Abstract
Aim:
Aim of the study was to investigate the prevalence, virulence gene profiles, and antimicrobial resistance pattern of Shiga toxigenic Escherichia coli (STEC) in diarrheic buffalo calves from Andhra Pradesh and Telangana States.
Materials and Methods:
A total of 375 fecal samples from diarrheic buffalo calves of 1-7, 8-30, 31-60, and 61-90 days age were collected from which STEC were isolated, and virulence genes were detected using multiplex polymerase chain reaction. The antimicrobial resistance of isolates was tested by disk diffusion method.
Results:
The prevalence of E. coli associated diarrhea in buffalo calves was 85.04%, of which 35.01% was STEC origin. In STEC, the combination of eaeA and, hlyA virulence genes was highest (42.45%) followed by stx1 (16.04%), stx1, stx2 and hlyA (13.21%), stx2 (12.64%), stx1, eae and hlyA (9.43%) and stx1 and hlyA (6.6%) genes were detected. Highest antimicrobial resistance was observed for tetracycline (63.21%) and ampicillin (48.11%), while chloramphenicol, gentamycin (96.33%) and imipenem (99.06%) antibiotics are susceptible. Multidrug resistance was detected in 69.81% of the STEC isolates from diarrheic buffalo calves.
Conclusion:
Higher prevalence of eaeA and hlyA genes carrying isolates of STEC may be a serious zoonotic threat and increased prevalence of multidrug resistance in E. coli may necessitate stringent selection of appropriate antimicrobial agent in treating buffalo calf diarrhea cases.
Keywords: antimicrobial resistance, buffalo calf diarrheia, Shiga toxigenic Escherichia coli, virulence genes
Introduction
Calf diarrhea is a complex syndrome with complex etiopathogenesis causing economic loss directly through mortality and indirectly through treatment costs and reduced growth rates in affected calves [1]. The mortality rate is high, particularly in buffalo calves of <3 months age in India [2]. Among all the etiological agents responsible for calf diarrhea, Escherichia coli are recognized as the leading cause [3]. Shiga toxigenic E. coli (STEC) is one of the pathogenic groups of E. coli that has zoonotic origin and cattle being recognized as the major reservoir for human infections.
In calves, STEC is the main cause of diarrhea and dysentery particularly in very young calves [4]. The pathogenicity of STEC is mediated mainly through Shiga toxins 1 and 2 encoded by stx1 and stx2 genes, respectively [5], intimin, an outer membrane surface adhesin encoded by eaeA gene [6] and enterohemolysin (ehly) which is encoded by the hlyA gene [7]. Antimicrobial therapy is an important tool in reducing the incidence and treatment of diarrhea in calves. However, widespread and indiscriminate use of antimicrobial agents leads to multi-drug resistant pathogenic bacteria in calves, resulting difficulty in treatment [8].
Therefore, the aim of this study was to investigate the diversity and distribution of virulence genes and to understand antimicrobial resistance epidemiology of STEC isolated from diarrheic buffalo calves in Andhra Pradesh and Telangana States.
Materials and Methods
Ethical approval
Ethical approval was not necessary for this study. However, samples were collected as per standard collection procedure without harming or giving stress to the animals.
Sample collection
A total of 375 fecal samples from diarrheic buffalo calves of 1-7, 8-30, 31-60, and 61-90 days age were collected randomly from organized dairy farms and individual farmers of Vizianagaram, Vishakapatnam, East Godavari, West Godavari, Krishna, Guntur, Prakasam, Districts of Andhra Pradesh State and Ranga Reddy and Khammam Districts of Telangana State during the period from May 2014 to November 2015. Geographical distribution and age of diarrheic calves were recorded during sampling. Fecal samples were collected using sterile rectal swabs. After collection, the swabs were immediately transported to the Department of Veterinary Microbiology, NTR College of Veterinary Science, Gannavaram in ice-cooled containers for E. coli isolation. All the samples were inoculated on MacConkey agar and incubated at 37°C for 24 h. The pink colonies obtained were again inoculated on EMB agar, and the colonies showing green metallic sheen were selected and confirmed as E. coli by standard biochemical tests [9] and by polymerase chain reaction (PCR) amplifying 16S rRNA gene [10].
