.1002/mbo3.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harmer NJ. 2010. The structure of sedoheptulose-7-phosphate isomerase from Burkholderia pseudomallei reveals a zinc binding site at the heart of the active site. J Mol Biol 400:379–392. doi: 10.1016/j.jmb.2010.04.058. [DOI] [PubMed] [Google Scholar]
- 57.Sandoval JM, Arenas FA, Vasquez CC. 2011. Glucose-6-phosphate dehydrogenase protects Escherichia coli from tellurite-mediated oxidative stress. PLoS One 6:e25573. doi: 10.1371/journal.pone.0025573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morse SA, Bartenstein L. 1974. Factors affecting autolysis of Neisseria gonorrhoeae. Proc Soc Exp Biol Med 145:1418–1421. doi: 10.3181/00379727-145-38025. [DOI] [PubMed] [Google Scholar]
- 59.Hebeler BH, Young FE. 1975. Autolysis of Neisseria gonorrhoeae. J Bacteriol 122:385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Elmros T, Burman LG, Bloom GD. 1976. Autolysis of Neisseria gonorrhoeae. J Bacteriol 126:969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elmros T, Sandstrom G, Burman L. 1976. Autolysis of Neisseria gonorrhoeae. Relation between mechanical stability and viability. Br J Vener Dis 52:246–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hebeler BH, Young FE. 1976. Mechanism of autolysis of Neisseria gonorrhoeae. J Bacteriol 126:1186–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sato H, Feix JB. 2006. Osmoprotection of bacterial cells from toxicity caused by antimicrobial hybrid peptide CM15. Biochemistry 45:9997–10007. doi: 10.1021/bi060979m. [DOI] [PubMed] [Google Scholar]
- 64.Lin L, Rosenberg M, Taylor KG, Doyle RJ. 1995. Kinetic analysis of ammonium sulfate-dependent aggregation of bacteria. Colloids Surf B 5:127–134. doi: 10.1016/0927-7765(95)01189-P. [DOI] [Google Scholar]
- 65.Wingfield PT. 2001. Protein precipitation using ammonium sulfate. Curr Protoc Protein Sci Appendix 3:Appendix 3F. doi: 10.1002/0471140864.psa03fs13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brittingham A, Wilson WA. 2014. The antimicrobial effect of boric acid on Trichomonas vaginalis. Sex Transm Dis 41:718–722. doi: 10.1097/OLQ.0000000000000203. [DOI] [PubMed] [Google Scholar]
- 67.Petty KJ. 2001. Metal-chelate affinity chromatography. Curr Protoc Mol Biol Chapter 10:Unit 10.11B. doi: 10.1002/0471142727.mb1011bs36. [DOI] [PubMed] [Google Scholar]
- 68.Zorko M, Jerala R. 2008. Alexidine and chlorhexidine bind to lipopolysaccharide and lipoteichoic acid and prevent cell activation by antibiotics. J Antimicrob Chemother 62:730–737. doi: 10.1093/jac/dkn270. [DOI] [PubMed] [Google Scholar]
- 69.Chawner JA, Gilbert P. 1989. Interaction of the bisbiguanides chlorhexidine and alexidine with phospholipid vesicles: evidence for separate modes of action. J Appl Bacteriol 66:253–258. doi: 10.1111/j.1365-2672.1989.tb02476.x. [DOI] [PubMed] [Google Scholar]
- 70.Katoh M, Hiratake J, Oda J. 1998. ATP-dependent inactivation of Escherichia coli gamma-glutamylcysteine synthetase by l-glutamic acid gamma-monohydroxamate. Biosci Biotechnol Biochem 62:1455–1457. doi: 10.1271/bbb.62.1455. [DOI] [PubMed] [Google Scholar]
- 71.Meister A, Anderson ME. 1983. Glutathione. Annu Rev Biochem 52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 72.Masip L, Veeravalli K, Georgiou G. 2006. The many faces of glutathione in bacteria. Antioxid Redox Signal 8:753–762. doi: 10.1089/ars.2006.8.753. [DOI] [PubMed] [Google Scholar]
