Abstract
Characterizing the proteome composition of organelles and subcellular regions of living cells can facilitate the understanding of cellular organization as well as protein interactome networks. Proximity labeling-based methods coupled with mass spectrometry (MS) offer a high-throughput approach for systematic analysis of spatially restricted proteomes. Proximity labeling utilizes enzymes that generate reactive radicals to covalently tag neighboring proteins with biotin. The biotinylated endogenous proteins can then be isolated for further analysis by MS. To analyze protein–protein interactions or identify components that localize to discrete subcellular compartments, spatial expression is achieved by fusing the enzyme to specific proteins or signal peptides that target to particular subcellular regions. Although these technologies have only been introduced recently, they have already provided deep insights into a wide range of biological processes. Here, we describe and compare current methods of proximity labeling as well as their applications. As each method has its own unique features, the goal of this review is to describe how different proximity labeling methods can be used to answer different biological questions.
INTRODUCTION
Specialized biological processes occur in different organelles and subcellular regions. In addition, protein functions correlate with their subcellular localizations and interactions. Understanding how cellular structures underlie specialized functions requires the comprehensive identification of proteins within spatially defined cellular domains. Furthermore, identification of interacting proteins is key to elucidating the mechanisms underlying complex cellular processes.
Mass spectrometry (MS) techniques have been used to systematically characterize the proteome of isolated organelles and protein interactors purified by affinity pull-down or following crosslinking. However, these approaches are limited by available purification methods, as it is not possible in many cases to obtain intact organelles of high purity. Moreover, even when purification is possible, contamination that results in false-positive identification is common. For example, false positives may be introduced by cellular disruption, as two proteins that normally localize in different subcellular regions may artificially interact when membranes are disrupted. In addition, false negatives often occur due to loss of components caused by disruption of isolated organelles or protein complexes. Additionally, a variety of discreet cellular regions cannot be purified by centrifugation, such as specialized endoplasmic reticulum (ER)–plasma membrane (PM) junctions that are critical for lipid metabolism and Ca2+ signaling.1–4 Similarly, transient or weak interactions may be lost during purification of a protein interactome due to stringent washes.
Recently, proximity-dependent biotin labeling methods have been developed and utilized for mapping compartmental proteome and protein interactomes. In this review, we compare proximity labeling techniques that utilize different enzymes and describe how they are used to address limitations of traditional methods.
OVERVIEW OF ENZYME-CATALYZED PROXIMITY LABELING FOR PROTEOMIC PROFILING
In general, proximity labeling relies on enzymes that convert a substrate into a reactive radical that covalently tags neighboring proteins with biotin. We will discuss three major enzymes utilized for proximity labeling: proximity-dependent biotin identification (BioID), horseradish peroxidase (HRP), and engineered ascorbate peroxidase (APEX).
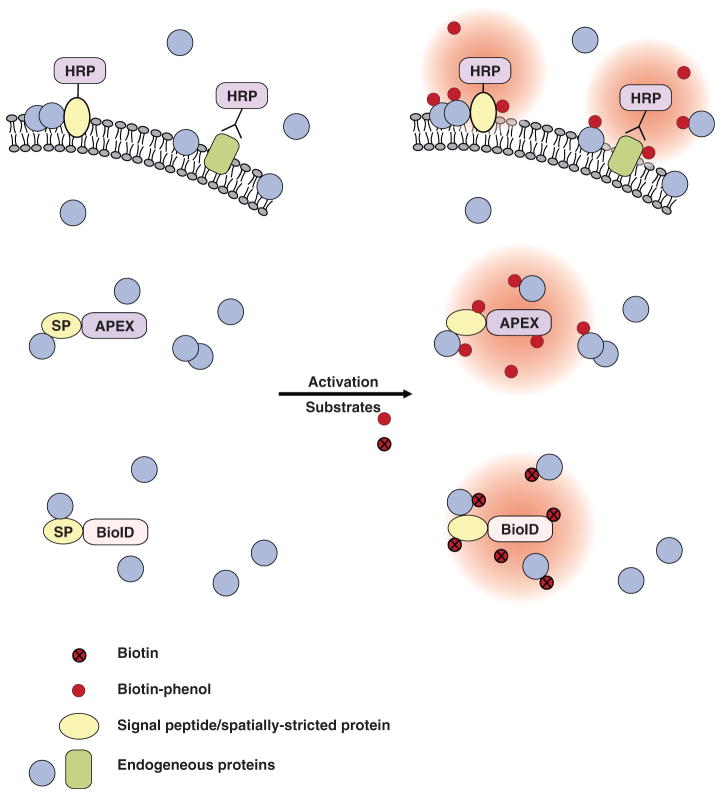
To achieve spatially restricted labeling, the enzymes are usually fused with a targeting signal peptide or a protein of interest. After performing proximity labeling in living cells, cells are then lysed and biotinylated endogenous proteins are isolated using streptavidin beads. Small peptides from enriched proteins are generated by trypsin digestion and subsequently ionized for MS analysis. The mass-to-charge (m/z) ratio of each peptide is then used to identify the peptide sequence, usually through computational comparison against an established database (Figure 1).
