ABSTRACT
Electron cryotomography (ECT) can reveal the native structure and arrangement of macromolecular complexes inside intact cells. This technique has greatly advanced our understanding of the ultrastructure of bacterial cells. We now view bacteria as structurally complex assemblies of macromolecular machines rather than as undifferentiated bags of enzymes. To date, our group has applied ECT to nearly 90 different bacterial species, collecting more than 15,000 cryotomograms. In addition to known structures, we have observed, to our knowledge, several uncharacterized features in these tomograms. Some are completely novel structures; others expand the features or species range of known structure types. Here, we present a survey of these uncharacterized bacterial structures in the hopes of accelerating their identification and study, and furthering our understanding of the structural complexity of bacterial cells.
IMPORTANCE Bacteria are more structurally complex than is commonly appreciated. Here we present a survey of previously uncharacterized structures that we observed in bacterial cells by electron cryotomography, structures that will initiate new lines of research investigating their identities and roles.
KEYWORDS: bacteria, electron cryotomography, bacterial ultrastructure, uncharacterized structures, electron microscopy
INTRODUCTION
The history of cell biology has been punctuated by advances in imaging technology. In particular, the development of electron microscopy (EM) in the 1930s produced a wealth of new information about the ultrastructure of cells (1). For the first time, the structure of cell envelopes, internal organelles, cytoskeletal filaments, and even large macromolecular complexes like ribosomes became visible. A further advance came in the 1980s and 1990s with the development of electron cryotomography (ECT) (2), which allows small cells to be imaged intact in 3D in a near-native, “frozen-hydrated” state to “macromolecular” (∼5 nm) resolution, without the limitations and artifacts of more traditional specimen-preparation methods (3).
ECT has helped reveal the previously unappreciated complexity of “simple” bacterial cells. Our group has been using ECT to study bacteria for more than a decade, generating more than 15,000 tomograms of 88 different species. These tomograms have revealed new insights into, among other things, the bacterial cytoskeleton (4–7), cell wall architecture (8, 9), morphogenesis (10), metabolism (11), motility (12–15), chemotaxis (16), sporulation (17), cell-cell interactions (18), and phage infection (19) (for a summary with more references from our and others' work, see reference 20).
A major hurdle in such studies is identifying the novel structures observed in tomograms. In some cases, we have identified structures by perturbing the abundance (either by knockout or overexpression) of candidate proteins (21). In others, we have used correlated light and electron microscopy (CLEM) to locate tagged proteins of interest (22, 23). In one striking example, we observed 12- and 15-nm-wide tubes in our tomograms of Vibrio cholerae cells. Ultimately, in collaboration with John Mekalanos' group, we identified them as type VI secretion systems (T6SS), which immediately led to the insight that the bacterial T6SS functions as a phage-tail-like, contractile molecular dagger (18).
Many other novel structures we have observed, though, remain unidentified. In some cases, we have published papers describing the novel structures seen in a particular species (e.g., references 24 and 25), but many have never been published. We therefore conducted a visual survey of the tomograms collected by our group, curated in the Caltech Tomography Database (26), as of 2015 and present here a catalog of previously undescribed bacterial structures. Some structures are, to our knowledge, completely novel; others belong to known types but present additional features or an expanded species range. We hope that sharing these images will help spur their identification and study, contributing to our expanding understanding of bacterial cell biology. In addition, we look forward to a future in which custom microbes are designed for diverse medical and industrial purposes; an expanded “parts list” of structures to be repurposed will aid in this effort.
RESULTS AND DISCUSSION
We performed a visual inspection of approximately 15,000 tomograms of intact frozen-hydrated cells belonging to 88 species and identified what we believed to be novel structures. A summary of the results of this survey is shown in Table S1 in the supplemental material, with features observed, species range, and frequency listed for each structure type. For full tomographic (3D) views of each feature, see reference 27; to view the figures in virtual reality, see reference 75.
Extracellular structures: external appendages.
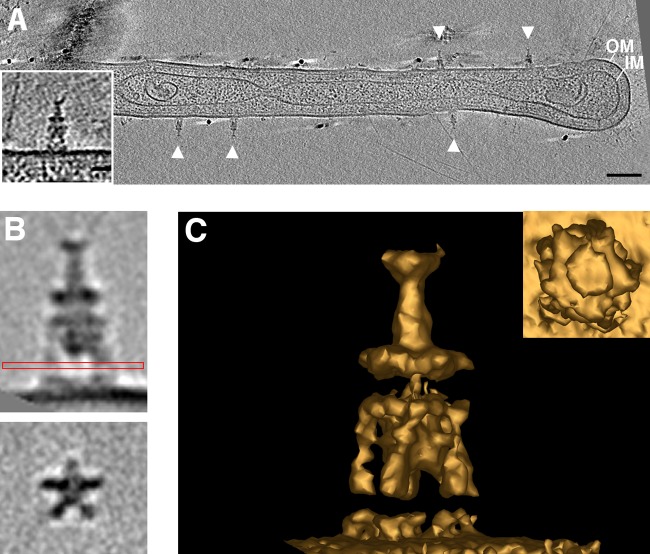
Prosthecobacter debontii is a bacterial species from the poorly studied phylum Verrucomicrobia. Each vibroid P. debontii cell possesses an appendage (prostheca), similar to the stalk of Caulobacter crescentus (28). In several tomograms of P. debontii, we observed novel extracellular appendages along the prosthecae, apparently attached to the cell membrane. Individual cells displayed up to 30 such appendages, which exhibited consistent sizes (∼20 nm wide and ∼50 nm long) and shapes (Fig. 1A). Subtomogram averaging of 105 particles revealed a distinctive structure. Extending outward from the cell membrane, five legs were attached to a disc, which in turn connected to a smaller disc and a long neck region (Fig. 1B and C). Individual particles showed that the structure culminated in two antenna-like filaments, which were likely lost in the average due to conformational variability. The appendages were observed in multiple cultures of the strain. While it remains unclear whether they originated intracellularly or extracellularly, no free-floating appendages were ever observed in the extracellular space. They may represent novel bacterial attachment organelles or appendages for nutrient acquisition, which has been proposed for a similar structure, the Caulobacter stalk. Under phosphate-limited conditions, Caulobacter grew elongated stalks (29). This increase in cell surface area with respect to cell volume was hypothesized to allow increased phosphate uptake (30). However, the presence of diffusion barriers challenges this view (31). The appendages could also be a novel secretion system (though we might expect a cell envelope spanning complex) or a novel bacteriophage (though there is a notable lack of a capsid-like density).
FIG 1.
Novel Prosthecobacter debontii appendages. Multiple external appendages (arrowheads) were observed by ECT on P. debontii prosthecae. (A) A central tomographic slice is shown, with a single appendage enlarged in the inset. Subtomogram averaging revealed the structure in more detail. (B) Side (top) and top (bottom) views show the characteristic disc-like densities and the five legs attaching to the cell surface. The red box shows which view was used to rotate the image 90° for the bottom image. (C) 3D isosurface of the average, seen from the side and top (inset). Bars, 50 nm (A) and 20 nm (inset).
