Abstract
Long non-coding RNAs (lncRNAs) are emerging as critical regulators of various biological processes and human diseases. The mechanisms of action involve their interactions with proteins, RNA and genomic DNA. Most lncRNAs display strong nuclear localization. Heterogeneous nuclear ribonucleoproteins (hnRNPs) are a large family of RNA-binding proteins that are important for multiple aspects of nucleic acid metabolism. hnRNPs are also predominantly expressed in the nucleus. This review discusses the interactions of lncRNAs and hnRNPs in regulating gene expression at transcriptional and post-transcriptional levels or by changing genomic structure, highlighting their involvements in glucose and lipid metabolism, immune response, DNA damage response, and other cellular functions. Toward the end, several techniques that are used to identify lncRNA binding partners are summarized. There are still many questions that need to be answered in this relatively new research area, which might provide novel targets to control the biological outputs of cells in response to different stimuli.
Keywords: epigenetics, eukaryotic gene expression, large intervening non-coding RNA, metabolism, ribonucleoproteins, signalling
Introduction
Long non-coding RNAs (lncRNAs) are RNA transcripts that do not code for protein. They are arbitrarily considered to be at least 200 nucleotides in length, on the basis of a convenient practical cut-off in RNA purification protocols, which distinguishes lncRNAs from short non-coding RNAs such as microRNAs [1–3]. Both human and mouse genomes are pervasively transcribed [4–6]. Protein-coding RNAs represent only a small fraction of the transcriptional output in higher eukaryotes. In humans, 19 000 proteins are coded for by less than 2% of the genome [7], while the vast majority is transcribed as non-coding RNAs. lncRNAs account for most of this pervasive transcription. More and more lncRNAs have been demonstrated to be functional molecules rather than transcriptional noise. Recently, 19 175 potentially functional lncRNAs were identified in the human genome [8]. They are expressed in many different cell types and tissues. They display strong cell- and tissue-specific expression, and poor conservation, at least at the primary sequence level among species. Based on the genomic location of their genes, lncRNAs are often placed into the following categories: (1) long intergenic non-coding RNAs (lincRNAs); (2) natural antisense transcripts; and (3) intronic lncRNAs.
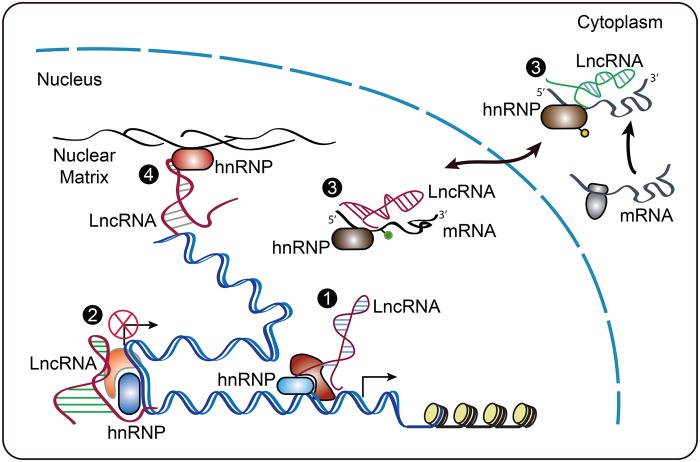
lncRNAs can be detected in the nucleus, cytoplasm, or both [9]. However, they are predominantly localized in the nucleus and are associated with chromatin [10]. These nuclear lncRNAs exert functions through their interaction with proteins, RNA or genomic DNAs. Heterogeneous nuclear ribonucleoproteins (hnRNPs) are a large family of RNA-binding proteins that are important for multiple aspects of nucleic acid metabolism [11,12]. hnRNPs consist of one or more RNA-binding domains including RNA recognition motifs, Arg-Gly-Gly repeats, or hnRNP K homology domains. The activity of hnRNPs is regulated by several post-translational modifications, including phosphorylation, SUMOylation, ubiquitination and methylation. Increasing evidence demonstrates that hnRNPs are involved in the development of human disease such as neurodegenerative diseases and cancer. In the review, we discuss how the interactions between hnRNPs and lncRNAs regulate gene expression and contribute to cellular homeostasis, participating in glucose metabolism, lipid metabolism, immune response, DNA damage response, and other cellular functions (Table 1). Based on the mechanisms of action, the interactions of lncRNAs and hnRNPs are categorized into four groups: (1) lncRNA interacts with hnRNP to induce gene expression; (2) lncRNA interacts with hnRNP to restrain gene expression; (3) lncRNA and hnRNP interaction controls mRNA stability and translation; and (4) lncRNA and hnRNP interaction regulates genome structure (Figure 1).
Table 1. The interaction of lncRNAs and hnRNPs in regulating cellular functions.
| lncRNAs | hnRNPs | Functions | References |
|---|---|---|---|
| lncLGR | hnRNP L | Regulation of hepatic glycogen storage | [13] |
| LeXis | Raly | Cholesterol level regulation, cholesterol biosynthesis gene expression | [15] |
| lincRNA-Cox2 | hnRNP A/B, A2/B1 | Macrophage gene expression after TLR signalling, NF-κB coactivator | [16,17] |
| lincRNA-EPS | hnRNP L | Restrains the expression of immune genes | [18] |
| THRIL | hnRNP L | TNF-α expression | [19] |
| lnc13 | hnRNP D | Regulation of inflammation in celiac disease patients | [20] |
| lincRNA-p21 | hnRNP K | Induces or suppresses p53 target genes | [14,21] |
| lnc00672 | hnRNP F,H,K | Endometrial carcinogenesis | [23] |
| linc-ROR | hnRNP I | Represses p53 expression, upregulation of cMyc | [24,25] |
| MYU | hnRNP K | CDK6 stabilization | [26] |
| EWSAT1 | hnRNP K | Gene suppression in Ewing sarcoma | [27] |
| treRNA | hnRNP K | Tumor metastsis | [28] |
| UCA1 | hnRNP A1, I | Promotes senescence in urothelial carcinoma, cell growth in breast cancer | [30,31] |
| Firre | hnRNP U | Adipogenesis regulation, cross-chromosomal interaction | [32,33] |
| lnc-BATE1 | hnRNP U | Adipogenesis of brown adipose tissue | [34] |
| Xist | hnRNP U | Inactivation of X-chromosome | [36] |
| H19 | hnRNP U | Transcriptional inhibition | [37] |
| BC200 | hnRNPv E1, E2 | Elimination of BC200 transcriptional inhibition | [40] |
Figure 1. The mechanisms of action by which lncRNA and hnRNP interactions regulate gene expression.