FIGURE 1.
Proximity labeling for proteomic profiling. To achieve regional protein labeling, the enzymes are usually fused with a targeting signal peptide or a spatially restricted protein (SP). The enzymes can also be fused with any protein of interest for protein interactome studies. After performing proximity labeling in living cells, the cells are lysed and the biotinylated endogenous proteins are isolated using steptavidin beads. Small peptides of enriched proteins are generated by trypsin digestion and subsequently ionized for mass spectrometry analysis. The mass-to-charge (m/z) ratio of each peptide is then used to identify peptide sequence usually through computational comparison against established databases.
To distinguish potential candidates from background, proteins with the highest abundance are usually chosen for further study as a semi-quantitative approach even though low-abundance candidates may potentially be biologically relevant. Alternatively, to generate a high-confidence and comprehensive list of candidates from MS data, proximity labeling has been coupled with quantitative MS. Quantitative MS can be achieved using metabolic labeling such as stable isotope labeling by amino acid in cell culture (SILAC).5 Cells or organisms are grown with supplement of arginine and lysine residues containing stable 13C and/or 15N to synthesize proteins with altered mass. In LC–MS, each trypsin-digested peptide contains altered amino acids that induce a small shift in the m/z ratio, thus allowing the relative quantification of peptides from different samples. Alternatively, quantitative MS can be done with in vitro chemical labeling, such as isobaric tags for relative and absolute quantification (iTRAQ)6 and tandem mass tags (TMT),7 whereby peptides from different samples are modified with chemical tags that can alter the m/z ratio to be detected by MS.
Importantly, with proximity labeling, cells remain intact when the proteome or interactome is labeled. Thus, the potential for false-positive identifications is minimized, as artificial interactions caused by disruption of cells and contaminants during purification steps no longer affect the results. Moreover, proximity labeling can be applied to bypass organelle purification steps, offering an alternative approach for systematic proteomic characterization in live cells. As proximity labeling is an emerging method that enables proteomic profiling of organelles and subcellular domains as well as protein interactomes, this review aims to provide an overview of the different methods to aid planning and execution of future experiments.
BioID-BASED PROXIMITY LABELING
BioID-based proximity labeling employs a mutant form of the biotin ligase BirA from E. coli.8–10 The biotin ligase BirA is a conserved enzyme that mediates the attachment of biotin to target proteins.11 In the presence of ATP, BirA biotinylates proteins by catalyzing the conversion of biotin to reactive biotinoyl-5′-AMP, which specifically tags a lysine residue of a subunit of the acetyl-CoA carboxylase.8,12 Wild-type BirA has a high affinity to biotinol-5′-AMP and keeps it in the active site until the acetyl-CoA carboxylase, or a short acceptor peptide, becomes available.13 As BirA has a high specificity for its target sequence, it has been used to study specific protein–protein interactions14: BirA is fused to a bait protein and biotin acceptor peptide (BAP) is fused to a prey protein. If the interaction occurs, the prey will be close enough to the bait to become biotinylated.
To achieve promiscuous labeling, the active site of BirA has been mutated, enabling random biotinylation of vicinity proteins without BAP.8,9 This method is named BioID and the mutated form of BirA for proximity labeling is called BioID or BirA* to be distinguished from the wild-type and other mutant forms of BirA10(Figure 2). When the active site of BirA is mutated (R118G), its affinity to biotin-5′-AMP is greatly reduced. The highly reactive biotinoyl-5′-AMP is released from the active site of BioID and nonspecifically reacts with nearby proteins. Therefore, BioID can covalently tag nearby endogenous proteins on lysine residues. Although the labeling radius of BioID may vary depending on the local environmental, the labeling radius of BioID is estimated to be around 10 nm using the structure of the nuclear pore complex as a ‘molecular ruler.’15
FIGURE 2.
Proximity labeling methods. BioID, a mutant form of the biotin ligase BirA, can convert biotin into radicals that can covalently tag neighboring proteins on lysine residues. HRP and APEX are peroxidases that, when activated by H2O2, are able to turn biotin-phenol substrates into highly reactive radicals that covalently tag neighboring proteins on electron-rich amino acids. In addition, fluorescein-aryl azide or biotin-aryl azide have been used for HRP-mediated proximity labeling (not shown in the figure). HRP is inactive in a reducing environment, such as the cytosol, but functions extracellularly. APEX, engineered ascorbate peroxidase; BioID, proximity-dependent biotin identification; HRP, horseradish peroxidase; SP, spatially restricted protein.
BioID has been used to map local interactomes and identify transient protein interactions, and thus provides a better understanding of cellular structures as well as interactions occurring during signal transduction. Recently, a new version of BioID, BioID2, has been generated with an R40G mutation in the reactive site of BirA from Aquifex aeolicus to allow promiscuous labeling.16 BioID2 lacks the DNA binding domain at the N-terminus and is thus smaller (233 a.a.) than the original BioID derived from E. coli (321 a.a.). BioID2 performs similar labeling chemistry as BioID but shows a higher activity and requires less biotin. As the application and impact of BioID have been extensively reviewed,17,18 we focus here on introducing the more recently developed APEX-mediated approach and on the comparisons between labeling methods.
