Abstract
Aim:
In Egypt as in many other countries, river water buffalo (Bubalus bubalis) is considered an important source of high-quality milk and meat supply. The objective of this study was to investigate serotypes, virulence genes, and antibiotic resistance determinants profiles of Escherichia coli isolated from buffalo at some places in Egypt; noticibly, this issue was not discussed in the country yet.
Materials and Methods:
A number of 58 rectal samples were collected from diarrheic buffalo calves in different regions in Egypt, and bacteriological investigated for E. coli existence. The E. coli isolates were biochemically, serologicaly identified, tested for antibiotic susceptibility, and polymerase chain reaction (PCR) analyzed for the presence of antibiotic resistance determinants and virulence genes.
Results:
Overall 14 isolates typed as E. coli (24.1%); 6 were belonged to serogroup O78 (10.3%), followed by O125 (4 isolates, 6.9%), then O158 (3 isolates, 5.2%) and one isolate O8 (1.7%), among them, there were 5 E. coli isolates showed a picture of hemolysis (35.7%). The isolates exhibited a high resistance to β lactams over 60%, followed by sulfa (50%) and aminoglucoside (42.8%) group, in the same time the isolates were sensitive to quinolone, trimethoprim-sulfamethoxazole, tetracycline (100%), and cephalosporine groups (71.4%). A multiplex PCR was applied to the 14 E. coli isolates revealed that all were carrying at least one gene, as 10 carried blaTEM (71.4%), 8 Sul1 (57.1%), and 6 aadB (42.8%), and 9 isolates could be considered multidrug resistant (MDR) by an incidence of 64.3%. A PCR survey was stratified for the most important E. coli virulence genes, and showed the presence of Shiga toxins in 9 isolates carried either one or the two Stx genes (64.3%), 5 isolates carried hylA gene (35.7%), and eae in 2 isolates only (14.3%), all isolates carried at least one virulence gene except two (85.7%).
Conclusion:
The obtained data displayed that in Egypt, buffalo as well as other ruminants could be a potential source of MDR pathogenic E. coli variants which have a public health importance.
Keywords: antibiotic resistance determinants, buffalo, Egypt, Escherichia coli, virulence
Introduction
Escherichia coli is a member of the genus Escherichia within the family Enterobacteriaceae. Members of this family are vastly extended in the environment. E. coli is ordinarily a non-pathogenic member of the animal intestinal flora. However, certain strains have developed virulence and antibiotic resistance factors, so may cause a variety of infections in humans and animals, strains of E. coli that cause enteric disease are stated enterovirulent or diarrheagenic E. coli (DEC) [1].
Ruminants represent a major reservoir for pathogenic E. coli access into the human communities through the food chain [2]. Diagnosing DEC in humans, food, and the environment has revealed that non-O157:H7 serotypes are more responsible for extreme infections in humans, and considered to be of greater clinical significance as causes of human disease than O157 strains in different countries [3].
Presently, the rising of antimicrobial resistance is a public health concern, in particular the use and misuse of antimicrobials, especially in animal population as growth promoters. The main factor in the emergence of multidrug-resistance (MDR) strains is the ability of bacteria to gain and disseminate exogenous genes through mobile genetic elements [4]. The upgrowth of E. coli isolates with MDR phenotypes has been formerly reported and may extend from one ecosystem to another by lateral gene transfer; integrons [5].
The pathogenic E. coli usually possess abundant virulence determinants that share in the disease pathogenesis; Shiga toxin-producing E. coli (STEC) represents a substantial emerging set of food-borne pathogens, as farm animals, particularly bovine, have been incriminated as natural reservoirs of STEC worldwide [6]. Attaching and effacing (A/E) E. coli are characterized by their ability to cause A/E lesions in the gut mucosa of human and animal hosts leading to diarrhea [7].
Recently, the studies concerned the virulence genes and serotypes of E. coli isolated from different animal and environmental sources in Egypt take a some attention [8-10], “but not yet in buffalo which is considered better than cattle in accommodation with the Egyptian climate conditions and distributed either in solitary populations or small holder farms [11].”