(1) lncRNAs recruit hnRNPs and turn on gene transcription; for example, lincRNA-p21 binds to hnRNP K and induces p21. (2) lncRNAs recruit hnRNPs, and their interactions restrain gene transcription; for example, lncRNA Lexis binds to Raly and inhibits cholesterol biosynthetic genes. (3) lncRNAs, hnRNPs and mRNA form a complex that controls mRNA stability or translation; for example, lincRoR and hnRNP I complex regulates p53 transalation. (4) lncRNAs regulate genome organization by interacting with nuclear matrix proteins; for example, Firre binds to the nuclear matrix protein hnRNP U.
lncRNA interacts with hnRNP to induce gene expression
The interactions of lncRNAs and hnRNPs can activate gene expression, although there are only a few examples. lncRNA THRIL (TNFα and hnRNP L related immunoregulatory lincRNA) also known as linc1992 is one of the lincRNAs that are differentially expressed following innate activation of THP1 macrophages [13]. THRIL forms a complex with hnRNP L and binds to the promoter of TNFα to induce its expression. Moreover, it was found that lincRNA-p21 acts in concert with hnRNP K as a coactivator for p53-dependent p21 transcription [14]. lincRNA-p21 was required for the recruitment of hnRNP K and p53 to the p21 promoter [14].
In addition to lncRNA THRIL and lincRNA-p21, there are several other lncRNAs including brown fat lncRNA-1 (Blnc1) and brown adipose tissue enriched long non-coding RNA 1 (lnc-BATE1) that can induce gene expression through interaction with hnRNPs [15–17]. Blnc1 expression is highly induced during brown adipocyte differentiation [16]. Blnc1 stimulates the thermogenic gene programme in brown adipocytes, and is required for brown adipocyte differentiation [16]. Blnc1 is conserved between mice and humans, and human Blnc1 also increases the expression of several transcriptional regulators of the thermogenic programme [17]. hnRNP U interacts with Blnc1 and facilitates the formation of a ribonucleoprotein complex that induces the expression of thermogenic genes [17]. By comparing transcriptomes of mouse brown, inguinal white, and epididymal white fat, lnc-BATE1 was found to be a brown adipose tissue-selective lncRNA required for the establishment and maintenance of the identity and thermogenic capacity of brown adipocytes [15]. lnc-BATE1 also interacts with hnRNP U, and both induce thermogenic gene expression and contribute to the adipogenesis of brown adipose tissue [15].
lncRNA interacts with hnRNP to inhibit gene expression
Although the interactions of lncRNAs and hnRNPs can induce gene expression, more studies have shown that the interactions repress gene expression. In this section, we will discuss how lncRNAs interact with hnRNPs to repress gene expression, and how they participate in glucose metabolism, lipid metabolism, immune response, p53 signaling and DNA damage response, and other cellular functions.
Glucose metabolism is tightly regulated in order to maintain blood glucose levels. Glycogen storage and degradation is one of the major mechanisms utilized to maintain glucose homeostasis between fasting and feeding conditions. The role of fasting-induced lncRNA-liver glucokinase repressor (lncLGR) in regulating the hepatic glycogen storage, along with hnRNP L, has been demonstrated by Ruan et al. [18]. Glucokinase (GCK) is one of the key regulators that oscillate the physiological state of the liver between fasting and feeding conditions. Over expression of lncLGR in the liver decreases GCK expression and glycogen storage while its depletion increases GCK expression and hepatic glycogen storage [18]. Similarly, knockdown of hnRNP L increases GCK expression at both the mRNA and protein levels. lncLGR facilitates the recruitment of hnRNP L to the GCK promoter and subsequently suppresses transcription of GCK [18]. In a fed state, activation of hepatic GCK activity shifts the liver from a glucose-utilizing organ to a glucose-storing organ. Fasting induces the expression of lncLGR, which binds to hnRNP L to form a functional complex that suppresses transcription of GCK, resulting in reduced glycogen storage [18]. These data suggest that the interaction of lncLGR and hnRNP L plays an important role in promoting glucose production from glycogen by inhibiting GCK expression during fasting. A previous study suggested that lncRNAs with very low copy numbers are usually involved in cis-regulation of gene expression [14]. In this study, lncLGR, with a very low copy number of 3.6 per cell, was demonstrated to be acting in trans by promoting the recruitment of hnRNP L to the GCK promoter [18]. It remains unknown how the lncLGR and hnRNP L complex is removed from the GCK promoter before the transition to fed state.
Cholesterol is essential for life, but higher cholesterol levels correlate with the risk of cardiovascular diseases. Therefore, cholesterol levels in cells and blood circulation are controlled at multiple levels. The liver X receptors (LXRs) are very important regulators of cellular and systemic cholesterol homeostasis. Activation of LXRs increases cholesterol efflux, inhibits cholesterol uptake, and promotes fatty acid synthesis. lncRNA LeXis (liver-expressed LXR-induced sequence) is the most robustly induced lncRNA in primary mouse hepatocytes in response to LXR activation with the selective synthetic agonist GW 3965 [19]. Adenovirus-mediated overexpression of LeXis decreased serum cholesterol, but not triglycerides, in chow-fed C57BL/6 mice. The expression of genes related to the cholesterol biosynthetic pathway was strongly downregulated in LeXis-expressing adenovirus-transduced livers [19]. The reduction of plasma cholesterol is independent of low-density lipoprotein receptor-mediated lipoprotein clearance. By contrast, knockdown of LeXis increases serum cholesterol levels and liver cholesterol contents, which is accompanied by increased expression of a number of cholesterol biosynthetic genes [19]. In LeXis-deficient mice, hepatic cholesterol content was markedly increased, while the expression of sterol synthesis genes in the liver was increased in chow-fed mice but deceased in mice maintained on a Western diet [19]. From an unbiased analysis of the LeXis interactome by mass spectrometry, the heterogeneous ribonucleoprotein Raly was identified as a binding partner of LeXis. Adenovirus-mediated knockdown of Raly in mouse liver reduced serum cholesterol, and inhibited the cholesterol biosynthetic pathway, effects that were independent of low-density lipoprotein receptor expression [19]. Raly knockdown also compromised the ability of LeXis to regulate serum cholesterol levels and hepatic gene expression, suggesting that the action of LeXis was dependent on Raly. The promoters of cholesterol biosynthetic genes were also associated with Raly [19]. These data demonstrate an important role of LeXis in mediating the inhibition of cholesterol biosynthesis upon LXR activation. A human ortholog of LeXis is thought to exist, but has not yet been characterized. How LeXis recruits Raly to specific genomic locations is also an open and interesting question.