HRP-BASED PROXIMITY LABELING
HRP is a peroxidase that, when activated by H2O2, is able to convert a substrate into a highly reactive radical that covalently tags neighboring proteins on electron-rich amino acids.19 HRP is inactive in a reducing environment, such as the cytosol, because the structure of HRP, which is maintained with four disulfide bonds and two Ca2+ ion-binding sites, is disrupted in reducing conditions.20 This has limited its use for determining intracellular interactomes, and motivated the development of APEX. Nevertheless, HRP is active in oxidizing environments, such as the lumen of the ER or the Golgi and the extracellular region. Thus, HRP has been used for proteomic mapping on the surface of living cells. In addition, HRP can also be used as an electron microscopy (EM) tag.21 With H2O2, HRP can catalyze the polymerization of 3,3′-diaminobenzidine (DAB), which precipitates and creates an EM contrast after OsO4 fixation.
Although HRP can catalyze a variety of substrates, for proximity labeling two in particular have been used: (1) the enzyme-mediated activation of radical source (EMARS) method uses fluorescein arylazide or biotin arylazide.22–30 Fluorescein arylazide reduces the cytosolic background generated by biotin-aryl azide,23 which is membrane permeable during the EMARS reaction and activated by endogenous enzymes22,24; and (2) the selective proteomic proximity labeling assay using tyramide (SPPLAT) method using biotin-tyramide, which is also known as biotin-phenol.19,31
HRP has been used extensively for other applications, such as ELISA and immunochemistry.32 Furthermore, antibody–HRP conjugates have been generated that can also be used for proximity labeling. However, this application is limited by the affinity of the antibody. Nevertheless, antibody–HRP conjugates have been successfully used to identify cell surface molecules such as the composition of the B-cell receptor cluster, as well as proteins that interact with Thy1, β1 integrin, CD20, and PrPC.19,22–30
APEX-BASED PROXIMITY LABELING
APEX, an engineered ascorbate peroxidase derived from plants, uses the same labeling chemistry and rapid kinetics as HRP to convert a substrate into a radical in the presence of H2O2.33,34 The key advantage of APEX over HRP, however, is that it remains active in the reducing environment of the cellular cytosol. Upon activation by H2O2, APEX catalyzes the conversion of its substrate biotin-phenol into short-lived (<1 ms) and highly reactive radicals, which can covalently attach to electron-rich amino acids such as tyrosine in nearby endogenous proteins.34,35 The labeling reaction can be stopped by the removal of H2O2 and the addition of quenching buffer, and the resulting biotinylated proteins can be subsequently isolated using streptavidin beads and further analyzed by MS. In addition, APEX can catalyze the polymerization and precipitation of DAB creating a contrast after OsO4 fixation,33 which can then be used for EM to visualize the structures where APEX is expressed.
Yeast display selection has been performed to screen for mutations that increase APEX activity.36 An improved version of APEX, called APEX2, has one additional mutation (A134P) and catalyzes the same chemistry as APEX but with higher activity and sensitivity for promiscuous labeling and EM.
APEX-mediated proximity labeling was first introduced by Rhee and colleagues to circumvent the limitations of traditional mitochondrial purification and to achieve spatial and temporal specificity of organelle proteome mapping.34 To examine the ability of APEX in proteomic labeling, a mitochondrial matrix-targeted APEX was used in human embryonic kidney (HEK) cells. Mitochondria are comprised of an outer membrane (OMM) and an inner membrane (IMM). The mitochondrial matrix is the most inner subcompartmental region surrounded by the IMM. The region located between the OMM and IMM is called the intermembrane space (IMS). To induce APEX-mediated proximity labeling, cells were preincubated with biotin-phenol followed by 1 min H2O2 treatment for APEX activation. To eliminate the background from endogenous biotinylated proteins and nonspecific candidates caused by the labeling procedure, a two-state SILAC labeling coupled with MS was used for relative quantification. Prior to APEX labeling, one group of cells expressing the mitochondrial matrix localized mito-APEX was cultured with heavy media (heavy arginine and lysine), and another group of cells without APEX, used as a negative control, was cultured with light medium (light arginine and lysine). After cell lysis, biotinylated endogenous proteins were enriched using streptavidin beads and then processed for MS analysis. Consequently, 495 proteins were identified as putative components of the mitochondrial matrix. Ninety-four percent of these proteins had prior mitochondrial annotation. This not only demonstrates that APEX-mediated proteomic mapping provides high specificity but also implies that the other 6% (31 proteins) are novel mitochondrial proteins. In addition, mito-APEX labeling provided a high coverage (85%) of known functional mitochondrial components.
The power of quantitative MS and the importance of appropriate controls are well illustrated by the study of mitochondrial IMS.35 As the outer mitochondrial membrane has pores that permit passive diffusion, highly reactive radicals generated by IMS-APEX diffuse within and out of the IMS to biotinylate cytosolic proteins. Simply using cells without APEX as a negative control is not sufficient to filter out the false-positively labeled cytosolic proteins. To solve this problem, an additional cytosolic APEX control, referred to as ratiometric APEX tagging, was used with a three-state SILAC experiment for quantitative MS: cells labeled with IMS-APEX, with cytosolic NES-APEX, or without APEX as a negative control, were cultured with heavy, medium, and light SILAC media. Using the biotinylated ratio among these three samples to define the IMS proteome, mitochondrial specificity was improved (from 40 to 80%) while the coverage of known IMS proteins dropped only very slightly (from 69 to 67%). The drop in coverage may represent false-negative results due to the removal of proteins localized both in the IMS and in the cytosol.