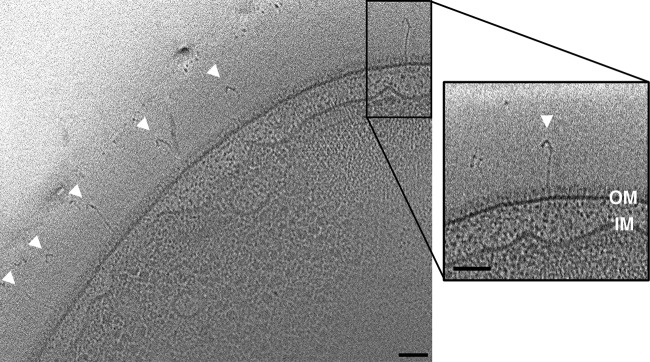
We observed a different novel extracellular appendage in tomograms of cell poles of Azospirillum brasilense, a plant growth-promoting bacterium of the Alphaproteobacteria class with a curved-rod morphology. Thin hooks were seen extending from the cell surface (Fig. 2). Individual cells exhibited dozens of hooks, each ∼3 nm wide and ∼75 nm long, associated with the outer membrane. The cells are large, and most tomograms included only the cell pole region, so while hooks were observed only at the pole region, they may also occur elsewhere on the cell. Hooks were seen in >90% of wild-type cells as well as in a strain in which the operon encoding the Che1 chemotaxis system was deleted. They were seen in ∼50% of cells in which the Che4 chemotaxis system operon was deleted, and none were seen in cells lacking both the Che1 and Che4 operons. A. brasilense is a well-studied plant growth-promoting bacterium. Cells attach to plant roots through a two-step process (32), a rapid reversible adsorption, thought to be mediated by the polar flagellum, and a slow irreversible anchoring, thought to be mediated by an as-yet unidentified surface polysaccharide (33). A recent study reported that mutants in components of the Che4 chemotaxis system are defective in this root colonization (34). A. brasilense cells also attach to conspecifics in the presence of elevated oxygen levels (35). Interestingly, it has been shown that mutants in components of the Che1 chemotaxis system form such attachments more rapidly than wild-type cells (36). The hooks we observed are vaguely reminiscent of the grappling hook-like structures that an archaeal species uses to anchor itself in biofilms (37), though those hooks were longer fibers with barbs. Those archaeal cells demonstrated very strong adhesion to a variety of surfaces as well as to each other. It is tempting, therefore, to speculate that the hooks shown here play a similar role in adhesion, to either other A. brasilense cells or plant roots.
FIG 2.
Novel Azospirillum brasilense hooks. Many hook-like structures were observed on the surface of A. brasilense cells. A central tomographic slice is shown, with arrowheads indicating hooks. A single hook is shown enlarged at right. Bars, 50 nm.
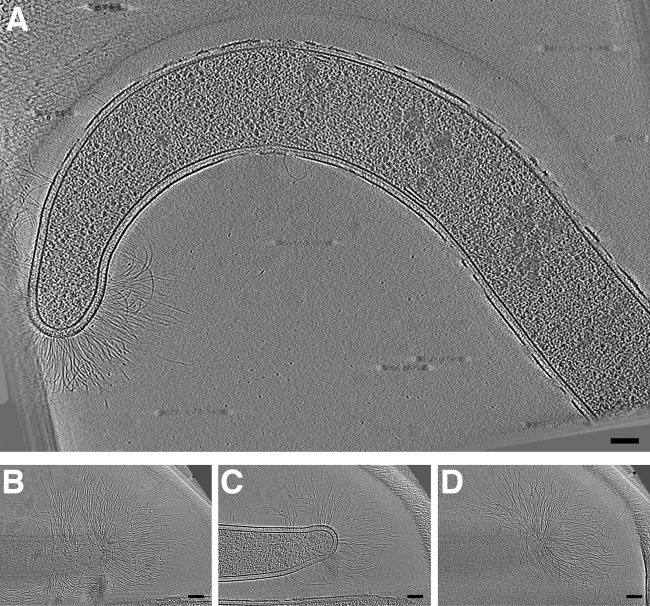
In cells of strain JT5 (a rod-shaped bacterium isolated from termite gut and related to the Dysgonomonas genus), we observed abundant fimbriae concentrated at the cell poles (Fig. 3). Fimbriae were also observed at the cell body but were much more concentrated at the cell poles. They were present in cells grown on cellulose or xylan, as well as under a starvation-inducing condition. Their width (∼4 nm), apparent flexibility, density on the cell envelope, and inhomogeneous distribution around the cell is consistent with curli, functional amyloids secreted by the type VIII secretion system that are involved in adhesion (38, 39). Curli systems are relatively divergent at the sequence level but are remarkably widespread phylogenetically, and the genes were reported to be present in Bacteroidetes (the phylum containing Dysgonomonas) (40). The appendages we observed in strain JT5 may therefore play a role in adhesion in the environment of the termite gut.
FIG 3.
Strain JT5 fimbriae. Examples are shown from two cells of strain JT5 (related to the Dysgonomonas genus) exhibiting abundant fimbrae at the cell pole. (A) Central slice revealing overall cell morphology; (B to D) slices at progressive z-heights through a cryotomogram of a cell of strain JT5 (related to the Dysgonomonas genus). Abundant fimbriae can be seen at the cell pole. Bars, 100 nm.
Intracellular structures. (i) Nanospheres.
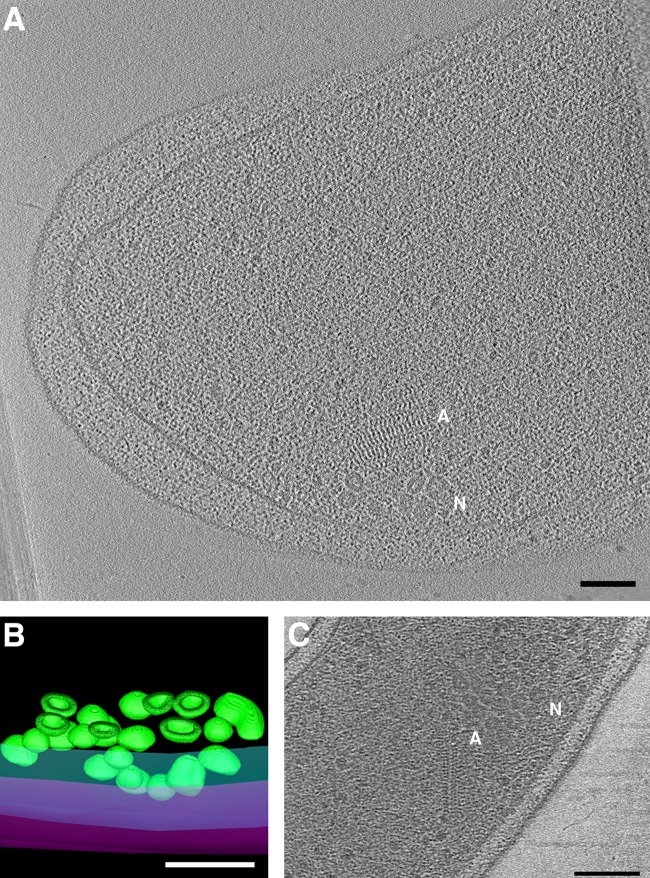
In two Vibrio cholerae cells (one from a C6706 lacZ mutant [41] and one from a ΔctxA ΔtcpB strain [15]) we observed clusters of nanospheres, hollow granules with thick walls (Fig. 4). The diameter of the nanospheres ranged from ∼18 to 37 nm, and the walls were ∼4 to 10 nm thick. They were pleomorphic; most were roughly spherical, but some were oblong or comma shaped. Each cluster contained about two dozen nanospheres. The clusters were observed at the cell periphery, near the inner membrane (although the clusters were large enough to extend to the center of the cell), and were always observed near a filament array structure (discussed below).
