Abstract
A novel mcr colistin resistance gene was identified in a strain of Salmonella enterica, monophasic variant of serovar Typhimurium (4,5,12:i:- ), isolated from a pig at slaughter in Italy in 2013, and in Escherichia coli strains collected during routine diagnostic of post-weaning diarrhoea in pigs from Spain and Belgium in 2015 and 2016. Immediate implementation of mcr-screening including this novel gene variant is required for Salmonella and E. coli from humans and food-producing animals in Europe.
Keywords: colistin, ColE, Salmonella, mcr
The wide use of colistin in veterinary medicine for the control of Enterobacteriaceae infections, and for prophylaxis purposes caused a significant increase in colistin resistance, especially in Escherichia coli from pigs [1].
The primary objective of this study was the identification of an unknown mechanism of colistin resistance in a Salmonellaenterica serovar Typhimurium (4,5,12:i:- ), of our collection [2]. The relevance of the new mcr-4 gene, for the first time here identified, was supported by its occurrence in numerous Escherichia coli of swine origin recently isolated from different European countries.
A novel mcr-gene in Salmonella with origins from Shewanella frigidimarina
In 2013, one colistin-resistant (minimum inhibitory concentration (MIC) = 8 mg/L, measured by broth microdilution) [3] monophasic variant of S. enterica serovar Typhimurium (4,5,12:i:– ) isolate R3445 was obtained from the caecal content of a pig at slaughter, during a study performed by the Istituto Zooprofilattico of Umbria and Marche in Italy [2]. The pig originated from a small finishing herd in central Italy [2].
Whole genome sequencing (WGS) of the strain was performed by an Illumina MiSeq (2 x 300 PE), with the objective to identify the mechanism of colistin resistance. The R3445 genome was negative for mcr-1, mcr-2 and mcr-3 genes. Genome comparative analysis, performed against the S. enterica Typhimurium LT2 genome (NC_003197.2) identified eight contigs > 1 kb that were not present in the reference genome. Three of them contained the chromosomally located resistance genes blaTEM-1B, strA-strB-sul2 and tetB, respectively (identified by ResFinder https://cge.cbs.dtu.dk/services/) [4]. In the remaining five divergent regions, PlasmidFinder (https://cge.cbs.dtu.dk/services/) detected ColE-like replicons [5]. Assembly and annotation analysis suggested the presence of five small extrachromosomal plasmids in the colistin-resistant R3445 Salmonella strain; large plasmids were not identified (Table 1).
Table 1. Characteristics of the mcr-4 positive strains analysed in this study, Italy 2013, Spain and Belgium, 2015 to 2016 (n = 4), and their transformants and conjugants.
| Strain | Country | MLST | Plasmid content | Plasmid-mediated colistin resistance | Additional resistance genes | Colistin MIC mg/L |
|---|---|---|---|---|---|---|
|
R3445
Salmonella enterica |
Italy | 34 | ColE10, ColRNAI_34, ColRNAI_36, ColRNAI_38, ColRNAI_46 |
mcr-4 | bla TEM-1B, strA, strB, sul2, tet(B) | 8 |
| 3445T (transformant in DH5-α) | ColE10 | mcr-4 | 2 | |||
| DH5-α | 0.25 | |||||
|
R4287
Escherichia coli |
Spain | 10 (CC10) | I1, I2, FII, FIB, FIC, HI2, Col156, ColRNAI_34, ColE10 | mcr-4 | bla TEM-1B, aph(4)-1a, aph(3’)-1c, aac(3)-IVa, aac(3)-IIa, strA, aadA1, floR, catA1, sul2, sul1, tet(B), dfrA1, mph(A), erm(B), mphB | 8 |
| 4287C (conjugant in CSH26 RifR) | ColE10, I2 | mcr-4 | 4 | |||
| CSH26 RifR | 0.25 | |||||
|
R4280
E. coli |
Belgium | 10 (CC10) |
I1, FII, FIB, FIC, HI2, Col(MG828), ColRNAI_34, ColE10 | mcr-4 | bla TEM-1B, aph4–1a, aac(3)-IVa, strB, aadA1, floR, catA1, sul1, tet(B), dfrA1, mph(B) | 8 |
|
R4278
E. coli |
Belgium | 7029 | I1, X1, FII(pCoo), ColE10 | mcr-4 | bla TEM-1B, strA, strB, aadA1, sul2, sul1, tet(A), dfrA1 | 16 |
MIC: minimum inhibitory concentration; MLST: multilocus sequence typing.
