ABSTRACT
We recently showed that mutations in the HIV-1 envelope (Env) destabilize the V3 loop, rendering neutralization-resistant viruses sensitive to V3-directed monoclonal antibodies (MAbs). Here, we investigated the propagation of this effect on other Env epitopes, with special emphasis on V2 loop exposure. Wild-type JR-FL and 19 mutant JR-FL pseudoviruses were tested for neutralization sensitivity to 21 MAbs specific for epitopes in V2, the CD4 binding site (CD4bs), and the CD4-induced (CD4i) region. Certain glycan mutants, mutations in the gp120 hydrophobic core, and mutations in residues involved in intraprotomer interactions exposed epitopes in the V2i region (which overlies the α4β7 integrin binding site) and the V3 crown, suggesting general destabilization of the distal region of the trimer apex. In contrast, other glycan mutants, mutations affecting interprotomer interactions, and mutations affecting the CD4bs exposed V3 but not V2i epitopes. These data indicate for the first time that V3 can move independently of V2, with V3 pivoting out from its “tucked” position in the trimer while apparently leaving the V2 apex intact. Notably, none of the mutations exposed V2 epitopes without also exposing V3, suggesting that movement of V2 releases V3. Most mutations increased sensitivity to CD4bs-directed MAbs without exposure of the CD4i epitope, implying these mutations facilitate the trimers' maintenance of an intermediate energy state between open and closed conformations. Taken together, these data indicate that several transient Env epitopes can be rendered more accessible to antibodies (Abs) via specific mutations, and this may facilitate the design of V1V2-targeting immunogens.
IMPORTANCE Many epitopes of the HIV envelope (Env) spike are relatively inaccessible to antibodies (Abs) compared to their exposure in the open Env conformation induced by receptor binding. However, the reduced infection rate that resulted from the vaccine used in the RV144 HIV-1 vaccine trial was correlated with the elicitation of V2- and V3-directed antibodies. Previously, we identified various mechanisms responsible for destabilizing the V3 loop; here, we determined, via mutation of numerous Env residues, which of these elements maintain the V1V2 loop in an inaccessible state and which expose V1V2 and/or V3 epitopes. Notably, our data indicate that V3 can move independently of V2, but none of the mutations studied expose V2 epitopes without also exposing V3. Additionally, V1V2 can be rendered more accessible to Abs via specific mutations, facilitating the development of engineered V2 immunogens.
KEYWORDS: HIV envelope, antibody, human immunodeficiency virus, immunogen design, protein dynamics, vaccines
INTRODUCTION
Numerous studies of the trimeric human immunodeficiency virus type 1 (HIV-1) envelope (Env) surface protein have demonstrated that the molecule exhibits significant flexibility and can assume several conformations regardless of whether it is bound by CD4 (1–4). When in the ground state, not bound by CD4, V3 is preferentially masked, and many epitopes of the Env spike are generally inaccessible to antibodies (Abs) relative to their accessibility in the open conformation induced by receptor binding (4–7). The dynamic movement of the Env trimer, with transient exposure of many parts of the molecule, along with gp120 shedding, helps to explain the robust immunogenicity of V3 and the presence of V3-specific Abs in the sera of essentially all HIV-infected individuals despite its occlusion in the ground state of the Env (8–10).
In a previous study, we investigated Env spike plasticity and the various mechanisms that keep V3 occluded when the Env trimer is in the closed ground state (11). We examined the effects of individual mutations in several regions of the gp120 monomer that forced the trimer into a more open state, rendering it highly interactive with V3-directed monoclonal Abs (MAbs). We determined that several interactions were responsible for the packing of the V3 loop in the Env spike and that many of these interactions, if disrupted, could “pop out” V3. These interactions included quaternary interprotomer interactions; hydrophobic packing of the core of gp120 in the bridging sheet; intraprotomer interactions of V3 with adjacent regions of gp120; and limiting flexibility of the V3 region, which can be mediated by glycans (11). Indeed, most primary HIV isolates are typically insensitive to V3 Abs, but many isolates that are sensitive to V3 Abs possess these types of mutations that release V3 (11). Whereas our previous study revealed the alterations that exposed V3 and rendered viruses susceptible to neutralization by V3 MAbs, here, we aimed to further understand the plasticity of Env and whether mutations that release V3 would propagate changes in Env structure and stability affecting exposure of V2 and other epitopes in the Env trimer.
Exposure of epitopes in the second variable loop (V2) of gp120 was of particular interest, since the RV144 clinical HIV vaccine trial data showed that vaccine-induced anti-V2 serum Abs were correlated with a reduced risk of infection (12–14). These data suggested that the V1V2 loop might be an important epitope for vaccine targeting and renewed interest in V2 MAbs. Given the generation and characterization of numerous V2-specific MAbs from infected patients, we sought to employ a large panel of these MAbs with specificities for various epitopes within V2 to determine whether the same structural features that affected V3 MAb potency affected the activities of V2 MAbs.
As V3 is highly flexible relative to other Env domains and V3 is among the most immunogenic regions of the Env glycoproteins (8–10, 15–17), we hypothesized that release of the V3 loop in various mutants from its occluded position in the Env spike occurs independently of substantial conformational alterations in the V1V2 loop. We also sought to determine if the same mutations that released V3 would affect the exposure of various epitopes presented by the V2 region of gp120, using the JR-FL pseudovirus, which exhibits a “closed” trimer conformation and is therefore relatively resistant to neutralization by V2i and V3 MAbs (18–23). The effects of these mutations were subsequently tested, as well, to determine how they affected the potency of MAbs specific for the CD4 binding site (CD4bs) and the CD4-induced (CD4i) epitopes. To address these questions, we measured the relative changes in neutralization sensitivity of wild-type (WT) and mutant JR-FL pseudoviruses to a large panel of human MAbs. The information provided by this study speaks to the overall plasticity of Env. Additionally, the data reveal how V2 Abs can impose immune pressure on the virus and how this information may be used to better elicit V2-specific Abs as part of a vaccine regimen.
RESULTS
Variable effects of JR-FL glycan mutations on neutralization by V2i MAbs.
The wild-type JR-FL pseudovirus is highly resistant to neutralization in the TZM-bl assay by virtually all V2i, V2p, and V2q MAbs tested, which is why it was selected for analysis (Table 1). Since glycosylation of Env shields and stabilizes Env from Abs of many specificities (5, 7, 24, 25), we first tested the effect on the neutralization sensitivity of JR-FL and its variants from which selected gp120 glycans had been deleted (Fig. 1 to 3). We had previously determined that the removal of the glycan in the N160K mutant destabilized V3 by significantly enhancing the flexibility of that region, causing the mutant to become highly sensitive to many V3 MAbs (Table 1) (11). We therefore tested the N160K mutant against the V2 MAb panel but found that the pseudovirus remained highly resistant to neutralization by essentially all V2 MAbs (Table 1). PGT145 neutralization was lost, as would be predicted by this MAb's binding requirements (Fig. 4) (26).
TABLE 1.
Sensitivities of mutant pseudoviruses to V2- and V3-specific MAbsa
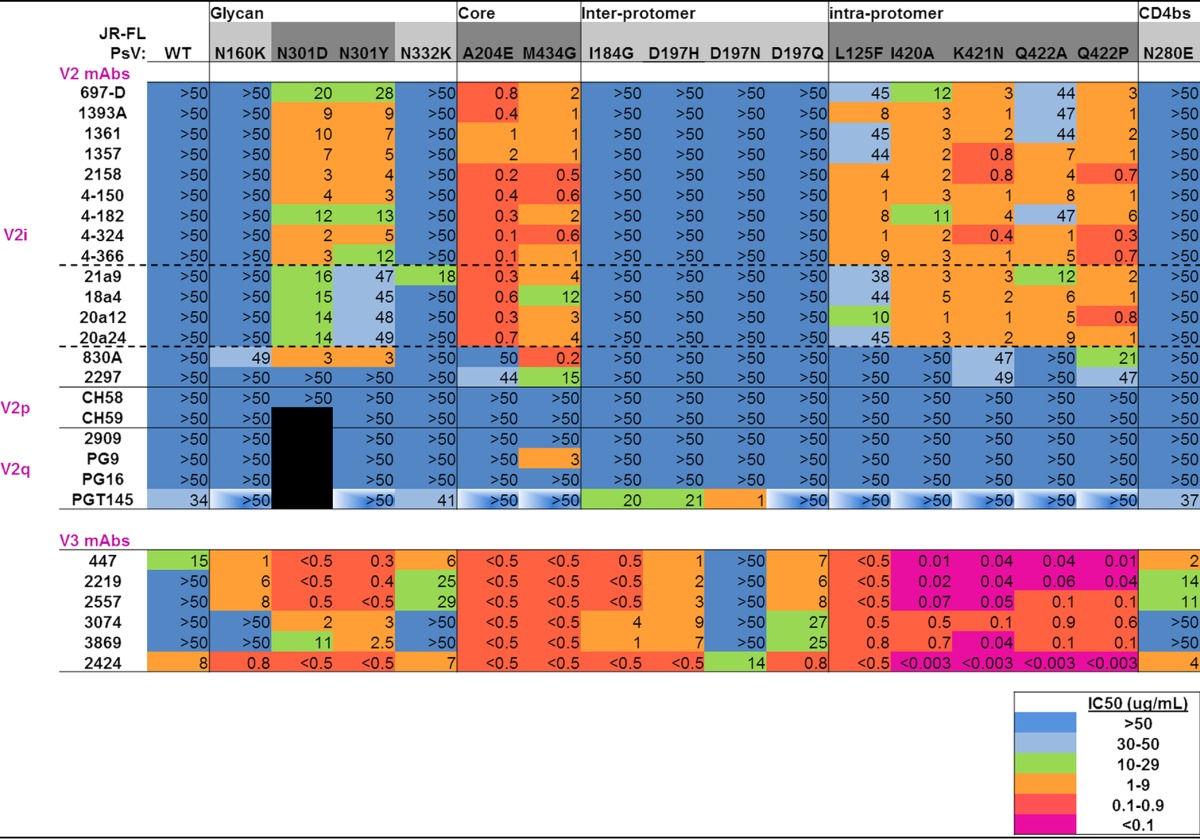
The titles at the top of the table indicate the types of interactions in the Env trimer affected by the mutations listed directly below. JR-FL WT and mutant pseudoviruses (PsV) are shown in the next row across the table. Mutants shaded in dark gray indicate neutralizing sensitivity to V2 and V3 MAbs compared to the WT, while mutants shaded in light gray show neutralization sensitivity to V3 MAbs but not V2 MAbs. IC50s are shown and were calculated by titration of MAbs in the TZM-bl assay, as detailed in Materials and Methods. “>50” (blue shading) indicates that 50% neutralization was not achieved even at the highest MAb concentration tested, 50 μg/ml. Increasing neutralization sensitivity is indicated by color coding, as shown in the color legend. The black boxes indicate that experiments were not done. The boxes with partial white shading indicate decreased neutralization sensitivity. The solid horizontal lines separate classes of MAbs, whereas the dashed horizontal lines indicate subdivisions with the V2i family of MAbs.