FIG 4.
Novel Vibrio cholerae nanospheres. Clusters of nanospheres were observed in two cryotomograms of V. cholerae cells (central slices shown in panels A and C). “N” indicates nanospheres; “A” indicates associated filament array. (B) Segmentation of the cluster seen in panel A, with outer and inner membranes in magenta and cyan, respectively, and nanospheres in green. A clipping plane cuts through the 3D segmentation revealing the thick walls and hollow centers of the nanospheres. Bars, 100 nm.
(ii) Filaments, bundles, arrays, chains and meshes.
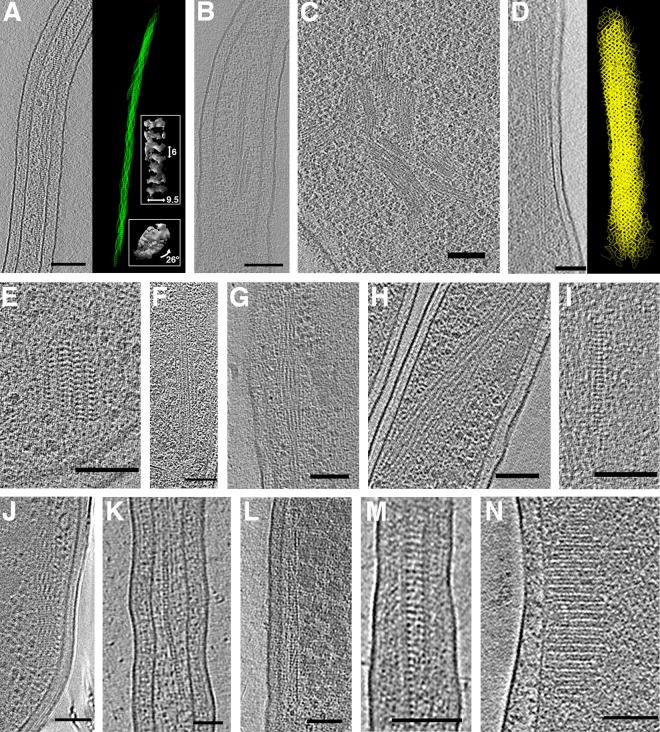
One of the strengths of ECT imaging is its power to resolve cytoskeletal elements in small bacterial cells. In addition to those we have already identified, we observed many novel filamentous structures in tomograms, including filament arrays, bundles, chains, and meshes (Fig. 5). In Hyphomonas neptunium, we observed long helical filament bundles in the prosthecae that connect dividing cells (Fig. 5A). The helix width was 9.5 ± 1.5 nm, the spacing between cross-densities 6.0 ± 0.3 nm, and the helical pitch was ∼26°. H. neptunium divides by asymmetric budding (42), and the genome of the parent cell is passed to the daughter cell through the narrow prostheca connecting the two cells (43). We observed that the helical structure was straightened in cells treated with ethidium bromide (an intercalator known to unwind DNA [44]) (Fig. 5B). We therefore propose that the helix is composed of supercoiled DNA, with each visible filament a DNA duplex connected to adjacent duplexes by cross-densities formed by an unidentified protein.
FIG 5.
Filament bundles, arrays, and chains. Hyphomonas neptunium division stalks contained helical bundles (A) that straightened when cells were treated with ethidium bromide (B). The right side of panel A shows a 3D segmentation of the helical bundle, with side and top views of subtomogram averaged insets. Labeled dimensions are in nanometers. (C) Large filament bundles in Helicobacter pylori. (D) A long mesh-like filament array in Vibrio cholerae, with segmentation at right. (E) A more typical V. cholerae filament array. (F to J) Filament arrays in Thiomonas intermedia (F), Hyphomonas neptunium (G), Hylemonella gracilis (H), Halothiobacillus neapolitanus c2 (I), and Mycobacterium smegmatis (J). (K) A chain in Prosthecobacter vanneervenii. (L to M) Filament arrays in Prosthecobacter debontii. (N) A filament array in a starved Campylobacter jejuni cell. Bars, 100 nm (A, B, D to J, L to N) and 50 nm (C and K).
In Helicobacter pylori cells, we observed extensive filament bundles. In one cell in an early stage of lysis, such bundles were observed throughout most of the cell (Fig. 5C). In V. cholerae we observed filament arrays resembling a honeycombed mesh (Fig. 5D). These arrays varied in length but were usually fairly short (∼100 nm in length and width), like the example shown in Fig. 5E. This is the structure we observed near the nanosphere clusters. Filament arrays exhibited different morphologies in other species. Thiomonas intermedia cells contained untwisted arrays that were ∼48 nm thick and ∼30 nm wide (Fig. 5F). In addition to the prosthecal helix described above, H. neptunium cells also contained a bundle of twisting filaments laddered by cross-densities (Fig. 5G). These bundles were ∼40 nm thick and ∼75 nm wide. In a Hylemonella gracilis cell we observed a helical bundle of filaments that varied in width and could be related to the nucleoid (Fig. 5H). In Halothiobacillus neapolitanus c2 cells grown in limited CO2 for several hours, we observed linear filament arrays with prominent cross-densities spaced 7 ± 0.8 nm apart (Fig. 5I). Mycobacterium smegmatis displayed straight arrays ∼80 nm thick and wide, comprising segments of pitched filaments (Fig. 5J). Filament arrays were also seen in multiple species of Prosthecobacter; P. vanneervenii contained linear chains (Fig. 5K), and one P. debontii cell contained a straight array similar to those observed in T. intermedia (Fig. 5L), as well as mesh-like arrays spanning the width of the prostheca (Fig. 5M).
In starving Campylobacter jejuni cells, we observed regular filament arrays (Fig. 5N). When subjected to environmental or cellular stress, some bacteria, including Escherichia coli, have been shown to reorganize their DNA into protective crystalline arrays (45). Since then, additional nucleoid associated proteins have been identified that organize DNA into higher order structures in stationary-phase or stress conditions (46, 47). The structures we observed in C. jejuni resemble those seen in E. coli cells overexpressing the protective DNA binding protein Dps (45) and may, therefore, represent such a nucleoprotein array.
Several other proteins have been shown to copolymerize with DNA into filaments for various functions, including RecA (homologous recombination) (48) and MuB from bacteriophage Mu (DNA transposition) (49). The width of such filaments in vitro (∼10 nm) is similar to widths we observed in cells; it is possible that some of the structures in Fig. 5 represent these DNA-related processes. Other bacterial proteins form filaments to regulate their function, and it has been suggested that this property may have been coopted in the evolution of the cytoskeleton (50). We previously observed such filaments of CTP synthase in tomograms of C. crescentus cells (21). Another protein, alcohol dehydrogenase, forms plaited filaments ∼10 nm wide, called spirosomes, in many bacteria capable of anaerobic metabolism (51, 52). It is possible that some of the filament arrays and chains that we observed in tomograms were filaments formed by these or other, yet uncharacterized, proteins.
