Abstract
Aim:
The aim was to characterize Salmonella enterica serovar Gallinarum isolated from backyard poultry by polymerase chain reaction (PCR) detection of virulence genes invasion (invA) and Salmonella plasmid virulence C (spvC).
Materials and Methods:
Two strains of Salmonella serovar Gallinarum isolates used in this study were obtained from an outbreak of fowl typhoid in backyard Vanaraja fowl. PCR technique was used for detection of invA and spvC genes using standard methodology. The invA PCR product from one representative isolate was sequenced and compared with other related Salmonella serovars in GenBank data.
Results:
Salmonella Gallinarum produced expected amplicons of invA and spvC gene products. Nucleotide sequence of 285 bp invA gene was deposited in GenBank with accession no. KX788214. Sequence analysis of invA gene was found conserved in Salmonella serovars and demonstrated 100% homology with closely related serovars of Salmonella.
Conclusion:
Invasion gene (invA) was found to be highly conserved in Salmonella Gallinarum and highly similar with closely related serovars. The isolates also contained plasmid-mediated spvC gene indicating possession of virulence plasmid.
Keywords: invA, polymerase chain reaction, Salmonella Gallinarum, Salmonella plasmid virulence C, virulence genes
Introduction
Fowl typhoid (FT) is disease of major economic significance in many countries of Asia, Africa, Central and South America [1]. It is an endemic disease of poultry in India with occasional outbreaks [2-4]. The pathogen Salmonella enterica serovar Gallinarum can colonize and cause disease in various domestic and wild birds. The pathogen can get transmitted by both horizontal and vertical routes. The majority of virulence genes of Salmonella are clustered in a region distributed over the chromosome, called Salmonella pathogenicity islands (SPI). A total of 19 SPI have been described with SPI-1 to SP-5 being present in most serovars and others were being less widely distributed [5]. Besides, one large plasmid of approximately 85 kb in Salmonella Gallinarum have the ability of strains to produce high mortality in chickens [6] and Salmonella plasmid virulence (spv) locus that carries the spv genes were reported to be present in Salmonella Gallinarum-Pullorum and few other non-typhoid Salmonella serovars, namely, Salmonella Abortusovis, Salmonella Choleraesuis, Salmonella Dublin, Salmonella Enteritidis, and Salmonella Typhimurium, and Salmonella Sendai [7] and absent in typhoid serovars Typhi and Paratyphi [8].
The chromosomally located invasion gene (invA) being thought to trigger the invasion of Salmonellae into cultured epithelial cells [9], while an operon (spvRABCD) in plasmid containing five genes, involved in intra-macrophage survival of Salmonella [10]. Characterization of Salmonella serovars has been carried out previously by various researchers by polymerase chain reaction (PCR) assay of different virulence factors [11-13], but the study was less reported with Salmonella Gallinarum particularly from backyard poultry.
The current study was aimed to characterize Salmonella Gallinarum obtained from backyard poultry by detection of virulence genes invA and spvC.
Materials and Methods
Ethical approval
As per the Committee for the Purpose of Control and Supervision on Experiments on Animals (CPCSEA) guidelines, this study does not require ethical approval from Institute Animal Ethics Committee.
Bacterial strains
Two isolates of Salmonella Gallinarum (WBSG-1, WBSG-2) obtained from the Department of Veterinary Microbiology, West Bengal University of Animal and Fishery Sciences, Kolkata, from an outbreak of FT in Vanaraja fowl were used. The isolates were serotyped with antigenic structure (9,12:-:-) at National Salmonella and Escherichia Centre, Kasauli, India.
Preparation of culture lysate
Bacterial culture lysate was prepared as described previously [14] with little modification. 1 ml of overnight broth culture of Salmonella Gallinarum was taken in a sterile 1.5 ml microcentrifuge tube (Tarsons, India) and centrifuged at 6000 rpm for 5 min. The pellet was washed twice with Tris-ethylenediaminetetraacetic acid (EDTA) buffer and was re-suspended in 1 ml Tris-EDTA buffer. Then, the culture was boiled for 10 min followed by chilling in ice. The cell debris was removed by centrifugation at 6000 rpm for 5 min. Then, the supernatant was stored at −20°C for further use as template DNA.
