Abstract
Extended-spectrum beta-lactamases (ESBLs), AmpC-type beta-lactamases (ACBLs) and carbapenemases are among the most important resistance mechanisms in Enterobacteriaceae. This study investigated the presence of these resistance mechanisms in consecutive non-replicate isolates of Escherichia coli (n = 2,352), Klebsiella pneumoniae (n = 697), and Proteus mirabilis (n = 275) from an Italian nationwide cross-sectional survey carried out in October 2013. Overall, 15.3% of isolates were non-susceptible to extended-spectrum cephalosporins but susceptible to carbapenems (ESCR-carbaS), while 4.3% were also non-susceptible to carbapenems (ESCR-carbaR). ESCR-carbaS isolates were contributed by all three species, with higher proportions among isolates from inpatients (20.3%) but remarkable proportions also among those from outpatients (11.1%). Most ESCR-carbaS isolates were ESBL-positive (90.5%), and most of them were contributed by E. coli carrying blaCTX-M group 1 genes. Acquired ACBLs were less common and mostly detected in P. mirabilis. ESCR-carbaR isolates were mostly contributed by K. pneumoniae (25.1% and 7.7% among K. pneumoniae isolates from inpatients and outpatients, respectively), with blaKPC as the most common carbapenemase gene. Results showed an increasing trend for both ESBL and carbapenemase producers in comparison with previous Italian surveys, also among outpatients.
Keywords: ESBL, carbapenemase, epidemiology, outpatients
Introduction
Enterobacteriaceae are the most common cause of healthcare associated infections, and beta-lactams are among the most used antibiotics in clinical practice for treatment of these infections [1,2]. During the last decades, Enterobacteriaceae with decreased susceptibility to beta-lactams have been increasingly reported worldwide, causing major problems [3-8].
The most important resistance mechanism to beta-lactams in Enterobacteriaceae is the production of beta-lactamases, and the most challenging enzymes of this family are the extended-spectrum beta-lactamases (ESBLs), the AmpC-type beta-lactamases (ACBLs), and the carbapenemases [9-12]. ESBLs are able to hydrolyse a wide range of beta-lactams, including penicillins, narrow- and extended-spectrum cephalosporins and monobactams, but not cephamycins and carbapenems. ESBLs are usually inhibited by conventional beta-lactamase inhibitors. CTX-M-type ESBLs, which emerged in the late 1980s, and demonstrated a high ability to disseminate in clinical settings, rapidly reaching a pandemic diffusion. Nowadays they are the most prevalent plasmid-encoded ESBLs overall [13,14]. Acquired ACBLs can confer resistance to penicillins and to most cephalosporins (including cephamycins) but are poorly or not active against monobactams and the zwitterionic oxyimino-cephalosporins, such as cefepime and cefpirome, and the carbapenems. ACBLs are generally less prevalent than ESBLs in Enterobacteriaceae but are nonetheless important for their contribution to beta-lactam resistance, which can be extended also to carbapenems when ACBLs are overproduced in combination with an impermeability defect [10,15,16]. Carbapenemase production is the leading resistance mechanism to carbapenems in Enterobacteriaceae. Acquired carbapenemases of the KPC-, VIM-, NDM- and OXA-48-types are the most prevalent, although with a notable geographical variability [6,17-20].
In Italy, the most recent data from the European Antimicrobial Resistance Surveillance Network (EARS-Net) reported very high proportions of resistance to extended-spectrum cephalosporins among Escherichia coli (30.1%) and Klebsiella pneumoniae (55.9%), and of resistance to carbapenems among K. pneumoniae (33.5%) [21]. However, the EARS-NET data neither cover other enterobacterial species nor describe the resistance mechanisms.
This survey, promoted by the Committee for Study of Antibiotics (CoSA) of the Italian Society of Clinical Microbiologists (AMCLI), was carried out to provide an updated picture of the molecular epidemiology of ESBL- and carbapenemase-producing Enterobacteriaceae circulating in Italy and to investigate, for the first time, the presence of ACBL producers on a nationwide scale.
Methods
Study design
Fourteen clinical microbiology laboratories from 13 Italian cities participated in the study (Figure 1).
Figure 1.
Distribution of the centers participating in the survey, Italy, October 2013 (n=14)
For each centre the presence of CTX-M extended-spectrum beta-lactamase and carbapenemase genes is also indicated.
Centers listed as follows: 1-Milano; 2-Lecco; 3-Novara; 4-San Remo; 5-Bolzano; 6-Udine; 7-Modena Bg; 8-Modena Pc; 9-Firenze; 10-Ancona; 11-Roma; 12-San Giovanni Rotondo; 13-Avellino; 14-Cosenza.
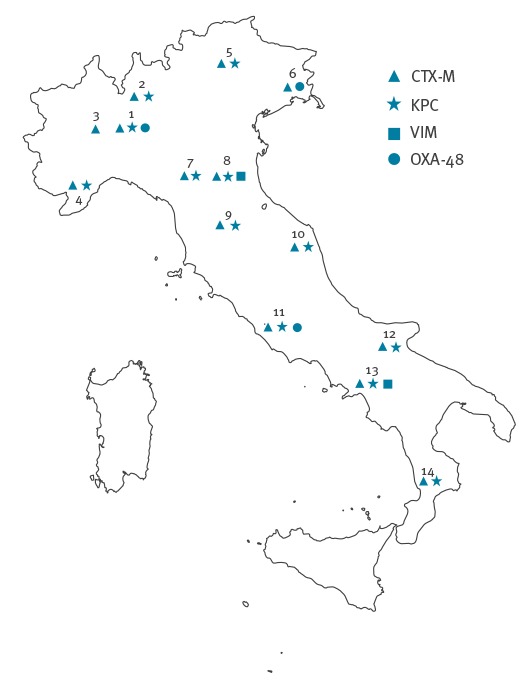
The laboratories were selected so as to provide a countrywide coverage by large laboratories associated with hospitals, representative of most Italian Regions. Twelve of the 14 laboratories had also been involved in the first Italian survey on carbapenemase-producing Enterobacteriaceae, carried out in 2011 [22]. The survey period was from 1 to 15 October 2013.
The laboratories consecutively collected all non-replicate (only first isolate from a patient included) clinical isolates of E. coli, K. pneumoniae and Proteus mirabilis, from any site of infection, showing a minimum inhibitory concentration (MIC) > 1 mg/L for extended-spectrum cephalosporins (ESC) (cefotaxime and/or ceftriaxone and/or ceftazidime and/or cefepime), and/or for ertapenem. Participating laboratories determined MICs of ESC and ertapenem by the automated systems routinely used in the respective laboratory: either Vitek 2 (bioMérieux, Marcy l’Etoile, France) or BD Phoenix (Becton, Dickinson and Co., New Jersey, United States (US)). The collected isolates were sent to reference laboratories for confirmation of species identification and characterisation of the resistance mechanisms.