The 8,749 bp ColE10 plasmid (named pMCR; Figure 1) encoded the RepB replicase, MobA/L mobilisation proteins and the toxin RelE, and showed 99% nucleotide identity, 65% coverage, with the pPSP_ee2 Pantoea spp. plasmid (CP009884).In pMCR an IS5 element (ISKpn6) flanked a region showing 99% nucleotide identity with the genome of Sh. frigidimarina (CP000447.1). The assembly of pMCR was confirmed by PCR-based closure, using the primers listed in Table 2.
Figure 1.
Plasmid content of the Salmonella isolate from swine, Italy 2013 (n = 1) and its transformant 3445T and map of the pMCR plasmid carrying the mcr-4 gene
Left panel: M: 1 Kb Plus DNA ladder (Invitrogen, Italy). EcoRV-restricted plasmids purified from the R3445 Salmonella strain and the pMCR plasmid purified from the 3445T strain obtained by transformation (indicated by the arrow). Among the five ColE-like plasmids predicted by WGS, the pMCR ColE10 was estimated to be 8.7 kb, three copies per cell, while the other four ColE1 ranged from 7.0 to 4.0 kb in size and were estimated at more than 10 copies per cell.
Right panel: map of pMCR in the R3445 genome (http://wolfe.ucd.ie/GenomeVx/). The sequence of this plasmid is released at the NCBI GenBank under accession number MF543359.
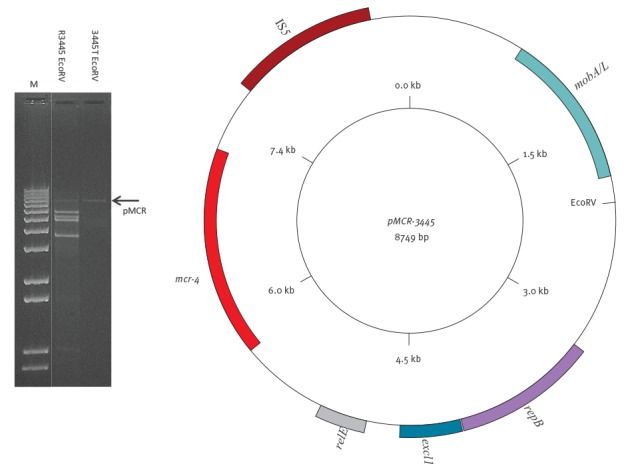
Table 2. Primers used in this study for the detection of the new mcr-4 gene.
| Primer name | Sequence | Target | Amplicon size |
|---|---|---|---|
| Mcr-4 FW | ATTGGGATAGTCGCCTTTTT | primers for the screening of the new mcr-4 gene | 487 bp |
| Mcr-4 RV | TTACAGCCAGAATCATTATCA | ||
| Mcr-4 ext FW | ATCTGTTAAGTTTGTTGGTGAC | external primers to amplify the complete mcr-4 gene | 1,820 bp |
| Mcr-4 ext RV | TGAGAGCTAAATGTAACAATAGA | ||
| ColE10_repBF | TAAAGCATCGTGTAGTGTTTT | primers for the detection of the repB of the ColE10 plasmid | 420 bp |
| ColE10_repBR | ATATAATCTGGAACATGTTAAG | ||
| IS5FW | CATGACCTCAATCAGCTGG | In combination with ColE10_repBF, used for PCR-based closure of pMCR | 4,588 bp |
| IS5 RV | TTTACTGAGATCTCTCCCAC | In combination with Mcr-4-RV, used for PCR-based closure of pMCR | 2,143 bp |
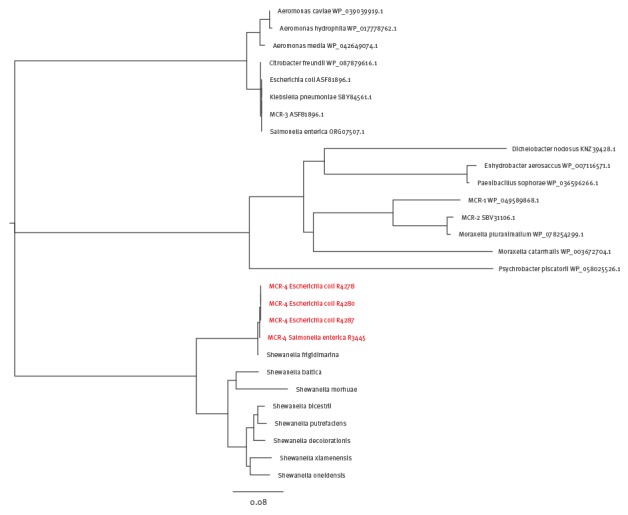
The Sh. frigidimarina region in pMCR encoded only one large protein that had an amino acid sequence with 82.0%–99.0% identity to phosphoethanolamine transferases found in Shewanella species and 34.0%, 35.0% and 49.0% amino acid sequence identity to MCR-1, MCR-2 and MCR-3 respectively (Figure 2).