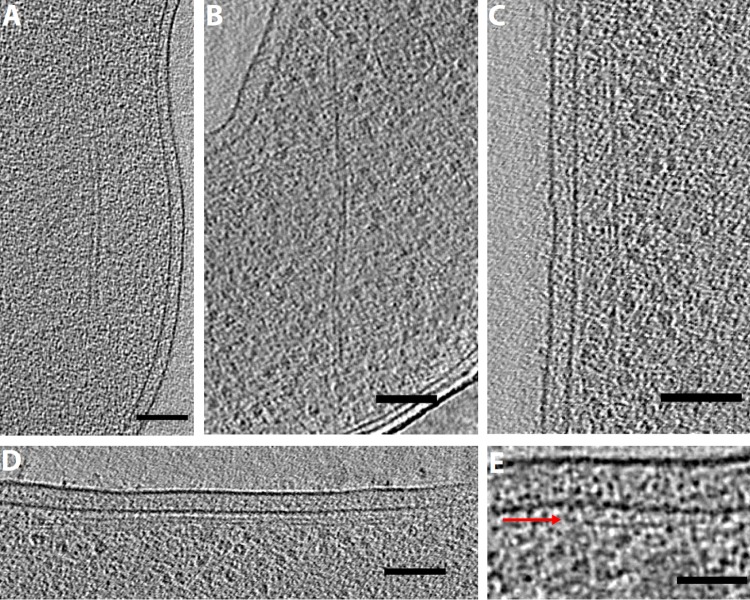
In addition to filament arrays and bundles, we observed individual or paired filaments in nearly every species imaged. Examples are shown in Fig. 6. (Note that due to their ubiquity, statistics are not included in Table S1 in the supplemental material.) Filaments were seen with various orientations in the cytoplasm (Fig. 6A to C), as well as running alongside the membrane (Fig. 6D and E). Consistent with our previous work (53), we did not observe any filaments immediately adjacent to the membrane as predicted by some studies of MreB (e.g., references 54 and 55). (Note that we did observe filaments corresponding to the known types of MamK [4, 56], FtsZ [5, 57], and bactofilins [58] but we do not show them here since they have already been characterized.) Paired filaments have been shown to function in plasmid segregation, so it is possible that some paired filaments that we observed were such ParM or TubZ structures (59, 60).
FIG 6.
Single and paired filaments. Tomographic slices showing paired filaments in Campylobacter jejuni (A) and Thiomicrospira crunogena (B) and membrane-aligned filaments in Shewanella putrefaciens (C), Prosthecobacter debontii (D), and Prosthecobacter fluviatilis (E) (the arrow shows a filament just under the inner membrane). Bars, 100 nm (A to D) and 50 nm (E).
(iii) Tubes.
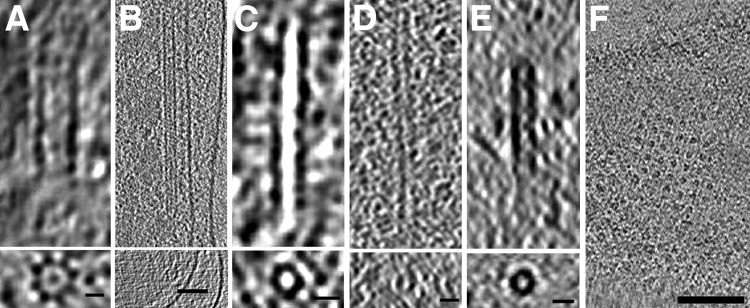
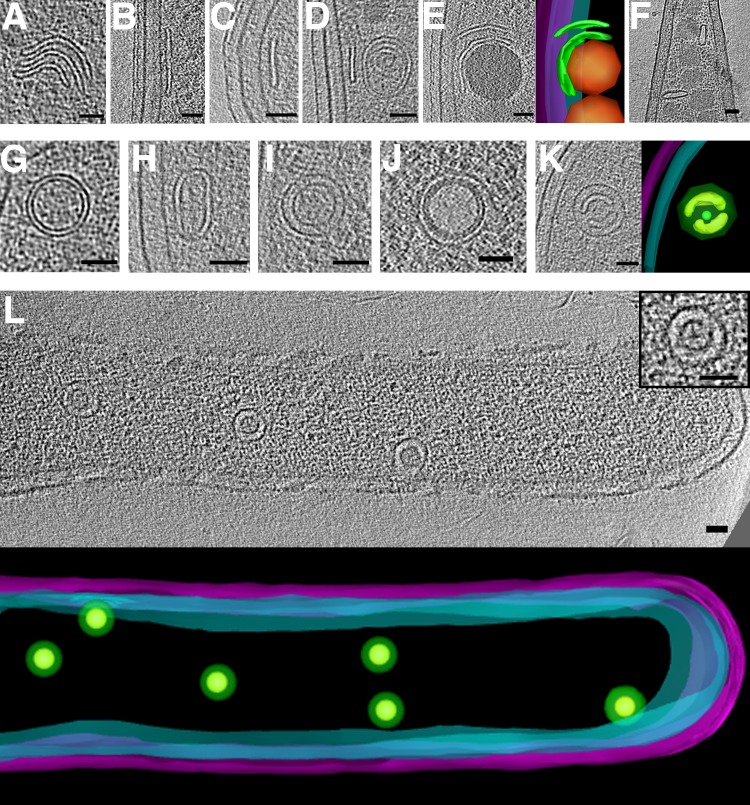
In addition to the known types of tubes we reported earlier, such as bacterial microtubules (6) and type VI secretion systems (18), we observed several novel tubular structures in bacterial cells (Fig. 7). In Thiomicrospira crunogena we found large tubes (18.6 ± 1.8 nm in diameter) containing eight outer protofilaments surrounding a central protofilament (Fig. 7A). H. neapolitanus c2 cells also contained large tubes (16.7 ± 0.7 nm in diameter) with a central filament (Fig. 7B). In several other species, we observed hollow tubes of varying dimensions: 8.9 ± 0.3 nm in diameter in Bdellovibrio bacteriovorus (Fig. 7C), 14.3 ± 1.7 nm in T. intermedia (Fig. 7D), and 8.3 ± 0.5 nm in H. neptunium (Fig. 7E). H. neptunium cells also contained many rings of similar diameter. In fact, we observed rings in many species, which could be assembly or disassembly intermediates of tubes.
FIG 7.
Tubes and rings. Tubes observed in Thiomicrospira crunogena (A), Halothiobacillus neapolitanus c2 (B), Bdellovibrio bacteriovorus (C), Thiomonas intermedia (D), and Hyphomonas neptunium (E). In each panel, tomographic slices show a side view (above) and a top view (bottom). (F) An array of rings observed in Helicobacter pylori. Bars, 10 nm (A, C, and E), 20 nm (B and D), and 100 nm (F).
In addition to isolated rings, in one case we observed an organized array of rings. One slightly lysed (a condition that flattens the cell and increases image quality) H. pylori cell contained a striking array of about two dozen evenly spaced rings near the cytoplasmic membrane (Fig. 7F). Each ring was ∼6 nm in diameter and ∼20 nm (center-to-center distance) from its neighbors in the square lattice.
(iv) Vesicles.