In Egypt, like many developing countries, there are commonly used antibiotics in human as well as the farm animal population as aminoglycosides (gentamicin, tobramycin, kanamycin, …etc.), β-lactam (ampicillin, amoxicillin, etc.), and sulfa groups due to their low costs, so investigation of their resistance genes is of public concern. Although there were many researches over the world implicated ruminants, especially cattle, as a principal reservoir of enterovirulent Escherichia coli, only a few reports on the role of buffalo as a reservoir of various E. coli pathotypes.
Materials and Methods
Ethical approval
As per CPCSEA guidelines, a study involving clinical samples does not require the approval of the Institute Animal Ethics Committee.
Bacterial isolation and identification
Rectal samples were collected from 58 diarrheic buffalo calves in sterile swabs, dipped into trypticase soya broth were and incubated aerobically overnight at 37°C.
Samples were streaked onto MacConkey sorbitol agar (oxoid); then, the lactose fermenting pink colonies were picked and cultured onto eosin methylene agar (oxoid) and incubated as described above. The green metallic colonies were picked and examined for their biochemical characters; API 20E kit (Bio Merieux) was performed according to manufacturer’s instruction to detect the biochemical profile of the isolated organisms.
Antimicrobial susceptibility testing
Phenotypical antibiotic susceptibility was tested applying the disk diffusion method on Mueller–Hinton agar plates (oxoid) according to the guidelines of Clinical and Laboratory Standards Institute [12]. A panel of 15 antibiotic discs was used as follows: Cefotaxime (30 μg), cefazolin (30 μg), trimethoprim/sulfamethoxazole (25 μg), sulphaprim (50 μg), ampicillin (10 μg), amoxicillin (10 μg), erythromycin (15 μg), amikacin (30 μg), kanamycin (30 μg), gentamycin (10 μg), ciprofloxacin (5 μg), norfloxacin (10 μg), cefadroxil (30 μg), chloramphenicol (10 μg), and tetracycline (10 μg).
Serological identification of E. coli isolates
The serological typing based on agglutination reactions of the somatic antigens (O). O antigens were identified as described by Guinée et al. [13]. All available somatic antigens were (O1 to O185) antisera which done in Animal Health Institute Laboratory, Dokki, Giza.
Hemolysis assay
E. coli isolates were propagated on blood agar base (oxoid) supplemented with 5% washed sheep erythrocytes. The plates were incubated at 37°C for 24 h, and the presence of a clear colorless zone surrounding the colonies indicated b-hemolytic activity [14].
DNA extraction
DNA extraction from the samples was performed using the QIAamp DNA Mini kit (Qiagen, Germany, GmbH) with modifications from the manufacturer’s recommendations. Briefly, 200 µl of the sample suspension was incubated with 10 µl of proteinase K and 200 µl of lysis buffer at 56°C for 10 min. After incubation, 200 µl of 100% ethanol was added to the lysate. The sample was then washed and centrifuged following the manufacturer’s recommendations. Nucleic acid was eluted with 100 µl of elution buffer provided in the kit.
Oligonucleotide primer
Primers used were supplied from Metabion (Germany) are listed in Tables-1 [15-17] and 2 [18-20].
Table-1.
Multiplex PCR: Primers sequences, target genes, amplicon sizes, and cycling conditions.
| Target gene | Primers sequences | Amplified segment (bp) | Primary denaturation | Amplification (35 cycles) | Final extension | Reference | ||
|---|---|---|---|---|---|---|---|---|
| Secondary denaturation | Annealing | Extension | ||||||
| aadB | GAGCGAAATCTGCCGCTCTGG | 319 | 94°C 5 min | 94°C 30 s | 55°C 45 s | 72°C 45 s | 72°C 10 min | [15] |
| CTGTTACAACGGACTGGCCGC | ||||||||
| blaTEM | ATCAGCAATAAACCAGC | 516 | [16] | |||||
| CCCCGAAGAAC GTTTTC | ||||||||
| Sul1 | CGGCGTGGGCTACCTGAA CG | 433 | [17] | |||||
| GCCGATCGCGTGAAGTTC CG | ||||||||
PCR=Polymerase chain reaction
Table-2.