- 73.Seib KL, Wu HJ, Kidd SP, Apicella MA, Jennings MP, McEwan AG. 2006. Defenses against oxidative stress in Neisseria gonorrhoeae: a system tailored for a challenging environment. Microbiol Mol Biol Rev 70:344–361. doi: 10.1128/MMBR.00044-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sköld O. 2000. Sulfonamide resistance: mechanisms and trends. Drug Resist Updat 3:155–160. doi: 10.1054/drup.2000.0146. [DOI] [PubMed] [Google Scholar]
- 76.Vree TB, Hekster YA. 1987. Clinical pharmacokinetics of sulfonamides and their metabolites: an encyclopedia. Karger, Basel, Switzerland. [PubMed] [Google Scholar]
- 77.Garénaux A, Guillou S, Ermel G, Wren B, Federighi M, Ritz M. 2008. Role of the Cj1371 periplasmic protein and the Cj0355c two-component regulator in the Campylobacter jejuni NCTC 11168 response to oxidative stress caused by paraquat. Res Microbiol 159:718–726. doi: 10.1016/j.resmic.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 78.Lu X, Roe F, Jesaitis A, Lewandowski Z. 1998. Resistance of biofilms to the catalase inhibitor 3-amino-1,2, 4-triazole. Biotechnol Bioeng 59:156–162. [PubMed] [Google Scholar]
- 79.Brown SD, Thompson MR, Verberkmoes NC, Chourey K, Shah M, Zhou J, Hettich RL, Thompson DK. 2006. Molecular dynamics of the Shewanella oneidensis response to chromate stress. Mol Cell Proteomics 5:1054–1071. doi: 10.1074/mcp.M500394-MCP200. [DOI] [PubMed] [Google Scholar]
- 80.Venier P, Montini R, Zordan M, Clonfero E, Paleologo M, Levis AG. 1989. Induction of SOS response in Escherichia coli strain PQ37 by 16 chemical compounds and human urine extracts. Mutagenesis 4:51–57. doi: 10.1093/mutage/4.1.51. [DOI] [PubMed] [Google Scholar]
- 81.Oda Y. 1987. Induction of SOS responses in Escherichia coli by 5-fluorouracil. Mutat Res 183:103–108. [DOI] [PubMed] [Google Scholar]
- 82.Gieringer JH, Wenz AF, Just HM, Daschner FD. 1986. Effect of 5-fluorouracil, mitoxantrone, methotrexate, and vincristine on the antibacterial activity of ceftriaxone, ceftazidime, cefotiam, piperacillin, and netilmicin. Chemotherapy 32:418–424. doi: 10.1159/000238445. [DOI] [PubMed] [Google Scholar]
- 83.Black CG, Fyfe JA, Davies JK. 1998. Absence of an SOS-like system in Neisseria gonorrhoeae. Gene 208:61–66. doi: 10.1016/S0378-1119(97)00653-7. [DOI] [PubMed] [Google Scholar]
- 84.Schook PO, Stohl EA, Criss AK, Seifert HS. 2011. The DNA-binding activity of the Neisseria gonorrhoeae LexA orthologue NG1427 is modulated by oxidation. Mol Microbiol 79:846–860. doi: 10.1111/j.1365-2958.2010.07491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rohde W, Mikelens P, Jackson J, Blackman J, Whitcher J, Levinson W. 1976. Hydroxyquinolines inhibit ribonucleic acid-dependent deoxyribonucleic acid polymerase and inactivate Rous sarcoma virus and herpes simplex virus. Antimicrob Agents Chemother 10:234–240. doi: 10.1128/AAC.10.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ooi N, Eady EA, Cove JH, O'Neill AJ. 2015. Redox-active compounds with a history of human use: antistaphylococcal action and potential for repurposing as topical antibiofilm agents. J Antimicrob Chemother 70:479–488. doi: 10.1093/jac/dku409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Worthington RJ, Melander C. 2013. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol 31:177–184. doi: 10.1016/j.tibtech.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Létoffé S, Audrain B, Bernier SP, Delepierre M, Ghigo JM. 2014. Aerial exposure to the bacterial volatile compound trimethylamine modifies antibiotic resistance of physically separated bacteria by raising culture medium pH. mBio 5(1):e00944-13. doi: 10.1128/mBio.00944-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thabaut A, Durosoir JL, Saliou P. 1981. Comparative in vitro activity of 8 cephalosporins on 109 strains of Neisseria gonorrhoeae and 60 strains of Neisseria meningitidis. Chemotherapy 27(Suppl 1):S19–S24. [DOI] [PubMed] [Google Scholar]