PCR assay
Salmonella specific primers described previously [15], the forward primer S139 and reverse primer S141 (Table-1) based on the invA gene of Salmonella were used. The amplification of the invA gene fragment was carried out as described earlier with little modifications [16]. The PCR was carried out with a 25 µl amplification mixture consisting of 3 µl template DNA, 5 µl of ×5 GoTaq® Flexi buffer, 0.5 µl of deoxynucleotide triphosphates (10 mM each), 1.6 µl of 25 mM MgCl2, 1 µl of 10 µM each primer and 0.3 µl of GoTaq® DNA polymerase (Promega, USA), and 12.6 µl nuclease free water. Amplification was conducted in a thermocycler (Mastercycler personal, Eppendorf, Germany). The cycle condition consisted an initial denaturation 94°C for 1 min followed by 35 cycles of denaturation at 94°C for 60 s, annealing at 64°C for 30 s, and elongation at 72°C for 30 s with 7 min final extension period at 72°C. The amplified products were visualized by agarose gel electrophoresis containing 1.5% w/v agarose (SRL, India) with ethidium bromide (0.5 µg/ml) and detected by gel documentation system (UVP, UK).
Table-1.
Oligonucleotides (primers) used for detection of virulence genes (invA and spvC) of Salmonella Gallinarum.
| Genes | Primer | Oligonucleotides (5’-3’) | Amplification product (bp) | References |
|---|---|---|---|---|
| invA | S139 | GTG AAA TTA TCG CCA CGT TCG GGC AA | 284 | [15] |
| S141 | TCATCGCACCGTCAAAGGAACC | |||
| spvC | SPV-1 | ACTCCTTGCACAACCAAATGCGGA | 571 | [7] |
| SPV-2 | TGTCTTCTGCATTTCGCCACCATCA |
spv=Salmonella plasmid virulence
For spvC gene, another set primer was used (Table-1) [7]. The amplification conditions for the spvC gene fragment being similar as described for invA gene except the annealing temperature was 58°C.
Nucleotide sequencing of invA gene
Positive amplification from a PCR reaction of invA gene from one representative isolate (WBSG1) was purified with DNA gel/PCR purification mini kit (Xcelris, India). Both strands of purified PCR product were sequenced with forward and reverse primers for invA gene in an ABI 3730 XL automated sequencer (Applied Biosystems) in custom sequencing facility of Xcelris, India. Sequence obtained was analyzed, and homology searches were conducted using the BLAST algorithm (www.ncbi.nlm.nih.gov/BLAST).
Results
Amplicons of invA and spvC virulence genes were observed in agarose gel as ~284 bp and ~571 bp products, respectively (Figure-1).
Figure-1.
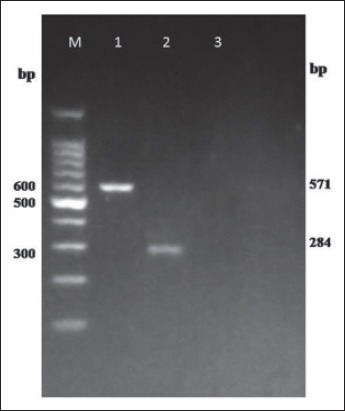
Polymerase chain reaction amplification of virulence genes invA and Salmonella plasmid virulence C (spvC) of Salmonella Gallinarum isolates. Lane M: 100 bp DNA ladder, Lane 1: invA gene (strain WBSG1), Lane 2: spvC gene (strain WBSG2), Lane 3: Negative control.