Figure 2.
Phylogenetic analysis of the entire MCR-4 protein sequence
Phylogenetic tree of the deduced amino acid sequences of putative phosphoethanolamine transferases from different bacterial species, in relation to the MCR-4-positive Salmonella isolate from swine, Italy 2013 and E. coli isolated in Europe in 2016, generated by bootstrap analysis using 1,000 replicates. The analysis was carried out using Geneious, version 10.1.3 [10]. Isolates in this study are highlighted in red.
Conjugation experiments failed to transfer the pMCR plasmid into an E. coli laboratory recipient, but purified plasmid DNA obtained from the R3445 Salmonella strain successfully transformed chemically competent DH5-alpha E. coli cells (Invitrogen, Italy), selecting the transformation on Luria-Bertani agar solid media plates containing 1 mg/L of colistin sulfate. The pMCR transformants showed a MIC of 2 mg/L for colistin, eightfold higher than the empty DH5-alpha recipient (MIC = 0.25 mg/L). pMCR plasmids in the transformants were analysed by restriction and PCR and found identical to that detected in the Salmonella strain (primers in Table 2). The phosphoethanolamine transferase identified on pMCR has been named MCR-4 (GenBank accession number: MF543359).
The novel mcr-4 gene in Escherichia coli strains from pigs in Europe
A total of 125 E. coli isolates were collected in Italy (34 strains), Spain (43 strains) and Belgium (48 strains) in 2015 and 2016, from specimens submitted for routine diagnosis of post-weaning diarrhoea in piglets. Fifty isolates were colistin-resistant with MIC > 2 mg/L [3]. A PCR screening for mcr was performed on the entire collection of 125 isolates: 32 strains were positive for mcr-1 [6], three for mcr-2 [7] and 11 for the novel mcr4 gene identified in the Salmonella isolate R3445 (primers used are shown in Table 2). Nine strains were from piglets from Spain and two from Belgium. The eleven mcr-4-positive strains were also positive for the ColE10 replicase and showed a MIC ≥ 4 mg/L for colistin.
WGSs of three E. coli positive for the new mcr-4 gene, were obtained (R4278, R4280 and R4287 in Table 1). The pMCR plasmid was detected in the three genomes, but several additional plasmids were also identified, belonging to the I1, I2, F, X1 and HI2 families, probably associated with the large repertoire of resistance genes in these E. coli strains (Table 1).
The pMCR plasmid was transferred by conjugation at 37 °C, at a 1 × 10−4 frequency (conjugants/donors colony-forming units), from strain R4287 to a rifampicin-resistant CSH26 E. coli recipient strain. Conjugants were screened by plating 10-fold serial dilutions of the mating mixture on Luria-Bertani agar solid media plates containing 2 mg/L of colistin sulfate and 100 mg/L rifampicin, as resistance markers for pMCR and CSH26, respectively.
All transconjugants contained the pMCR and a co-resident I2 plasmid that acted as the helper plasmid promoting conjugation. The R4287 pMCR conjugants showed MIC = 4 mg/L for colistin, 16-fold higher than the empty CSH26 recipient (Table 1). From strain R4280, pMCR was not transferred by conjugation. Interestingly, this strain did not carry the I2 helper plasmid that was present in R4287.