Diplex PCR: Primers sequences, target genes, amplicon sizes, and cycling conditions.
| Target gene | Primers sequences | Amplified segment (bp) | Primary denaturation | Amplification (35 cycles) | Final extension | Reference | ||
|---|---|---|---|---|---|---|---|---|
| Secondary denaturation | Annealing | Extension | ||||||
| Stx1 | ACACTGGATGATCTCAGT GG | 614 | 95°C 3 min | 94°C 60 s | 59°C 45 s | 72°C 90 s | 72°C 10 min | [18-20] |
| CTGAATCCCCCTCCATTA TG | ||||||||
| Stx2 | CCATGACAACGGACAGCAGTT | 779 | ||||||
| CCTGTCAACTGAGCAGCACTTTG | ||||||||
| hylA | ACGATGTGGTTTATTCTG GA | 165 | 53°C 45 s | |||||
| CTTCACGTGCCATACATAT | ||||||||
| eae A | GACCCGGCA ACAAGCATA | 384 | ||||||
| AGC | ||||||||
| CCACCTGCAGCAACAAGAGG | ||||||||
PCR=Polymerase chain reaction
Polymerase chain reaction (PCR) amplification
Primers were utilized in a 50-µl, PCR reaction containing 25 µl of EmeraldAmp Max PCR Master Mix (Takara, Japan), 1 µl of each primer of 20 pmol concentrations, 9 µl of water, and 10 µl of DNA template. The reactions were performed either multiplex (aadB, blaTEM, and Sul1) or diplex (Stx1 and Stx2) or (hylA and eae A) as described in the Tables-1 and 2 in an applied biosystem 2720 thermal cycler.
Analysis of the PCR products
The products of PCR were separated by electrophoresis on 1.5% agarose gel (Applichem, Germany, GmbH) in ×1 Tris/Borate/ethylenediaminetetraacetic acid buffer at room temperature using gradients of 5 V/cm. For gel analysis, 30 µl of the products was loaded in each gel slot. A 100 bp ladder (Qiagen, Germany, GmbH) was used to determine the fragment sizes. The gel was photographed by a gel documentation system (Alpha Innotech, Biometra) and the data were analyzed through computer software.
Reference E. coli strain ATCC35150 (O157:H7, stx1, stx2, eae, hylA), and E. coli HB 101 inv+ were used as a positive control and S. aureus ATCC29737 as a negative control.
Results and Discussion
The isolation and serotyping of E. coli
Of 58 collected rectal samples, there were 14 typed isolates as E. coli (24.1%); 6 were belonged to serogroup O78 (10.3%), followed by O125 (4 isolates, 6.9%), then O158 (3 isolates, 5.2%) and one isolate O8 (1.7%), only 5 E. coli isolates showed a picture of hemolysis (35.7%). These results were some extent close to that obtained by Beraldo et al. [6] who determined the prevalence of enteropathogenic E. coli and STEC E. coli among buffalo and found that 76 out of 256 samples (29.7%) were positive, somewhat approach another study applied on fecal samples from 174 slaughter buffalo in Dhaka, Bangladesh, as 37.9% [21]. On the other side, there were other studies showed a higher incidences; Oliveira et al. [22] reported the presence of E. coli in water buffaloes in Brazil “as the first survey in South America” ranged from 0 to 64%, depending on the farm. However, in another study in Italy, the incidence of isolation reached 70% (220/314) among Mediterranean water buffalo calves <4 weeks old affected by severe diarrhea with a lethal outcome [23]; moreover, the same incidence was recorded in buffalo farms, in the central region of Vietnam [24].
The serotyping of the study isolates showed three serogroups O8, O125, and O158, which also detected among 36 different O groups in another study [21], the fourth serotype represented O78 which was commonly found in cattle population in Egypt [9,10].