Nucleotide sequence of invA gene of Salmonella Gallinarum strainWBSG1 obtained in this study was analyzed and 285 bp sequences deposited with NCBI under GenBank accession number KX788214. Sequence alignment with BLAST revealed that invA gene of Salmonella Gallinarum strain WBSG1 was highly similar (100%) with some other poultry serovars such as Salmonella enterica serovar Gallinarum strain 9184 (accession no. CP019035.1) and Salmonella Enteritidis strain OLF 00D 98987-1 (accession no. CP011942.1) isolated elsewhere.
Discussion
Detection of invasion gene of Salmonella by PCR-based assays may be useful for rapid pathogen identification as well. Molecular identification of Salmonella sp. with invA gene primer set S139-S141 conforms to be international standard [17-19] with very high specificity [15]. However, choosing suitable primers are important as the primer sets targeting different sequences within invA gene [7], often resulted in non-specific amplification with the fecal and gut-associated bacteria [20]. In one study, Salmonella isolates belonging to serotypes Anatum, Enteritidis and Amsterdam were also reported negative for the invA gene using those primers [21].
High prevalence of invA virulence gene in Salmonella serovars has also been reported by other workers [22,23]. We found invA gene was 100% similar with other Salmonella serovars. Other studies also reported similar results [24], which were expected since the invasion gene (invA) is conserved among Salmonella serovars. Serovar Enteritidis, Dublin, and Gallinarum were reported to be closely related where serovar Dublin and Gallinarum diverging independently from an Enteritidis-like ancestor [25].
In this study, both Salmonella Gallinarum isolates were positive to spvC gene. This finding was similar with a study in Kashmir where all isolates of Salmonella from poultry harbored virulence genes invA and spvC [26]. However, less prevalence of spv genes was noticed in Salmonella serovars by several workers [9,27,28]. In a study with 37 Salmonella comprising serovar Enteritidis (n=12) and Typhimurium (n=24) originated from pork and slaughterhouse environment, all have produced 284 bp invA gene, but no spvC gene [23]. In another study, a high prevalence (88.6%) of spvA, spvB, and spvC genes was observed in S. Enteritidis from poultry source [13]. One main function of the spv operon is to potentiate the systemic spread of the pathogen [29], and these genes can restore pathogenicity for systemic spread in plasmid-cured strains [30]. The spv region contains three genes required for the virulence phenotype in mice; the positive transcriptional regulator spvR and two structural genes spvB and spvC [8]. Mutations in spvC and spvD genes cause various (allele-specific) defects in Salmonella virulence [31].
Conclusion
Invasion gene (invA) was found to be highly conserved in Salmonella Gallinarum and highly similar with closely related serovars. The isolates also contained spvC gene indicating possession of plasmid virulence.
Authors’ Contributions
SP, KB, and SD planned and designed the study. The experiment was conducted by SP, AB, and IS, data analysis was performed by SNJ, SD, and DPI. All authors participated in the draft and revision of the manuscript. All authors read and approved the final manuscript.
Acknowledgments
The authors are grateful to the Vice-Chancellor, Director of Research, West Bengal University of Animal and Fishery Sciences and Dean, Faculty of Veterinary Science for providing necessary facilities and ICAR for financial assistance (Development Grant 7.1).