In contrast to eukaryotic cells, relatively little is known about membrane remodeling in bacteria. Compartmentalized cells in the Planctomycetes-Verrucomicrobia-Chlamydiae (PVC) superphylum have been shown to contain homologs of eukaryotic membrane trafficking proteins (61) and exhibit endocytosis-like protein uptake (62). An additional potential membrane-remodeling system based on FtsZ homologues is more widespread across bacteria, but its function remains unknown (63).
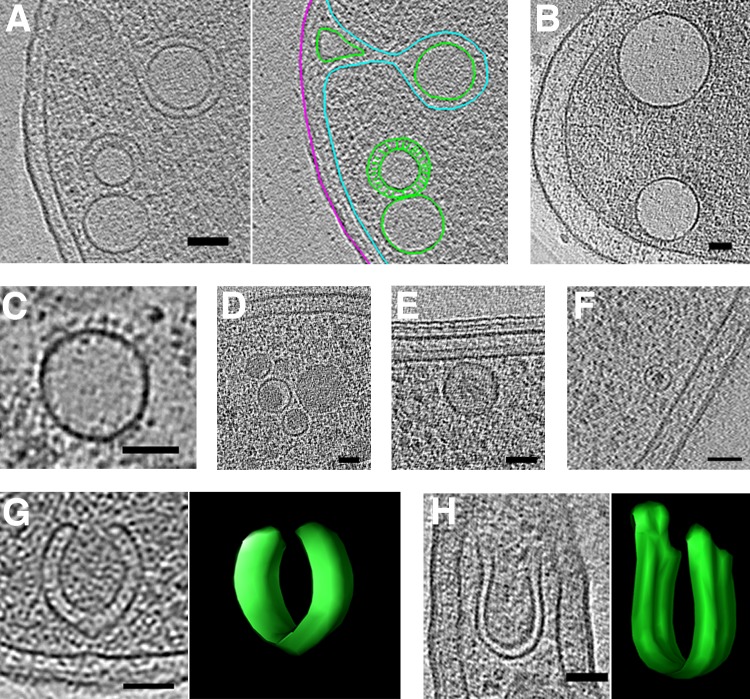
Despite this limited evidence for membrane remodeling in bacteria, we observed intracellular vesicles in nearly every species imaged. They exhibited various sizes, shapes, membrane layers, and contents and were frequently found near the cytoplasmic membrane. Figure 8 shows examples of round and horseshoe-shaped vesicles. Round vesicles were found in nearly every species imaged; therefore, no statistics for them are compiled in Table S1 in the supplemental material. Most round vesicles were empty (density similar to background [e.g., Fig. 8A to C]). One of these vesicles, observed in a lysed cell (improving clarity by reducing cytoplasmic crowding), exhibited regularly spaced protein densities around its exterior (Fig. 8C). Others were at least partially filled with denser material (e.g., Fig. 8D to F). In two Myxococcus xanthus cells overexpressing a fluorescent fusion of a periplasmic protein (PilP-sfGFP), we observed round vesicles containing a dense amorphous core (Fig. 8F). These could be a novel form of membrane-bound inclusion bodies, perhaps packaged from the periplasm. In eight species, we observed horseshoe-shaped vesicles (Fig. 8G and H).
FIG 8.
Round and horseshoe-shaped vesicles. Tomographic slices showing examples of round vesicles in Escherichia coli (A) (segmentation shown at right), Helicobacter pylori (B), Helicobacter hepaticus (C), Myxococcus xanthus (D), Caulobacter crescentus (E), and Myxococcus xanthus overexpressing PilP-sfGFP (F). Examples of horseshoe-shaped vesicles in Ralstonia eutropha (G) and Prosthecobacter fluviatilis (H), with 3D segmentations shown at right. In the segmentation in panel A, outer and inner membranes are in magenta and cyan, respectively, and vesicles are in green. Bars, 50 nm.
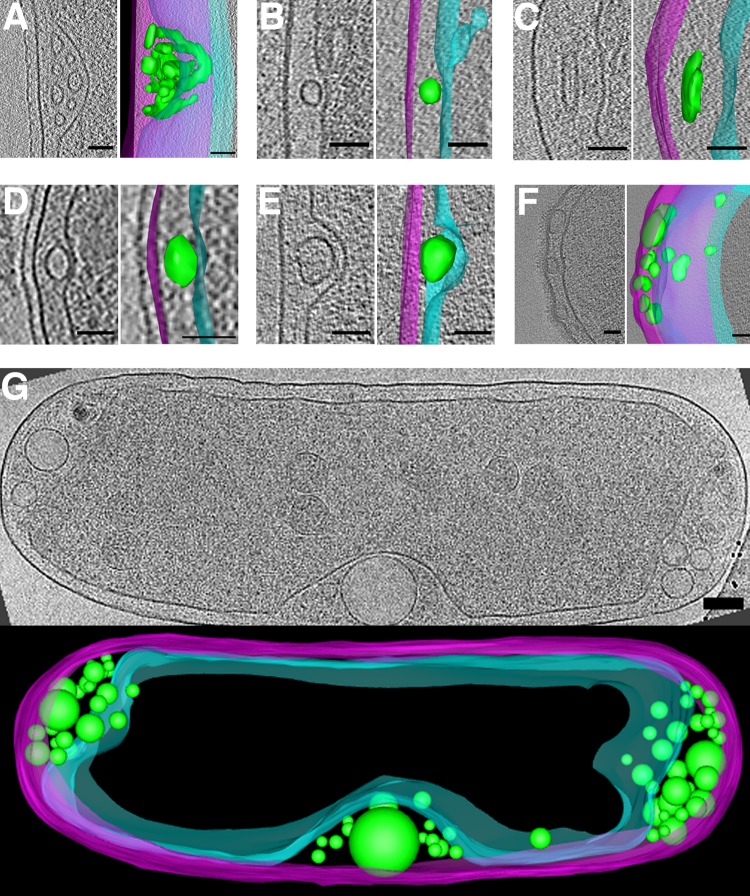
Flattened vesicles (Fig. 9A to F) were less common than round vesicles, and were usually observed near membranes or wrapping around storage granules (Fig. 9E), suggesting a possible functional relationship. Flattened vesicles were usually empty. One T. intermedia cell contained a stack of flattened vesicles (Fig. 9A). Flattened vesicles were particularly prevalent in C. crescentus cells (Fig. 9B to E). P. debontii cells contained flattened vesicles that neither ran along the membrane nor wrapped around granules (Fig. 9F). Since the lowest energy shape of a liposome is a sphere, it is likely that the vesicles were flattened by cytoplasmic pressure or some other constraint such as an associated protein.
FIG 9.
Flattened and nested vesicles. Examples of flattened vesicles in Thiomonas intermedia (A), Caulobacter crescentus (B to E), and Prosthecobacter debontii (F). Note storage granules in panels E and F, shown in orange in the segmentation panel E (G to L). Examples of nested vesicles in Serpens flexibilis (G), Caulobacter crescentus (H), Borrelia burgdorferi (I), Vibrio cholerae (J), Caulobacter crescentus with segmentation (K), and strain JT5 (L). (L, inset) Shows an enlargement of central vesicle, and a 3D segmentation of the visible portion of the cell is shown below. In segmentations, outer and inner membranes are shown in magenta and cyan, respectively, and vesicles are in green. Bars, 50 nm.
Many cells contained nested vesicles, with diverse sizes and shapes, as well as subcellular locations (Fig. 9G to L). In some nested vesicles, densities were observed bridging the inner and outer membranes (Fig. 8A and 9G and H). Cells of strain JT5 exhibited multiple nested vesicles of uniform shape and size (Fig. 9L).