The antibiotic sensitivity assay showed that the E. coli isolates were more frequently resistant to ampicillin (71.4%), amoxicillin (64.3%), followed by intermediate resistance to sulphaprim (50%) then gentamicin and kanamycin (42.8%), while the isolates were found to be highyly sensitive to ciprofloxacin, norfloxacin, trimethoprim-sulfamethoxazole, and tetracycline (100%), then less sensitive to amikacin and cephalosporine group (71.4%).
Results of PCR for the detection of certain antibiotic resistance genes
The 14 E. coli isolates were screened for presence certain antibiotic resistance determinant genes, the results obtained from Figure-1 revealed that all 14 isolated E. coli serotypes which were used in mPCR, carried at least one gene, as 10 carried blaTEM (71.4%), 8 Sul1 (57.1%), and 6 aadB (42.8%), one carried the 3 genes (7.1%) and 8 carried 2 genes (57.1%). These results lead until there were 9 isolates could be considered MDR by an incidence of 64.3%. The previous investigations stated the isolated E. coli strains were resistant most frequent to erythromycin (95.83%), cephalothin (62.5%), amikacin (54.17%), kanamycin (45.83%), and gentamicin (41.67%) [25]. While in another one the majority of E. coli strains were susceptible to all antimicrobials tested. Resistance to one drug (nalidixic acid, streptomycin, or ampicillin) and resistance to two antimicrobials (ampicillin plus streptomycin and nalidixic acid plus ampicillin) were found in 10 (17.8%) and 2 (3.6%) of the strains, respectively [22].
Figure-1.
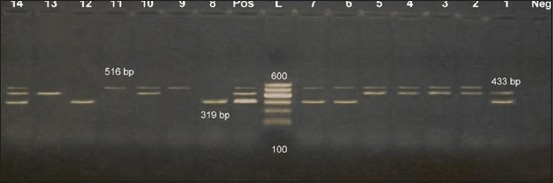
Multiplex polymerase chain reaction detection of antibiotic resistant genes in Escherichia coli isolates showing: L: 100 bp DNA ladder. Lanes 1-14: E. coli isolates. Lane Pos.: Positive control; amplification of 319 bp represented aadB, 433 bp represented Sul1, and 516 bp represented blaTEM. Lane Neg.: Negative control.
Results of PCR for the detection of certain virulence genes
Shiga toxins play a major role in intense inflammatory response and may explain the ability of STEC strains to cause severe illness in affected human and animals. These toxins were expressed by Stx genes which constitute two major subfamilies; Stx1 and Stx2 [26]. Besides that there are other virulent determinants included in E. coli pathogenicity; as intimin coded by eae gene, which responsible for the attachment and invasion of the organism to mucosa, and hylA gene which expresses the hemolytic activity [27].
The results obtained from Figure-2 recognized Stx1 gene in 6 isolates (42.8%), two were belonged to both serotypes O125 and O78, while O8 and O158 represented one for each, while Stx2 was present in 4 isolates (28.6%), belonged to both O125 and O78. One O125 isolate harbored the two genes, overall, 9 isolates carried either one or the two Stx genes (64.3%).
Figure-2.
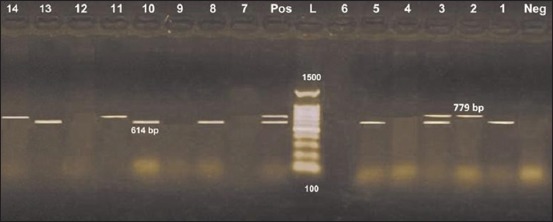
Diplex polymerase chain reaction detection of Shiga toxins genes in Escherichia coli isolates showing: L: 100 bp DNA ladder. Lanes 1-14: E. coli isolates. Lane Pos.: Positive control; amplification of 614 bp represented Stx1, 779 bp represented Stx2. Lane Neg.: Negative control.