For each isolate, information on the type of clinical specimen and of patient (inpatient or outpatient) were provided. Isolates from patients from nursing homes or other long-term care facilities, and isolates from surveillance specimens, were excluded. Each laboratory also provided information on the total number of non-replicate clinical isolates of Enterobacteriaceae of the same species observed during the collection period from inpatients and outpatients.
Bacterial identification, antimicrobial susceptibility testing and phenotypic characterisation of resistance mechanisms
At the reference laboratories, identification of collected isolates was confirmed by MALDI-TOF mass spectrometry (Vitek MS, bioMérieux), and susceptibility to ESC (cefotaxime, ceftazidime, cefepime), carbapenems (ertapenem, imipenem, and meropenem), aztreonam, aminoglycosides (amikacin and gentamicin), ciprofloxacin, trimethoprim/sulfamethoxazole was determined by disc diffusion on Mueller-Hinton agar (for P. mirabilis imipenem was not tested) according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) methodology [23]. MICs of tigecycline and colistin for carbapenemase producers were determined using reference broth microdilution [24] with custom lyophilised plates (TREK Diagnostic Systems, Cleveland, Ohio, US). All results were interpreted according to the EUCAST breakpoints [25]. ESBL production was investigated using the double disk method testing the synergistic activity between amoxicillin-clavulanate and cefotaxime, ceftazidime, cefepime, and aztreonam [26].
Molecular characterisation of resistance genes
The presence of carbapenemase genes was investigated by multiplex real time-PCR (mRT-PCR) targeting blaKPC-type,blaVIM-type, blaOXA-48-type and blaNDM-type genes, using primers and conditions reported in Table 1. The presence of blaCTX-M ESBL genes was investigated by a mRT-PCR, able to distinguish among different blaCTX-M-type variants of groups 1, 2, 8/25 and 9, using primers and conditions reported in Table 1. The presence of ACBL genes was investigated by a multiplex PCR targeting genes encoding ACBLs, as described previously [27]. For E. coli, specific primers designed to distinguish between acquired and chromosomal ACBL genes were used [27]. The presence of mcr-1-like genes was investigated by a novel real time-PCR (Table 1).
Table 1. Sequence of primers and probes used in nationwide surveillance survey of the molecular epidemiology of ESBL- and carbapenemase-producing Enterobacteriaceae, Italy, October 2013.
| Target | Primer name | Sequence (5’-3’)a | Reference | Positive control |
|---|---|---|---|---|
| OXA-48-like-rt-F | GTAGCAAAGGAATGGCAAGAAA | [44] |
Escherichia coli ECBZ-1 (blaOXA-48) [45] |
|
| bla OXA-48-like genes | OXA-48-like-rt-R | GATGCGGGTAAAAATGCTTG | ||
| OXA-48-like-rt-P | HEX-CTCTGGAATGAGAATAAGCAGCAAGG-BHQ-1 | |||
| bla KPC genes | KPC-rt-F | GATACCACGTTCCGTCTGG | [46] |
Klebsiella pneumoniae FIPP-1 (blaKPC-3) [47] |
| KPC-rt-R | GCAGGTTCCGGTTTTGTCTC | |||
| KPC-rt-P | FAM-AGCGGCAGCAGTTTGTTGATTG-BHQ-1 | |||
| bla VIM genes | VIM-rt-F | TGGTCTCATTGTCCGTGATG | [48] |
K. pneumoniae VA-416/02 (blaVIM-4) [49] |
| VIM-rt-R | CATGAAAGTGCGTGGAGA | |||
| VIM-rt-P | ROX-AAGCAAATTGGACTTCCCGTAACGC-BHQ-2 | |||
| bla NDM genes | blaNDM1_F | CGCAACACAGCCTGACTTT | [50] |
E. coli CVB-1 (blaNDM-1) [51] |
| blaNDM1_R | TCGATCCCAACGGTGATATT | |||
| blaNDM1_P | CY5-CAACTTTGGCCCGCTCAAGGTATTT-BHQ-3 | |||
| bla CTX-M group 1 genes | CTX-M-group-1_FW | AAAAATCACTGCGCCAGTTC | [52] |
E. coli V460a (blaCTX-M-15), in-house control |
| CTX-M-group-1_REV | AGCTTATTCATCGCCACGTT | |||
| CTX-M-group1-P | HEX-TGGCGACGGCAACCGTCACGCTGTT-BHQ-1 | This study | ||
|
bla
CTX-M group 2 genes |
CTX-M-group-2_FW | CGACGCTACCCCTGCTATT | [52] |
E. coli C277a (blaCTX-M-2), in-house control |
| CTX-M-group-2_REV | CCAGCGTCAGATTTTTCAGG | |||
| CTX-M-group2-P | FAM-TATTGAGCGTGGGCTCGGTTCTGTCCAG-BHQ-1 | This study | ||
| bla CTX-M group 8/25 genes | CTX-M-group-8/25_ FW | CGATACCACCACGCCATTAG | This study |
E. coli M26a (blaCTX-M-8), in-house control |
| CTX-M-group-8/25_REV | AACCCACGATGTGGGTAGC | [52] | ||
| CTX-M-group8/25-P | CY5-CCTGAATGCTGGCAGCGCCGGTG-BHQ-3 | This study | ||
| bla CTX - M group 9 genes | CTX-M-group-9_FW | CAAAGAGAGTGCAACGGATG | [52] |
E. coli V404a (blaCTX-M-14) [53] |
| CTX-M-group-9_REV | ATTGGAAAGCGTTCATCACC | |||
| CTX-M-group9-P | ROX-CGTGCATTCCGCTGCTGCTGGGCA-BHQ-2 | This study | ||
| All blaCTX-M genes |
U-CTX-M- FW | ATYRAYACMGCVGATAAYWCGCA | This study | E. coli V460a (blaCTX-M-15), E. coli C277a (blaCTX-M-2), E. coli M26a (blaCTX-M-8), E. coli V404a (blaCTX-M-14) |
| U-CTX-M- REV | CSGCAATSGGRTTRTAGTTAAC | This study | ||
| U-CTX-M-P | CY5.5-ATGTGCAGYACCAGTAARGTKATGGC-BHQ-3 | This study | ||
| PhHV (internal control) | PhHV-267s | GGGCGAATCACAGATTGAATC | [54] | PhHV DNA cloned in pGEM-T-easy E. coli DH5α |
| PhHV-337as | GCGGTTCCAAACGTACCAA | |||
| PhHV-305tq | Cy5.5 -TTTTATGTGTCCGCCACCATCTGGATC-BHQ-3 | |||
|
mcr-1-like genes |
Mcr-1-rt-fwd | ATCAGCCAAACCTATCCCATC | This study |
E. coli FI-4531 (mcr-1) [55], K. pneumoniae KP-6884 (mcr-1.2) [33] |
| Mcr-1-rt-rev | ACACAGGCTTTAGCACATAGC | |||
| Mcr-1-rt-p | Cy5-GACAATCTCGGCTTTGTGCTGACGATC-BHQ-3 |
ESBL: extended-spectrum beta-lactamases.
a The amplification programme consisted of 35 two-step cycles of 15s at 95 °C and 60s at 60 °C.