We also observed periplasmic vesicles in many species (Fig. 10). They were typically empty and exhibited great variability in size, shape, and abundance. In some cases, they were even seen to form branching networks (Fig. 10A). As with cytoplasmic vesicles, they were most abundant in cells showing signs of stress (rupture of inner or outer membrane, separation of inner and outer membrane, lysis, or membrane blebbing).
FIG 10.
Periplasmic vesicles. Examples of periplasmic vesicles in Caulobacter crescentus (A), Helicobacter pylori (Β), Brucella abortus (C), Thiomonas intermedia (D), Hyphomonas neptunium (E), Myxococcus xanthus (F), and Halothiobacillus neapolitanus c2 (G). In each panel, a central tomographic slice is shown, as well as a segmentation with outer and inner membranes in magenta and cyan, respectively, and vesicles are in green. Bars, 50 nm (A to F) and 100 nm (G).
Conclusions.
Here we present the results of a survey of, to our knowledge, uncharacterized bacterial structures that we have observed in our work over the past 10 or more years. We hope that further study will identify them and their functions. Already, they signal the wealth of complexity still to be discovered in bacterial cells.
MATERIALS AND METHODS
Strains and growth.
Unless otherwise noted, bacterial strains were wild type and grown in species-standard medium and conditions to mid-log or early stationary phase. Azospirillum brasilense cultures were switched to nitrogen-free medium for ∼16 h prior to imaging to induce nitrogen fixation and digestion of storage granules that decrease image quality. Predatory Bdellovibrio bacteriovorus cells were cocultured with Vibrio cholerae strain MKW1383. Helicobacter pylori cells were cultured with human gastric carcinoma cells. Vibrio cholerae and Borrelia burgdorferi were grown according to conditions in references 64 and 65). E. coli were grown according to conditions in references 16 and 66). Caulobacter crescentus were grown according to conditions in reference 67. Prosthecobacters were all grown according to conditions in reference 6. Hyphomonas neptunium was grown according to conditions in reference 68.
In all cases, samples of cells in growth medium were mixed with BSA-treated 10 nm colloidal gold fiducials (Sigma), applied to glow-discharged EM grids (Quantifoil), and plunge-frozen in a liquid ethane-propane mixture (69). Grids were maintained at liquid nitrogen temperature throughout storage, transfer, and imaging.
Electron cryotomography.
The references to growing conditions above also provide specific data collection settings. Generally, plunge-frozen samples were imaged using either a Polara or Titan Krios 300 kV FEG transmission electron microscope (FEI Company) equipped with an energy filter (Gatan). Images were recorded using either a lens-coupled 4,000 by 4,000 UltraCam CCD (Gatan) or a K2 Summit direct electron detector (Gatan). Tilt-series were recorded from −60° to +60° in 1 to 2° increments, with defoci of ∼6 to 12 μm and a cumulative dose of ∼100 to 200 e−/Å2. Tilt-series were acquired automatically using either Leginon (70) or UCSF Tomography (71) software. Tomographic reconstructions were calculated using either the IMOD software package (72) or Raptor (73). 3D segmentations and movies were produced with IMOD (72). Subtomogram averages were calculated using PEET software (74).
Supplementary Material
ACKNOWLEDGMENTS
We thank our collaborators who provided strains for imaging: Andrew Camilli (Streptococcus pneumoniae), Eric Matson (strain JT5), Gladys Alexandre (Azospirillum brasilense mutants), Lotte Søgaard-Andersen, Simon Ringgaard, and Matthew K. Waldor (Vibrio cholerae wild type and mutants), Michael Marletta (Shewanella putrefaciens), and Gordon Cannon and Sabine Heinhorst (Halothiobacillus neapolitanus and Thiomonas intermedia). We also thank members of the Jensen lab for their helpful discussions.
We declare no competing interests.
M.J.D. and G.J.J. conceived the idea for the study; M.J.D., C.M.O., A.P., J.C., K.G., T.J., J.T., J.D., Y.P., A.K., A.I.J., M.P., S.C., E.I.T., Y.-W.C., A.B., J.S., Z.L., P.S., C.V.I., B.A.S., and A.W.M. performed formal analysis and investigation; M.J.D. and C.M.O. wrote and prepared the original article draft; M.J.D., C.M.O., and G.J.J. wrote, reviewed, and edited the final article draft; M.J.D. and G.J.J. acquired funding; G.J.J. provided resources; and M.J.D. and G.J.J. supervised the study.
This work was supported by the Hampshire College Dr. Lucy fund and the Collaborative Modeling Center, NIH grant R01 AI27401 (to G.J.J.), the Beckman Institute at Caltech, the Gordon and Betty Moore Foundation, the Human Frontier Science Program, the Howard Hughes Medical Institute, and the John Templeton Foundation as part of the Boundaries of Life project.
The opinions expressed in this publication are those of the authors and do not necessarily reflect the views of the John Templeton Foundation.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00100-17.
REFERENCES
- 1.Ruska E. 1987. Nobel lecture. The development of the electron microscope and of electron microscopy. Biosci Rep 7:607–629. [DOI] [PubMed] [Google Scholar]
- 2.Koster AJ, Grimm R, Typke D, Hegerl R, Stoschek A, Walz J, Baumeister W. 1997. Perspectives of molecular and cellular electron tomography. J Struct Biol 120:276–308. doi: 10.1006/jsbi.1997.3933. [DOI] [PubMed] [Google Scholar]
- 3.Pilhofer M, Ladinsky MS, McDowall AW, Jensen GJ. 2010. Bacterial TEM: new insights from cryo-microscopy. Methods Cell Biol 96:21–45. doi: 10.1016/S0091-679X(10)96002-0. [DOI] [PubMed] [Google Scholar]
- 4.Komeili A, Li Z, Newman DK, Jensen GJ. 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- 5.Li Z, Trimble MJ, Brun YV, Jensen GJ. 2007. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J 26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pilhofer M, Ladinsky MS, McDowall AW, Petroni G, Jensen GJ. 2011 Microtubules in bacteria: ancient tubulins build a five-protofilament homolog of the eukaryotic cytoskeleton. PLoS Biol 9:e1001213. doi: 10.1371/journal.pbio.1001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swulius MT, Jensen GJ. 2012. The helical MreB cytoskeleton in Escherichia coli MC1000/pLE7 is an artifact of the N-Terminal yellow fluorescent protein tag. J Bacteriol 194:6382–6386. doi: 10.1128/JB.00505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gan L, Chen S, Jensen GJ. 2008. Molecular organization of Gram-negative peptidoglycan. Proc Natl Acad Sci U S A 105:18953–18957. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beeby M, Gumbart JC, Roux B, Jensen GJ. 2013. Architecture and assembly of the Gram-positive cell wall. Mol Microbiol 88:664–672. doi: 10.1111/mmi.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. 2008. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell 134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, Martino A, Jensen GJ. 2007. The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J Mol Biol 372:764–773. doi: 10.1016/j.jmb.2007.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy GE, Leadbetter JR, Jensen GJ. 2006. In situ structure of the complete Treponema primitia flagellar motor. Nature 442:1062–1064. doi: 10.1038/nature05015. [DOI] [PubMed] [Google Scholar]