In the R4278 genome, the pMCR was located on the chromosome integrated within a Type I methylation gene. Two IS5 copies in direct orientation flanked the complete pMCR plasmid sequence, suggesting that a transposition event has driven the chromosomal integration of the plasmid.
Conclusions
The chromosomal mechanism of colistin resistance in Enterobacteriaceae involves mutations in several genes involved in decoration and structural modifications of lipopolysaccharides (LPS) [8]. Plasmid-borne mcr genes encoding phosphoethanolamine transferases, modifying the lipid A have been described as an important mechanism conferring decreased susceptibility to colistin. Since its discovery, the mcr-1 gene has been widely detected in enterobacteria isolated from animals and humans in all the world [8]. In contrast, the mcr-2 variant has been rarely identified, and only in E. coli strains of swine and bovine origin in Belgium [7]. The recently discovered mcr-3 gene has been identified in E. coli, Klebsiella pneumoniae and Salmonella from Asia and the United States [9].
Our findings demonstrate the presence of a new plasmid-mediated colistin mcr-4 gene on a small, not self-conjugative plasmid. ColE are plasmids with a broad host range that can be expected to be able to replicate in different bacterial species and genera. However, the mobilisation of the plasmid needs a helper plasmid to promote conjugation.
We found the mcr-4 gene in a monophasic variant of Salmonella Typhimurium isolated from a pig at slaughter in Italy and in 11 of 125 (9%) E. coli isolates from piglets. Nine of 43 E. coli from Spanish piglets were positive, 37% (9/24) of strains from Spain with a colistin MIC > 2 mg/L. Among the Belgian strains, two positive E. coli among 15 colistin-resistant isolates were found. These findings suggest considerable dissemination of the novel gene in Europe. Screening for this mcr-4 variant should be immediately implemented in zoonotic bacteria, including Salmonella and E. coli from food-producing animals and human sources to understand its diffusion in Europe, and to evaluate the risk for human health posed by this novel plasmid-mediated colistin resistance determinant.
Acknowledgements
We are grateful to Dr. Luca Busani and Dr. Giovanni Rezza for critical reading of the manuscript and to Mr. Sergio Arena for helpful technical assistance. This work was supported by a project to AC financed by the Ministry of Health of Italy (grant number RF-2011-02346987).
Conflict of interest: None declared.
Authors’ contributions: This study was designed by AC and CFM. Isolates were collected by AL, GP and CFM. Experimental work was done by AC, LV, CF, LC and SO. Data was analysed and interpreted by AC, LV, AL, GP and CFM. The manuscript was drafted by AC and LV, and was reviewed by all authors.
References
- 1. Rhouma M, Beaudry F, Letellier A. Resistance to colistin: what is the fate for this antibiotic in pig production? Int J Antimicrob Agents. 2016;48(2):119-26. 10.1016/j.ijantimicag.2016.04.008 [DOI] [PubMed] [Google Scholar]
- 2. Pesciaroli M, Cucco L, De Luca S, Massacci FR, Maresca C, Medici L, et al. Association between pigs with high caecal Salmonella loads and carcass contamination. Int J Food Microbiol. 2017;242:82-6. 10.1016/j.ijfoodmicro.2016.11.021 [DOI] [PubMed] [Google Scholar]
- 3.European Committee on Antimicrobial Susceptibility Testing (EUCAST). Clinical Breakpoints. Växjö: EUCAST. Jun 2017. Available from: http://eucast.org/clinical_breakpoints
- 4. Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640-4. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895-903. 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161-8. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 7. Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016;21(27):30280. 10.2807/1560-7917.ES.2016.21.27.30280 [DOI] [PubMed] [Google Scholar]
- 8. Poirel L, Jayol A, Nordmann P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin Microbiol Rev. 2017;30(2):557-96. 10.1128/CMR.00064-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, et al. Novel Plasmid-Mediated Colistin Resistance Gene mcr-3 in Escherichia coli. MBio. 2017;8(3):e00543-17. 10.1128/mBio.00543-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647-9. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]