Spectrophotometric assay for carbapenemase activity
In carbapenem non-susceptible isolates that tested negative for known carbapenemase genes, carbapenemase production was further investigated by measuring the imipenem-hydrolysing specific activity in bacterial crude extracts as described previously [28], using a Cary 100 UV-Vis spectrophotometer (Varian, Walnut Creek, California, US).
Statistical analysis
Statistical evaluation of differences between resistance rates observed in this study and previous surveillance studies was carried out with the chi-squared test with Yates correction, using the Stata Statistical Software (release 13, College Station, Texas, US). Since the surveillance studies were overall similar by design but there were a few sampling differences, which could act as confounding factors, the tests were treated rather as exploratory analyses, which may indicate trends.
Results
Prevalence of Enterobacteriaceae with resistance mechanisms to extended-spectrum cephalosporins and/or carbapenems
During the study period, a total of 3,324 consecutive non-replicate clinical isolates of Enterobacteriaceae of the target species were isolated in the 14 laboratories participating in the survey, including 2,352 (70.7%) E. coli, 697 (21.0%) K. pneumoniae, and 275 (8.3%) P. mirabilis. Of these, 1,509 isolates (45.4%) were from inpatients and 1,815 (54.6%) from outpatients (Table 2).
Table 2. Proportions of ESCR-carbaS and ESCR-carbaR of Enterobacteriaceae by species included and origin of isolate, nationwide surveillance survey, Italy, October 2013 (n=3,324 isolates).
| Species | Isolates from inpatients | Isolates from outpatients | All isolates | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | ESCR | % | ESCR‑carbaS | % | ESCR‑carbaR | % | Total | ESCR | % | ESCR‑carbaS | % | ESCR‑carbaR | % | Total | ESCR | % | ESCR‑carbaS | % | ESCR‑carbaR | % | |
| Escherichia coli | 920 | 230 | 25.0 | 219 | 23.8 | 11 | 1.2 | 1,432 | 162 | 11.3 | 159 | 11.1 | 3 | 0.2 | 2,352 | 392 | 16.7 | 378 | 16.1 | 14 | 0.6 |
| Klebsiella pneumoniae | 437 | 159 | 36.4 | 49 | 11.2 | 110 | 25.1 | 260 | 36 | 13.8 | 16 | 6.2 | 20 | 7.7 | 697 | 195 | 28.0 | 65 | 9.3 | 130 | 18.7 |
| Proteus mirabilis | 152 | 39 | 25.7 | 39 | 25.7 | 0 | NA | 123 | 26 | 21.1 | 26 | 21.1 | 0 | NA | 275 | 65 | 23.6 | 65 | 23.6 | 0 | NA |
| Total target species | 1,509 | 428 | 28.4 | 309 | 20.3 | 121 | 8.0 | 1,815 | 224 | 12.3 | 201 | 11.1 | 23 | 1.3 | 3,324 | 652 | 19.6 | 508 | 15.3 | 144 | 4.3 |
ESCR: non-susceptible to extended-spectrum cephalosporins; ESCR-carbaS: non-susceptible to extended-spectrum cephalosporins but susceptible to carbapenems; ESCR-carbaR: isolates non-susceptible to extended-spectrum cephalosporins and non-susceptible to carbapenems; NA: not applicable.
Overall, 508 isolates were confirmed to be non-susceptible to ESC but susceptible to carbapenems (ESCR-carbaS phenotype), and 144 isolates to be non-susceptible to both ESC and carbapenems (ESCR-carbaR). Isolates susceptible to ESC and non-susceptible to carbapenems were not detected.
The ESCR-carbaS phenotypes were contributed by all three species, with higher proportions among P. mirabilis (23.6%) and E. coli (16.1%) than among K. pneumoniae (9.3%), and with a higher proportion among isolates from inpatients (20.3%) but a remarkable proportion (11.1%) also among those from outpatients (Table 2).
The ESCR-carbaR phenotypes were mostly contributed by K. pneumoniae, with higher proportion among isolates from inpatients (25.1% of K. pneumoniae) but also a notable proportion among those from outpatients (7.7% of K. pneumoniae) (Table 2).
Considering the nature of resistant isolates, the majority of those from outpatients (n=224) were from urine (206/224, 91.9%), while those from inpatients (n=428) were from different specimens (urine: 239/428, 55.8%; blood: 55/428, 12.8%; respiratory tract samples: 54/428, 12.6%).
Beta-lactamase phenotypes and genotypes of the ESCR and carbapenem-resistant isolates
Of the ESCR-carbaS isolates, 460 (90.5%) were ESBL-positive and 384 of them (83.4%) carried a blaCTX-M ESBL gene (Figure 2 and Table 3).
Figure 2.
Distribution of Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis isolates according to resistance phenotypes and genotypes, nationwide surveillance survey, Italy, October 2013 (n=652 isolates)
ACBL: isolates producing acquired class C beta-lactamase; CARBA: carbapenemase producing isolates; E. coli: Escherichia coli; ESBL: isolates producing extended spectrum beta-lactamase; ESCR-carbaS: isolates non-susceptible to extended-spectrum cephalosporins but susceptible to carbapenems; ESCR-carbaR: isolates non-susceptible to extended-spectrum cephalosporins and non-susceptible to carbapenems; K. pneumoniae: Klebsiella pneumoniae; P. mirabilis: Proteus mirabilis.
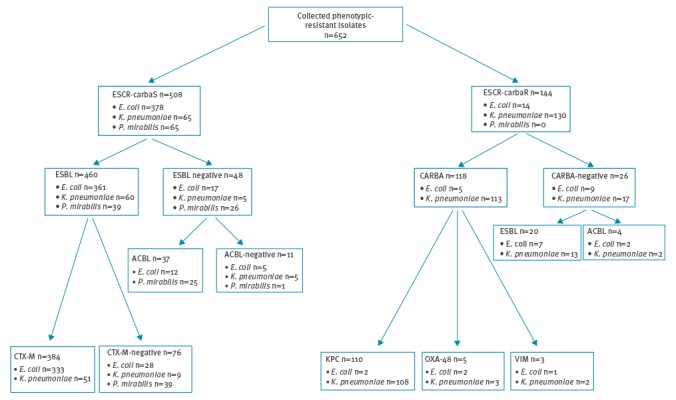
Table 3. Resistance mechanisms detected in the investigated phenotypic resistant isolates by species included, nationwide surveillance survey, Italy, October 2013 (n=652 isolates).