As biotin-phenoxyl radicals are not membrane-permeable,34 APEX is excellent for proteomic profiling of membrane-enclosed subcellular compartments, such as the mitochondrial matrix. Nevertheless, APEX is not limited to analysis of membrane-enclosed organelles, and has been used successfully to profile the proteome of nonmembrane enclosed organelles.37 Using cilia-localized APEX, signaling molecules such as the kinases PKA, AMPK, and LKB1 were identified in primary cilia. Rather than using quantitative MS, enrichment was estimated by comparing spectral counts from the labeled cilia-APEX sample to those from unlabeled cilia-APEX and a labeled control-APEX sample. Moreover, a proteomic profile of dysfunctional cilia was obtained by comparing cilia-APEX-based profiling of wild-type and lft27 mutant cells, and identified abnormal accumulation of factors. In addition to successful use of APEX for organelle profiling, APEX also provides a good tool for identification of protein–protein interactions. For example, APEX2 was fused with the ER-resident Ca2+ sensor STIM1 to map the proteome of the ER–PM junction in living cells, leading to the identification of the STIM-activating enhancer TMEM110 (STIMATE).38
The APEX-based approach has also been used successfully in vivo.39 APEX was shown to be active in different Drosophila tissues when expressed in multiple subcellular domains. A mito-APEX construct was used to characterize the mitochondrial matrix proteome in live Drosophila muscle cells. Muscle tissue with or without mito-APEX expression was dissected and labeled with biotin-phenol, and biotinylated proteins were subsequently isolated using streptavidin beads. After on-bead tryptic digestion, peptides from control muscles that do not express APEX and muscles expressing mitochondrial APEX were chemically labeled with 4-plex iTRAQ and simultaneously analyzed by MS. iTRAQ enables comparison between samples of muscle with and without mito-APEX expression, allowing one to distinguish between proteins specifically biotinylated by APEX and endogenously biotinylated proteins. Use of an APEX-based method for mitochondrial matrix mapping in Drosophila achieved an excellent specificity (83% compared with 49–57% for the isolation-based approach) with high coverage.
The APEX2 approach has been further adapted to yeast cells.40 Additional steps that boost biotin-phenol delivery are required to achieve optimal APEX2 labeling in yeast.40,41 In addition, an increase in osmolarity has been shown to promote biotin-phenol uptake or retention, significantly increasing APEX2 labeling in the fission yeast Schizosaccharomyces pombe. Interestingly, high osmolarity alone is insufficient to achieve proper APEX2 labeling in the budding yeast Saccharomyces cerevisiae. Disruption of cell wall integrity using Zymolyase dramatically improved the labeling efficiency in budding yeast, even though the general composition of the cell wall in these two species of yeast is similar. Using the optimized protocol for APEX2 labeling in yeast, a binding protein of the Golgi-localized protease Rbd2 was identified and shown to function in the ability of Rbd2 to recognize its targets.42
COMPARISON BETWEEN BIOTIN LIGASE-BASED AND PEROXIDASE-BASED APPROACHES
The major differences between biotin ligase-based and peroxidase-based labeling approaches are the substrates, the targeted amino acid(s), the kinetics, and the working conditions (Table 1). In addition to differences in proteomic labeling, APEX, like HRP, can be used for EM, thus allowing confirmation of fine subcellular localization. However, the proper expression and localization of BioID can only be verified by other methods like immunostaining and/or Western blotting to rule out the possibility of false positive from mis-localization of the fusion proteins or slow translation of the fusion protein.
TABLE 1.
Comparison of the Different Proximity Labeling Methods
| Enzyme | BioID | HRP | APEX |
|---|---|---|---|
| Enzymatic activity | Biotin ligase based | Peroxidase based | Peroxidase based |
| Labeling target | Lysine | Electron-rich amino acids | Tyrosine and potentially other electron-rich amino acids |
| Size (kD) | 35 | 44 | 27 |
| Labeling time | 15–24 h | 5–10 min | 1 min |
| Incubation time with substrate | 15–24 h | 5–10 min | 30–60 min |
| Activation by H2O2 | No | Yes | Yes |
| Substrates for protein labeling | Biotin | Biotin-phenol (and biotin- or fluorescein-acylazide) | Biotin-phenol |
| Half-life of generated radicals | Mins | <1 ms | <1 ms |
| Active region | Intracellular | Extracellular, secretory pathway (inactive in cytosol) | Intracellular |
| Available variants | BioID2 (27 kD) | Split HRP | APEX2 |
| Note | Reduced activity below 37 degrees | Can be used as an EM tag; HRP- conjugated antibodies available | Can be used as an EM tag |
| Organisms | Mammalian cells, xenograft tumors in mice, Trypanosoma brucei, Toxoplasma gondii, Dictyostelium discoideum, Plasmodium berghei | Mammalian cells | Mammalian cells, Schizosaccharomyces pombe, Saccharomyces cerevisiae, Drosophila melanogaster |
APEX, engineered ascorbate peroxidase; BioID, proximity-dependent biotin identification; EM, electron microscopy; HRP, horseradish peroxidase.