FIG 1.
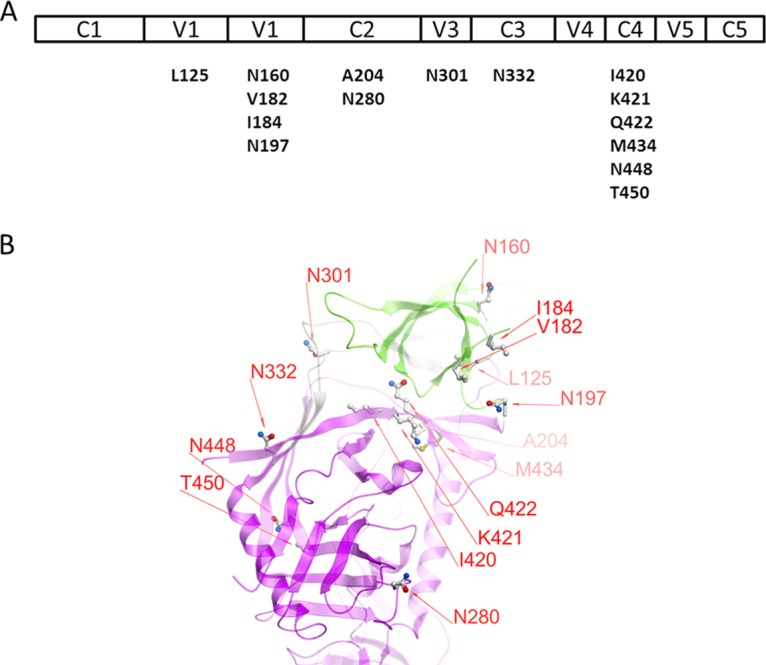
Mutated residues of the JR-FL-JB pseudoviruses used in neutralization assays. (A) The various regions of gp120 are shown above the residues and positions in the wild-type virus that were mutated for this study. (B) Diagram of the positions of the mutations made in gp120. The V1V2 domain is shown in green, the gp120 core in magenta, and the V3 loop in gray ribbon representation. The image is based on PDB entry 4TVP, the crystal structure of the BG505 SOSIP trimer complexed with MAbs PGT122 and 35O22 (27). The numbering is based on the HxB2 reference sequence.
FIG 2.
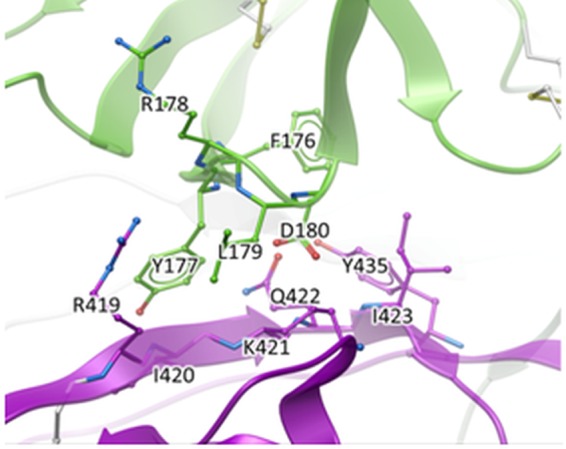
Closeup of the interface between the V1V2 domain (green) and the gp120 core (magenta). Extensive intraprotomer interactions between the region in V1V2 that includes the putative integrin binding motif (179LDI181) and the R419-to-I423 segment in C4 of gp120 are shown. The image is based on PDB entry 4TVP.
FIG 3.
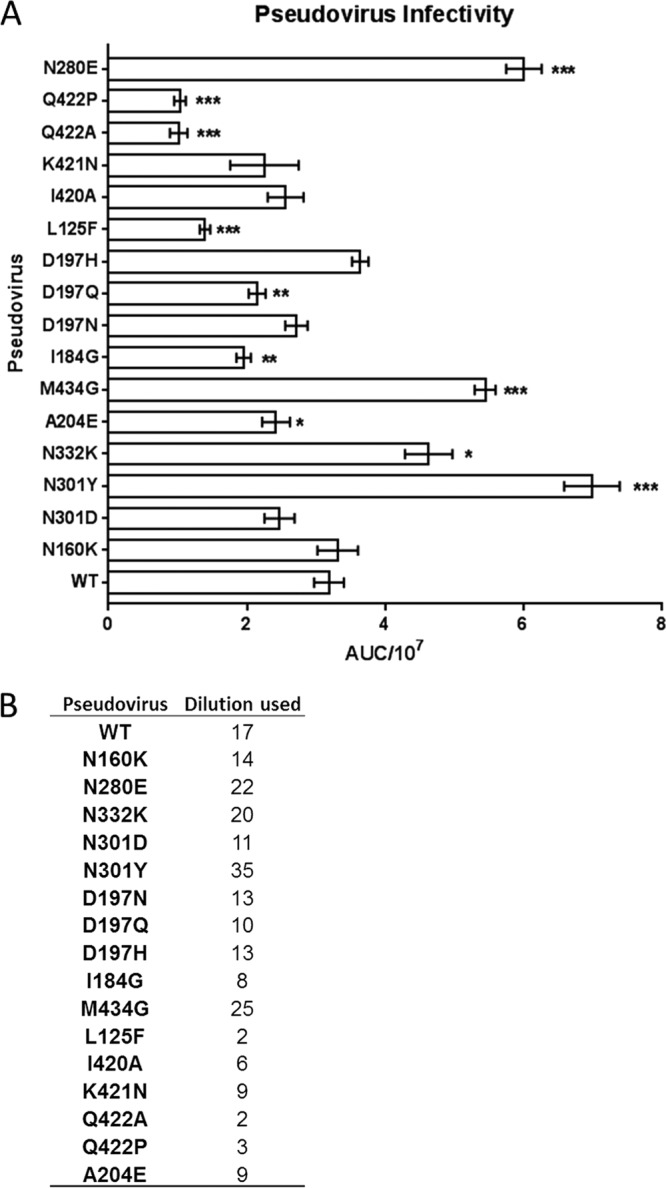
Pseudovirus infectivity. (A) Pseudoviruses were titrated on TZM-bl cells as described previously (61). Replicate titration curves were used to calculate the area under the concentration/time curve (AUC) values using GraphPad Prism. The values were divided by 107. The asterisks indicate increasing degrees of significant (*, 0.05 > P > 0.01; **, 0.01 > P > 0.001; ***, P < 0.001) changes from the WT as determined by 2-tailed Student's t test. (B) Using the titration curves, the appropriate pseudovirus dilutions needed to achieve ∼150,000 RLU were interpolated.
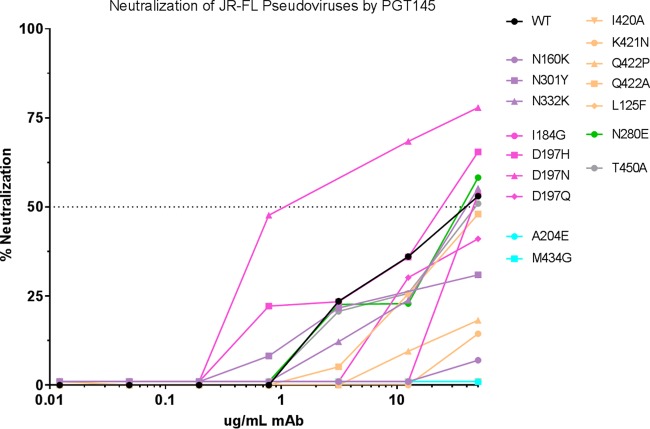
FIG 4.