lncRNAs are important regulators of immune response. lincRNA-Cox2 is amongst the most highly induced lncRNAs in macrophages during the innate immune response after the activation of Toll-like receptor signaling [20]. The expression of lincRNA-Cox2 decreases or increases the expression of more than one thousand genes [20]. Many genes related to the immune response are upregulated when lincRNA-Cox2 is silenced. HnRNP A/B and hnRNP A2/B1 were identified as lincRNA-Cox2 binding partners. lincRNA-Cox2 is present in both the cytosol and nucleus, and forms a complex with hnRNP A/B and A2/B1 in both compartments [20]. Many genes repressed by lincRNA-Cox2 overlap with the genes repressed by the two hnRNP proteins. Recruitment of RNA Pol II to the promoters of Ccl5 and Irf7 but not Il6 was increased when lincRNA-Cox2 or the hnRNPs were knocked down [20]. These data confirm that hnRNP A/B and hnRNP A2/B1 form a complex with lincRNA-Cox2 to restrain the transcription of a subset of immune genes. lincRNA-EPS (erythroid prosurvival) is also one of lncRNAs that restrain the expression of immune genes [21]. Its expression is downregulated in macrophages upon stimulation by Toll-like receptor ligands. Genetic deletion of lincRNA-EPS potentiates the expression of immune response genes in macrophages [21]. Interestingly, many of these genes are often clustered within specific chromosomal locations. By contrast, ectopic expression of lincRNA-EPS inhibits the expression of a number of immune response genes [21]. lincRNA-EPS is predominantly expressed in the nucleus and is associated with chromatin in resting macrophages. This association maintains a heterochromatic (repressive) chromatin state at many genomic loci of immune response genes [21]. Absence of lincRNA-EPS leads to changes in nucleosome positioning and an increase in chromatin accessibility. The authors found lincRNA-EPS specifically interacts with hnRNP L both in vitro and in vivo [21]. Knockdown of hnRNP L recapitulates the effect of lincRNA-EPS deficiency on gene expression of Cxcl10, Il6, Ifit1, Ccl5, and increased H3K4me3 levels at the promoters of some of these genes [21]. Importantly, lincRNA-EPS also restrains lethal inflammation in mice injected with lipopolysaccharides (LPS). In the study by Atianand et al., it remains to be fully defined how lincRNA-EPS and hnRNP L localize to the genomic loci of specific genes [21]. Further studies are needed to examine whether the interaction of lincRNA-EPS and hnRNP L participates in the regulation of immune response genes in other cell types such as B cells, T cells, and endothelial cells. The barrier needs to be removed in response to stimuli to allow gene expression. Genetic variations of lncRNA are associated with human disease. Castellanos-Rubio et al. uncovered the significance of a lncRNA named lnc13 in restraining a subset of inflammatory genes associated with celiac disease (CeD) [22]. They demonstrated that disease-associated single-nucleotide polymorphisms can affect lncRNA expression, and thereby the pathogenesis of human disease. Expression of lnc13 is found to be reduced in the small intestines of CeD patients [22]. lnc13 is primarily located in the nucleus, and exclusively associated with the chromatin fraction in macrophages. Its expression was significantly reduced upon LPS stimulation. In non-treated macrophages, lnc13 is constitutively expressed, and forms complexes with the p42 isoform of hnRNP D to recruit Hdac1 on to the promoters of certain inflammatory genes, which results in their repression. In LPS-treated macrophages, an active NF-κB pathway elevates decapping protein 2 (Dcp2) [22]. Decapping of lnc13 by Dcp2 causes its degradation, leaving the inflammatory genes unrestrained and active. Dcp2 expression was also increased in biopsies of CeD patients, which explains the reduction of lnc13. In lnc13-deficient cells, hnRNP D and Hdac1 enrichment was found to be reduced at the transcription start site of lnc13 target genes [22]. Similarly, in hnRNP D knockout cells, binding of Hdac1 to the lnc13-regulated promoters was less efficient, which indicates the role of hnRNP D in recruiting Hdac1 to lnc13-regulated promoters [22]. Wild type lnc13 binds more efficiently to hnRNP D relative to disease-associated counterparts harboring the mutation due to the CeD-associated single-nucleotide polymorphism. This demonstrates that genetic variation of lnc13 results in modifications of its secondary structure that weakens its binding with hnRNP D, which interferes with its suppressive effect on inflammatory genes [22]. The results demonstrate the significant role of lnc13 and hnRNP D in regulating intestinal immune homeostasis by restraining the expression of inflammatory genes. NF-κB signaling is often divided into canonical and non-canonical pathways [23,24]; however, it remains unknown how these lncRNAs specifically regulate different NF-κB pathways and target gene expression in different cell types.
lncRNAs play a key regulatory role in DNA damage and p53 transcriptional response. Many lncRNAs are p53 transcriptional targets. One of the p53-induced lincRNAs, lincRNA-p21, mediates the transcriptional repression of p53 in response to DNA damage, and triggers cell apoptosis [25]. lincRNA-p21 interacts with hnRNP K, and the interaction co-represses genes in the p53 transcriptional response. lincRNA-p21 is required to direct the binding of hnRNP K to the promoters of these repressed genes, while hnRNP K was also identified as a transcriptional coactivator of p53, promoting gene expression in response to DNA damage [26]. linc00672 is a direct transcriptional target of p53. Overexpression of linc00672 caused a significant accumulation of cells in the G2/M phases, and reduced cell proliferation in the human endometrial carcinoma cell line [27]. Bioinformatics and RNA-pulldown were performed to identify the binding partners of linc00672. linc00672 binds to hnRNP F and H, and is required to recruit hnRNPs to promote p53-dependent suppression of the LIM and SH3 protein 1. In addition to hnRNP F and H, hnRNP K was also detected at the p53 binding site at the LIM and SH3 protein 1 promoter. These data suggest that linc00672 is an important regulator of p53 protein-mediated gene suppression and the oncogenesis of endometrial carcinoma.