DNA isolation
Isolation of DNA from E. coli was carried out by conventional boiling and snap chilling method [11] with slight modifications. A single colony was inoculated in 1 ml tryptic soy broth and incubated at 37°C for 24 h. The cells were harvested by centrifugation at 5000 rpm for 10 min. The pellet was washed with phosphate buffer saline by centrifuging at 500 rpm for 10 min for twice. Then, the pellet was resuspended in 500 µl nuclease free water and boiled for 5-10 min at 100°C and snap chilled on ice, after centrifugation at 1000 rpm for 5 min; supernatant was used as template DNA.
Detection of Shiga toxigenic E. coli
The primers used in the present study for the detection of Shiga toxin producing E. coli were as described [5] (Table-1). Multiplex PCR for amplification of the stx1, stx2, eaeA, and hlyA genes was set up in 25 µl reaction contained 3 µl of bacterial lysate, 2.5 µl of 10× PCR buffer with 1.5 mM of MgCl2, 250 nM of each forward primer and reverse primer, 0.5 µl of 10 Mm dNTP’S and 0.5 units of Taq DNA polymerase, and autoclaved milli-Q water to make volume to 25 µl. Samples were subjected to 35 PCR cycles, each consisting of 1 min of denaturation at 95°C; 2 min of annealing at 65°C for the first 10 cycles, decrementing to 60°C by cycle 15, and 1.5 min of elongation at 72°C, incrementing to 2.5 min from cycles 25 to 35. PCR reaction mixtures were electrophoresed on 2% agarose gels and stained with ethidium bromide.
Table-1.
Details of the primers used for the detection of stχ1, stχ2, eaeA and hly A genes.
| S. No | Primer | Sequence (5’---- 3’) | Target gene | Expected amplicon size (bp) |
|---|---|---|---|---|
| 1 | Stχ1F | ATAAATCGCCATTCGTTGACTAC | stχ1 | 180 |
| Stχ2 R | AGAACGCCCACTGAGATCATC | |||
| 2 | Stχ2 F | GGCACTGTCTGAAACTGCTCC | stχ2 | 254 |
| Stχ2 R | TCGCCAGTTATCTGACATTCTG | |||
| 3 | eaeA F | GACCCGGCACAAGCATAAGC | eaeA | 384 |
| eaeA R | CCACCTGCAGCAACAAGAGG | |||
| 4 | hlyA F | GCATCATCAAGCGTACGTTCC | hlyA | 534 |
| hlyA R | AATGAGCCAAGCTGGTTAAGCT |
Antibiotic susceptibility testing
Antimicrobial resistance against 18 different antibiotics, i.e., for ampicillin, cefotaxime, ceftazidime, amoxicillin clavulanic acid, gentamycin, kanamycin, streptomycin, sulfisoxazole, cotrimoxazole, nalidixic acid, ciprofloxacin, aztreonam piperacillintazobactem, tetracyclin, chloramphenicol, meropenem, imipenem, and nitrofurantoin was studied by disk diffusion method [12]. The diameter of the zone of inhibition was compared with the standard known value against each specific antimicrobial agent as suggested in the product information (interpretation guideline) from manufacturer.
Results and Discussion
This study detected 80.53% prevalence of E. coli in diarrheic buffalo calves in AP and TS states (Table-2), and the prevalence (88.20%) was highest in commercial diaries than in buffalo calves maintained by individual farmers (72.86%). This might be due to unhygienic housing, poor feeding, and management of buffalo calves in commercial diaries than at individual farmers. The prevalence rate reported in this study was higher than prevalence of E. coli reported in diarrheic buffalo calves in Jabalpur, India (59.37%) [13], Egypt (66 and 72%) [14,15], Pakistan (14.6%) [16], and Bangladesh (45) [17]. The differences of the prevalence rates of E. coli in diarrheic calves may be attributed to the geographical locations and management practices as well as hygienic measures which influence the susceptibility of calves to E. coli infection [18,19].
Table-2.
Prevalence of E. coli in the fecal samples of diarrheic calves obtained from different ages.
| Age (days) | Number of samples collected | Number positive for E. coli | Prevalence (%) |
|---|---|---|---|
| 1-7 | 127 | 108 | 85.04 |
| 8-30 | 187 | 157 | 83.96 |
| 31-60 | 49 | 31 | 63.27 |
| 61-90 | 12 | 6 | 50.00 |
| Total | 375 | 302 | 80.53 |
E. coli: Escherichia coli
This study also observed the highest prevalence of E. coli (85.04%) associated diarrhea in 1-7 days age buffalo calves followed by 83.96% and 63.27% prevalence in 8-30 and 31-60 day age groups, while lowest (50.0%) prevalence was observed in 61-90 days age buffalo calves (Table-2). Higher prevalence in younger calves may be due to increased susceptibility to E. coli infection [20-22] and predisposing factors like overcrowding and malnutrition, which are supposed to be a primary cause of immunosuppression [14]. Further, E. coli is a commensal organism and is responsible for diarrhea in calves, particularly calves receiving less or no maternal antibodies through colostrum where milk is mainly used for commercial purposes [23].