Neutralization curves of MAb PGT145 against mutant pseudoviruses. Neutralization was performed using the TZM-bl assay, and the percent neutralization was calculated as detailed in Materials and Methods. The curves are colored according to the type of mutation: glycan deletion, lavender; gp120 core disruption, cyan; interprotomer disruption, pink; intraprotomer disruption, tan; CD4bs abrogation, green. The wild type is colored black.
Removal of the glycan at position 332 in the N332K mutant was previously shown to have a relatively small effect on JR-FL V3 MAb sensitivity, as it is part of the secondary and tertiary structures of the outer domain and does not significantly disturb the space that is potentially accessible for local V3 flexing (11). Similarly, the N332K mutant was found to have essentially no effect on sensitivity to any of the V2 MAbs, with the exceptions of V2i MAb 21a9, which became only moderately effective against N332K, exhibiting a 50% inhibitory concentration (IC50) neutralization value of 18 μg/ml (Table 1).
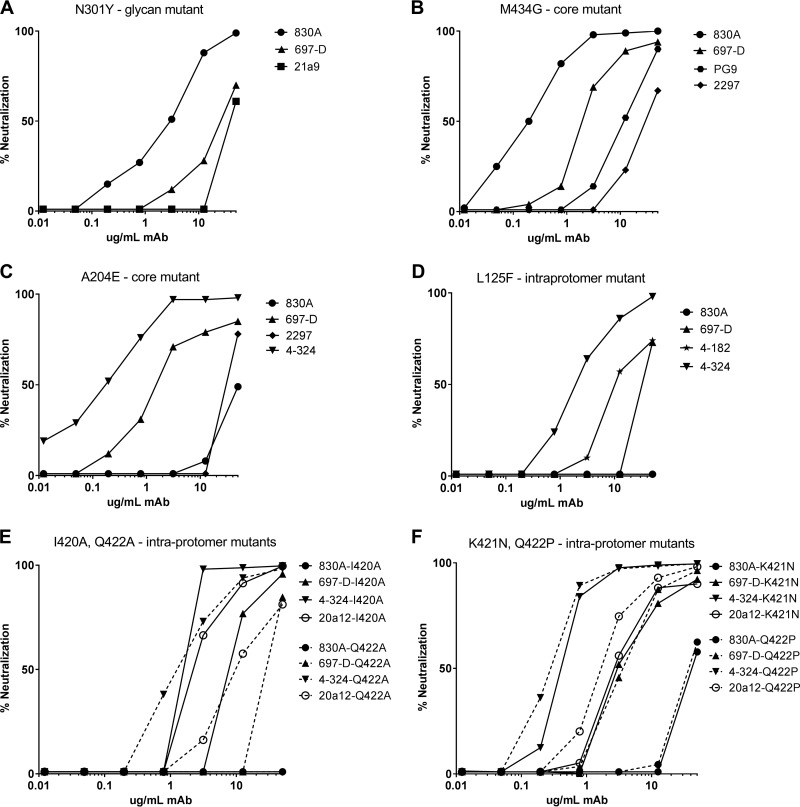
In contrast to the minimal effects elicited by the elimination of glycans at 160 and 332, we found that the elimination of the N301 glycan had significant effects on sensitivity to V2i MAbs. We had previously demonstrated through modeling that the N301 glycan plays a critical role in limiting the flexibility of the crown and stem of V3 (roughly between residues 300 and 328), thus affecting the packing of V3, and that removal of this glycan in the N301Y and N301D mutants resulted in profound increases in sensitivity to V3 MAbs (Table 1) (11). When the V2i MAb panel was tested against these mutants, it was found that the relatively flexible trimer apex of the pseudoviruses allowed access to V2i MAbs compared to JR-FL WT. Notably, this increased sensitivity was not as pronounced as was previously shown for V3 MAbs, as the IC50s for V2i MAbs generally ranged from 2 to 28 μg/ml, values that were on average more than an order of magnitude higher than those for V3 MAbs (<0.05 to 11 μg/ml) (Table 1) (11). However, one group of V2i MAbs (21a9, 18a4, 20a12, and 20a24) was distinct in barely achieving 50% neutralization of N301Y (IC50 = 47 to 49 μg/ml), and MAb 2297 remained unable to neutralize either of the N301 mutants (Table 1). Sample titration curves of the neutralizing activities of three V2i MAbs active against the N301Y mutant JR-FL are shown in Fig. 5A. The V2p and V2q MAb potencies appeared unaffected by the N301 glycan mutants of JR-FL, with the exception of PGT145, whose potency against N301Y was ablated (Fig. 4 and Table 1).
FIG 5.
Neutralization curves of V2 MAbs against V2-destabilizing mutant pseudoviruses. Neutralization was performed using the TZM-bl assay, and the percent neutralization was calculated as detailed in Materials and Methods.
Profound effects of mutations in the gp120 hydrophobic core on neutralization sensitivity to V2i-specific MAbs.
Residues A204 and M434 in the V2 and C4 regions of JR-FL, respectively, form a part of the hydrophobic core underlying the “prebridging sheet” in each Env monomer, and we previously predicted from modeling that alteration of this core would elicit generalized Env-destabilizing effects (11). Indeed, V3 MAb sensitivity profoundly increased for pseudoviruses with either of these mutations, consistent with previous studies of a virus isolated from a patient with a core mutant at residue 424 that exhibited increased V3 MAb sensitivity and with mutants of simian-human immunodeficiency virus (SHIV) (27, 28). Similarly, it was predicted that the structural instability induced by mutations in the hydrophobic core would propagate structural changes throughout gp120, and indeed, it was found that both the A204E and M434G pseudovirus mutants became highly sensitive to V2i MAb neutralization, with IC50s ranging from 0.1 to 2.0 μg/ml (A204E) and 0.2 to 15.0 μg/ml (M434G) for most V2i MAbs tested, although both mutants remained relatively resistant to neutralization by the two V2i MAbs (830A and 2297) with distinct characteristics, as described in Materials and Methods (Table 1 and Fig. 5B and C). While the neutralizing activity of V2p MAbs was unaffected by mutations affecting core stability, the activity of V2q MAbs was impacted. The M434G mutant was rendered sensitive to PG9 and neutralized with an IC50 of 2.9 μg/ml; in contrast, neutralization by PGT145 was ablated (Table 1 and Fig. 4 and 5B).
Destabilization of intraprotomer interactions induces sensitivity to V2i-specific MAbs.
Within each protomer, the hydrophobic face of V3 also reacts with the gp120 core via the V1V2 stem. Previously, we found that mutation of the V2 stem residue L125 dramatically increased sensitivity to V3 MAbs (11). The L125F mutant was also found to be sensitized to V2i MAb neutralization, although the effect was less striking than with V3 MAbs: the IC50s for 7 of 15 V2i MAbs ranged from 1 to 9.9 μg/ml, while for V3 MAbs, the IC50s were consistently ≤0.8 μg/ml (Table 1 and Fig. 5D) (11).
The intraprotomer interface between V1V2 and the gp120 core shown in Fig. 2 was analyzed to identify side chains that contribute significantly to the interactions between the two domains. Because mutations on the V1V2 domain side could affect binding of V1V2 MAbs, we focused on the gp120 side of the interface, specifically residues I420, K421, and Q422 in the C4 region. Two mutations to alanine were chosen to eliminate stabilizing interactions: I420A eliminates lipophilic contacts, and Q422A eliminates several hydrogen bonds. Two other mutations were designed to create steric clashes (repulsion): K421N and Q422P. Specific substitutions were selected using multiple alignments of 4,173 gp120 sequences from the LANL database (2014 data set; https://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/2014/hiv1prot.pdf) to identify rare but naturally observed substitutions. Thus, four mutations not previously tested for V3 release were identified and predicted to disrupt the intraprotomer interface between V1V2 and the gp120 core. Since the V1V2 domain normally forms one of the walls of the pocket in which the V3 loop sits, when V1V2 is released from its interaction with the gp120 core, V3 would also be predicted to be released. Residues 420, 421, and 422 are directly in contact with the V2 YKLDV kink/integrin binding motif of the same protomer (29, 30). I420A and Q422A mutations rendered these mutant pseudoviruses susceptible to neutralization by most V2i MAbs with IC50s ranging from 1.3 to 12 μg/ml (Table 1 and Fig. 5E). Replacement of these C4 residues with more repulsive substitutions (K421N and Q422P) yielded greater release of the V1V2 loop: V2i MAbs exhibited neutralization of these pseudoviruses, with most IC50s between 0.7 and 5.5 μg/ml (Table 1 and Fig. 5F). All V3 MAbs tested with these novel mutants exhibited increases in neutralization potency by 2 or 3 orders of magnitude (Table 1). In contrast to the enhanced sensitivity of intraprotomer mutants to V2i MAbs, these mutations resulted in a loss of neutralization ability by the V2q MAb PGT145, consistent with the disruption of the trimer association domain (Table 1 and Fig. 4). As with the WT and all the tested mutants of JR-FL, V2p MAbs remained unable to neutralize (Table 1).