| Phenotypic resistance | Resistance mechanism | Escherichia coli | Klebsiella pneumoniae | Proteus mirabilis | Total | ||||
|---|---|---|---|---|---|---|---|---|---|
| N | % | n | % | n | % | n | % | ||
| Total | 2,352 | 100 | 697 | 100 | 275 | 100 | 3,324 | 100 | |
| Resistant | 392 | 16.7 | 195 | 28.0 | 65 | 23.6 | 652 | 19.6 | |
| ESCR-carbaS | Total | 378 | 96.4 | 65 | 33.3 | 65 | 100.0 | 508 | 77.9 |
| ESBL | CTX-M-1 | 264 | 69.8 | 50 | 76.9 | 0 | NA | 314 | 61.8 |
| CTX-M-9 | 61 | 16.1 | 0 | NA | 0 | NA | 61 | 12.0 | |
| CTX-M-1 + 9 | 8 | 2.1 | 1 | 1.5 | 0 | NA | 9 | 1.8 | |
| Other ESBL | 28 | 7.4 | 9 | 13.8 | 39 | 60.0 | 76 | 15.0 | |
| ACBL | CMY/LAT/ACT/MIR | 12 | 3.2 | 0 | NA | 24 | 36.9 | 36 | 7.1 |
| DHA | 0 | NA | 0 | NA | 1 | 1.5 | 1 | 0.2 | |
| Other | Other resistance mechanism | 5 | 1.3 | 5 | 7.7 | 1 | 1.5 | 11 | 2.1 |
| ESCR-carbaR | Total | 14 | 3.6 | 130 | 66.7 | 0 | 0.0 | 144 | 22.1 |
| CARBA | KPC | 2 | 14.3 | 108 | 83.2 | 0 | NA | 110 | 76.4 |
| VIM | 1 | 7.1 | 2 | 1.5 | 0 | NA | 3 | 2.1 | |
| OXA-48 | 2 | 14.3 | 3 | 2.3 | 0 | NA | 5 | 3.5 | |
| ESBL | CTX-M-1 | 4 | 28.6 | 9 | 6.9 | 0 | NA | 13 | 9.0 |
| CTX-M-9 | 3 | 21.4 | 0 | NA | 0 | NA | 3 | 2.1 | |
| CTX-M-1 + 9 | 0 | NA | 1 | 0.8 | 0 | NA | 1 | 0.7 | |
| ACBL | CMY/LAT/ACT/MIR | 2 | 14.3 | 0 | NA | 0 | NA | 2 | 1.4 |
| DHA | 0 | NA | 2 | 1.5 | 0 | NA | 2 | 1.4 | |
| Other | Other resistance mechanism | 0 | NA | 5 | 3.8 | 0 | NA | 5 | 3.5 |
| Total | Total | 392 | 100 | 195 | 100 | 65 | 100 | 652 | 100 |
| CARBA | KPC | 2 | 0.5 | 108 | 55.4 | 0 | NA | 110 | 16.9 |
| VIM | 1 | 0.3 | 2 | 1.0 | 0 | NA | 3 | 0.5 | |
| OXA-48 | 2 | 0.5 | 3 | 1.5 | 0 | NA | 5 | 0.8 | |
| ESBL | CTX-M-1 | 268 | 68.4 | 59 | 30.3 | 0 | NA | 327 | 50.2 |
| CTX-M-9 | 64 | 16.3 | 0 | NA | 0 | NA | 64 | 9.8 | |
| CTX-M-1 + 9 | 8 | 2.0 | 2 | 1.0 | 0 | NA | 10 | 1.5 | |
| ACBL | CMY/LAT/ACT/MIR | 14 | 3.6 | 0 | NA | 24 | 36.9 | 38 | 5.8 |
| DHA | 0 | NA | 2 | 1.0 | 1 | 1.5 | 3 | 0.5 | |
| Other | Other resistance mechanism | 33 | 8.4 | 19 | 9.7 | 40 | 61.5 | 92 | 14.1 |
ACBL: acquired class C beta-lactamase-producing; CARBA: carbapenemase-producing; CTX-M-1: CTX-M group 1 producers; CTX-M-9: CTX-M group 9 producers; CTX-M-1+9: CTX-M group 1 and group 9 producers; ESBL: extended-spectrum beta-lactamase-producing; ESCR-carbaR: non-susceptible to extended-spectrum cephalosporins and non-susceptible to carbapenems; ESCR-carbaS: non-susceptible to extended-spectrum cephalosporins but susceptible to carbapenems; NA: not applicable.
The proportion of blaCTX-M genes was 92.2% among ESBL-positive E. coli and 84.7% among ESBL-positive K. pneumoniae, while such genes were never detected among ESBL-positive isolates of P. mirabilis (Figure 2). Among CTX-M-positive isolates, most carried blaCTX-M genes of group 1 (81.5%), while a minority carried blaCTX-M genes of group 9 (16,0%) and a small proportion carried blaCTX-M genes of both CTX-M groups (2.5%) (Table 3). CTX-M-positive isolates were detected from all centers (Figure 1).
Comparison of these data with results from the earlier Italian nationwide survey on producing ESBL-producing Enterobacteriaceae, carried out in 2003 [29], revealed a notable increase in the proportion of ESBL-producing isolates (from 6.4% to 14.4%; p < 0.001, considering the three target species). Among outpatients, the proportion of ESBL producers increased from 3.4% to 11.0% (p < 0.001), and the increase was especially large among E. coli (from 1.9% to 10.9%; p < 0.001).
The majority of the ESCR-carbaS isolates that were ESBL-negative were positive for an acquired ACBL gene (n = 37, 79%) which, in most cases, belonged to the CMY/LAT/ACT/MIR lineage. Most of the ACBL-positive isolates were P. mirabilis (Figure 2, Table 3).
Concerning the ESCR-carbaR isolates, most (81.9%) were positive for carbapenemase genes, including blaKPC- (93.2%), blaOXA-48- (4.2%), and blaVIM-type (2.6%) carbapenemase genes. The majority of carbapenemase-producers were K. pneumoniae, while only five E. coli produced a carbapenemase (Figure 2). Interestingly, some KPC-positive K. pneumoniae (KPC-KP) (6.5%) and most OXA-48-positive isolates also harbored a blaCTX-M gene (Table 3). The 26 ESCR-carbaR isolates that tested negative for carbapenemase genes were confirmed to be carbapenemase non-producers by spectrophotometric assay. Most of these isolates carried blaCTX-M genes (65.3%) while a minority was positive for an ACBL gene (blaDHA or blaCMY/LAT/ACT/MIR), suggesting that in these cases the carbapenem resistance mechanism was related with production of an ESBL or ACBL in combination with reduced permeability [30].