Whereas the biotin ligase-based method uses biotin as a substrate, the peroxidase-based approaches use biotin-phenol as the substrate for proteomic analysis. Delivery of the substrate to the region of interest is a critical factor. Biotin is actively imported into mammalian cells and other organisms though distinct mechanisms.43 Even though biotin-phenol can be simply incubated with mammalian cells for cytosolic and mitochondrial protein labeling, a number of studies have shown that biotin-phenol may not effectively penetrate membranes.19,31 Moreover, high osmolarity and perturbation of the cell wall are required for efficient delivery of biotin-phenol and optimal proximity labeling in yeast.40 Therefore, optimizing biotin-phenol delivery to a region of interest in a specific cell type may be required to achieve successful protein labeling. In addition, the half-life of BioID generated biotin-5′-AMP radicals is on the order of minutes in aqueous solutions,44 which is longer than that of APEX-generated biotin-phenoxyl radicals (<1 ms).34,35 The shorter half-life of unstable radicals may result in a smaller labeling radius, which is also determined by other factors, such as local intracellular environments. Unfortunately, the labeling radius of BioID and APEX has been estimated by different methods and in different cellular regions.
BioID labels lysine residues of nearby proteins whereas APEX and HRP tag electron-rich tyrosine residues. Generally, the estimated amount of lysine present in proteins is higher than that of tyrosine.45,46 Thus, when the number of available tyrosine residues is limited, potential target proteins may not be identified using APEX and HRP.
Importantly, BioID shows much slower kinetics than APEX or HRP. The optimal labeling time for APEX (~1 min) is shorter than that for HRP (5–10 min) and much shorter than for BioID (15–24 h). Although biotin is not toxic, biotinylation of proteins over the long BioID labeling period may perturb protein function or lead to artificial interactions. In addition to the unique chemical attributes of each type of reaction, there is a dramatic difference in the temporal landscape of the proteomes obtained by the two methods. While BioID is useful in capturing entire changes in protein complexes during a longer period of time, APEX is excellent for characterizing rapid dynamic changes in proteomes that can only be achieved with a short labeling window, such as acute responses to drug treatment.
Notably, the activity of BioID or BioID2 is greatly reduced at temperatures below 37°C.16 For model systems that need to be maintained under 37°C, BioID cannot be easily used. Nevertheless, BioID has been successfully applied to several organisms in addition to mammalian cells,47–54 as summarized in Table 1. However, APEX has been shown to be active in Drosophila cultured cells at 25°C and in yeast cultured at room temperature, in addition to showing good activity in mammalian cells that are cultured at 37°C. This temperature range allows APEX to be broadly suitable for studies in a variety of model organisms.
COMPARISON BETWEEN APEX AND HRP-BASED APPROACHES
In contrast to BioID, both APEX and HRP catalyze the same proximity labeling chemistry. The key parameter that one should consider for their usage is the environment to which the enzyme will be exposed. As mentioned above, HRP is inactive in the cytosol; however, it is functional when it faces outside the cell on the cell surface and has been successfully used to identify membrane proteins.19,22–30 Notably, many previous studies used antibody-conjugated HRP.19,22–24,26–30 A key advantage of the HRP-mediated approach is that many antibody–HRP conjugates are currently available. As noted previously, however, the use of antibody-conjugated HRP in proximity labeling is limited by the affinity of the antibody.
Interestingly, a bimolecular complementation version of HRP has recently been reported.55 This split HRP has been generated to characterize intercellular protein–protein interactions and visualize synapses. The two split HRP fragments were fused with neurexin and neuroligin, which bind to each other across the synaptic cleft. When the split fragments are brought together as a result of the neurexin–neuroligin interaction, they reconstitute a functional form of HRP that allows proximity labeling. This binary system offers another level of control to the HRP system, making it useful for finer spatial restriction. Although split HRP has not yet been used for proteomics, its potential use for proteomic mapping of cell–cell interactions is very promising.
CONCLUSION/PERSPECTIVES
Since the recent introduction of proximity labeling, the method has made significant contributions to the mapping of local interactomes relevant to a wide range of biological processes. By tagging regional proteomes, proximity labeling overcomes issues associated with traditional approaches of organelle purification and allows proteomic analysis of other types of subcellular regions. A disadvantage that all proximity labeling-based methods have in common is that they cannot distinguish direct binding of two proteins from proximity of two adjacent proteins. Thus, these methods serve as discovery methods that require detailed follow-up studies. Nevertheless, as proximity labeling does not require disruption of cells for complex isolation, these methods not only preserve evidence of weak or transient interactions that are not detectable using traditional approaches but also minimizes false discovery by eliminating false positives generated during lysis or disruption. Importantly, as proximity labeling can be performed in living cells, researchers can study protein–protein interactions and proteomic alterations in physiologically relevant conditions. Proximity labeling has been adapted to several model systems, making this technology available to study diverse biological problems in a wide range of organisms.