Mutation of residues critical for interprotomer interactions produces little effect on neutralization sensitivity to V2i-specific MAbs.
To test for the effects of mutations that disturb interprotomer interactions within the Env spike, several mutants were made that had previously been shown to disrupt the stabilizing interactions between the V1V2 stem and the tip of V3 from the neighboring protomer (11, 31). JR-FL has an aspartate at position 197 (D197); however, numerous strains possess a glycosylated asparagine at this position, which has been shown to have a stabilizing effect on the trimer (11, 31). Restoration of the glycan at position 197 was previously shown not to affect V3 sensitivity, while the D197H and D197Q mutants were highly sensitive to V3 neutralization (Table 1) (11). This suggested that D197 itself forms critical contacts that stabilize V3. In contrast to these data, the present study found none of the D197 mutants tested to be sensitive to V2i neutralization (Table 1). Moreover, disruption of the I184 residue, which putatively contributes to the quaternary interaction of the V1V2 regions of neighboring protomers (Fig. 6 shows the van der Waals interactions between the only partially solvent-accessible surfaces of I184 and L165 from the neighboring protomer), had no effect on V2i neutralization (Table 1) despite being previously shown to render JR-FL exceptionally sensitive to V3 MAbs (11).
FIG 6.
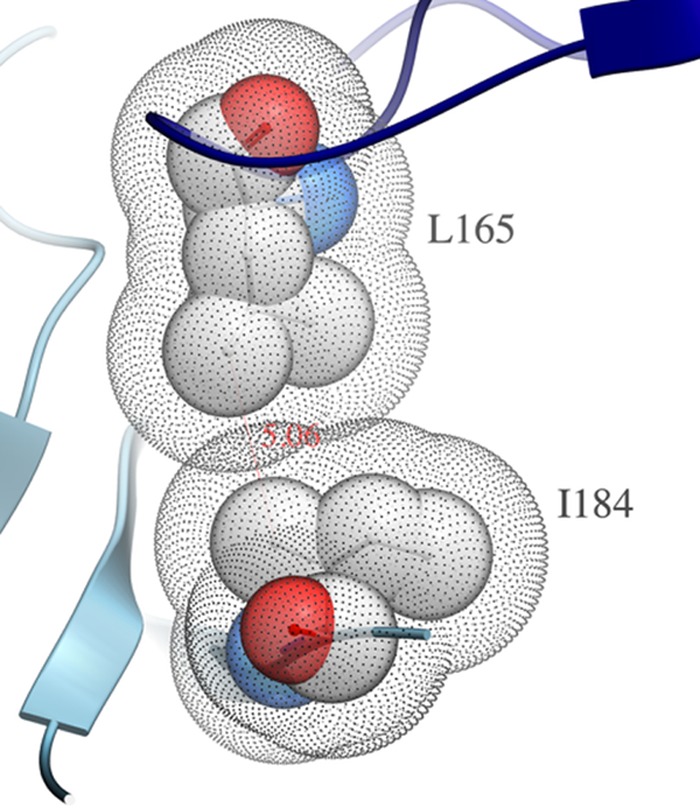
I184 lipophilic contact between protomers. This illustration was generated from PDB entry 4TVP, the X-ray structure of SOSIP BG505 (29). The dot mesh shading represents the solvent-accessible surfaces (SAS) (the van der Waals surfaces expanded by the water molecule radius) of the two residues from adjacent protomers (73, 74). Where the SAS overlap, the two residues exclude solvent access to each other.
Most V2p and V2q MAbs were also unaffected by mutation of residues critical for interprotomer interactions, with the exception of PGT145, whose neutralizing IC50s for D197N decreased by an order of magnitude, again suggesting the requirement for a closed trimer apex in order for the MAb to bind and function (Table 1 and Fig. 4).
Mutation of the CD4bs does not affect neutralization by V2 MAbs.
Next, we investigated the effect of a mutation in the CD4bs on V1V2 loop exposure. Previously, we had determined that mutation of the CD4bs moderately affected V3 stability, as the N280E JR-FL mutant became sensitive to most V3 MAbs at concentrations ranging from 2 to 28 μg/ml whereas WT JR-FL was resistant to most V3 MAbs (Table 1) (11). Here, we tested this CD4bs mutant for V2 MAb-mediated neutralization sensitivity and found the N280E mutant pseudovirus to be unchanged in its sensitivity to any of the V2 MAbs tested.
V1V2 domain motion simulations in the trimer context.
To gain insight into how V2i MAbs might acquire access to their epitopes, we performed stochastic Monte Carlo simulations of the V1V2 domain movements within the context of the trimer. The X-ray structures of complexes of V2i MAbs with the V1V2 domain showed that the internal tertiary β-barrel structure of the domain is largely preserved between the closed Env trimer state and the Ab-bound state (7). Therefore, we set up simulations in such a way as to specifically explore the movements that involve the flexible V1V2 stems, radial movements of whole protomers with respect to the trimer axis, and V3 “flickering” (with two discrete V3 states) (Fig. 7).
FIG 7.
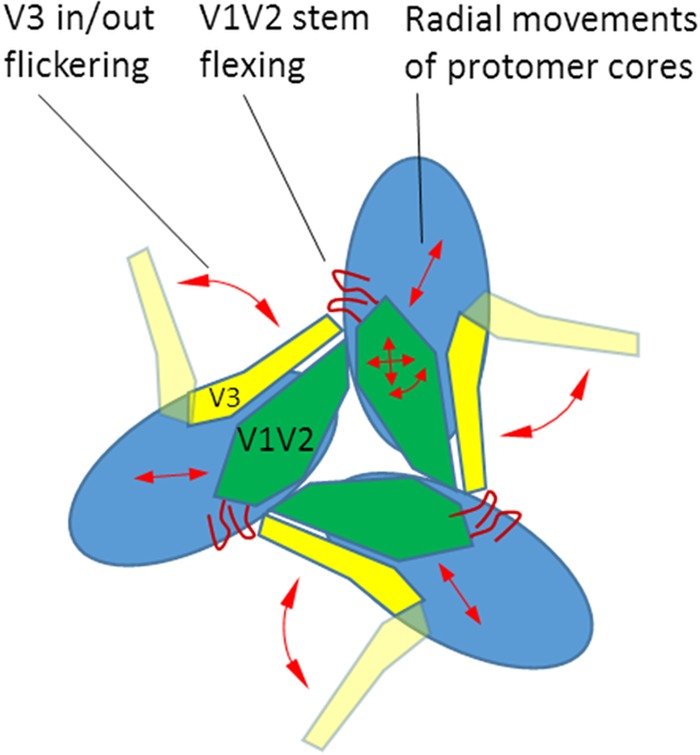
Types of stochastic movements in the Monte Carlo simulations of the radial movements of the protomer cores, as well as the mobility of the V3 and V1V2 domains within the Env trimer.
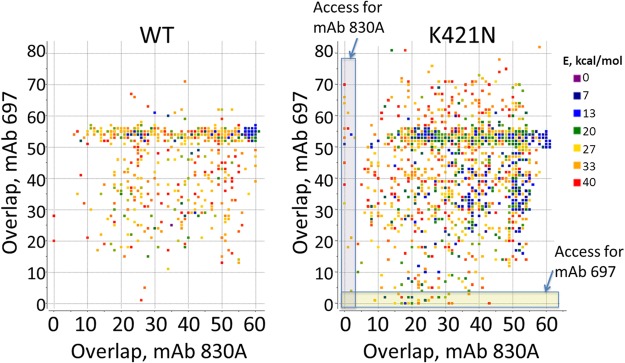
Ensembles of conformations were generated for the WT and the K421N mutant, which affects the packing of the intraprotomer core. In order to detect events of V2i exposure, each conformation was analyzed for compatibility with binding of V2i MAbs 697 and 830A: the Fab fragments were placed in their bound positions with respect to the V1V2 domain, and the number of backbone atoms that would overlap other parts of the spike was determined. Lack of overlap would thus be indicative of unobstructed epitope access. The original closed conformation exhibited overlap of 55 to 60 atoms, i.e., epitopes were clearly highly sterically obstructed in the closed WT conformation. In order to visualize how the system sampled states with different degrees of epitope exposure, we plotted the points representing two overlaps for each conformation (Fig. 8). For WT simulations, low-energy conformations largely clustered around the original experimental crystallographic structure. Some higher-energy states had reduced overlaps (between 5 and 50) (Fig. 8), but few if any had unobstructed V2i epitope access (<5 overlaps). In contrast, simulations of the K421N mutant showed that the low-energy states now sampled a much broader range of conformations, all the way down to fully V2i-exposed states with <5 overlaps (Fig. 8). The ensembles also showed relatively limited correlation between accesses for V2i MAbs 830A and 697, i.e., most of the conformers that allowed access for one antibody were not compatible with the other, and vice versa. This is likely related to the significantly different angles of access of the two MAbs with respect to the V1V2 domain (32).