Comparison of these data with results from the first Italian nationwide survey on carbapenem-resistant Enterobacteriaceae (CRE), carried out in 2011 [22], revealed an increase in the proportion of carbapenem-resistant K. pneumoniae isolates (from 11.9% to 18.7%) although the difference was not statistically significant (p = 0.5). KPC-KP remained the major contributors (83.1% of carbapenem-resistant K. pneumoniae in 2013 vs. 87.2% in 2011). Among outpatients, the proportion of KPC-KP increased from 2.2% in 2011 to 4.6% (p < 0.05).
A detailed distribution of the resistance genes in the Italy is reported in Figure 2, and the distribution by species and type of patients is reported in Table 2.
Antimicrobial susceptibility
All ESBL-producing isolates that were carbapenemase-negative remained susceptible to imipenem and meropenem, while ertapenem susceptibility was 98.1% among E. coli and 82.2% among K. pneumoniae (Table 4).
Table 4. Antimicrobial susceptibility testing results for the investigated isolates, nationwide surveillance survey, Italy, October 2013 (n=652 isolates).
| Species | Phenotypic resistance | n | Susceptible (%) |
| AK | GM | CIP | CAZ | CTX | FEP | IMI | MEM | ERT | AZT | TRIM/SUL | COLa | TIGa | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Escherichia coli | ESBL | 368 | 94.8 | 61.7 | 9.8 | 10.6 | 0.8 | 14.9 | 100.0 | 100.0 | 98.1 | 6.3 | 43.8 | NA | NA |
| CARBA | 5 | 80.0 | 60.0 | 20.0 | 20.0 | 0.0 | 20.0 | 80.0 | 60.0 | 0.0 | 20.0 | 40.0 | 100.0 | 100.0 | |
| Others | 19 | 100.0 | 89.5 | 52.6 | 0.0 | 0.0 | 94.7 | 100.0 | 100.0 | 89.5 | 5.3 | 42.1 | NA | NA | |
| Total | 392 | 94.9 | 63.0 | 12.0 | 10.2 | 0.8 | 18.9 | 99.7 | 99.5 | 96.4 | 6.4 | 43.6 | NA | NA | |
| Klebsiella pneumoniae | ESBL | 73 | 91.8 | 39.7 | 21.9 | 4.1 | 6.8 | 6.8 | 100.0 | 100.0 | 82.2 | 8.2 | 26.0 | NA | NA |
| CARBA | 113 | 16.8 | 48.7 | 0.9 | 0.0 | 0.0 | 0.0 | 3.5 | 3.5 | 0.0 | 0.0 | 25.7 | 61.9 | 94.4 | |
| Others | 9 | 66.7 | 88.9 | 44.4 | 0.0 | 66.7 | 77.8 | 100.0 | 88.9 | 55.6 | 77.8 | 55.6 | NA | NA | |
| Total | 195 | 47.2 | 47.2 | 10.8 | 1.5 | 5.6 | 6.2 | 44.1 | 43.6 | 33.3 | 6.7 | 27.2 | NA | NA | |
| Proteus mirabilis | ESBL | 39 | 92.3 | 0.0 | 30.8 | 43.6 | 7.7 | 61.5 | NA | 100.0 | 100.0 | 87.2 | 25.6 | NA | NA |
| CARBA | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
| Others | 26 | 96.2 | 57.7 | 7.7 | 0.0 | 0.0 | 88.5 | NA | 100.0 | 100.0 | 100.0 | 7.7 | NA | NA | |
| Total | 65 | 93.8 | 23.1 | 21.5 | 26.2 | 4.6 | 72.3 | NA | 100.0 | 100.0 | 92.3 | 18.5 | NA | NA |
AK: amikacin; AZT: aztreonam; CARBA: carbapenemase production; CAZ: ceftazidime; CIP: ciprofloxacin; COL: colistin ; CTX: ceftriaxone; ERT: ertapenem; ESBL: extended spectrum beta-lactamase production; FEP: cefepime; GM: gentamicin; IMI: imipenem; MEM: meropenem; NA: not applicable; TIG: tigecycline; TRIM/SUL: trimethoprim/sulfamethoxazole .
a COL and TIG were tested only for carbapenemase producers.
Among the carbapenemase producers, all were non-susceptible to ertapenem, while most carbapenemase-producing E. coli were susceptible to imipenem and meropenem (80.0% and 60.0%, respectively). Carbapenemase-producing K. pneumoniae were more susceptible to gentamicin than to amikacin (48.7% vs. 16.8%). Broth microdilution assays, carried out with the 113 carbapenemase-producing K. pneumoniae, revealed that 61.7% and 94.4% were susceptible to colistin and tigecycline, respectively, while all five carbapenemase-producing E. coli were susceptible to both drugs (Table 4). These results were overall similar to those reported from the European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) survey in 2015 [31], and underscore the remarkable rate of colistin resistance among carbapenemase-producing K. pneumoniae circulating in Italy. No significant differences in susceptibility to these two drugs were detected among inpatients and outpatients (data not shown).
Screening for mcr-1-like genes in colistin-resistant isolates
The 43 colistin-resistant carbapenemase-positive K. pneumoniae isolates were screened for the presence of mcr-1-like genes, encoding transferable colistin resistance [32,33]. All the tested isolates yielded negative results, revealing that colistin resistance was caused by different mechanisms.
Discussion
This study provides an updated picture of the prevalence, distribution, beta-lactamase profiles and susceptibilities of ESBL-, carbapenemase- and ACBL-producing E. coli, K. pneumoniae and P. mirabilis circulating in Italy.
Compared with the earlier Italian nationwide survey on ESBL-producing Enterobacteriaceae, carried out in 2003 [29], the proportion of ESBL-producing isolates (considering the three target species) showed a notable increase, especially among outpatients and among E. coli. At the level of resistance mechanisms, this epidemiological evolution was associated with a remarkable increase in the prevalence of CTX-M-type enzymes, which, in 2013, represented by far the most common type of ESBL in E. coli and in K. pneumoniae. Interestingly, despite their ability to spread, the genes encoding these resistance mechanisms have not disseminated in P. mirabilis, where the ESCR phenotype was found to be relatively common (23.6% of isolates) but due to other mechanisms, i. e. ESBLs other than CTX-M-type or ACBLs, mostly of the CMY/LAT/ACT/MIR lineage. A similar epidemiological evolution was observed in other European Countries including Belgium (with 77% vs 46% of CTX-M-positive isolates, among ESBL-producing E. coli, in 2008 vs 2006) [34], Spain (with 72% vs 52% of CTX-M- positive isolates among ESBL-producing E. coli in 2006 vs 2000) [35].