On the other hand, the results obtained from Figure-3 recognized hylA gene in 5 isolates (35.7%), the same of what presented in Figure-2, two were belonged to both serotypes O125 and O78, while one was O8 and O158 represented one for each, while eae was present in 2 isolates only (14.3%), belonged to both O8 and O78. One O8 isolate harbored the two genes. These results were near that obtained Oliveira et al. [22] who stated that out of 109 STEC isolates, 42 (38.5%) carried stx2, 43 (39.5%) carried stx1 and stx2 sequences, and only 24 (22%) harbored the stx1 sequence and no intimin. Furthermore, the prevalence of virulence genes in 49 non-O157 E. coli strains, 41 showed detection of stx1 (83.1%), 4 stx2 (8.2%), 4 both (8.2%), 3 showed detection of hylA (6.1%), and no intimin was detected, Beraldo et al. [6], Islam et al. [21] characterize stx1, stx2 in 28.8% and eae was found in 14.4% in tested isolates. While in Vu-Khac and Cornick [24] stx-positive strains were recovered from 27% of buffaloes and eae gene was not detected in buffalo isolates.
Figure-3.
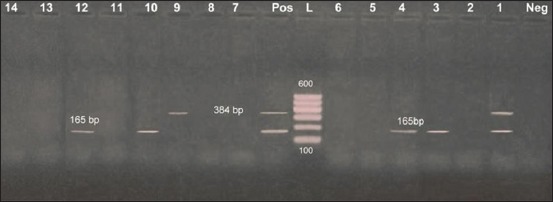
Diplex polymerase chain reaction detection of hylA and eae genes in Escherichia coli isolates showing: L: 100 bp DNA ladder. Lanes 1-14: E. coli isolates. Lane Pos.: Positive control; amplification of 165 bp represented hylA, 384 bp represented eae. Lane Neg.: Negative control.
Moreover, “E. coli” that harboring any gene for Shiga toxins (stx1 or stx2) was detected in 24 (6.61%) of the 363 E. coli isolates [25]. On the opposite manner, there were high records as, Borriello et al. [23] characterized the 120 E. coli isolates for the presence of the virulence factors Stx1, Stx2, haemolysins, eae, and found that Stx1 (80%) more frequent than Stx2 (27%) while the eae and hylA were positive in all isolates.
Conclusion
The obtained data of our study revealed that the Egyptian buffalo may play role in colonization and dissemination of pathogenic, diarrhegenic E. coli and may constitute a prospective human threatening as a source of certain zenotic serotypes infection through milk and meat consumption. The problem is more exacerbating with the marked incidence of multi-antimicrobial resistance isolates, so the attention should be directed to hygienic precautions, as well as careful use of antibiotics in farm animal husbandry.
Authors’ Contributions
ASH supervised the experiment. STO and ASH shared in the isolation and the antibiotic susceptibility performance, EAF collected the samples and help in antibiotic susceptibility, ASH and EAF identified the isolates by API 20E kit. STO, SMS, and ASH performed PCR, ASH prepared and reviewed the manuscript. All authors read and approved the final manuscript.