FIG 8.
Motion simulations of V2i MAb access to the V1V2 domain. Ensembles of conformations from the simulations of JR-FL WT and from the K421N mutant Env trimers were analyzed to determine binding accessibility for two V2i-specific MAbs: 697 and 830A. Each dot represents a conformation, colored according to its energy (E) relative to the lowest energy conformer in the ensemble. The x and y coordinates show the numbers of atoms that would overlap other parts of the spike if the MAb were bound to its epitope on the V1V2 domain. Access to an epitope is unobstructed if the overlap is close to zero.
Mutations affecting V2 and V3 exposure enhance access to CD4bs- but not CD4i-specific MAbs.
Mutations that affected V3 or V2 and V3 epitope exposure also potently enhanced the availability of epitopes recognized by CD4bs MAbs b12, 654, and 559. They included glycan mutations, particularly removal of the N301 glycan, as well as mutations affecting the gp120 core and interprotomer and intraprotomer interactions (Table 2). The potency of these CD4bs MAbs increased by 2 to 3 orders of magnitude, even inducing neutralization in the case of MAbs 559 and 654, which do not neutralize WT JR-FL. VRC01 similarly displayed increased potency against many of the mutants, though sensitivity to this MAb was reduced ∼10-fold in the cases of the A204E and M434G core mutants and the more disruptive K421N and Q422P intraprotomer mutants (Table 2). In contrast, most of the same mutations had little effect on the neutralization potencies of four CD4i MAbs.
TABLE 2.
Sensitivities of mutant pseudoviruses to CD4bs and CD4i MAbsa
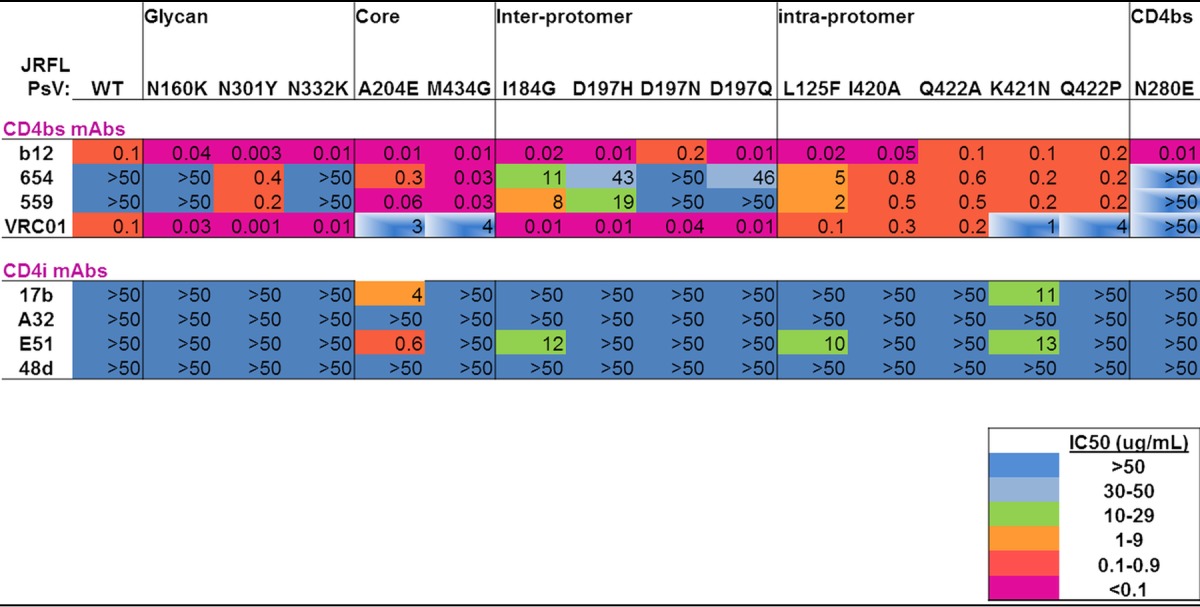
See footnote a to Table 1 for an explanation of the colors and symbols.
Mutant JR-FL variants that were not expected to have structurally disrupted Env spikes were included as controls in all of the MAb neutralization experiments; they included the C4 mutations N448Q and T450A and the V2 mutation V182Q. The neutralization sensitivities of these mutants to all V2 MAbs were unchanged from those of the WT (data not shown). Similarly, the mutant pseudoviruses, as well as the WT, were not neutralized by the negative-control anti-parvovirus human MAb 1418.
DISCUSSION
The data presented here demonstrate that selected individual mutations in gp120 expose both V2 and V3 epitopes that are generally occluded in circulating viruses. Thus, mutations that eliminate the N301 glycan, that disrupt the gp120 hydrophobic core, and that alter intraprotomer packing expose both V2 and V3, suggesting general destabilization of the distal region of the trimer. Interestingly, a complementary study showed that mutants in V1V2 render epitopes elsewhere in the trimer more accessible, also highlighting the dynamic nature of the molecule (33). In contrast, several mutants studied exposed V3 but had no effect on V2 epitope accessibility. Thus, mutants that eliminated the N160K glycan, disrupted the CD4bs, or interfered with interprotomer contacts exposed V3 only. These data indicate for the first time that V3 can move independently of V2, pivoting out from its “tucked” position in the trimer while apparently leaving the V2 apex intact. These data are consistent with previous work by Upadhyay et al. showing that engagement of the CD4 receptor with soluble CD4 prior to subsequent incubation with MAbs and pseudovirus strongly affected V3 MAb potency but had only a mild effect on V2 neutralization (1). Similarly, production of pseudoviruses in the presence of kifunensine, which inhibits complex glycan processing, resulted in increased neutralization potency of V3 MAbs but little change in the potency of V2 MAbs (1). It is noteworthy, however, that Lyumkis et al. described the V3 loop as being situated directly beneath the V1V2 hairpin, using their cryo-electron microscopy (EM) structure of the Env trimer in complex with MAb PGV04, and suggested it was unlikely that there could be disruption of V3 without V1V2 reorganization (34). The data described above clearly show that V3 is not “locked” in a cryptic configuration, as predicted by the cryo-EM technique, which provides a depiction of the most prevalent trimer structure at only one time point. Thus, MAb neutralization of the mutants studied demonstrated exposure of V3 alone or V2 and V3, but none exposed V2 epitopes without also exposing V3, indicating that the movement of V2 releases V3, but not vice versa.
These studies, along with many others cited, indicate that intricate contacts throughout the trimer are required to maintain the metastable, closed state of the Env trimer and that disruption of any one of these contacts can release V3 or both V1V2 and V3. Moreover, the closed state of the unliganded Env trimer is only one configuration that can be adopted by the unliganded Env spike. Indeed, several studies have now shown that the unliganded trimer undergoes shifts between a closed ground state (state 1), a functional intermediate (state 2), and an open conformation (state 3) in which the chemokine receptor binding site (CD4i epitope) is exposed (5, 6, 29, 33). Importantly, the SOSIP structure, such as that published by Lyumpkis et al. bound to PGV04, illustrates the most prevalent state of the trimer rather than the only possible state. The data presented in our paper underscore the dynamic nature of Env; there is no single unbound conformation of the trimer. Our V1V2 mobility simulations were performed using a SOSIP trimer in order to show how fluctuations around that predominant state explain the observed data. Studies by both Spurrier et al. (35) and Herschhorn et al. (33) identified “restricting” amino acid residues in the hydrophobic core of V2; these lead to lowered activation barriers between the various states of the Env trimer. In the current study, we identify additional residues within, as well as outside, V2 that behave similarly, causing conformational changes that release previously occluded epitopes and rendering viruses sensitive to the neutralizing effects of MAbs to which they were previously resistant.
The spontaneous transitions between these states of the trimer may provide an explanation for how V2i Abs, which were identified as the only independent correlate of reduced infection risk in the RV144 clinical vaccine trial, could play a protective role and impose immunologic pressure on infecting and circulating viruses (16, 36). In the Herschhorn et al. study (33) of the tier 2 JR-FL virus, the majority of viruses (53%) were present in the closed state 1, while 19% were present in the intermediate state 2 and 28% were present in the open state 3. The majority of the viruses with a closed trimer are resistant to V3 and V2i MAbs under standard neutralizing assay conditions when the V3 and V2i epitopes are occluded. However, assuming that a significant portion of the viruses are present in vivo in, or can spontaneously transition into, the intermediate or open state, these viruses should be sensitive to the V3 and V2i Abs. In fact, in the data shown above, the mutant JR-FL viruses appear to be in the intermediate state, i.e., with V2i and/or V3 epitopes accessible to MAbs but with an occluded CD4i site, as indicated by their general resistance to CD4i MAbs (Tables 1 and 2). These findings appear to be highly relevant in explaining how V2i and V3 Abs were effective in RV144, reducing the rate of infection and imposing immune pressure on the virus in vaccinees (13, 15, 16, 36): if a significant proportion of the trimers on circulating viruses or infected cells are, like JR-FL, in the intermediate or open conformation, then these viruses and infected cells could be affected by the vaccine-induced V2i and V3 Abs.