- 90.Tucker AN, Lillich TT. 1974. Effect of the systemic fungicide carboxin on electron transport function in membranes of Micrococcus denitrificans. Antimicrob Agents Chemother 6:572–578. doi: 10.1128/AAC.6.5.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shin SJ, Collins MT. 2008. Thiopurine drugs azathioprine and 6-mercaptopurine inhibit Mycobacterium paratuberculosis growth in vitro. Antimicrob Agents Chemother 52:418–426. doi: 10.1128/AAC.00678-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Falagas ME, Kasiakou SK. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant Gram-negative bacterial infections. Clin Infect Dis 40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 93.Khan A, Sarkar S, Sarkar D. 2008. Bactericidal activity of 2-nitroimidazole against the active replicating stage of Mycobacterium bovis BCG and Mycobacterium tuberculosis with intracellular efficacy in THP-1 macrophages. Int J Antimicrob Agents 32:40–45. doi: 10.1016/j.ijantimicag.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 94.Myers K, Cannon J, Montoya D, Dickson J, Lonergan S, Sebranek J. 2013. Effects of high hydrostatic pressure and varying concentrations of sodium nitrite from traditional and vegetable-based sources on the growth of Listeria monocytogenes on ready-to-eat (RTE) sliced ham. Meat Sci 94:69–76. doi: 10.1016/j.meatsci.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 95.Major TA, Panmanee W, Mortensen JE, Gray LD, Hoglen N, Hassett DJ. 2010. Sodium nitrite-mediated killing of the major cystic fibrosis pathogens Pseudomonas aeruginosa, Staphylococcus aureus, and Burkholderia cepacia under anaerobic planktonic and biofilm conditions. Antimicrob Agents Chemother 54:4671–4677. doi: 10.1128/AAC.00379-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ogawa T, Usui M, Yatome C, Idaka E. 1989. Influence of chromium compounds on microbial growth and nucleic acid synthesis. Bull Environ Contam Toxicol 43:254–260. doi: 10.1007/BF01701756. [DOI] [PubMed] [Google Scholar]
- 97.Knowles TJ, Browning DF, Jeeves M, Maderbocus R, Rajesh S, Sridhar P, Manoli E, Emery D, Sommer U, Spencer A, Leyton DL, Squire D, Chaudhuri RR, Viant MR, Cunningham AF, Henderson IR, Overduin M. 2011. Structure and function of BamE within the outer membrane and the beta-barrel assembly machine. EMBO Rep 12:123–128. doi: 10.1038/embor.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tzeng YL, Ambrose KD, Zughaier S, Zhou X, Miller YK, Shafer WM, Stephens DS. 2005. Cationic antimicrobial peptide resistance in Neisseria meningitidis. J Bacteriol 187:5387–5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ryan KR, Taylor JA, Bowers LM. 2010. The BAM complex subunit BamE (SmpA) is required for membrane integrity, stalk growth and normal levels of outer membrane β-barrel proteins in Caulobacter crescentus. Microbiology 156:742–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shaw KJ, Rather PN, Hare RS, Miller GH. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev 57:138–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tso W-W, Fung W-P. 1981. Correlation between the antibacterial activity and alkali metal ion transport efficiency of crown ether. Inorg Chim Acta 55:129–134. doi: 10.1016/S0020-1693(00)90794-1. [DOI] [Google Scholar]