RNA-binding protein EWS and transcription factor FLI1 (EWS-FLI1) can both activate and repress genes that are critical for cell transformation. lncRNA Ewing sarcoma-associated transcript 1 (EWSAT1) is induced by EWS-FLI1 in bone marrow-derived mesenchymal progenitor cells that were isolated from pediatric patients [28]. Examination of EWSAT1 expression in tumor samples revealed that EWSAT1 is an lncRNA that is upregulated in Ewing sarcoma. EWSAT1 is an important molecule that mediates repression of genes downstream of EWS-FLI1. Using protein array, the authors found EWSAT1 binds to 22 proteins. An interaction between hnRNP K and EWSAT1 was validated using RNA immunoprecipitation. Interestingly, 20 out of 302 genes repressed by hnRNP K were also repressed by EWSAT1, suggesting that EWSAT1 and hnRNP K act together to repress the expression of a subset of genes in the context of Ewing sarcoma. Since not all EWSAT1-repressed genes were changed after hnRNP K downregulation, EWSAT1 probably represses many genes through interaction with other proteins.
lncRNA and hnRNP interaction controls mRNA stability and translation
lncRNAs control gene expression at not only the transcriptional but also the post-transcriptional level. In this section, we will discuss a few lncRNAs that regulate mRNA stability and translation. lncRNA regulator of reprogramming (linc-ROR) was first discovered in induced pluripotent stem cells. In a more recent study, linc-RoR was identified as a negative regulator of p53 through interaction with hnRNP I [29]. hnRNP I can bind to the internal ribosome entry site of p53 mRNA and promote its translation. The interaction of linc-RoR with hnRNP I occurs predominantly in the cytoplasm with phosphorylated hnRNP I, which prevents or interrupts the interaction of hnRNP I with p53 mRNA, and suppresses DNA-damage-induced p53 translation. The expression of linc-RoR is induced by p53, thus forming a negative feedback loop. linc-RoR also aggravates oncogenesis by enhancing c-Myc mRNA and protein levels [30]. Both c-Myc and linc-RoR expression are up-regulated in cancer tissues. Overexpression of linc-RoR in HCT-116 cells significantly promotes its proliferation and tumorigenesis characteristics. Conversely, linc-RoR knockout reduces the cellular c-Myc mRNA and protein levels [30]. Further investigation of lncRNA-mediated c-Myc regulation uncovered the significant role of hnRNP I and hnRNP D (also known as AUF1) in fine-tuning c-Myc mRNA stability. In tumor cells, linc-RoR facilitates the interaction between hnRNP I and c-Myc, which results in the increased stability of c-Myc mRNA. Consequently, the interaction of linc-RoR with hnRNP I prevents the binding of hnRNP D to c-Myc mRNA, whereas binding of hnRNP D to c-Myc mRNA results in decay of c-Myc mRNA in non-cancerous cells. In this condition, linc-RoR renders specificity in regulating the stability of c-Myc mRNA by determining its binding with hnRNP D or hnRNP I. The findings emphasize the role of lncRNAs acting as molecular anchors that fine-tune cell signaling by recruiting different types of multifunctional hnRNPs, resulting in repression or activation of specific genes. Transcription factor c-Myc plays a critical role in the development of colorectal cancer. c-Myc is known to induce the expression of an lncRNA named MYU [31]. MYU interacts with hnRNP K to stabilize cyclin-dependent kinase 6 (CDK6) expression and thereby promote the G1-S transition of the cell cycle. Mechanistically, MYU, hnRNP K, and CDK6 mRNA form a complex in which hnRNP K inhibits the interaction of miR-16 with its binding site in the 3′UTR of CDK6 mRNA.
We have discussed that lncRNAs linc-RoR and MYU control mRNA stability. Next, the translation inhibition by lncRNA-hnRNP interactions are summarized.
Human translational regulatory lncRNA (treRNA) can function as an enhancer and regulate Snail transcription in cis (the regulation of Snail depends on the site of transcription of treRNA) [32]. In lymph-node metastatic human breast cancer samples, treRNA expression is higher than in primary breast-cancer carcinoma. Enforced expression of treRNA increases cell migration and cell invasion through Matrigel™ in non-invasive and non-metastatic breast cancer MCF7 cells. Knockdown of endogenous treRNA suppressed tumor metastasis in vivo. From mass spectrometry analysis, three RNA-binding proteins among the top hits were identified, including hnRNP K, FXR1 and FXR2. HnRNP K and FXR2 indeed mediate the effect of treRNA on both E-cadherin suppression and promotion metastasis in vivo. TreRNA expression promotes binding of the PUF60–SF3B3 complex to hnRNP K, FXR1 and FXR2 to form a treRNA RNP complex, which suppresses translation by binding to translation initiation factor eIF4G1 [33]. lncRNA urothelial carcinoma associated 1 (UCA1) inhibits cell proliferation and promotes senescence [34]. UCA1 binds and sequesters hnRNP A1, preventing it from destabilizing p16INK mRNA. lncRNA UCA1 is a direct target of the complex of CAPERα (Coactivator of AP1 and Estrogen Receptor) and T-box3, mediating their effects on cell proliferation during development and disease. By contrast, UCA1 promotes breast cancer cell growth both in vitro and in vivo [35]. The expression of tumor suppressor p27 is suppressed by UCA1 through its interaction with hnRNP I. In addition to UCA1, an additional 18 lncRNAs were identified in RNA immunoprecipitation assays with hnRNP I antibodies. Ectopic expression of hnRNP I increased UCA1 RNA stability and its level. Doxorubicin, a p53 activator, induces hnRNP I cytoplasmic localization and phosphorylation, and its interaction with UCA1. This interaction competes with hnRNP1 for p27 binding, thus preventing hnRNP I from enhancing the translation of p27, a tumor suppressor. Brain cytoplasmic RNA 200 nucleotides (BC200 RNA) inhibit translation in human cells [36]. HnRNP E1 and hnRNP E2 were identified as binding proteins of BC200 RNA using a yeast three-hybrid screening [37]. HnRNP E1 and E2 compete with eIF4A for binding to BC200 RNA. HnRNP E2 forms a ternary complex with BC200 RNA and eIF4A, while the binding of hnRNP E1 to BC200 RNA causes an alteration of the RNA secondary structure. The results suggest that hnRNP E1 and E2 probably restore BC200 RNA-mediated translation inhibition through different mechanisms [37].