Generally, the various mutants studied here did not induce exposure of V2q epitopes in JR-FL, which is resistant to MAbs 2909, PG9, and PG16 (Table 1). This was not unexpected, since these MAbs do not bind JR-FL well, as key residues in the C β-strand of V2 that are required for engagement are not present in the JR-FL sequence (37). In order for the antibody to recognize the epitope it should (i) be in the right conformation, (ii) be exposed, and (iii) contain correct residues. One can only detect changes in the first two prerequisites on the condition that the third is satisfied. Our data do confirm that these V2q epitopes cannot be efficiently exposed merely by opening the trimer and exposing V2. PG9 is somewhat unusual within this class, as it binds to selected gp120 monomers (38, 39). Consistent with this feature of PG9, the M434G mutant tested here was sensitive to PG9 neutralization, suggesting that the significant structural displacements induced by disrupting the hydrophobic core of gp120 allowed unusual contacts with the MAb to form. In contrast, JR-FL is moderately sensitive to PGT145 neutralization, and this MAb is trimer dependent, requiring an intact, closed trimer apex for binding (26, 34, 37). The loss of PGT145 neutralizing activity for the core mutants highlights the significant reorganization of Env, where no closed trimer contacts could be formed. Similarly, all trimer-opening mutations abolished PGT145 neutralization, including the intraprotomer and the N160 and N301 glycan mutations. The D197Q interprotomer mutant appears to have also ablated the trimer contacts required for PGT145 binding, though no V2i epitopes were made available. Intriguingly, replacement of D197 with a glycosylated asparagine, which is present in most strains at this position and has been shown to have a stabilizing effect on the trimer, seems to significantly enhance PGT145 neutralization, suggesting the N197 glycan either contributes directly to or helps stabilize the PGT145 epitope (11, 31).
For expanded studies, we created a novel family of mutants expected to expose the V1V2 loop. They were identified by analyzing the crystal structures of BG505.SOSIP and identifying residues forming the interface between the V1V2 domain and the gp120 core (29, 30). In the C4 region of gp120, the I420, K421, and Q422 side chains interact extensively with the V1V2 domain, in particular with the 177YKLDV181 motif, which forms a “kink” connecting the C and C′ strands of the domain (Fig. 1 and 2) (32). As expected, mutation of these C4 residues caused the pseudovirus mutants to be susceptible to neutralization by V2i MAbs (Table 1). In addition, mutants carrying the C4 mutations were also generally rendered more sensitive to CD4bs-specific MAbs. Indeed, two CD4bs MAbs, 654 and 559, that do not neutralize most primary isolates and did not neutralize WT JF-FL could effectively neutralize the C4 intraprotomer mutants (Table 2). While structural data for complexes of these MAbs with gp120 are not available, it is likely that poor neutralization of primary isolates is due to the restriction of access to the CD4bs by the adjacent protomer or by the V1V2 domain and/or its glycans in the context of the tightly closed trimer. Stringent requirements for the angle of approach of broadly neutralizing MAbs are illustrated by crystal structures of SOSIP with the CD4bs-specific MAbs PGV04 and VRC01 (34, 40). However, mutation-induced liberation of V1V2 movement and increased separation of the protomers in mutant spikes would relax these restrictions, allowing binding and neutralization by normally nonneutralizing CD4bs Abs. The CD4bs was also rendered accessible by the more globally destabilizing mutations to N301, A204, and M434 (Table 2).
Interestingly, MAb VRC01 displayed increased potency for N301Y by more than 2 orders of magnitude (Table 2). The crystal structure of the SOSIPJR-FL-VRC01 complex demonstrates that substantial contacts exist between the back of the VRC01 heavy-chain Fab and the N301 glycan of the adjacent gp120 protomer (40). Our results suggest that this interaction is not favorable and that VRC01 has to push aside the N301 glycan to access its epitope on the closed trimer. In contrast to the several mutations that increased sensitivity to neutralization by various MAbs, the potency of VRC01 was decreased by mutations in the core (A204E and M343G) and by intraprotomer mutations at K421N and Q422P (Table 2), documenting the unique nature of this CD4bs MAb and its dependence on the metastable packing of the Env spike.
As opposed to the effects of Env mutations on the various classes of MAbs studied here, most mutations had weak or no effect on the neutralization potencies of four CD4i MAbs. We had previously suggested that the A204E mutation promotes transition from a mixed β-sheet conformation to an all antiparallel conformation of the bridging sheet/coreceptor binding site (11). CD4i MAbs 17b and E51 target this site; emergence of neutralization by these MAbs is in agreement with the expected structural effects of A204E (41–43). The specificity of MAb A32, while phenomenologically classified as CD4i, in fact targets a cluster A epitope located in the inner domain of gp120, far from the coreceptor binding site, and requires a much larger opening of the trimer (44–46). Accordingly, none of our mutant viruses were neutralized by A32 (Table 2). Generally, these results indicate that exposure of many V1V2 and V3 epitopes requires only a limited degree of trimer opening, significantly smaller than the major transition induced by CD4 binding (4, 47).
While Abs to V2q are present in a very small proportion of HIV-infected individuals (48, 49) and the V2p class of Abs has not yet been studied extensively in natural infection, Abs directed at the V2i epitope of gp120 are found in greater than 80% of naturally infected subjects, indicating that V1V2 is highly immunogenic (references 22 and 50–52 and M. K. Gorny, unpublished data), and many human MAbs specific for this highly conformational region of V2 have been generated and characterized in detail (18, 22, 32, 35, 53, 54). Indeed, the present study was mainly focused on reactivity to V2i MAbs and the mutations that would change the exposure of the V2i epitope. Many V2q-specific MAbs do not bind well to JR-FL, as the key residues in the C β-strand of V2 that are required for engagement are not present in the JR-FL sequence. Our data confirm that the V2q epitope is not present in WT JR-FL and cannot be efficiently exposed or formed by opening the trimer and exposing V2 and V3.
MAbs with V2i-directed specificities are now sufficiently plentiful and well studied to allow recognition of subtle differences in this Ab category. For example, in contrast to the effect of the intraprotomer mutants on most V2i MAbs, these mutants did not render V2i MAbs 830A and 2297 more potent (Table 1). Similarly, MAbs 21a9, 18a4, 20a12, and 20a24 were generally much less effective against the N301D, N301Y, and L125 mutants than were the nine V2i MAbs shown at the top of Table 1. A further example of the subtle differences within the V2i MAb family is revealed by examining the disparate activities of MAbs 697 and 830A. MAbs 697 and 830A have distinct angles of approach to the V1V2 domain (22, 32). It seems (and simulations give some support to this) that the epitope of MAb 697 is in general more easily accessed by V1V2 domain movements than is that of MAb 830A. One can speculate that M434G introduces more flexibility in the bridging sheet region, facilitating the flexion of the V1V2 stem and allowing V1V2 rotation that exposes the 830A epitope.
Overall, these data speak to the plasticity of the HIV-1 Env. Beyond illuminating the fine points of the structural dynamics of HIV Env, the data provide a road map for creating Env constructs and engineered immunogens designed to target the V1V2 loop. In addition, the data address how Env plasticity provides a plausible explanation for the efficacy of V2 Abs in reducing infection rates of HIV, SHIV, and simian immunodeficiency virus (SIV) (13, 36, 55–60) and the potential value of vaccines that target V1V2.
MATERIALS AND METHODS
Envelope clones and site-directed mutagenesis.
Mutant plasmids of pCAGGS_JR-FL.JB gp160 (originally supplied by John Mascola) were created using QuikChange II XL or QuikChange Lightning kits (Stratagene, La Jolla, CA, USA) according to the manufacturer's instructions or by Genscript (Piscataway, NJ). All constructs were sequenced completely to confirm their identities before use. Mutations were selected from those that had been shown previously to release V3 from its occluded state in the Env trimer, and control mutants that had been shown not to affect the exposure of V3 were used (11). These mutations were in various regions of gp120: V1 (L125F), V2 (N160K, V182Q, I184G, and D197H), C2 (A204E and N280E), V3 (N301D and N301Y), C3 (N332K), and C4 (M434G, N448Q, and T450A) (Fig. 1). Additionally, mutations in the C4 region not previously tested for V3 release and predicted to expose the V2i epitope were also tested: I420A, K421N, Q422P, and Q422A. These sites are directly in contact with the V2 YKLDV kink/integrin binding motif (Fig. 2) (32).
Pseudovirus production and neutralization assays.
Pseudoviruses were produced as previously described (61) in 293T cells transfected with 2 μg of envelope plasmid and 10 μg of an env-deficient subtype B backbone plasmid, pSG3Δenv (NIH AIDS Research and Reference Reagent Program). Pseudoviruses in supernatants were harvested at 48 h posttransfection. Pseudovirus was titrated in TZM-bl cells as previously described (61). The titrations found that, compared to the WT, 5 mutants exhibited increased infectivity, 6 mutants exhibited decreased infectivity, and 7 exhibited no change in infectivity (Fig. 3A). These differences in infectivity were normalized by using a dilution of pseudoviral supernatant shown by titration to yield 150,000 relative light units (RLU) (Fig. 3B). Neutralization assays were performed in TZM-bl cells as previously described. Briefly, after incubation of titrated MAb and pseudovirus for 1 h, cells were added, and the plates were incubated for 48 h and read using Britelite Plus reagent (Perkin-Elmer) according to the manufacturer's protocol on a Biotek Cytation3 imaging reader. The percent neutralization was determined by comparison to pseudovirus-only control wells. The average percent neutralization at each MAb concentration from at least two independent experiments was used to determine the IC50 of the MAb in GraphPad Prism.