Among the E. coli isolates, 35.10% were detected as STEC by multiplex PCR (Figure-1) which was higher (6.8%) and lower (47.30%) than STEC reported from fecal samples of diarrheic buffalo calves in Italy [24] and diarrheic calves in Iran [25]. Studies carried out in different countries by several researchers revealed that 10-80% of cattle may carry STEC [26]. These differences might be due to variation in environment which has an influence on the shedding of STEC in calves [27].
Figure-1.
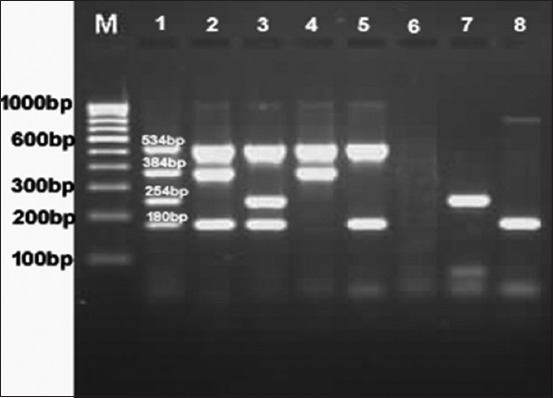
Multiplex polymerase chain reaction detecting the virulence genes. Lane M: 100 bp DNA ladder, Lane 1: Standard stx1 (180 bp), stx2 (254 bp), eaeA (384 bp), hlyA (534 bp), Lane 6: Negative, Lane 2-5,7 and 8: Escherichia coli isolates from diarrheic buffalo calves.
The virulence gene profile of the STEC isolates from diarrheic calves was found in diverse combinations (Table-3 and Figure-1).
Table-3.
Distribution of virulence genes among STEC isolates in diarrheic buffalo calves.
| Total E. coli isolates | STEC | Virulence gene | Number of isolates | % in STEC |
|---|---|---|---|---|
| 302 | 106 (35.10%) | Stχ1 | 17 | 16.04 |
| Stχ2 | 13 | 12.26 | ||
| Stχ1stχ2HlyA | 14 | 13.21 | ||
| Stχ1EaeAHlyA | 10 | 9.43 | ||
| Stχ1HlyA | 7 | 6.60 | ||
| EaeA, HlyA | 45 | 42.45 |
E. coli: Escherichia coli, STEC: Shiga toxigenic Escherichia coli
The STEC isolates carrying eaeA and hlyA genes were most prevalent (42.45%). Similar prevalence rate of eaeA and hlyA genes harboring isolates from diarrheic calves was also reported in other studies [6,28]. Several investigators have reported that the strong association between the carriage of eaeA gene, and the capacity of STEC to cause severe human disease [6,29]. Therefore, higher prevalence of eaeA and hlyA genes in STEC isolates from diarrheic buffalo calves detected in the present study may be a serious zoonotic threat in this geographic region. The STEC isolates carrying 16.04% of Stx1 and 12.26% of Stx2 genes in the present study was lower than reported in Iran [30] but higher than reported in India [31], Poland [32], and Turkey [33]. However, higher prevalence of stx1 gene than stx2 gene observed from diarrheic calves in the present study is comparable to the observations in Argentina [34] and in Austria [35].
The antimicrobial susceptibility testing revealed 69.81% of the STEC isolates were resistant to three or more of the antimicrobial agents tested. Among the STEC isolates, highest percentage of antimicrobial resistance was observed for tetracycline (63.21), followed by ampicillin (48.11%), aztreonam (36.79%), cefotaxime, ceftazidime, and streptomycin, (31.13%), nalidixic acid (29.25%), sulfisoxazole (28.30%), cotrimoxazole (26.42%), amoxicillin clavulanic acid (20.75%), piperacillintazobactem (18.87%) meropenem (17.92%) kanamycin and nitrofurantoin (12.26%) ciprofloxacin (4.72), chloramphenicol and gentamycin (3.77%) while lowest % of 0.94 was observed for imipenem antibiotics. The present findings were corroborated with findings of 100% multidrug resistance in STEC isolates from diarrheic calves in Brazil [36].