Concerning carbapenem resistance, a notable increase was observed among K. pneumoniae in comparison with data from the first Italian nationwide survey on CRE, carried out in 2011 [22], with KPC-KP remaining the major contributors of CRE endemicity in Italy and other types of carbapenemases remaining uncommon. These findings underscore the ability of carbapenemase-producing Enterobacteriaceae (CPE) to rapidly disseminate and establish conditions of high-level endemicity, and the notion that CPE are associated with a heightened epidemiological risk and deserve special attention, as readily acknowledged by the European Centre for Disease Prevention and Control (ECDC) [36]. Indeed, the data from the EARS-NET surveillance system indicate that there is an overall increasing spread of CRE in some European countries [21], and results from the EuSCAPE project have shown that some regions are experiencing a worrisome prevalence of CPE per 10,000 hospital admissions, ranging from four to six for Italy, Greece and Spain [37].
Interestingly, KPC-KP isolates were also found from outpatients, at a rate that was approximately double compared with that found in the 2011 survey (4.6% vs 2.2%) [22]. Since a significant proportion of these outpatients could have been recently hospitalised or otherwise have had contact with healthcare services, this figure may not reflect accurately the prevalence of KPC-KP in the community, in Italy. However, this finding points to an increasing dissemination of CPE also outside the hospital setting, likely reflecting the ability of KPC-KP to establish persistent intestinal colonisation even after hospital discharge [38,39]. It might herald a possible dissemination in the community like the one previously witnessed with CTX-M-producing E. coli [40]. Indeed, recent reports from the US and Spain have described the emergence of CPE infections also in the community, underscoring this possibility [41,42]. Such a scenario should be avoided, and suitable infection control measures should be enforced to address this phenomenon in settings of high CRE endemicity like Italy and some other European countries [37].
In this grim scenario, the good news was that (i) overall carbapenem susceptibility was retained by P. mirabilis, with no carbapenemase-positive isolates detected in this species despite the high endemicity of CPE; and that (ii) no mcr-1-like transferable colistin resistance genes were detected among the colistin-resistant CPE isolates, suggesting that this worrisome resistance mechanism remains uncommon among colistin-resistant CPE circulating in Italy.
Updated molecular surveillance data on resistance genes to beta-lactams are of increasing importance due to the introduction on the market of new antibiotics for resistant Gram-negatives that are based on new beta-lactamase inhibitors (e. g. avibactam), which will be useful for treatment of infections caused by ESBL and carbapenemase producers [43]. The new inhibitors, however, are not active against all beta-lactamases e. g. avibactam is able to inhibit class A but not class B beta-lactamases. Thus the possibility of using these new drugs will depend on the specific mechanism of resistance, and rapid testing for these resistance mechanisms will become an essential step in antibiotic stewardship programs. In this perspective, this survey underscores the major role of beta-lactamase genes as resistance determinants in Enterobacteriaceae in the Italian epidemiological setting, and provides relevant information for the selection of the most suitable diagnostic strategies based on molecular detection of beta-lactam resistance mechanisms. A limitation of this study is that, due to the study design, laboratory-based surveillance, information about risk factors for developing antibiotic resistance was not available. Future studies are warranted to investigate these aspects, which are relevant to understand the reasons for the epidemiological differences observed in different European countries and to contextualise infection control policies.
Acknowledgements
Results of this work were partially presented at the 25th ECCMID in Copenhagen (2015) as an oral presentation, O143.
This work was partially supported by a research grant from Pfizer to AMCLI, and from EvoTAR (no. HEALTH-F3-2011-2011-282004) to GMR.
AMCLI-CoSA Survey participants: Milan, Niguarda Ca’ Granda Hospital – C. Vismara; Lecco, A. Manzoni Hospital – B. Pini; Novara, Maggiore Hospital – S. Andreoni; San Remo, Azienda Sanitaria Imperiese Hospital – P. A. Dusi; Bolzano, Azienda Sanitaria dell’Alto Adige Hospital – R. Aschbacher; Udine, Azienda Ospedaliera Santa Maria della Misericordia – C. Scarparo; Modena, Baggiovara Hospital – M. Sarti; Modena, Modena University Hospital –C. Venturelli; Florence, Careggi University Hospital – P. Pecile; Ancona, ‘Torrette’ University Hospital – E. Manso; Rome, Policlinico Gemelli – T. Spanu; San Giovanni Rotondo, IRCCS Casa Sollievo della Sofferenza Hospital – M. Labonia; Avellino, San Giuseppe Moscati Hospital – G. Buonopane; Cosenza, Annunziata Hospital – C. Giraldi.
Conflict of interest: None declared.
Authors’ contributions: TG, FA, AP, FL, LP, and GMR contributed to the design of the study; TG, AA, FL and GMR contributed to draft and finalise the manuscript; CM, SB, and FL performed the phenotypic characterisation of the isolates; AA, JN, MC performed the detection of the ESBL and carbapenemase genes by PCR; AA, JN performed the detection of the class C beta-lactamase genes by PCR; AA, TG, FL and GMR entered and analysed data; TG and FL planned and coordinated the activities of the AMCLI-CoSA Survey Participants; the AMCLI-CoSA Survey participants collected the isolates and the denominator data; TG, AA and GMR wrote the manuscript; all authors revised and approved the final manuscript.