Notably, while improved second-generation BioID and APEX are now available, further improvements are likely to be made in the near future. In particular, variants of BioID with faster kinetics and higher activity at lower temperatures would be more suitable for a broad range of model organisms. In addition, a split APEX2 that could reconstitute a functional APEX would also find many applications. For mapping specific subcellular regions, such as organelle–organelle interaction sites, split APEX2 may provide a way to further fine-tune spatial restrictions.
Importantly, the ease of applying genetically encoded enzymes will benefit greatly from the powerful genome editing using CRISPR technology,56,57 as these enzymes can now be easily fused to any gene of interest via a knock-in approach. In addition, numerous genetic engineering tools already available for organisms such as Drosophila facilitate a wide range of proximity-labeling applications. For example, the existing library of MiMICs, a transposon insertion resource for engineering Drosophila genes, allows for rapid tagging of genes.58,59 Altogether, a broad range of proximity-labeling applications that build on existing tools are now possible and likely to provide deep insights into various biological questions.
Acknowledgments
We thank Alice Ting and members of her laboratory for stimulating discussions of APEX-based methods over the past few years. We are grateful to Raghuvir Viswanatha, Justin Bosch, Stephanie Mohr, and Richard Binari for helpful comments on the manuscript. This work was supported in part by P01-CA120964, R21-ES025615, and R01-DK088718 to N.P. and the HCIA Program from HHMI. N.P. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Conflict of interest: The authors have declared no conflicts of interest for this article.
References
- 1.Carrasco S, Meyer T. STIM proteins and the endoplasmic reticulum-plasma membrane junctions. Annu Rev Biochem. 2011;80:973–1000. doi: 10.1146/annurev-biochem-061609-165311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hogan PG, Lewis RS, Rao A. Molecular basis of calcium signaling in lymphocytes: STIM and ORAI. Annu Rev Immunol. 2010;28:491–533. doi: 10.1146/annurev.immunol.021908.132550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stefan CJ, Manford AG, Emr SD. ER-PM connections: sites of information transfer and inter-organelle communication. Curr Opin Cell Biol. 2013;25:434–442. doi: 10.1016/j.ceb.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elbaz Y, Schuldiner M. Staying in touch: the molecular era of organelle contact sites. Trends Biochem Sci. 2011;36:616–623. doi: 10.1016/j.tibs.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 6.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 8.Choi-Rhee E, Schulman H, Cronan JE. Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Sci. 2004;13:3043–3050. doi: 10.1110/ps.04911804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cronan JE. Targeted and proximity-dependent promiscuous protein biotinylation by a mutant Escherichia coli biotin protein ligase. J Nutr Biochem. 2005;16:416–418. doi: 10.1016/j.jnutbio.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 10.Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakravartty V, Cronan JE. Altered regulation of Escherichia coli biotin biosynthesis in BirA superrepressor mutant strains. J Bacteriol. 2012;194:1113–1126. doi: 10.1128/JB.06549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapman-Smith A, Cronan JE., Jr Molecular biology of biotin attachment to proteins. J Nutr. 1999;129:477S–484S. doi: 10.1093/jn/129.2.477S. [DOI] [PubMed] [Google Scholar]
- 13.Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez-Suarez M, Chen TS, Ting AY. Protein-protein interaction detection in vitro and in cells by proximity biotinylation. J Am Chem Soc. 2008;130:9251–9253. doi: 10.1021/ja801445p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci USA. 2014;111:E2453–E2461. doi: 10.1073/pnas.1406459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim DI, Jensen SC, Noble KA, Kc B, Roux KH, Motamedchaboki K, Roux KJ. An improved smaller biotin ligase for BioID proximity labeling. Mol Biol Cell. 2016;27:1188–1196. doi: 10.1091/mbc.E15-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim DI, Roux KJ. Filling the void: proximity-based labeling of proteins in living cells. Trends Cell Biol. 2016;26:804–817. doi: 10.1016/j.tcb.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varnaite R, MacNeill SA. Meet the neighbors: mapping local protein interactomes by proximity-dependent labeling with BioID. Proteomics. 2016;16:2503–2518. doi: 10.1002/pmic.201600123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li XW, Rees JS, Xue P, Zhang H, Hamaia SW, Sanderson B, Funk PE, Farndale RW, Lilley KS, Perrett S, et al. New insights into the DT40 B cell receptor cluster using a proteomic proximity labeling assay. J Biol Chem. 2014;289:14434–14447. doi: 10.1074/jbc.M113.529578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopkins C, Gibson A, Stinchcombe J, Futter C. Chimeric molecules employing horseradish peroxidase as reporter enzyme for protein localization in the electron microscope. Methods Enzymol. 2000;327:35–45. doi: 10.1016/s0076-6879(00)27265-0. [DOI] [PubMed] [Google Scholar]