Several investigators [36-38] around the world also detected highest antibiotic resistance for E. coli isolates from diarrheic calves to tetracycline antibiotic. Highest sensitivity of the STEC isolates to chloramphenicol and gentamycin antibiotics observed in the present study was comparable with findings of Wani et al. [31] and Rehman et al. [39] who reported higher sensitivity for STEC isolates from diarrheic calves to chloramphenicol and gentamycin antibiotics in Jammu and Kashmir of India.
The present results concluded higher prevalence of eaeA and hlyA genes in STEC from diarrheic buffalo calves may be a serious zoonotic threat in this geographical region. Further, the multidrug resistance of STEC isolates may necessitates stringent selection of appropriate antimicrobial agent and judicious use in treating buffalo calf diarrhea cases.
Authors’ Contributions
MS carried out the research work. YNR and KVS designed and supervised the experiment. TSR helped in carried out PCR analysis. MRR did the data analysis. All authors read and approved the final manuscript.
Acknowledgments
The authors are thankful to the Sri Venkateswara Veterinary University, Tirupati, for providing necessary funds (grant number; 1851/B4/B1 dated 15.06.2015 of comptroller, SVVU, Tirupati to the department of Veterinary Microbiology, NTR CVSc, Gannavaram) and facilities to carry out the research work.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Radostits O.M, Gay C.C, Hinchcliff K.W, Constable P.D, editors. Veterinary Medicine. 10th ed. Philadelphia, PA: Saunders; 2007. pp. 847–888. [Google Scholar]
- 2.Tiwari R, Sharma M.C, Singh B.P. Buffalo calf health care in commercial dairy farms: A field study in Uttar Pradesh (India) Livest. Res. Rural Dev. 2007;19(3):38. [Google Scholar]
- 3.Abubaker A, Ayis E.I, Ali A, Elgaddal Y, Almofti A. Isolation, identification and enterotoxin detection of Escherichia coli isolated from calf diarrhea and their virulence characteristics. J. Appl. Ind. Sci. 2015;3(4):141–149. [Google Scholar]
- 4.Sandhu K.S, Gyles C.L. Pathogenic shiga toxin-producing Escherichia coli in the intestine of calves. Can. J. Vet. Res. 2002;66:65–72. [PMC free article] [PubMed] [Google Scholar]
- 5.Paton A.W, Paton J.C. Detection and characterization of shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA rfbO111, and rfbO157. J. Clin. Microbiol. 1998;36:598–602. doi: 10.1128/jcm.36.2.598-602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanco M, Blanco J.E, Mora A, Dahbi G, Alonso M.P, Gonzalez E.A, Bernardez M.I, Blanco J. Serotypes virulence genes, and intimin types of shiga toxin (verotoxin)-producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eae-ξ) J. Clin. Microbiol. 2004;42:645–651. doi: 10.1128/JCM.42.2.645-651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt H, Beutin L, Karch H. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157: H7 strain EDL 933. Infect. Immun. 1995;63:1055–1061. doi: 10.1128/iai.63.3.1055-1061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar A, Verma A.K, Malik S, Gupta M.K, Sharma A, Rahal A. Occurrence of extended spectrum beta-lactamases producing alpha hemolytic Escherichia coli in neonatal diarrhea. Pak. J. Biol. Sci. 2013;10(3):923–929. doi: 10.3923/pjbs.2014.109.113. [DOI] [PubMed] [Google Scholar]
- 9.Cruickshank R. Medical Microbiology. 11th ed. Edinburgh: The English Language Book Society E and Livingston Ltd; 1970. [Google Scholar]
- 10.Sundong-Bo S, Rui W.U, Xian-Jing H.E, Shuang W, Yun-Cheng L, Xu H.A, Yue-Qiang W, Ting-Ting G.U.O, Guo-Jun W.U, Ke-Li Y. Development of a multiplex PCR for diagnosis of Staphylococcus aureus, Escherichia coli and Bacillus cereus from cows with endometritis. Agric. Sci. China. 2011;10(10):1624–1629. [Google Scholar]