lncRNA and hnRNP interaction regulates genome structure
The role of lncRNAs in regulating chromatin structure and nuclear architecture has been reviewed recently [38]. Chromosomes are organized into domains, and linearly distant genomic loci can be associated to yield chromosome loops and interchromosomal interactions [39]. The location of a gene relative to certain chromatin domains or nuclear structures can determine its activation [40]. Emerging evidence has demonstrated that lncRNAs participate in the regulation of nuclear architecture. For example, as a physical scaffold, lncRNA Xist can organize a repressive chromosome compartment [38]. hnRNP U is a nuclear matrix- or scaffold-attachment region-associated protein [41]. In a study by Hasegawa et al., depletion of hnRNP U resulted in the detachment of Xist from the inactive X-chromosome, suggesting hnRNP U is required for Xist RNA localization on the inactive X-chromosome [42]. The initial phase of transcription by RNA polymerase II involves the interaction of hnRNP with actin and their association with RNA polymerase II [43]. McHugh et al. [44] adapted an RNA antisense purification method (reference) to purify an lncRNA complex, followed by quantitative mass spectrometry to identify the interacting proteins. They identified 10 proteins that were enriched for Xist relative to control non-coding RNA. These proteins include HDAC associated repressor protein (SHARP), hnRNP M, hnRNP U, hnRNP C, Raly, Ptbp1, Lamin B receptor and others. In addition to hnRNP U, it was demonstrated that SHARP and Lamin B receptor are also required for Xist-mediated transcriptional silencing of the X-chromosome [44]. An lncRNA termed functional intergenic repeating RNA element (Firre, previously referred to as linc-RAP-1) is required for proper adipogenesis [45]. Firre binds to the nuclear matrix protein hnRNP U. This physical interaction is required for the proper focal and nuclear localization of Firre in order to maintain the multi-chromosomal nuclear interactions [46]. Firre along with hnRNP U probably impart specificity in organizing chromatin domains and nuclear architecture [46]. In addition to the lncRNA Firre, there are several other lncRNAs including Blnc1 [16,17], lnc-BATE1 [15], and H19 [47] that interact with hnRNP U. It remains unknown whether lncRNAs including Blnc1, lnc-BATE1 and H19 are involved in organizing trans-chromosomal associations and nuclear structure.
Techniques to interrogate lncRNA–protein interactions
RNA immunoprecipitation
RNA immunoprecipitation (RIP) is a technique used to detect the interaction between an individual protein and specific RNA molecules [48]. An antibody against a specific protein is used to immunoprecipitate the protein and associated RNAs. Then the associated RNAs including lncRNAs can be detected with real-time PCR or RNA sequencing [49]. RIP has limitations. It can produce false-positive results that do not reflect in vivo interaction, or false-negative results because the interaction cannot survive the immunoprecipitation procedures. Cross-linked RIP has been developed to address the limitation, but it also introduces caveats [50].
Chromatin isolation by RNA purification
Chromatin isolation by RNA purification (ChIRP) was first described to map genome-wide lncRNA occupancy at chromatin in 2011 [51]. Later on, it was also successfully used to identify lncRNA-binding proteins [52]. Briefly, chromatin is extracted from cross-linked cells, then biotin-conjugated antisense DNA oligos that tile the full length of lncRNA (1 probe/100 bp length of lncRNA) are added for hybridization [53]. The biotinylated DNA probes and associated binding partners are isolated using magnetic beads. DNA and RNA are isolated from the beads sample for sequencing, while proteins are eluted from the beads for mass spectrometry. So ChIRP can be used to identify DNA, RNA or protein-binding partners of lncRNA. Antisense DNA probes are conjugated with biotin at the 3′-end with 20 nucleotides in length. LncRNA Xist interacts with 81 proteins identified by ChIRP, and hnRNP K is one of the proteins that participates in Xist-mediated gene silencing and histone modifications [52].
Capture hybridization analysis of RNA targets and RNA affinity purification
Capture hybridization analysis of RNA targets (CHART) was initially developed to determine where lncRNAs bind to chromatin [54]. Antisense oligonucleotides 24–25 nucleotides in length were designed where the RNA molecule is accessible with high RNase-H sensitivity. This is different from how DNA probes are designed in the ChIRP assay. CHART was successfully adapted to identify the proteins that are associated with lncRNAs by using mass spectrometry [55]. RNA affinity purification (RAP) is also used to identify a lncRNA of interest and its associated proteins using antisense biotinylated DNA oligos [56]. RAP utilizes 120-nucleotide antisense RNA probes to form extremely strong hybrids with the target RNA for high specificity. A pool of overlapping probes tiled across the full length of the target RNA is used for robustly capturing the endogenous lncRNA. RAP was developed further to purify an lncRNA complex and identify the interacting protein by quantitative mass spectrometry [44].
Psoralen analysis of RNA interactions and structures
Psoralen analysis of RNA interactions and structures (PARIS) is a method for mapping RNA duplexes on a transcriptome-wide scale [57]. After UV-mediated psoralen crosslinking of duplex regions in cells, the isolated cross-linked fragments are analyzed by reverse transcription and next generation DNA sequencing. Sequencing results are mapped to the reference genome and analyzed to collect the information for RNA interactions and structures. PARIS is a great tool to map lncRNA structures and explore its functional roles.
RNA pulldown assay
lncRNA pulldown is a relatively easier method and is frequently used to identify various proteins that interact with an lncRNA of interest. lncRNA molecules are generated using in vitro transcription assay. Biotin-UTP is incorporated along the sequence of the RNA. The biotinylated lncRNA molecules are used for pulldown of its protein-binding partners identified by mass spectrometry.