References
- 1.European Centre for Disease Prevention and Control (ECDC). Point prevalence survey of healthcare- associated infections and antimicrobial use in European acute care hospitals. Stockholm: ECDC; 2013. Available from: http://ecdc.europa.eu/en/publications/publications/healthcare-associated-infections-antimicrobial-use-pps.pdf
- 2. Van Boeckel TP, Gandra S, Ashok A, Caudron Q, Grenfell BT, Levin SA, et al. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect Dis. 2014;14(8):742-50. 10.1016/S1473-3099(14)70780-7 [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization (WHO). Strategies for global surveillance of antimicrobial resistance: report of a technical consultation. Geneva: WHO; Oct 2013. Available from: http://www.who.int/drugresistance/publications/surveillance-meeting2012/en/index.html
- 4. Falagas ME, Karageorgopoulos DE, Nordmann P. Therapeutic options for infections with Enterobacteriaceae producing carbapenem-hydrolyzing enzymes. Future Microbiol. 2011;6(6):653-66. 10.2217/fmb.11.49 [DOI] [PubMed] [Google Scholar]
- 5. Maragakis LL, Perencevich EN, Cosgrove SE. Clinical and economic burden of antimicrobial resistance. Expert Rev Anti Infect Ther. 2008;6(5):751-63. 10.1586/14787210.6.5.751 [DOI] [PubMed] [Google Scholar]
- 6. Jean S-S, Lee W-S, Lam C, Hsu C-W, Chen R-J, Hsueh P-R. Carbapenemase-producing Gram-negative bacteria: current epidemics, antimicrobial susceptibility and treatment options. Future Microbiol. 2015;10(3):407-25. 10.2217/fmb.14.135 [DOI] [PubMed] [Google Scholar]
- 7. Borer A, Saidel-Odes L, Riesenberg K, Eskira S, Peled N, Nativ R, et al. Attributable mortality rate for carbapenem-resistant Klebsiella pneumoniae bacteremia. Infect Control Hosp Epidemiol. 2009;30(10):972-6. 10.1086/605922 [DOI] [PubMed] [Google Scholar]
- 8. Falagas ME, Lourida P, Poulikakos P, Rafailidis PI, Tansarli GS. Antibiotic treatment of infections due to carbapenem-resistant Enterobacteriaceae: systematic evaluation of the available evidence. Antimicrob Agents Chemother. 2014;58(2):654-63. 10.1128/AAC.01222-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bush K. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr Opin Microbiol. 2010;13(5):558-64. 10.1016/j.mib.2010.09.006 [DOI] [PubMed] [Google Scholar]
- 10. Philippon A, Arlet G, Jacoby GA. Plasmid-determined AmpC-type β-lactamases. Antimicrob Agents Chemother. 2002;46(1):1-11. 10.1128/AAC.46.1.1-11.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev. 2007;20(3):440-58. 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med. 2012;18(5):263-72. 10.1016/j.molmed.2012.03.003 [DOI] [PubMed] [Google Scholar]
- 13. D’Andrea MM, Arena F, Pallecchi L, Rossolini GM. CTX-M-type β-lactamases: a successful story of antibiotic resistance. Int J Med Microbiol. 2013;303(6-7):305-17. 10.1016/j.ijmm.2013.02.008 [DOI] [PubMed] [Google Scholar]
- 14. Cantón R, Coque TM. The CTX-M β-lactamase pandemic. Curr Opin Microbiol. 2006;9(5):466-75. 10.1016/j.mib.2006.08.011 [DOI] [PubMed] [Google Scholar]
- 15. Jacoby GA. AmpC β-lactamases. Clin Microbiol Rev. 2009;22(1):161-82. 10.1128/CMR.00036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arena F, Giani T, Becucci E, Conte V, Zanelli G, D’Andrea MM, et al. Large oligoclonal outbreak due to Klebsiella pneumoniae ST14 and ST26 producing the FOX-7 AmpC β-lactamase in a neonatal intensive care unit. J Clin Microbiol. 2013;51(12):4067-72. 10.1128/JCM.01982-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rossolini GM. Extensively drug-resistant carbapenemase-producing Enterobacteriaceae producing carbapenemases: an emerging challenge for clinicians and healthcare systems. J Intern Med. 2015;277(5):528-31. 10.1111/joim.12350 [DOI] [PubMed] [Google Scholar]
- 18. Albiger B, Glasner C, Struelens MJ, Grundmann H, Monnet DL, European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) working group Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries, May 2015. Euro Surveill. 2015;20(45):30062. 10.2807/1560-7917.ES.2015.20.45.30062 [DOI] [PubMed] [Google Scholar]
- 19. Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother. 2012;67(7):1597-606. 10.1093/jac/dks121 [DOI] [PubMed] [Google Scholar]
- 20. Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791-8. 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.European Center for Disease Prevention and Control (ECDC). Antimicrobial resistance interactive database (EARS-Net). Stockholm: ECDC; [Accessed 07/12/2017]. Available from: http://atlas.ecdc.europa.eu/public/index.aspx
- 22. Giani T, Pini B, Arena F, Conte V, Bracco S, Migliavacca R, et al. AMCLI-CRE Survey Participants Epidemic diffusion of KPC carbapenemase-producing Klebsiella pneumoniae in Italy: results of the first countrywide survey, 15 May to 30 June 2011. Euro Surveill. 2013;18(22):20489. [PubMed] [Google Scholar]
- 23.European Committee on Antimicrobial Susceptibility Testing (EUCAST). Antimicrobial susceptibility testing. EUCAST Disk Diffusion-Manual. Version 5.0. Växjö: EUCAST; Jan 2015. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/Manual_v_5.0_EUCAST_Disk_Test.pdf
- 24.Clinical and Laboratory Standards Institute (CLSI). 2015. Methods for dilution antimicrobial susceptibility. Tests for bacteria that grow aerobically; approved standards. Tenth edition. CLSI document M07-A10. Available from: https://clsi.org
- 25.European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Version 6.0. Växjö: EUCAST; 1 Jan 2016. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_6.0_Breakpoint_table.pdf
- 26.European Committee on Antimicrobial Susceptibility Testing (EUCAST). EUCAST guideline for the detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. Version 1.0. Växjö: EUCAST; Dec 2016. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_v1.0_20131211.pdf
- 27. D’Andrea MM, Nucleo E, Luzzaro F, Giani T, Migliavacca R, Vailati F, et al. CMY-16, a novel acquired AmpC-type β-lactamase of the CMY/LAT lineage in multifocal monophyletic isolates of Proteus mirabilis from northern Italy. Antimicrob Agents Chemother. 2006;50(2):618-24. 10.1128/AAC.50.2.618-624.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, et al. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother. 1999;43(7):1584-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luzzaro F, Mezzatesta M, Mugnaioli C, Perilli M, Stefani S, Amicosante G, et al. Trends in production of extended-spectrum β-lactamases among enterobacteria of medical interest: report of the second Italian nationwide survey. J Clin Microbiol. 2006;44(5):1659-64. 10.1128/JCM.44.5.1659-1664.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jacoby GA, Mills DM, Chow N. Role of β-lactamases and porins in resistance to ertapenem and other β-lactams in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48(8):3203-6. 10.1128/AAC.48.8.3203-3206.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Monaco M, Giani T, Raffone M, Arena F, Garcia-Fernandez A, Pollini S, et al. Network EuSCAPE-Italy Colistin resistance superimposed to endemic carbapenem-resistant Klebsiella pneumoniae: a rapidly evolving problem in Italy, November 2013 to April 2014. Euro Surveill. 2014;19(42):20939. 10.2807/1560-7917.ES2014.19.42.20939 [DOI] [PubMed] [Google Scholar]