Acknowledgments
The authors acknowledge Serology Department, Animal Health Institute Laboratory, Dokki, Giza, for serological identification performance. The research was conducted by personal finance.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Stenutz R, Weintraub A, Widmalm G. The structures of Escherichia coli O-polysaccharide antigens. FEMS Microbiol. Rev. 2006;30:382–403. doi: 10.1111/j.1574-6976.2006.00016.x. [DOI] [PubMed] [Google Scholar]
- 2.Pradel N, Boukhors K.Y, Forestier C, Martin C, Livrelli V. Heterogeneity of Shiga toxin-producing Escherichia coli strains isolated from hemolytic-uremic syndromepatients, cattle, and food samples in central France. Appl. Environ. Microbiol. 2001;67:2460–2468. doi: 10.1128/AEM.67.6.2460-2468.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Majalija S, Segal H, Ejobi F, Elisha B. Shiga toxin gene-containing Escherichia coli from cattle and diarrheic children in the pastoral systems of southwestern Uganda. J. Clin. Microbiol. 2008;46:352–354. doi: 10.1128/JCM.01995-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jianyu S, Shi L, Liansheng Y, Zenghuang X, Li X, Shinji Y. Analysis of integrons inclinical isolates of Escherichia coli in China during the last 6 years. FEMS Microbiol. Lett. 2006;254:75–80. doi: 10.1111/j.1574-6968.2005.00025.x. [DOI] [PubMed] [Google Scholar]
- 5.Canal N, Meneghetti K.L, Almeida C.P.D, Bastos M.D.R, Otton L.M, Corc G. Characterization of the variable region in the class 1 integron of antimicrobial-resistant Escherichia coli isolated from surface water. Braz. J. Microbiol. 2016;4(7):337–344. doi: 10.1016/j.bjm.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beraldo L.G, Borges C.A, Maluta R.P, Cardozo M.V, Rigobelo E.C, de Ávila F.A. Detection of Shiga toxigenic (STEC) and enteropathogenic (EPEC)Escherichia coli in dairy buffalo. Vet. Microbiol. 2014;170(1-2):162–166. doi: 10.1016/j.vetmic.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 7.Fröhlicher E, Krause G, Zweifel C, Beutin L, Stephan R. Characterization of attaching and effacing Escherichia coli (AEEC) isolated from pigs and sheep. BMC Microbiol. 2008;11(8):144. doi: 10.1186/1471-2180-8-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osman K.M, Mustafa A.M, Aly M.A.K, El-Hamed G.S.A. Serotypes, virulencegenes, and intimin types of shiga toxin-producing Escherichia coli and enteropathogenic Escherichia coli isolated from mastitic milk relevant to human health in Egypt. Vector BorneZoonotic Dis. 2012;12(4):297–305. doi: 10.1089/vbz.2010.0257. [DOI] [PubMed] [Google Scholar]
- 9.Galal H.M, Hakim A.S, Sohad M.D. Phenotypic and virulence genes screening of Escherichia coli strains isolated from different sources in delta Egypt. Life Sci. J. 2013;10(2):352. [Google Scholar]
- 10.Osman K.M, Mustafa A.M, Elhariri M, El-Hamed G.S.A. The distribution of Escherichia coli serovars, virulence genes, gene association and combinations and virulence genes encoding serotypes in pathogenic E. coli recovered from diarrhoeic calves, sheep and goat. Transbound Emerg. Dis. 2013;60(1):69–78. doi: 10.1111/j.1865-1682.2012.01319.x. [DOI] [PubMed] [Google Scholar]
- 11.FAO World Livestock. Livestock in Food Security. Rome: FAO; 2011. [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Second Informational Supplement M100-S21. Wayne: CLSI; 2012. [Google Scholar]
- 13.Guinée P.A.M, Jansen W.H, Wadström T, Sellwood R. In: Laboratory Diagnosis in Neonatal Calf and Pig Diarrhea: Current Topics in Veterinary and Animal Science. 13th ed. Netherlands: Martinus-Nijhoff, The Hague; 1981. Escherichia coli associated with neonatal diarrhea in piglets and calves; pp. 126–162. [Google Scholar]
- 14.Vaishnavi C, Kaur S, Beutin L, Krueger U. Phenotypic and molecular characterization of clinically isolated Escherichia coli. Indian J. Pathol. Microbiol. 2010;53:503–508. doi: 10.4103/0377-4929.68298. [DOI] [PubMed] [Google Scholar]