Concluding remarks and future prospects
In this review, we summarized the role of lncRNA and hnRNP interactions in regulating gene expression and cellular functions. The mechanisms of action can mainly be categorized into the following groups: (1) lncRNAs recruit hnRNPs and turn on gene transcription by binding to gene promoters; (2) lncRNAs recruit hnRNPs, and their interactions at specific genomic locations restrain gene transcription; (3) lncRNAs, hnRNPs and mRNA form a complex that controls mRNA stability or translation; and (4) lncRNAs regulate genome organization by interacting with nuclear matrix proteins, indirectly affecting gene expression (Figure 1). More and more lncRNAs will be identified as critical regulators of biological processes and human diseases.
There are several areas that warrant further investigation. Firstly, a general framework has been proposed that the act of transcription of lncRNAs would mark the spots for nuclear proteins such as hnRNPs to pull the genomic DNA, thus affecting the spatial organization of the genome and transcription output [58]. How hnRNPs contribute to the interplay between genome organization and the act of transcription of lncRNAs is important to investigate. With new technology available to assess how genome structure influences whether genes are switched on or off [59], the role of more and more lncRNAs in controlling gene expression by affecting genome structure will be revealed. Secondly, the function–structure relationship should be further investigated. Protein crystallization has been very important for understanding the structure–function relationship of proteins. This reminds us that crystallization of RNA molecules or RNA–protein complexes could greatly facilitate lncRNA functional studies. Post-transcriptional modifications such as methylation at the 6 position of adenosine (m6A) alters the stability of RNA secondary structure, thus affecting function [60]. For example, an m6A site in the lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) induces a local change in structure and increases recognition and binding by hnRNP C [61]. These studies provide a new direction for studying how post-transcriptional modifications such as m6A regulate the structure of lncRNAs and their interactions with protein-binding partners. Thirdly, lncRNA conservation between species warrants further study. lncRNA conservation should be examined at different dimensions including the primary sequence, structure, function, and expression from syntenic loci [62]. Using a protein-centric approach to probing hnRNP-lncRNA interactions in mouse and human may help to identify functionally conserved lncRNAs. Fourthly, the regulation of hnRNP-lncRNA interaction should be further investigated. The location and magnitude of the interaction for a specific lncRNA and hnRNP must be regulated to accommodate the change of gene expression and cellular function. As an example, the mechanisms that regulate the nucleo-cytoplasmic shuttling of hnRNP and lncRNA complexes are important to investigate. Finally, many of the hnRNPs are important players in neurodegenerative disease [11]. lncRNAs display very high tissue-specific expression patterns, and a large fraction of tissue-specific lncRNAs are expressed in the brain [10]. Studying the interaction of hnRNPs and lncRNAs in neurodegenerative disease will increase our knowledge about the pathogenesis of the disease, and provide novel therapeutic targets.
Abbreviations
- BC200 RNA
brain cytoplasmic RNA 200 nucleotides
- CDK6
cyclin-dependent kinase 6
- CHART
capture hybridization analysis of RNA targets
- CeD
celiac disease
- ChIRP
chromatin isolation by RNA purification
- Cox-2
cyclooxygenase-2
- Dcp2
decapping protein 2
- EWS-FLI1
EWS and transcription factor FLI1
- EWSAT1
Ewing sarcoma-associated transcript 1
- Firre
functional intergenic repeating RNA element
- GCK
glucokinase
- hnRNPs
heterogeneous nuclear ribonucleoproteins
- linc-ROR
lncRNA regulator of reprogramming
- lincRNA-EPS
lincRNA erythroid prosurvival
- lincRNAs
long intergenic non-coding RNAs
- lnc-BATE1
Brown adipose tissue enriched long non-coding RNA 1
- lncLGR
lncRNA-liver glucokinase repressor
- lncRNAs
long non-coding RNAs
- LPS
lipopolysaccharides
- LXRs
liver X receptors
- LeXis
liver-expressed LXR-induced sequence
- PARIS
psoralen analysis of RNA interactions and structures
- RAP
RNA affinity purification
- RIP
RNA immunoprecipitation
- THRIL
TNFα and hnRNP L related immunoregulatory LincRNA
- TNF
tumor necrosis factor
- TreRNA
translational regulatory lncRNA
- UCA1
urothelial carcinoma associated 1
- m6A
methylation at the 6 position of adenosine
Funding
This work was supported by the University of Nebraska – Lincoln - Layman award (to X.H.S.), American Heart Association [SDG#15SDG25400012 to X.H.S.], and a NIH-funded COBRE grant to X.H.S. [under NIH 1P20GM104320].