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Barrow P.A, Freitas Neto O.C. Pullorum disease and fowl typhoid-new thoughts on old diseases: A review. Avian Pathol. 2011;40:1–13. doi: 10.1080/03079457.2010.542575. [DOI] [PubMed] [Google Scholar]
- 2.Rajagopal R, Mini M. Outbreaks of salmonellosis in three different poultry farms of Kerala, India. Asian Pac. J. Trop. Biomed. 2013;3:496–500. doi: 10.1016/S2221-1691(13)60103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumari D, Mishra S.K, Lather D. Pathomicrobial studies on Salmonella Gallinarum infection in broiler chickens. Vet. World. 2013;6(10):725–729. [Google Scholar]
- 4.Gupta R, Jindal N, Arora D, Singh M, Kapoor P.K. Detection of Salmonella Typhimurium Salmonella Enteritidis and Salmonella Gallinarum from suspected cases of fowl typhoid in poultry in Haryana. Indian Vet. J. 2016;93(7):44–47. [Google Scholar]
- 5.Foley S.L, Lynne A.M, Nayak R. Salmonella challenges: Prevalence in swine and poultry and potential pathogenicity of such isolates. J. Anim. Sci. 2008;86:E149–E162. doi: 10.2527/jas.2007-0464. [DOI] [PubMed] [Google Scholar]
- 6.Barrow P.A, Simpson J.M, Lovell M.A, Binns M.M. Contribution of Salmonella Gallinarum large plasmid toward virulence in fowl typhoid. Infect. Immun. 1987;55:388–392. doi: 10.1128/iai.55.2.388-392.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu C.H, Ou J.T. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes inv A and spv C, by an enrichment broth culture-multiplex PCR combination assay. J. Clin. Microbiol. 1996;34(10):2619–2622. doi: 10.1128/jcm.34.10.2619-2622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guiney D.G, Fierer J. The role of the spv genes in Salmonella pathogenesis. Front. Microbiol. 2011;129:1–10. doi: 10.3389/fmicb.2011.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swamy S.C, Barnhart H.M, Lee M.D, Dreesen D.W. Virulence determinants inv A and spv C in Salmonellae isolated from poultry products, wastewater, and human sources. Appl. Environ. Microbiol. 1996;62:3768–3771. doi: 10.1128/aem.62.10.3768-3771.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rychlik I, Gregorova D, Hradecka H. Distribution and function of plasmids in Salmonella enterica. Vet. Microbiol. 2006;112:1–10. doi: 10.1016/j.vetmic.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 11.Oliveira S.D, Rodenbusch C.R, Michael G.B, Cardoso M.I.R, Canal C.W, Brandelli A. Detection of virulence genes in Salmonella Enteritidis isolated from different sources. Braz. J. Microbiol. 2003;34(Suppl 1):123–124. [Google Scholar]
- 12.Okamoto A.S, Filho R.L.A, Rocha T.S, Menconi A, Marietto-Gonçalves G.A. Relation between the spv Cand inv Avirulence genes and resistance of Salmonella enterica serotype Enteritidis isolated from avian material. Int. J. Poult. Sci. 2009;8(6):579–582. [Google Scholar]
- 13.Kaushik P, Anjay K.S, Bharti S.K, Dayal S. Isolation and prevalence of Salmonella from chicken meat and cattle milk collected from local markets of Patna, India. Vet. World. 2014;7(2):62–65. [Google Scholar]
- 14.Shanmugasamy M, Velayutham T, Rajeswar J. Inv A gene specific PCR for detection of Salmonella from broilers. Vet. World. 2011;4(12):562–564. [Google Scholar]
- 15.Rahn K, De Grandis S.A, Clarke R.C, McEwen S.A, Galan J.E, Ginocchio C, Curtiss R, 3rd, Gyles C.L. Amplification of an inv A gene sequence of Salmonella Typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes. 1992;6(4):271–279. doi: 10.1016/0890-8508(92)90002-f. [DOI] [PubMed] [Google Scholar]
- 16.Dey S, Mahanti A, Batabyal K, Joardar S.N, Samanta I, Isore D.P, Pakhira M.C. Identification and antimicrobial susceptibility of Salmonella Gallinarum isolated from fowl typhoid outbreak in backyard poultry. Explor. Anim. Med. Res. 2016;6(1):63–67. [Google Scholar]