- 15.Frana T.S, Carlson S.A, Griffith R.W. Relative distribution and conservation of genes encoding aminoglycoside-modifying enzymes in Salmonella enterica serotype Typhimurium phage type DT104. Appl. Environ. Microbiol. 2001;67:445–448. doi: 10.1128/AEM.67.1.445-448.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colom K, Pèrez J, Alonso R, Fernández-Aranguiz A, Lariňo E, Cisterna R. Simple and reliable multiplex PCR assay for detection of bla TEM, blaSHV and blaOXA-1 genes in Enterobacteriaceae. FEMS Microbiol. Lett. 2003;223:147–151. doi: 10.1016/S0378-1097(03)00306-9. [DOI] [PubMed] [Google Scholar]
- 17.Ibekwe A.M, Murinda S.E, Graves A.K. Genetic diversity and antimicrobial resistance of Escherichia coli from human and animal sources uncovers multiple resistances from human sources. PLoS One. 2011;6(6):e20819. doi: 10.1371/journal.pone.0020819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Márcia R.S, Geórgio F.V, Domingos S.L, Jesús B, Tomomasa Y. Virulence factors of Escherichia coli isolated from calves with diarrhea in Brazil. Braz. J. Microbiol. 2003;34(3):230–235. [Google Scholar]
- 19.Sheng H, Davis M.A, Knecht J. Characterization of a Shiga toxin, intimin and enterotoxin hemolysin producing E. coli ONT: H25 strain commonly isolated from healthy cattle. J. Clin. Microbiol. 2005;43:3213–3220. doi: 10.1128/JCM.43.7.3213-3220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leyla G, Kadri G. Virulence properties of Escherichia coli isolated from clinical bovine mastitis. Turk. J. Vet. Anim. Sci. 2007;31(5):361–365. [Google Scholar]
- 21.Islam M.A, Mondol A.S, de Boer E, Beumer R.R, Zwietering M.H, Talukder K.A, Heuvelink A.E. Prevalence and genetic characterization of shiga toxin-producing Escherichia coli isolates from slaughtered animals in Bangladesh. Appl. Environ. Microbiol. 2008;74(17):5414–5421. doi: 10.1128/AEM.00854-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliveira M.G, Brito J.R.F, Carvalho R.R, Guth B.E.C, Gomes T.A.T, Vieira M.A.M, Kato M.A.M, Ramos I.I, Vaz T.M.I, Irino K. Water buffaloes (Bubalus bubalis) identified as an important reservoir of Shiga toxin-producing Escherichia coli in Brazil. Appl. Environ. Microbiol. 2007;73(18):5945–5948. doi: 10.1128/AEM.00929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borriello G, Lucibelli M.G, De Carlo E, Auriemma C, Cozza D, Ascione G, Scognamiglio F, Iovane G, Galiero G. Characterization of enterotoxigenic E. coli (ETEC), Shiga-toxin producing E. coli (STEC) and necrotoxigenic E. coli (NTEC) isolated from diarrhoeic Mediterranean water buffalo calves (Bubalus bubalis) Res. Vet. Sci. 2012;93(1):18–22. doi: 10.1016/j.rvsc.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vu-Khac H, Cornick N.A. Prevalence and genetic profiles of Shiga toxin-producing Escherichia coli strains isolated from buffaloes, cattle, and goats in central Vietnam. Vet. Microbiol. 2008;126(4):356–363. doi: 10.1016/j.vetmic.2007.07.023. [DOI] [PubMed] [Google Scholar]
- 25.Mahanti A, Samanta I, Bandyopadhyay S, Joardar S.N. Molecular characterization and antibiotic susceptibility pattern of caprine Shiga toxin producing-Escherichia coli (STEC) isolates from India. Iran. J. Vet. Res. 2015;16(1):31–35. [PMC free article] [PubMed] [Google Scholar]
- 26.Gyles C.L. Shiga toxin-producing Escherichia coli: An overview. J. Anim. Sci. 2007;85(13):E45–E62. doi: 10.2527/jas.2006-508. [DOI] [PubMed] [Google Scholar]
- 27.Lorusso V, Dambrosio A, Quaglia N.C, Parisi A, LaSalandra G, Lucifora G, Mula G, Virgilio S, Carosielli L, Rella A, Dario M, Normanno G. Verocytotoxin-producing Escherichia coli O26 in raw water buffalo (Bubalus bubalis) milk products in Italy. J. Food Prot. 2009;72(8):1705–1708. doi: 10.4315/0362-028x-72.8.1705. [DOI] [PubMed] [Google Scholar]