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Ponting C.P., Oliver P.L. and Reik W. (2009) Evolution and functions of long non-coding RNAs. Cell 136, 629–641 doi: 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 2.Mercer T.R., Dinger M.E. and Mattick J.S. (2009) Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159 doi: 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- 3.Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T. et al. (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488 doi: 10.1126/science.1138341 [DOI] [PubMed] [Google Scholar]
- 4.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A. et al. (2012) Landscape of transcription in human cells. Nature 489, 101–108 doi: 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hangauer M.J., Vaughn I.W., McManus M.T. and Rinn J.L. (2013) Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic non-coding RNAs. PLoS Genet. 9, e1003569 doi: 10.1371/journal.pgen.1003569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yue F., Cheng Y., Breschi A., Vierstra J., Wu W., Ryba T. et al. (2014) A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355–364 doi: 10.1038/nature13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ezkurdia I., Juan D., Rodriguez J.M., Frankish A., Diekhans M., Harrow J. et al. (2014) Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum. Mol. Genet. 23, 5866–5878 doi: 10.1093/hmg/ddu309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hon C.C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J.L., Gough J. et al. (2017) An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543, 199–204 doi: 10.1038/nature21374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhatt D.M., Pandya-Jones A., Tong A.J., Barozzi I., Lissner M.M., Natoli G. et al. (2012) Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 150, 279–290 doi: 10.1016/j.cell.2012.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H. et al. (2012) The GENCODE v7 catalog of human long non-coding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 doi: 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geuens T., Bouhy D. and Timmerman V. (2016) The hnRNP family: insights into their role in health and disease. Hum. Genet. 135, 851–867 doi: 10.1007/s00439-016-1683-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han S.P., Tang Y.H. and Smith R. (2010) Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 430, 379–392 doi: 10.1042/BJ20100396 [DOI] [PubMed] [Google Scholar]
- 13.Li Z., Chao T.-C., Chang K.-Y., Lin N., Patil V.S., Shimizu C. et al. (2014) The long non-coding RNA THRIL regulates TNF expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. U.S.A. 111, 1002–1007 doi: 10.1073/pnas.1313768111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dimitrova N., Zamudio J.R., Jong R.M., Soukup D., Resnick R., Sarma K. et al. (2014) LincRNA-p21 activates p21 In cis to promote polycomb target gene expression and to enforce the G1/S checkpoint. Mol. Cell 54, 777–790 doi: 10.1016/j.molcel.2014.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvarez-Dominguez J.R., Bai Z., Xu D., Yuan B., Lo K.A., Yoon M.J. et al. (2015) De novo reconstruction of adipose tissue transcriptomes reveals long non-coding RNA regulators of brown adipocyte development. Cell Metab. 21, 764–776 doi: 10.1016/j.cmet.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao X.-Y., Li S., Wang G.-X., Yu Q. and Lin J.D. (2014) A long non-coding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol. Cell 55, 372–382 doi: 10.1016/j.molcel.2014.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mi L., Zhao X.-Y., Li S., Yang G. and Lin J.D. (2017) Conserved function of the long non-coding RNA Blnc1 in brown adipocyte differentiation. Mol. Metab. 6, 101–110 doi: 10.1016/j.molmet.2016.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruan X., Li P., Cangelosi A., Yang L. and Cao H. (2016) A long non-coding RNA, lncLGR, regulates hepatic glucokinase expression and glycogen storage during fasting. Cell Rep. 14, 1867–1875 doi: 10.1016/j.celrep.2016.01.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sallam T., Jones M.C., Gilliland T., Zhang L., Wu X., Eskin A. et al. (2016) Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature 534, 124–128 doi: 10.1038/nature17674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carpenter S., Aiello D., Atianand M.K., Ricci E.P., Gandhi P., Hall L.L. et al. (2013) A long non-coding RNA mediates both activation and repression of immune response genes. Science 341, 789–792 doi: 10.1126/science.1240925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atianand M.K., Hu W., Satpathy A.T., Shen Y., Ricci E.P., Alvarez-Dominguez J.R. et al. (2016) A long non-coding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 165, 1672–1685 doi: 10.1016/j.cell.2016.05.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castellanos-Rubio A., Fernandez-Jimenez N., Kratchmarov R., Luo X., Bhagat G., Green P.H.R. et al. (2016) A long non-coding RNA associated with susceptibility to celiac disease. Science 352, 91–95 doi: 10.1126/science.aad0467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oeckinghaus A., Hayden M.S. and Ghosh S. (2011) Crosstalk in NF-κB signaling pathways. Nat. Immunol. 12, 695–708 doi: 10.1038/ni.2065 [DOI] [PubMed] [Google Scholar]
- 24.Cildir G., Low K.C. and Tergaonkar V. (2016) Noncanonical NF-κB signaling in health and disease. Trends Mol. Med. 22, 414–429 doi: 10.1016/j.molmed.2016.03.002 [DOI] [PubMed] [Google Scholar]
- 25.Huarte M., Guttman M., Feldser D., Garber M., Koziol M.J., Kenzelmann-Broz D. et al. (2010) A large intergenic non-coding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419 doi: 10.1016/j.cell.2010.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moumen A., Masterson P., O'Connor M.J. and Jackson S.P. (2005) hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell 123, 1065–1078 doi: 10.1016/j.cell.2005.09.032 [DOI] [PubMed] [Google Scholar]
- 27.Li W., Li H., Zhang L., Hu M., Li F., Deng J. et al. (2017) Long non-coding RNA LINC00672 contributes to p53 protein-mediated gene suppression and promotes endometrial cancer chemosensitivity. J. Biol. Chem. 292, 5801–5813 doi: 10.1074/jbc.M116.758508 Epub 2017 Feb 23. PubMed PMID: 28232485; PubMed Central PMCID: PMC5392574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marques Howarth M., Simpson D., Ngok S.P., Nieves B., Chen R., Siprashvili Z. et al. (2014) Long non-coding RNA EWSAT1-mediated gene repression facilitates Ewing sarcoma oncogenesis. J. Clin. Invest. 124, 5275–5290 doi: 10.1172/JCI72124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang A., Zhou N., Huang J., Liu Q., Fukuda K., Ma D. et al. (2013) The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 23, 340–350 doi: 10.1038/cr.2012.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang J., Zhang A., Ho T.-T., Zhang Z., Zhou N., Ding X. et al. (2016) Linc-RoR promotes c-Myc expression through hnRNP I and AUF1. Nucleic Acids Res. 44, 3059–3069 doi: 10.1093/nar/gkv1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawasaki Y., Komiya M., Matsumura K., Negishi L., Suda S., Okuno M. et al. (2016) MYU, a target lncRNA for Wnt/c-Myc signaling, mediates induction of CDK6 to promote cell cycle progression. Cell Rep. 16, 2554–2564 doi: 10.1016/j.celrep.2016.08.015 [DOI] [PubMed] [Google Scholar]