- 102.Alfonso I, Quesada R. 2013. Biological activity of synthetic ionophores: ion transporters as prospective drugs? Chem Sci 4:3009–3019. doi: 10.1128/JB.187.15.5387-5396.2005. [DOI] [Google Scholar]
- 103.Bezrukov SM, Krasilnikov OV, Yuldasheva LN, Berezhkovskii AM, Rodrigues CG. 2004. Field-dependent effect of crown ether (18-crown-6) on ionic conductance of alpha-hemolysin channels. Biophys J 87:3162–3171. doi: 10.1529/biophysj.104.044453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jensen KK, Javor GT. 1981. Inhibition of Escherichia coli by thioglycerol. Antimicrob Agents Chemother 19:556–561. doi: 10.1128/AAC.19.4.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Javor GT. 1983. Inhibition of respiration of Escherichia coli by thioglycerol. Antimicrob Agents Chemother 24:868–870. doi: 10.1128/AAC.24.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Javor GT. 1983. Depression of adenosylmethionine content of Escherichia coli by thioglycerol. Antimicrob Agents Chemother 24:860–867. doi: 10.1128/AAC.24.6.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hammoudeh DI, Zhao Y, White SW, Lee RE. 2013. Replacing sulfa drugs with novel DHPS inhibitors. Future Med Chem 5:1331–1340. doi: 10.4155/fmc.13.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sandlie I, Lossius I, Sjastad K, Kleppe K. 1983. Mechanism of caffeine-induced inhibition of DNA synthesis in Escherichia coli. FEBS Lett 151:237–242. doi: 10.1016/0014-5793(83)80077-5. [DOI] [PubMed] [Google Scholar]
- 109.Kang TM, Yuan J, Nguyen A, Becket E, Yang H, Miller JH. 2012. The aminoglycoside antibiotic kanamycin damages DNA bases in Escherichia coli: caffeine potentiates the DNA-damaging effects of kanamycin while suppressing cell killing by ciprofloxacin in Escherichia coli and Bacillus anthracis. Antimicrob Agents Chemother, 56:3216–3223. doi: 10.1128/AAC.00066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sideropoulos AS, Shankel DM. 1968. Mechanism of caffeine enhancement of mutations induced by sublethal ultraviolet dosages. J Bacteriol 96:198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kuismanen E, Jantti J, Makiranta V, Sariola M. 1992. Effect of caffeine on intracellular transport of Semliki Forest virus membrane glycoproteins. J Cell Sci 102:505–513. [DOI] [PubMed] [Google Scholar]
- 112.Nicholson BH, Peacocke AR. 1966. The inhibition of ribonucleic acid polymerase by acridines. Biochem J 100:50–58. doi: 10.1042/bj1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maltzman JS, Koretzky GA. 2003. Azathioprine: old drug, new actions. J Clin Invest 111:1122–1124. doi: 10.1172/JCI200318384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ohnishi M, Saika T, Hoshina S, Iwasaku K, Nakayama S, Watanabe H, Kitawaki J. 2011. Ceftriaxone-resistant Neisseria gonorrhoeae, Japan. Emerg Infect Dis 17:148–149. doi: 10.3201/eid1701.100397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ohnishi M, Golparian D, Shimuta K, Saika T, Hoshina S, Iwasaku K, Nakayama S, Kitawaki J, Unemo M. 2011. Is Neisseria gonorrhoeae initiating a future era of untreatable gonorrhea? Detailed characterization of the first strain with high-level resistance to ceftriaxone. Antimicrob Agents Chemother 55:3538–3545. doi: 10.1128/AAC.00325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Spence JM, Wright L, Clark VL. 2008. Laboratory maintenance of Neisseria gonorrhoeae. Curr Protoc Microbiol Chapter 4:Unit 4A.1. doi: 10.1002/9780471729259.mc04a01s8. [DOI] [PubMed] [Google Scholar]
- 117.Mehr IJ, Long CD, Serkin CD, Seifert HS. 2000. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics 154:523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.