Human monoclonal antibodies.
A large panel of human HIV MAbs made in house or obtained from the AIDS Reagent Repository were tested in this study, including representatives of all three classes of V2 MAbs. V2i-specific MAbs included 697-D, 1393A, 1361, 1357, 2158, 4-150, 4-182, 4-324, 4-366, 21a9, 18a4, 20a12, 20a24, 2297, and 830A (produced in house) (18–22). As previously described, V2i Abs recognize a conformation-dependent epitope on V2 that includes the α4β7 integrin binding site and are commonly elicited during infection (22, 35). It is noteworthy that MAb 2297 was previously shown to have a slightly different pattern of binding reactivity based on competition with other V2i MAbs for binding to V1V2-gp70 and was unable to neutralize any pseudoviruses, including tier 1A and 1B pseudoviruses (22). In addition, the structure of the paratope of MAb 830A is different than that of other V2i MAbs, like 697 (22) and 2158 (35); 830A has a narrower neutralizing breadth against tier 1 viruses than other V2i MAbs, and it was shown to have a different angle of approach to the V1V2 region of gp120 than the well-characterized V2i MAb 697 (32). These characteristics of MAbs 2297 and 830A may account for their patterns of reactivity, which distinguish them from other V2i MAbs. In addition, it is noteworthy that V2i MAbs 21a9, 18a4, 20a12, and 20a24 were selected from patient samples with a scaffolded V1V2 (V1V2-gp70) rather than with gp120 (reference 22 and Liuzhe Li, Lily Liu, Susan Zolla-Pazner, Xiang-Peng Kong, and Miroslaw K. Gorny, unpublished data).
The V2p-specific MAbs CH58 and CH59 and the V2q (or V2 apex)-specific MAbs 2909, PG9, PG16, and PGT145 (20, 62–67) were prepared in house or obtained through the AIDS Reagent Repository. V2p Abs are glycan independent and bind to peptides representing a V2 linear epitope on the C strand of V1V2 (65). V2q Abs target a quaternary epitope, including regions of V2 as represented in the Env trimer; these V2q MAbs are glycan dependent, are rarely elicited by infection, are extensively hypermutated, and preferentially bind to trimeric Env (35, 66). It should be noted that the specificity of PG9 for quaternary epitopes is not absolute, while the PG145 epitope exists only on the trimer, and PG16 binding is greatly enhanced on the trimer (38). CD4bs MAbs b12, 654, 559, and VRC01, as well as CD4i MAbs 17b, A32, E51, and 48d, were also tested (64, 68–71). The human anti-parvovirus MAb 1418 was used as a negative control, and CD4 Ig, obtained from the AIDS Reagent Repository, served as a positive control (72).
V1V2 mobility simulations.
Structure preparation, Monte Carlo simulations, and analysis of the resulting conformational ensembles were performed in the ICM molecular-modeling package (Molsoft, San Diego, CA). As a starting point for the simulations, the X-ray structure of the JR-FL SOSIP.559 trimer (PDB accession no. 5FYK) was used (40). Stochastic Monte Carlo perturbations were applied to the internal variables of the two loops comprising the V1V2 stem (amino acid residues V119 to L124 and N197 to A204), to the radial positions of the gp120 protomer cores with respect to the 3-fold symmetry axis of the trimer, and to a V3 conformation. Flickering of V3 was sampled in a discrete manner, i.e., a single Monte Carlo step would switch the entire loop between the tucked-in conformation observed in the X-ray structure and a fully exposed (popped-out) conformation. (A representative conformation was taken from the V3 simulations we previously reported [11].) Radial movements were restrained by a soft (0.25-kcal/Å2) harmonic constraint, as well as hard boundaries beyond −2 and +18 Å. Ensembles of conformations were collected from 100 independent simulation runs performed for both WT and K421N mutant structures. Antibody epitope access for each conformation in the ensemble was evaluated by Fab overlap analysis: the X-ray structure of the Fab-V1V2 complex was superimposed onto each of the V1V2 domains in the given trimer conformation from the simulations, and the number of Fab backbone atoms within a strong clash cutoff (1.5 Å) of the Env spike backbone was determined.
ACKNOWLEDGMENTS
We thank Xiang-Peng Kong and Catarina Hioe for insightful discussions that enhanced the design and interpretation of these studies.
The NIH/NIAID provided funding to all of us under grant number P01 AI100151. In addition, S. Zolla-Pazner was supported by funds from the Department of Medicine, Division of Infectious Diseases, Icahn School of Medicine at Mount Sinai.
REFERENCES
- 1.Upadhyay C, Mayr LM, Zhang J, Kumar R, Gorny MK, Nadas A, Zolla-Pazner S, Hioe CE. 2014. Distinct mechanisms regulate exposure of neutralizing epitopes in the V2 and V3 loops of HIV-1 envelope. J Virol 88:12853–12865. doi: 10.1128/JVI.02125-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Totrov M. 2014. Estimated secondary structure propensities within V1/V2 region of HIV gp120 are an important global antibody neutralization sensitivity determinant. PLoS One 9:e94002. doi: 10.1371/journal.pone.0094002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korkut A, Hendrickson WA. 2012. Structural plasticity and conformational transitions of HIV envelope glycoprotein gp120. PLoS One 7:e52170. doi: 10.1371/journal.pone.0052170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guttman M, Cupo A, Julien JP, Sanders RW, Wilson IA, Moore JP, Lee KK. 2015. Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat Commun 6:6144. doi: 10.1038/ncomms7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB III, Kwong PD, Blanchard SC, Mothes W. 2014. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munro JB, Mothes W. 2015. Structure and dynamics of the native HIV-1 Env trimer. J Virol 89:5752–5755. doi: 10.1128/JVI.03187-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, Soto C, Terry DS, Yang Y, Zhou T, Ahlsen G, Bailer RT, Chambers M, Chuang GY, Doria-Rose NA, Druz A, Hallen MA, Harned A, Kirys T, Louder MK, O'Dell S, Ofek G, Osawa K, Prabhakaran M, Sastry M, Stewart-Jones GB, Stuckey J, Thomas PV, Tittley T, Williams C, Zhang B, Zhao H, Zhou Z, Donald BR, Lee LK, Zolla-Pazner S, Baxa U, Schon A, Freire E, Shapiro L, Lee KK, Arthos J, Munro JB, Blanchard SC, Mothes W, Binley JM, McDermott AB, Mascola JR, Kwong PD. 2015. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol 22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zolla-Pazner S. 2005. Improving on nature: focusing the immune response on the V3 loop. Hum Antibodies 14:69–72. [PubMed] [Google Scholar]
- 9.Tomaras GD, Yates NL, Liu P, Qin L, Fouda GG, Chavez LL, Decamp AC, Parks RJ, Ashley VC, Lucas JT, Cohen M, Eron J, Hicks CB, Liao HX, Self SG, Landucci G, Forthal DN, Weinhold KJ, Keele BF, Hahn BH, Greenberg ML, Morris L, Karim SS, Blattner WA, Montefiori DC, Shaw GM, Perelson AS, Haynes BF. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol 82:12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis KL, Gray ES, Moore PL, Decker JM, Salomon A, Montefiori DC, Graham BS, Keefer MC, Pinter A, Morris L, Hahn BH, Shaw GM. 2009. High titer HIV-1 V3-specific antibodies with broad reactivity but low neutralizing potency in acute infection and following vaccination. Virology 387:414–426. doi: 10.1016/j.virol.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zolla-Pazner S, Cohen SS, Boyd D, Kong XP, Seaman M, Nussenzweig M, Klein F, Overbaugh J, Totrov M. 2015. Structure/function studies involving the V3 region of the HIV-1 envelope delineate multiple factors that affect neutralization sensitivity. J Virol 90:636–649. doi: 10.1128/JVI.01645-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH, MOPH-TAVEG Investigators. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 13.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung AW, Kumar MP, Arnold KB, Yu WH, Schoen MK, Dunphy LJ, Suscovich TJ, Frahm N, Linde C, Mahan AE, Hoffner M, Streeck H, Ackerman ME, McElrath MJ, Schuitemaker H, Pau MG, Baden LR, Kim JH, Michael NL, Barouch DH, Lauffenburger DA, Alter G. 2015. Dissecting polyclonal vaccine-induced humoral immunity against HIV using systems serology. Cell 163:988–998. doi: 10.1016/j.cell.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zolla-Pazner S, Edlefsen PT, Rolland M, Kong XP, deCamp A, Gottardo R, Williams C, Tovanabutra S, Sharpe-Cohen S, Mullins JI, deSouza MS, Karasavvas N, Nitayaphan S, Rerks-Ngarm S, Pitisuttihum P, Kaewkungwal J, O'Connell RJ, Robb ML, Michael NL, Kim JH, Gilbert P. 2014. Vaccine-induced human antibodies specific for the third variable region of HIV-1 gp120 impose immune pressure on infecting viruses. EBioMedicine 1:37–45. doi: 10.1016/j.ebiom.