- 32.Ørom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G. et al. (2010) Long non-coding RNAs with enhancer-like function in human cells. Cell 143, 46–58 doi: 10.1016/j.cell.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gumireddy K., Li A., Yan J., Setoyama T., Johannes G.J., Ørom U.A. et al. (2013) Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J. 32, 2672–2684 doi: 10.1038/emboj.2013.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar P.P., Emechebe U., Smith R., Franklin S., Moore B., Yandell M. et al. (2014) Coordinated control of senescence by lncRNA and a novel T-box3 co-repressor complex. eLife 3, e02805 doi: 10.7554/eLife.02805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang J., Zhou N., Watabe K., Lu Z., Wu F., Xu M. et al. (2014) Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. 5, e1008 doi: 10.1038/cddis.2013.541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kondrashov A.V., Kiefmann M., Ebnet K., Khanam T., Muddashetty R.S. and Brosius J. (2005) Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly(A)-binding protein (PABP). J. Mol. Biol. 353, 88–103 doi: 10.1016/j.jmb.2005.07.049 [DOI] [PubMed] [Google Scholar]
- 37.Jang S., Shin H., Lee J., Kim Y., Bak G. and Lee Y. (2017) Regulation of BC200 RNA-mediated translation inhibition by hnRNP E1 and E2. FEBS Lett. 591, 393–405 doi: 10.1002/1873-3468.12544 [DOI] [PubMed] [Google Scholar]
- 38.Tomita S., Abdalla M.O.A., Fujiwara S., Yamamoto T., Iwase H., Nakao M. et al. (2017) Roles of long non-coding RNAs in chromosome domains. Wiley Interdiscip. Rev. RNA 8, e1384 doi: 10.1002/wrna.1384 [DOI] [PubMed] [Google Scholar]
- 39.Dekker J., Marti-Renom M.A. and Mirny L.A. (2013) Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 14, 390–403 doi: 10.1038/nrg3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A. et al. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 doi: 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romig H., Fackelmayer F.O., Renz A., Ramsperger U. and Richter A. (1992) Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J. 11, 3431–3440 PubMed PMID: 1324173; PubMed Central PMCID: PMC556878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasegawa Y., Brockdorff N., Kawano S., Tsutui K., Tsutui K. and Nakagawa S. (2010) The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev. Cell 19, 469–476 doi: 10.1016/j.devcel.2010.08.006 [DOI] [PubMed] [Google Scholar]
- 43.Kukalev A., Nord Y., Palmberg C., Bergman T. and Percipalle P. (2005) Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat. Struct. Mol. Biol. 12, 238–244 doi: 10.1038/nsmb904 [DOI] [PubMed] [Google Scholar]
- 44.McHugh C.A., Chen C.-K., Chow A., Surka C.F., Tran C., McDonel P. et al. (2015) The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236 doi: 10.1038/nature14443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun L., Goff L.A., Trapnell C., Alexander R., Lo K.A., Hacisuleyman E. et al. (2013) Long non-coding RNAs regulate adipogenesis. Proc. Natl. Acad. Sci. U. S. A. 110, 3387–3392 doi: 10.1073/pnas.1222643110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hacisuleyman E., Goff L.A., Trapnell C., Williams A., Henao-Mejia J., Sun L. et al. (2014) Topological organization of multichromosomal regions by the long intergenic non-coding RNA Firre. Nat. Struct. Mol. Biol. 21, 198–206 doi: 10.1038/nsmb.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bi H.-S., Yang X.-Y., Yuan J.-H., Yang F., Xu D., Guo Y.-J. et al. (2013) H19 inhibits RNA polymerase II-mediated transcription by disrupting the hnRNP U–actin complex. Biochim. Biophys. Acta, Gen. Subj. 1830, 4899–4906 doi: 10.1016/j.bbagen.2013.06.026 [DOI] [PubMed] [Google Scholar]
- 48.Gilbert C. and Svejstrup J.Q. (2006) RNA immunoprecipitation for determining RNA-protein associations in vivo. Curr. Protoc. Mol. Biol. Chapter 27, Unit 27.4 doi: 10.1002/0471142727.mb2704s75 Review. PubMed PMID: 18265380 [DOI] [PubMed] [Google Scholar]
- 49.Selth L.A., Close P. and Svejstrup J.Q. (2011) Methods in molecular biology. Methods Mol. Biol. 791, 253–264 doi: 10.1007/978-1-61779-316-5_19 [DOI] [PubMed] [Google Scholar]
- 50.Gagliardi M. and Matarazzo M.R. (2016) Methods in molecular biology. Methods Mol. Biol. 1480, 73–86 doi: 10.1007/978-1-4939-6380-5_7 [DOI] [PubMed] [Google Scholar]
- 51.Chu C., Qu K., Zhong F.L., Artandi S.E. and Chang H.Y. (2011) Genomic maps of long non-coding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 44, 667–678 doi: 10.1016/j.molcel.2011.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chu C., Zhang Q.C., da Rocha S.T., Flynn R.A., Bharadwaj M., Calabrese J.M. et al. (2015) Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416 doi: 10.1016/j.cell.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chu C., Quinn J. and Chang H.Y. (2012) Chromatin isolation by RNA purification (ChIRP). J. Vis. Exp. (61), pii: 3912 doi: 10.3791/3912 PubMed PMID: 22472705; PubMed Central PMCID: PMC3460573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simon M.D., Wang C.I., Kharchenko P.V., West J.A., Chapman B.A., Alekseyenko A.A. et al. (2011) The genomic binding sites of a non-coding RNA. Proc. Natl. Acad. Sci. U. S. A. 108, 20497–20502 doi: 10.1073/pnas.1113536108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I. et al. (2014) The long non-coding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell 55, 791–802 doi: 10.1016/j.molcel.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Engreitz J.M., Pandya-Jones A., McDonel P., Shishkin A., Sirokman K., Surka C. et al. (2013) The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, 1237973 doi: 10.1126/science.1237973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu Z., Zhang Q.C., Lee B., Flynn R.A., Smith M.A., Robinson J.T. et al. (2016) RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165, 1267–1279 doi: 10.1016/j.cell.2016.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melé M. and Rinn J.L. (2016) “Cat's Cradling” the 3D genome by the act of LncRNA transcription. Mol. Cell 62, 657–664 doi: 10.1016/j.molcel.2016.05.011 [DOI] [PubMed] [Google Scholar]
- 59.Stevens T.J., Lando D., Basu S., Atkinson L.P., Cao Y., Lee S.F. et al. (2017) 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64 doi: 10.1038/nature21429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pan T. (2013) N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem. Sci. 38, 204–209 doi: 10.1016/j.tibs.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu N., Dai Q., Zheng G., He C., Parisien M. and Pan T. (2015) N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 518, 560–564 doi: 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Diederichs S. (2014) The four dimensions of non-coding RNA conservation. Trends Genet. 30, 121–123 doi: 10.1016/j.tig.2014.01.004 [DOI] [PubMed] [Google Scholar]