- 13.Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, Li Z, Shi J, Tocheva EI, Muller A, Dobro MJ, Jensen GJ. 2011. Structural diversity of bacterial flagellar motors. EMBO J 30:2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abrusci P, Vergara-Irigaray M, Johnson S, Beeby MD, Hendrixson DR, Roversi P, Friede ME, Deane JE, Jensen GJ, Tang CM, Lea SM. 2013. Architecture of the major component of the type III secretion system export apparatus. Nat Struct Mol Biol 20:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang YW, Rettberg LA, Treuner-Lange A, Iwasa J, Sogaard-Andersen L, Jensen GJ. 2016. Architecture of the type IVa pilus machine. Science 351:aad2001. doi: 10.1126/science.aad2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. 2012. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci U S A 109:3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tocheva EI, Matson EG, Morris DM, Moussavi F, Leadbetter JR, Jensen GJ. 2011. Peptidoglycan remodeling and conversion of an inner membrane into an outer membrane during sporulation. Cell 146:799–812. doi: 10.1016/j.cell.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerrero-Ferreira RC, Viollier PH, Ely B, Poindexter JS, Georgieva M, Jensen GJ, Wright ER. 2011. Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus. Proc Natl Acad Sci U S A 108:9963–9968. doi: 10.1073/pnas.1012388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oikonomou CM, Jensen GJ. 2016. A new view into prokaryotic cell biology from electron cryotomography. Nat Rev Microbiol 14:205–220. doi: 10.1038/nrmicro.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. 2010. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat cell biol 12:739–746. doi: 10.1038/ncb2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Briegel A, Ding HJ, Li Z, Werner J, Gitai Z, Dias DP, Jensen RB, Jensen GJ. 2008. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol Microbiol 69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang YW, Chen S, Tocheva EI, Treuner-Lange A, Lobach S, Sogaard-Andersen L, Jensen GJ. 2014. Correlated cryogenic photoactivated localization microscopy and cryo-electron tomography. Nat Methods 11:737–739. doi: 10.1038/nmeth.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy GE, Matson EG, Leadbetter JR, Berg HC, Jensen GJ. 2008. Novel ultrastructures of Treponema primitia and their implications for motility. Mol Microbiol 67:1184–1195. doi: 10.1111/j.1365-2958.2008.06120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller A, Beeby M, McDowall AW, Chow J, Jensen GJ, Clemons WM Jr. 2014. Ultrastructure and complex polar architecture of the human pathogen Campylobacter jejuni. Microbiologyopen 3:702–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding HJ, Oikonomou CM, Jensen GJ. 2015. The caltech tomography database and automatic processing pipeline. J Struct Biol 192:279–286. doi: 10.1016/j.jsb.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobro MJ, Piper A. 2017. Uncharacterized bacterial structures revealed by electron cryotomography. figshare https://figshare.com/s/782461843c3150d27cfa Retrieved 23 June 2017. [DOI] [PMC free article] [PubMed]
- 28.Staley JT, Bont JA, Jonge K. 1976. Prosthecobacter fusiformis nov. gen. et sp., the fusiform caulobacter. Antonie Van Leeuwenhoek 42:333–342. doi: 10.1007/BF00394132. [DOI] [PubMed] [Google Scholar]
- 29.Gonin M, Quardokus EM, O'Donnol D, Maddock J, Brun YV. 2000. Regulation of stalk elongation by phosphate in. Caulobacter crescentus. J Bacteriol 182:337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner JK, Setayeshgar S, Sharon LA, Reilly JP, Brun YV. 2006. A nutrient uptake role for bacterial cell envelope extensions. Proc Natl Acad sci U S A 103:11772–11777. doi: 10.1073/pnas.0602047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlimpert S, Klein EA, Briegel A, Hughes V, Kahnt J, Bolte K, Maier UG, Brun YV, Jensen GJ, Gitai Z, Thanbichler M. 2012. General protein diffusion barriers create compartments within bacterial cells. Cell 151:1270–1282. doi: 10.1016/j.cell.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Troch P, Vanderleyden J. 1996. Surface Properties and motility of rhizobium and azospirillum in relation to plant root attachment. Microb Ecol 32:149–169. doi: 10.1007/BF00185885. [DOI] [PubMed] [Google Scholar]
- 33.Steenhoudt O, Vanderleyden J. 2000. Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects. FEMS Microbiol Rev 24:487–506. doi: 10.1111/j.1574-6976.2000.tb00552.x. [DOI] [PubMed] [Google Scholar]
- 34.Mukherjee T, Kumar D, Burriss N, Xie Z, Alexandre G. 2016. Azospirillum brasilense chemotaxis depends on two signaling pathways regulating distinct motility parameters. J Bacteriol 198:1764–1772. doi: 10.1128/JB.00020-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bible AN, Khalsa-Moyers GK, Mukherjee T, Green CS, Mishra P, Purcell A, Aksenova A, Hurst GB, Alexandre G. 2015. Metabolic adaptations of Azospirillum brasilense to oxygen stress by cell-to-cell clumping and flocculation. Appl Environ Microbiol 81:8346–8357. doi: 10.1128/AEM.02782-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bible A, Russell MH, Alexandre G. 2012. The Azospirillum brasilense Che1 chemotaxis pathway controls swimming velocity, which affects transient cell-to-cell clumping. J Bacteriol 194:3343–3355. doi: 10.1128/JB.00310-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moissl C, Rachel R, Briegel A, Engelhardt H, Huber R. 2005. The unique structure of archaeal ‘hami’, highly complex cell appendages with nano-grappling hooks. Mol Microbiol 56:361–370. doi: 10.1111/j.1365-2958.2005.04294.x. [DOI] [PubMed] [Google Scholar]
- 38.Epstein EA, Reizian MA, Chapman MR. 2009. Spatial clustering of the curlin secretion lipoprotein requires curli fiber assembly. J Bacteriol 191:608–615. doi: 10.1128/JB.01244-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Gerven N, Klein RD, Hultgren SJ, Remaut H. 2015. Bacterial amyloid formation: Structural insights into curli biogensis. Trends Microbiol 23:693–706. doi: 10.1016/j.tim.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dueholm MS, Albertsen M, Otzen D, Nielsen PH. 2012. Curli functional amyloid systems are phylogenetically widespread and display large diversity in operon and protein structure. PLoS One 7:e51274. doi: 10.1371/journal.pone.0051274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cameron DE, Urbach JM, Mekalanos JJ. 2008. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc Natl Acad Sci U S A 105:8736–8741. doi: 10.1073/pnas.0803281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiner RM, Melick M, O'Neill K, Quintero E. 2000. Hyphomonas adhaerens sp. nov., Hyphomonas johnsonii sp. nov. and Hyphomonas rosenbergii sp. nov., marine budding and prosthecate bacteria. Int J Syst Evol Microbiol 50 Part 2:459–469. [DOI] [PubMed] [Google Scholar]
- 43.Zerfas PM, Kessel M, Quintero EJ, Weiner RM. 1997. Fine-structure evidence for cell membrane partitioning of the nucleoid and cytoplasm during bud formation in Hyphomonas species. J Bacteriol 179:148–156. doi: 10.1128/jb.179.1.148-156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pommier Y, Covey JM, Kerrigan D, Markovits J, Pham R. 1987. DNA unwinding and inhibition of mouse leukemia L1210 DNA topoisomerase I by intercalators. Nucleic Acids Res 15:6713–6731. doi: 10.1093/nar/15.16.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolf SG, Frenkiel D, Arad T, Finkel SE, Kolter R, Minsky A. 1999. DNA protection by stress-induced biocrystallization. Nature 400:83–85. doi: 10.1038/21918. [DOI] [PubMed] [Google Scholar]