- 17.Malorny B, Hoorfar J, Bunge C, Helmuth R. Multicenter validation of the analytical accuracy of Salmonella PCR: Towards an international standard. Appl. Environ. Microbiol. 2003;69:290–296. doi: 10.1128/AEM.69.1.290-296.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ezzat M.E, Shabana I.I, Esawy A.M, Elsotohy M.E. Detection of virulence genes in Salmonella serovars isolated from broilers. Anim. Vet. Sci. 2014;2(6):189–193. [Google Scholar]
- 19.El-Tawwab A.A.A, Ammar A.M, Ali A.R, El-Hofy F.I, Sayed A.M.E. Detection of common (inv A) gene in salmonellae isolated from poultry using polymerase chain reaction technique. Benha Vet. Med. J. 2013;25(2):70–77. [Google Scholar]
- 20.Ziemer C.J, Steadham S.R. Evaluation of the specificity of Salmonella PCR primers using various intestinal bacterial species. Lett. Appl. Microbiol. 2003;37:463–469. doi: 10.1046/j.1472-765x.2003.01430.x. [DOI] [PubMed] [Google Scholar]
- 21.Turki Y, Mehr I, Ouzari H, Khessairi A, Hassen A. Molecular typing, antibiotic resistance, virulence gene and biofilm formation of different Salmonella enterica serotypes. J. Gen. Appl. Microbiol. 2014;60(4):123–130. doi: 10.2323/jgam.60.123. [DOI] [PubMed] [Google Scholar]
- 22.Karmi M. Detection of virulence gene (inv A) in Salmonella isolated from meat and poultry products. Int. J. Genet. 2013;3(2):7–12. [Google Scholar]
- 23.Chaudhary J.H, Nayak J.B, Brahmbhatt M.N, Makwana P.P. Virulence genes detection of Salmonella serovars isolated from pork and slaughter house environment in Ahmedabad, Gujarat. Vet. World. 2015;8(1):121–124. doi: 10.14202/vetworld.2015.121-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samanta I, Joardar S.N, Das P.K, Sar T.K, Bandyopadhyay S, Dutta T.K, Sarkar U. Prevalence and antibiotic resistance profiles of Salmonella serotypes isolated from backyard poultry flocks in West Bengal, India. J. Appl. Poult. Res. 2014;23:536–545. [Google Scholar]
- 25.Porwollik S, Santiviago C.A, Cheng P, Florea L, Jackson S, McClelland M. Differences in gene content between Salmonella enterica serovar Enteritidis isolates and comparison to closely related serovars Gallinarum and Dublin. J. Bacteriol. 2005;187:6545–6555. doi: 10.1128/JB.187.18.6545-6555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mir I.A, Wani S.A, Hussain I, Qureshi S.D, Bhat M.A, Nishikawa Y. Molecular epidemiology and in vitro antimicrobial susceptibility of Salmonella isolated from poultry in Kashmir. Rev. Sci. Tech. Off. Int. Epiz. 2010;29(3):677–686. doi: 10.20506/rst.29.3.2011. [DOI] [PubMed] [Google Scholar]
- 27.Ammar A.M, Mohamed A.A, El-Hamid M.I.A, El-Azzouny M.M. Virulence genotypes of clinical Salmonella serovars from broilers in Egypt. J. Infect. Dev. Ctries. 2016;10(4):337–346. doi: 10.3855/jidc.7437. [DOI] [PubMed] [Google Scholar]
- 28.Purkayastha A, Borah P, Sharma R.K, Borah P.P. Multiplex PCR for detection of virulence gene profiles of Salmonella isolates from animals and man. Indian J. Appl. Res. 2015;5(12):142–144. [Google Scholar]
- 29.Heithoff D.M, Shimp W.R, Lau P.W, Badie G, Enioutina E.Y, Daynes R.A, Byrne B.A, House J.K, Mahan M.J. Human Salmonella clinical isolates distinct from those of animal origin. Appl. Environ. Microbiol. 2008;10:1757–1766. doi: 10.1128/AEM.02740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gulig P.A, Danbara H, Guiney D.G, Lax A.J, Norel F, Rhen M. Molecular analysis of spv virulence genes of the Salmonella virulence plasmid. Mol. Microbiol. 1993;6:825–830. doi: 10.1111/j.1365-2958.1993.tb01172.x. [DOI] [PubMed] [Google Scholar]
- 31.Rotger R, Casadesús J. The virulence plasmids of Salmonella. Int. Microbiol. 1999;2:177–184. [PubMed] [Google Scholar]