- 21.Ellisman MH, Deerinck TJ, Shu X, Sosinsky GE. Picking faces out of a crowd: genetic labels for identification of proteins in correlated light and electron microscopy imaging. Methods Cell Biol. 2012;111:139–155. doi: 10.1016/B978-0-12-416026-2.00008-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honke K, Kotani N. Identification of cell-surface molecular interactions under living conditions by using the enzyme-mediated activation of radical sources (EMARS) method. Sensors. 2012;12:16037–16045. doi: 10.3390/s121216037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang S, Kotani N, Ohnishi T, Miyagawa-Yamguchi A, Tsuda M, Yamashita R, Ishiura Y, Honke K. A proteomics approach to the cell-surface interactome using the enzyme-mediated activation of radical sources reaction. Proteomics. 2012;12:54–62. doi: 10.1002/pmic.201100551. [DOI] [PubMed] [Google Scholar]
- 24.Kotani N, Gu J, Isaji T, Udaka K, Taniguchi N, Honke K. Biochemical visualization of cell surface molecular clustering in living cells. Proc Natl Acad Sci USA. 2008;105:7405–7409. doi: 10.1073/pnas.0710346105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyagawa-Yamaguchi A, Kotani N, Honke K. Expressed glycosylphosphatidylinositol-anchored horse-radish peroxidase identifies co-clustering molecules in individual lipid raft domains. PLoS One. 2014;9:e93054. doi: 10.1371/journal.pone.0093054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyagawa-Yamaguchi A, Kotani N, Honke K. Each GPI-anchored protein species forms a specific lipid raft depending on its GPI attachment signal. Glycoconj J. 2015;32:531–540. doi: 10.1007/s10719-015-9595-5. [DOI] [PubMed] [Google Scholar]
- 27.Iwamaru Y, Kitani H, Okada H, Takenouchi T, Shimizu Y, Imamura M, Miyazawa K, Murayama Y, Hoover EA, Yokoyama T. Proximity of SCG10 and prion protein in membrane rafts. J Neurochem. 2016;136:204–1218. doi: 10.1111/jnc.13488. [DOI] [PubMed] [Google Scholar]
- 28.Hashimoto N, Hamamura K, Kotani N, Furukawa K, Kaneko K, Honke K. Proteomic analysis of ganglioside-associated membrane molecules: substantial basis for molecular clustering. Proteomics. 2012;12:3154–3163. doi: 10.1002/pmic.201200279. [DOI] [PubMed] [Google Scholar]
- 29.Ishiura Y, Kotani N, Yamashita R, Yamamoto H, Kozutsumi Y, Honke K. Anomalous expression of Thy1 (CD90) in B-cell lymphoma cells and proliferation inhibition by anti-Thy1 antibody treatment. Biochem Biophys Res Commun. 2010;396:329–334. doi: 10.1016/j.bbrc.2010.04.092. [DOI] [PubMed] [Google Scholar]
- 30.Yamashita R, Kotani N, Ishiura Y, Higashiyama S, Honke K. Spatiotemporally-regulated interaction between beta1 integrin and ErbB4 that is involved in fibronectin-dependent cell migration. J Biochem. 2011;149:347–355. doi: 10.1093/jb/mvq148. [DOI] [PubMed] [Google Scholar]
- 31.Rees JS, Li XW, Perrett S, Lilley KS, Jackson AP. Selective proteomic proximity labeling assay using tyramide (SPPLAT): a quantitative method for the proteomic analysis of localized membrane-bound protein clusters. Curr Protoc Protein Sci. 2015;80:11–18. doi: 10.1002/0471140864.ps1927s80. [DOI] [PubMed] [Google Scholar]
- 32.Ryan BJ, Carolan N, O’Fagain C. Horseradish and soybean peroxidases: comparable tools for alternative niches? Trends Biotechnol. 2006;24:355–363. doi: 10.1016/j.tibtech.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, Ting AY. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hung V, Zou P, Rhee HW, Udeshi ND, Cracan V, Svinkina T, Carr SA, Mootha VK, Ting AY. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol Cell. 2014;55:332–341. doi: 10.1016/j.molcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, Ting AY. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods. 2015;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mick DU, Rodrigues RB, Leib RD, Adams CM, Chien AS, Gygi SP, Nachury MV. Proteomics of primary cilia by proximity labeling. Dev Cell. 2015;35:497–512. doi: 10.1016/j.devcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jing J, He L, Sun A, Quintana A, Ding Y, Ma G, Tan P, Liang X, Zheng X, Chen L, et al. Proteomic mapping of ER-PM junctions identifies STIMATE as a regulator of Ca(2)(+) influx. Nat Cell Biol. 2015;17:1339–1347. doi: 10.1038/ncb3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen CL, Hu Y, Udeshi ND, Lau TY, Wirtz-Peitz F, He L, Ting AY, Carr SA, Perrimon N. Proteomic mapping in live Drosophila tissues using an engineered ascorbate peroxidase. Proc Natl Acad Sci USA. 2015;112:12093–12098. doi: 10.1073/pnas.1515623112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hwang J, Espenshade PJ. Proximity-dependent biotin labelling in yeast using the engineered ascorbate peroxidase APEX2. Biochem J. 2016;473:2463–2469. doi: 10.1042/BCJ20160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hung V, Udeshi ND, Lam SS, Loh KH, Cox KJ, Pedram K, Carr SA, Ting AY. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat Protoc. 2016;11:456–475. doi: 10.1038/nprot.2016.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang J, Ribbens D, Raychaudhuri S, Cairns L, Gu H, Frost A, Urban S, Espenshade PJ. A Golgi rhomboid protease Rbd2 recruits Cdc48 to cleave yeast SREBP. EMBO J. 2016;35:2332–2349. doi: 10.15252/embj.201693923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Azhar A, Booker GW, Polyak SW. Mechanisms of biotin transport. Biochem Anal Biochem. 2015;4:8. [Google Scholar]
- 44.Demoss JA, Genuth SM, Novelli GD. The enzymatic activation of amino acids via their acyl-adenylate derivatives. Proc Natl Acad Sci USA. 1956;42:325–332. doi: 10.1073/pnas.42.6.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Echols N, Harrison P, Balasubramanian S, Luscombe NM, Bertone P, Zhang Z, Gerstein M. Comprehensive analysis of amino acid and nucleotide composition in eukaryotic genomes, comparing genes and pseudogenes. Nucleic Acids Res. 2002;30:2515–2523. doi: 10.1093/nar/30.11.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tourasse NJ, Li WH. Selective constraints, amino acid composition, and the rate of protein evolution. Mol Biol Evol. 2000;17:656–664. doi: 10.1093/oxfordjournals.molbev.a026344. [DOI] [PubMed] [Google Scholar]
- 47.Dingar D, Kalkat M, Chan PK, Srikumar T, Bailey SD, Tu WB, Coyaud E, Ponzielli R, Kolyar M, Jurisica I, et al. BioID identifies novel c-MYC interacting partners in cultured cells and xenograft tumors. J Proteomics. 2015;118:95–111. doi: 10.1016/j.jprot.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 48.Hu H, Zhou Q, Li Z. SAS-4 protein in Trypanosoma brucei controls life cycle transitions by modulating the length of the flagellum attachment zone filament. J Biol Chem. 2015;290:30453–30463. doi: 10.1074/jbc.M115.694109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McAllaster MR, Ikeda KN, Lozano-Nunez A, Anrather D, Unterwurzacher V, Gossenreiter T, Perry JA, Crickley R, Mercadante CJ, Vaughan S, et al. Proteomic identification of novel cytoskeletal proteins associated with TbPLK, an essential regulator of cell morphogenesis in Trypanosoma brucei. Mol Biol Cell. 2015;26:3013–3029. doi: 10.1091/mbc.E15-04-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morriswood B, Havlicek K, Demmel L, Yavuz S, Sealey-Cardona M, Vidilaseris K, Anrather D, Kostan J, Djinovic-Carugo K, Roux KJ, et al. Novel bilobe components in Trypanosoma brucei identified using proximity-dependent biotinylation. Eukaryot Cell. 2013;12:356–367. doi: 10.1128/EC.00326-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Q, Hu H, Li Z. An EF-hand-containing protein in Trypanosoma brucei regulates cytokinesis initiation by maintaining the stability of the cytokinesis initiation factor CIF1. J Biol Chem. 2016;291:14395–14409. doi: 10.1074/jbc.M116.726133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kehrer J, Frischknecht F, Mair GR. Proteomic analysis of the Plasmodium berghei gametocyte egressome and vesicular bioID of osmiophilic body proteins identifies merozoite TRAP-like protein (MTRAP) as an essential factor for parasite transmission. Mol Cell Proteomics. 2016;15:2852–2862. doi: 10.1074/mcp.M116.058263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen AL, Kim EW, Toh JY, Vashisht AA, Rashoff AQ, Van C, Huang AS, Moon AS, Bell HN, Bentolila LA, et al. Novel components of the Toxoplasma inner membrane complex revealed by BioID. mBio. 2015;6:e02357–14. doi: 10.1128/mBio.02357-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Batsios P, Ren X, Baumann O, Larochelle DA, Graf R. Src1 is a protein of the inner nuclear membrane interacting with the dictyostelium lamin NE81. Cells. 2016:5. doi: 10.3390/cells5010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martell JD, Yamagata M, Deerinck TJ, Phan S, Kwa CG, Ellisman MH, Sanes JR, Ting AY. A split horse-radish peroxidase for the detection of intercellular protein-protein interactions and sensitive visualization of synapses. Nat Biotechnol. 2016;34:774–780. doi: 10.1038/nbt.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Housden BE, Perrimon N. Cas9-mediated genome engineering in Drosophila melanogaster. Cold Spring Harb Protoc. 2016 doi: 10.1101/pdb.top086843. [DOI] [PubMed] [Google Scholar]
- 57.Komor AC, Badran AH, Liu DR. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods. 2011;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nagarkar-Jaiswal S, Lee PT, Campbell ME, Chen K, Anguiano-Zarate S, Gutierrez MC, Busby T, Lin WW, He Y, Schulze KL, et al. A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. Elife. 2015;4:e05338. doi: 10.7554/eLife.05338. [DOI] [PMC free article] [PubMed] [Google Scholar]