- 46.Teramoto J, Yoshimura SH, Takeyasu K, Ishihama A. 2010. A novel nucleoid protein of Escherichia coli induced under anaerobiotic growth conditions. Nucleic Acids Res 38:3605–3618. doi: 10.1093/nar/gkq077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim CJ, Lee SY, Teramoto J, Ishihama A, Yan J. 2013. The nucleoid-associated protein Dan organizes chromosomal DNA through rigid nucleoprotein filament formation in E. coli during anoxia. Nucleic Acids Res 41:746–753. doi: 10.1093/nar/gks1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Egelman EH, Stasiak A. 1986. Structure of helical RecA-DNA complexes. Complexes formed in the presence of ATP-gamma-S or ATP. J Mol Biol 191:677–697. [DOI] [PubMed] [Google Scholar]
- 49.Mizuno N, Dramicanin M, Mizuuchi M, Adam J, Wang Y, Han YW, Yang W, Steven AC, Mizuuchi K, Ramon-Maiques S. 2013. MuB is an AAA+ ATPase that forms helical filaments to control target selection for DNA transposition. Proc Natl Acad Sci U S A 110:E2441–E2450. doi: 10.1073/pnas.1309499110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barry RM, Gitai Z. 2011. Self-assembling enzymes and the origins of the cytoskeleton. Curr Opin Microbiol 14:704–711. doi: 10.1016/j.mib.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matayoshi S, Oda H, Sarwar G. 1989. Relationship between the production of spirosomes and anaerobic glycolysis activity in Escherichia coli B. J Gen Microbiol 135:525–529. [DOI] [PubMed] [Google Scholar]
- 52.Laurenceau R, Krasteva PV, Diallo A, Ouarti S, Duchateau M, Malosse C, Chamot-Rooke J, Fronzes R. 2015. Conserved Streptococcus pneumoniae spirosomes suggest a single type of transformation pilus in competence. PLoS Pathog 11:e1004835. doi: 10.1371/journal.ppat.1004835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swulius MT, Chen S, Jane Ding H, Li Z, Briegel A, Pilhofer M, Tocheva EI, Lybarger SR, Johnson TL, Sandkvist M, Jensen GJ. 2011. Long helical filaments are not seen encircling cells in electron cryotomograms of rod-shaped bacteria. Biochem Biophys Res Commun 407:650–655. doi: 10.1016/j.bbrc.2011.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones LJ, Carballido-Lopez R, Errington J. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913–922. doi: 10.1016/S0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 55.Shih YL, Le T, Rothfield L. 2003. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci U S A 100:7865–7870. doi: 10.1073/pnas.1232225100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schuler D. 2006. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 440:110–114. doi: 10.1038/nature04382. [DOI] [PubMed] [Google Scholar]
- 57.Szwedziak P, Wang Q, Bharat TA, Tsim M, Lowe J. 2014. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife 9:e04601. doi 10.7554/elife.04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuhn J, Briegel A, Morschel E, Kahnt J, Leser K, Wick S, Jensen GJ, Thanbichler M. 2010. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J 29:327–339. doi: 10.1038/emboj.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aylett CH, Wang Q, Michie KA, Amos LA, Lowe J. 2010. Filament structure of bacterial tubulin homologue TubZ. Proc Natl Acad Sci U S A 107:19766–19771. doi: 10.1073/pnas.1010176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bharat TA, Murshudov GN, Sachse C, Lowe J. 2015. Structures of actin-like ParM filaments show architecture of plasmid-segregating spindles. Nature 523:106–110. doi: 10.1038/nature14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santarella-Mellwig R, Franke J, Jaedicke A, Gorjanacz M, Bauer U, Budd A, Mattaj IW, Devos DP. 2010. The compartmentalized bacteria of the Planctomycetes-Verrucomicrobia-Chlamydiae superphylum have membrane coat-like proteins. PLoS biology 8:e1000281. doi: 10.1371/journal.pbio.1000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lonhienne TG, Sagulenko E, Webb RI, Lee KC, Franke J, Devos DP, Nouwens A, Carroll BJ, Fuerst JA. 2010. Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci U S A 107:12883–12888. doi: 10.1073/pnas.1001085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Makarova KS, Koonin EV. 2010. Two new families of the FtsZ-tubulin protein superfamily implicated in membrane remodeling in diverse bacteria and archaea. Biol Direct 5:33. doi: 10.1186/1745-6150-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Müller A, Iancu CV, Murphy GE, Dobro MJ, Zhulin IB, Jensen GJ. 2009. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci U S A 106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Briegel A, Ortega DR, Mann P, Kjær A, Ringgaard S, Jensen GJ. 2016. Chemotaxis cluster 1 proteins form cytoplasmic arrays in Vibrio cholerae and are stabilized by a double signaling domain receptor DosM. Proc Natl Acad Sci U S A 113:10412–10417. doi: 10.1073/pnas.1604693113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Briegel A, Ames P, Gumbart JC, Oikonomou CM, Parkinson JS, Jensen GJ. 2013. The mobility of two kinase domains in the Escherichia coli chemoreceptor array varies with signalling state. Mol Microbiol 89:831–841. doi: 10.1111/mmi.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Briegel A, Beeby M, Thanbichler M, Jensen GJ. 2011. Activated chemoreceptor arrays remain intact and hexagonally packed. Mol Microbiol 82:748–757. doi: 10.1111/j.1365-2958.2011.07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cserti E, Rosskopf S, Chang YW, Eisheuer S, Selter L, Shi J, Regh C, Koert U, Jensen GJ, Thanbichler M. 2017. Dynamics of the peptidoglycan biosynthetic machinery in the stalked budding bacterium Hyphomonas neptunium. Mol Microbiol 103:875–895. doi: 10.1111/mmi.13593. [DOI] [PubMed] [Google Scholar]
- 69.Tivol WF, Briegel A, Jensen GJ. 2008. An improved cryogen for plunge freezing. Microsc Microanal 14:375–379. doi: 10.1017/S1431927608080781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suloway C, Shi J, Cheng A, Pulokas J, Carragher B, Potter CS, Zheng SQ, Agard DA, Jensen GJ. 2009. Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol 167:11–18. doi: 10.1016/j.jsb.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng SQ, Keszthelyi B, Branlund E, Lyle JM, Braunfeld MB, Sedat JW, Agard DA. 2007. UCSF tomography: an integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. J Struct Biol 157:138–147. doi: 10.1016/j.jsb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 72.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 73.Amat F, Moussavi F, Comolli LR, Elidan G, Downing KH, Horowitz M. 2008. Markov random field based automatic image alignment for electron tomography. J Struct Biol 161:260–275. doi: 10.1016/j.jsb.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 74.Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. 2006. The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 75.Piper AT. 2017. Cell visualization VR. https://play.google.com/store/apps/details?id=com.BishopVisual.Mk2&rdid=com.BishopVisual.Mk2 Retrieved 23 June 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.