- 11.Wani S.A, Bhat M.A, Samanta I, Nishikawa Y, Buchh A.S. Isolation and characterization of shiga toxin-producing Escherichia coli (STEC) and enteropathogenic Escherichia coli (EPEC) from calves and lambs with diarrhea in India. Lett. Appl. Microbiol. 2003;37:121–126. doi: 10.1046/j.1472-765x.2003.01364.x. [DOI] [PubMed] [Google Scholar]
- 12.Bauer A.W, Kirly W.M.M, Sherris J.C, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- 13.Deepti N, Madhu S, Shrivastav A.B. Detection of verotoxin producing strain of E. coli in buffalo calves. Buffalo Bull. 2015;34(2):227–229. [Google Scholar]
- 14.Abdulgayeid M, Hazem S, Seham F, Madiha S.I. Molecular characterization of Escherichia coli isolated from buffalo calves in El-Behera Governorate. Alex. J. Vet. Sci. 2015;47(1):90–96. [Google Scholar]
- 15.Helal A.D, Hanan E.N, Abdel-Samea M.E, Abdel-Fattaiah S.H.M. Using of traditional and quantitative cytochmecal methods of identification and enzyme characterization of some E. coli serogroups using entiritis in buffaloes. Assiut Vet. Med. J. 2014;60(142):121–131. [Google Scholar]
- 16.Anwarullah M, Khan J.A, Khan M.S.K, Ashraf K, Avais M. Prevalence of Salmonella and Escherichia coli associated diarrhea in buffalo and cow calves. Buffalo Bull. 2014;33(3):332–336. [Google Scholar]
- 17.Islam M.A, Mondol A.S, de Boer E, Beumer R.R, Zwietering M.H, Talukder K.A, Heuvelink A.E. Prevalence and genetic characterization of shiga toxin-producing Escherichia coli isolates from slaughtered animals in Bangladesh. Appl. Environ. Microbiol. 2008;74(17):5414–5421. doi: 10.1128/AEM.00854-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho Y, Yoon K.J. An overview of calf diarrhea-infectious etiology, diagnosis, and intervention. J. Vet. Sci. 2014;15(1):1–17. doi: 10.4142/jvs.2014.15.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Içen H, Arserim N.B, Işik N, Özkan C, Kaya A. Prevalence of four enteropathogens with immunochromatographic rapid test in the feces of diarrheic calves in east and southeast of Turkey. Pak. Vet. J. 2013;33(4):496–499. [Google Scholar]
- 20.Islam K.M.A, Rahman M, Nahar A, Khair A, Alam M.M. Investigation of pathogenic Escherichia coli from diarrheic calves in selective area of Bangladesh. Bang. J. Vet. Med. 2015;13(1):45–51. [Google Scholar]
- 21.Shahrani M, Dehkordi F.S, Momtaz H. Characterization of Escherichia coli virulence genes, pathotypes and antibiotic resistance properties in diarrheic calves in Iran. Biol. Res. 2014;47:28. doi: 10.1186/0717-6287-47-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paul S.K, Khan M.S.R, Rashid M.A, Hassan J, Mahmud S.M.S. Isolation and characterization of Escherichia coli from buffalo calves in some selected areas of Bangladesh. Bang. J. Vet. Med. 2010;8(1):23–26. [Google Scholar]
- 23.Mailk S, Amit K, Amit K.V, Manoj K.G, Som D.S, Arvind K.S, Anu R. Incidence and drug resistance pattern of collibacillosis in cattle and buffalo calves in Western Utter Pradesh in India. J. Anim. Health Prod. 2013;1(2):15–19. [Google Scholar]
- 24.Borriello G.M.G, Lucibelli E.D.E, Carlo C, Auriemma D, Cozza G, Ascione F, Scognamiglio G, Iovane G, Galiero G. Characterization of enterotoxigenic E. coli (ETEC), shiga-toxin producing E. coli (STEC) and necrotoxigenic E. coli (NTEC) isolated from diarrhoeic Mediterranean water buffalo calves (Bubalus bubalis) Res. Vet. Sci. 2012;93:18–22. doi: 10.1016/j.rvsc.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Askari B.M, Zahraei S.T, Rabbani K.M, Tadjbakhsh H, Nikbakht B.G.M, Nadalian M.G. Virulence gene profiles and intimin subtypes of shiga toxin-producing Escherichia coli isolated from healthy and diarrhoeic calves. Vet. Rec. 2010;167:858–861. doi: 10.1136/vr.c4009. [DOI] [PubMed] [Google Scholar]
- 26.Cobbold R, Desmarchelier P. A longitudinal study of shiga toxigenic Escherichia coli (STEC) prevalence in three Australian dairy herds. Vet. Microbiol. 2000;71:125–137. doi: 10.1016/s0378-1135(99)00173-x. [DOI] [PubMed] [Google Scholar]