- 32. Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161-8. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 33. Di Pilato V, Arena F, Tascini C, Cannatelli A, Henrici De Angelis L, Fortunato S, et al. MCR-1.2: a new MCR variant encoded by a transferable plasmid from a colistin-resistant KPC carbapenemase-producing Klebsiella pneumoniae strain of sequence type 512. Antimicrob Agents Chemother. 2016;60(9):5612-5. 10.1128/AAC.01075-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rodriguez-Villalobos H, Bogaerts P, Berhin C, Bauraing C, Deplano A, Montesinos I, et al. Trends in production of extended-spectrum β-lactamases among Enterobacteriaceae of clinical interest: results of a nationwide survey in Belgian hospitals. J Antimicrob Chemother. 2011;66(1):37-47. 10.1093/jac/dkq388 [DOI] [PubMed] [Google Scholar]
- 35. Díaz MA, Hernández-Bello JR, Rodríguez-Baño J, Martínez-Martínez L, Calvo J, Blanco J, et al. Spanish Group for Nosocomial Infections (GEIH) Diversity of Escherichia coli strains producing extended-spectrum β-lactamases in Spain: second nationwide study. J Clin Microbiol. 2010;48(8):2840-5. 10.1128/JCM.02147-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.European Centre for Disease Prevention and Control (ECDC). Rapid risk assessment: Carbapenem-resistant Enterobacteriaceae. Stockholm: ECDC; 8 Apr 2016. Available from: http://ecdc.europa.eu/en/publications/Publications/carbapenem-resistant-enterobacteriaceae-risk-assessment-april-2016.pdf
- 37. Grundmann H, Glasner C, Albiger B, Aanensen DM, Tomlinson CT, Andrasević AT, et al. European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Working Group Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect Dis. 2017;17(2):153-63. 10.1016/S1473-3099(16)30257-2 [DOI] [PubMed] [Google Scholar]
- 38. Feldman N, Adler A, Molshatzki N, Navon-Venezia S, Khabra E, Cohen D, et al. Gastrointestinal colonization by KPC-producing Klebsiella pneumoniae following hospital discharge: duration of carriage and risk factors for persistent carriage. Clin Microbiol Infect. 2013;19(4):E190-6. 10.1111/1469-0691.12099 [DOI] [PubMed] [Google Scholar]
- 39. Zimmerman FS, Assous MV, Bdolah-Abram T, Lachish T, Yinnon AM, Wiener-Well Y. Duration of carriage of carbapenem-resistant Enterobacteriaceae following hospital discharge. Am J Infect Control. 2013;41(3):190-4. 10.1016/j.ajic.2012.09.020 [DOI] [PubMed] [Google Scholar]
- 40. Cantón R, González-Alba JM, Galán JC. CTX-M enzymes: origin and diffusion. Front Microbiol. 2012;3:110. 10.3389/fmicb.2012.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guh AY, Bulens SN, Mu Y, Jacob JT, Reno J, Scott J, et al. Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012-2013. JAMA. 2015;314(14):1479-87. 10.1001/jama.2015.12480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paño-Pardo JR, López Quintana B, Lázaro Perona F, Ruiz Carrascoso G, Romero-Gómez MP, Loeches Yagüe B, et al. Community-Onset Bloodstream and Other Infections, Caused by Carbapenemase-Producing Enterobacteriaceae: Epidemiological, Microbiological, and Clinical Features. Open Forum Infect Dis. 2016;3(3):ofw136. 10.1093/ofid/ofw136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bush K. A resurgence of β-lactamase inhibitor combinations effective against multidrug-resistant Gram-negative pathogens. Int J Antimicrob Agents. 2015;46(5):483-93. 10.1016/j.ijantimicag.2015.08.011 [DOI] [PubMed] [Google Scholar]
- 44. Antonelli A, Di Palo DM, Galano A, Becciani S, Montagnani C, Pecile P, et al. Intestinal carriage of Shewanella xiamenensis simulating carriage of OXA-48-producing Enterobacteriaceae. Diagn Microbiol Infect Dis. 2015;82(1):1-3. 10.1016/j.diagmicrobio.2015.02.008 [DOI] [PubMed] [Google Scholar]
- 45. Giani T, Conte V, Di Pilato V, Aschbacher R, Weber C, Larcher C, et al. Escherichia coli from Italy producing OXA-48 carbapenemase encoded by a novel Tn1999 transposon derivative. Antimicrob Agents Chemother. 2012;56(4):2211-3. 10.1128/AAC.00035-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hindiyeh M, Smollen G, Grossman Z, Ram D, Davidson Y, Mileguir F, et al. Rapid detection of blaKPC carbapenemase genes by real-time PCR. J Clin Microbiol. 2008;46(9):2879-83. 10.1128/JCM.00661-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Giani T, D’Andrea MM, Pecile P, Borgianni L, Nicoletti P, Tonelli F, et al. Emergence in Italy of Klebsiella pneumoniae sequence type 258 producing KPC-3 Carbapenemase. J Clin Microbiol. 2009;47(11):3793-4. 10.1128/JCM.01773-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Antonelli A, Arena F, Giani T, Colavecchio OL, Valeva SV, Paule S, et al. Performance of the BD MAX™ instrument with Check-Direct CPE real-time PCR for the detection of carbapenemase genes from rectal swabs, in a setting with endemic dissemination of carbapenemase-producing Enterobacteriaceae. Diagn Microbiol Infect Dis. 2016;86(1):30-4. 10.1016/j.diagmicrobio.2016.06.002 [DOI] [PubMed] [Google Scholar]
- 49. Luzzaro F, Docquier JD, Colinon C, Endimiani A, Lombardi G, Amicosante G, et al. Emergence in Klebsiella pneumoniae and Enterobacter cloacae clinical isolates of the VIM-4 metallo-β-lactamase encoded by a conjugative plasmid. Antimicrob Agents Chemother. 2004;48(2):648-50. 10.1128/AAC.48.2.648-650.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ong DC, Koh TH, Syahidah N, Krishnan P, Tan TY. Rapid detection of the blaNDM-1 gene by real-time PCR. J Antimicrob Chemother. 2011;66(7):1647-9. 10.1093/jac/dkr184 [DOI] [PubMed] [Google Scholar]
- 51. D’Andrea MM, Venturelli C, Giani T, Arena F, Conte V, Bresciani P, et al. Persistent carriage and infection by multidrug-resistant Escherichia coli ST405 producing NDM-1 carbapenemase: report on the first Italian cases. J Clin Microbiol. 2011;49(7):2755-8. 10.1128/JCM.00016-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum (β)-lactamases. J Antimicrob Chemother. 2006;57(1):154-5. 10.1093/jac/dki412 [DOI] [PubMed] [Google Scholar]
- 53. Riccobono E, Di Pilato V, Villagran AL, Bartoloni A, Rossolini GM, Pallecchi L. Complete sequence of pV404, a novel IncI1 plasmid harbouring blaCTX-M-14 in an original genetic context. Int J Antimicrob Agents. 2014;44(4):374-6. 10.1016/j.ijantimicag.2014.06.019 [DOI] [PubMed] [Google Scholar]
- 54. van Doornum GJ, Guldemeester J, Osterhaus AD, Niesters HG. Diagnosing herpesvirus infections by real-time amplification and rapid culture. J Clin Microbiol. 2003;41(2):576-80. 10.1128/JCM.41.2.576-580.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cannatelli A, Giani T, Antonelli A, Principe L, Luzzaro F, Rossolini GM. First detection of the mcr-1 colistin resistance gene in Escherichia coli in Italy. Antimicrob Agents Chemother. 2016;60(5):3257-8. 10.1128/AAC.00246-16 [DOI] [PMC free article] [PubMed] [Google Scholar]