- 27.Shaw D.J, Jenkins C, Pearce M.C, Cheasty T, Gunn G.J, Dougan G. Shedding patterns of verocytotoxinproducing Escherichia coli strains in a cohort of calves and their dams on a Scottish beef farm. Appl. Environ. Microbiol. 2004;70:7456–765. doi: 10.1128/AEM.70.12.7456-7465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aidar-Ugrinovich L, Blanco J, Blanco M, Blanco J.E, Leomil L, Dahbi G. Serotypes, virulence genes, and intimin types of Shiga toxin-producing Escherichia coli (STEC) and enteropathogenic E. coli (EPEC) isolated from calves in Sao Paulo, Brazil. Int. J. Food Microbiol. 2007;115:297–306. doi: 10.1016/j.ijfoodmicro.2006.10.046. [DOI] [PubMed] [Google Scholar]
- 29.Beutin L, Krause G, Zimmermann S, Kaulfuss S, Gleifer K. Characterization of shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3 years period) J. Clin. Microbiol. 2004;42:1099–1108. doi: 10.1128/JCM.42.3.1099-1108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dastmalchi S.H, Ayremlou N. Characterization of shiga toxin-producing Escherichia coli (STEC) in feces of healthy and diarrheic calves in Urmia region, Iran. Iran. J. Microbiol. 2012;4(2):63–69. [PMC free article] [PubMed] [Google Scholar]
- 31.Wani S.A, Hussain I, Beg S.A, Rather M.A, Kabli Z.A, Mir M.A, Nishikawa Y. Diarrhoeagenic Escherichia coli and salmonellae in calves and lambs in Kashmir: Absence, prevalence and antibiogram. Sci. Tech. Rev. 2013;32(3):1–17. doi: 10.20506/rst.32.2.2213. [DOI] [PubMed] [Google Scholar]
- 32.Osek J, Gallien P, Protz D. Characterization of shiga toxin-producing Escherichia coli strains isolated from calves in Poland. Comp. Immunol. Microbiol. Infect. Dis. 2000;23:267–276. doi: 10.1016/s0147-9571(00)00008-4. [DOI] [PubMed] [Google Scholar]
- 33.Guler L, Gunduz K, Ok U. Virulence factors and antimicrobial susceptibility of Escherichia coli isolated from calves in Turkey. Zoonoses Public Health. 2008;55:249–257. doi: 10.1111/j.1863-2378.2008.01121.x. [DOI] [PubMed] [Google Scholar]
- 34.Mercado E.C, Gioffre A, Rodriguez S.M, Cataldi A, Irino K, Elizondo A.M. NonO157 shiga toxin producing Escherichia coli isolated from diarrheic calves in Argentina. J. Vet. Med. 2004;51:82–88. doi: 10.1111/j.1439-0450.2004.00729.x. [DOI] [PubMed] [Google Scholar]
- 35.Luna C.H, Klein D, Lapan G, Revilla-Fernandez S, Haschek B, Sommerfeld-Stur I, Moestl K, Baumgartner W. Characterization of virulence factors in Escherichia coli isolated from diarrheic and healthy calves in Austria shedding various enteropathogenic agents. Vet. Med. 2009;54(1):1–11. [Google Scholar]
- 36.Rigobelo E.C, Gamez H.J, Marin J.M, Macedo C, Ambrosin J.A, Ávila F.A. Virulence factors of Escherichia coli isolated from diarrheic calves. Braz. J. Vet. Res. Anim. Sci. 2006;58(3):305–310. [Google Scholar]
- 37.Alberto M, Pilar H, Sonia J, de la Fuente R, Ruiz-Santa-Quiteria J.A, Gustavo D.B, Orden J.A. Phenotypic and genotypic characterization of antimicrobial resistance in enterohemorrhagic Escherichia coli and atypical enteropathogenic E. coli strains from ruminants. J. Vet. Diagn. Invest. 2011;23:91–95. doi: 10.1177/104063871102300114. [DOI] [PubMed] [Google Scholar]
- 38.Carl M.S, Cuiwei Z, Chitrita D.R, Jocelyn T, Shaohua Z, White D.G, Wagner D.D, McDermott P.F, Walker R.D, Meng J. Antimicrobial resistance of Escherichia coli O157 isolated from humans, cattle, swine, and food. Appl. Environ. Microbiol. 2002;68(2):576–581. doi: 10.1128/AEM.68.2.576-581.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rehman M.U, Rashid M, Sheikh J.A, Wani S.A, Farooq S. Multi-drug resistance among shiga toxin producing Escherichia coli isolated from bovines and their handlers in Jammu region, India. Vet. World. 2014;6(9):655–658. [Google Scholar]