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottardo R, Bailer RT, Korber BT, Gnanakaran S, Phillips J, Shen X, Tomaras GD, Turk E, Imholte G, Eckler L, Wenschuh H, Zerweck J, Greene K, Gao H, Berman PW, Francis D, Sinangil F, Lee C, Nitayaphan S, Rerks-Ngarm S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Robb ML, Michael NL, Kim JH, Zolla-Pazner S, Haynes BF, Mascola JR, Self S, Gilbert P, Montefiori DC. 2013. Plasma IgG to linear epitopes in the V2 and V3 regions of HIV-1 gp120 correlate with a reduced risk of infection in the RV144 vaccine efficacy trial. PLoS One 8:e75665. doi: 10.1371/journal.pone.0075665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaine M, Wang S, Liu Q, Arthos J, Montefiori D, Goepfert P, McElrath MJ, Lu S. 2010. Profiles of human serum antibody responses elicited by three leading HIV vaccines focusing on the induction of Env-specific antibodies. PLoS One 5:e13916. doi: 10.1371/journal.pone.0013916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorny MK, Moore JP, Conley AJ, Karwowska S, Sodroski J, Williams C, Burda S, Boots LJ, Zolla-Pazner S. 1994. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of HIV-1. J Virol 68:8312–8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorny MK, VanCott TC, Williams C, Revesz K, Zolla-Pazner S. 2000. Effects of oligomerization on the epitopes of the Human Immunodeficiency Virus Type 1 envelope glycoproteins. Virology 267:220–228. doi: 10.1006/viro.1999.0095. [DOI] [PubMed] [Google Scholar]
- 20.Nyambi PN, Mbah HA, Burda S, Williams C, Gorny MK, Nadas A, Zolla-Pazner S. 2000. Conserved and exposed epitopes on intact, native, primary human immunodeficiency virus type 1 virions of group M. J Virol 74:7096–7107. doi: 10.1128/JVI.74.15.7096-7107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinter A, Honnen WJ, He Y, Gorny MK, Zolla-Pazner S, Kayman SC. 2004. The V1/V2 domain of gp120 is a global regulator of sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J Virol 78:5205–5215. doi: 10.1128/JVI.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorny MK, Pan R, Williams C, Wang XH, Volsky B, O'Neal T, Spurrier B, Sampson JM, Li L, Seaman MS, Kong XP, Zolla-Pazner S. 2012. Functional and immunochemical cross-reactivity of V2-specific monoclonal antibodies from human immunodeficiency virus type 1-infected individuals. Virology 427:198–207. doi: 10.1016/j.virol.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar R, Pan R, Upadhyay C, Mayr L, Cohen S, Wang XH, Balasubramanian P, Nadas A, Seaman MS, Zolla-Pazner S, Gorny MK, Kong XP, Hioe CE. 2015. Functional and structural characterization of human V3-specific monoclonal antibody 2424 with neutralizing activity against HIV-1 JRFL. J Virol 89:9090–9102. doi: 10.1128/JVI.01280-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Rourke SM, Sutthent R, Phung P, Mesa KA, Frigon NL, To B, Horthongkham N, Limoli K, Wrin T, Berman PW. 2015. Glycans flanking the hypervariable connecting peptide between the A and B strands of the V1/V2 domain of HIV-1 gp120 confer resistance to antibodies that neutralize CRF01_AE viruses. PLoS One 10:e0119608. doi: 10.1371/journal.pone.0119608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong L, Wilson IA, Kwong PD. 2015. Crystal structure of a fully glycosylated HIV-1 gp120 core reveals a stabilizing role for the glycan at Asn262. Proteins 83:590–596. doi: 10.1002/prot.24747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrabi R, Voss JE, Liang CH, Briney B, McCoy LE, Wu CY, Wong CH, Poignard P, Burton DR. 2015. Identification of common features in prototype broadly neutralizing antibodies to HIV envelope V2 apex to facilitate vaccine design. Immunity 43:959–973. doi: 10.1016/j.immuni.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ringe R, Sharma D, Zolla-Pazner S, Phogat S, Risbud A, Thakar M, Paranjape R, Bhattacharya J. 2011. A single amino acid substitution in the C4 region in gp120 confers enhanced neutralization of HIV-1 by modulating CD4 binding sites and V3 loop. Virology 418:123–132. doi: 10.1016/j.virol.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyd DF, Peterson D, Haggarty BS, Jordan AP, Hogan MJ, Goo L, Hoxie JA, Overbaugh J. 2015. Mutations in HIV-1 envelope that enhance entry with the macaque CD4 receptor alter antibody recognition by disrupting quaternary interactions within the trimer. J Virol 89:894–907. doi: 10.1128/JVI.02680-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Cleveland B, Klots I, Travis B, Richardson BA, Anderson D, Montefiori D, Polacino P, Hu SL. 2008. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J Virol 82:638–651. doi: 10.1128/JVI.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan R, Gorny MK, Zolla-Pazner S, Kong XP. 2015. The V1V2 region of HIV-1 gp120 forms a five-stranded beta barrel. J Virol 89:8003–8010. doi: 10.1128/JVI.00754-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herschhorn A, Ma X, Gu C, Ventura JD, Castillo-Menendez L, Melillo B, Terry DS, Smith AB III, Blanchard SC, Munro JB, Mothes W, Finzi A, Sodroski J. 2016. Release of gp120 restraints leads to an entry-competent intermediate state of the HIV-1 envelope glycoproteins. mBio 7:e01598–. doi: 10.1128/mBio.01598-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spurrier B, Sampson J, Gorny MK, Zolla-Pazner S, Kong XP. 2014. Functional implications of the binding mode of a human conformation-dependent V2 monoclonal antibody against HIV. J Virol 88:4100–4112. doi: 10.1128/JVI.03153-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zolla-Pazner S, deCamp A, Gilbert PB, Williams C, Yates NL, Williams WT, Howington R, Fong Y, Morris DE, Soderberg KA, Irene C, Reichman C, Pinter A, Parks R, Pitisuttithum P, Kaewkungwal J, Rerks-Ngarm S, Nitayaphan S, Andrews C, O'Connell RJ, Yang ZY, Nabel GJ, Kim JH, Michael NL, Montefiori DC, Liao HX, Haynes BF, Tomaras GD. 2014. Vaccine-induced IgG antibodies to V1V2 regions of multiple HIV-1 subtypes correlate with decreased risk of HIV-1 infection. PLoS One 9:e87572. doi: 10.1371/journal.pone.0087572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doria-Rose NA, Georgiev I, O'Dell S, Chuang GY, Staupe RP, McLellan JS, Gorman J, Pancera M, Bonsignori M, Haynes BF, Burton DR, Koff WC, Kwong PD, Mascola JR. 2012. A short segment of the HIV-1 gp120 V1/V2 region is a major determinant of resistance to V1/V2 neutralizing antibodies. J Virol 86:8319–8323. doi: 10.1128/JVI.00696-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffenberg S, Powell R, Carpov A, Wagner D, Wilson A, Kosakovsky Pond S, Lindsay R, Arendt H, Destefano J, Phogat S, Poignard P, Fling SP, Simek M, Labranche C, Montefiori D, Wrin T, Phung P, Burton D, Koff W, King CR, Parks CL, Caulfield MJ. 2013. Identification of an HIV-1 clade A envelope that exhibits broad antigenicity and neutralization sensitivity and elicits antibodies targeting three distinct epitopes. J Virol 87:5372–5383. doi: 10.1128/JVI.02827-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, Bylund T, Choi CW, Davison JR, Georgiev IS, Joyce MG, Kwon YD, Pancera M, Taft J, Yang Y, Zhang B, Shivatare SS, Shivatare VS, Lee CC, Wu CY, Bewley CA, Burton DR, Koff WC, Connors M, Crispin M, Baxa U, Korber BT, Wong CH, Mascola JR, Kwong PD. 2016. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell 165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol 67:3978–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salzwedel K, Smith ED, Dey B, Berger EA. 2000. Sequential CD4-coreceptor interactions in human immunodeficiency virus type 1 Env function: soluble CD4 activates Env for coreceptor-dependent fusion and reveals blocking activities of antibodies against cryptic conserved epitopes on gp120. J Virol 74:326–333. doi: 10.1128/JVI.74.1.326-333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, Wyatt R, Choe H, Farzan M, Kwong PD. 2004. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci U S A 101:2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferrari G, Pollara J, Kozink D, Harms T, Drinker M, Freel S, Moody MA, Alam SM, Tomaras GD, Ochsenbauer C, Kappes JC, Shaw GM, Hoxie JA, Robinson JE, Haynes BF. 2011. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J Virol 85:7029–7036. doi: 10.1128/JVI.00171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richard J, Pacheco B, Gohain N, Veillette M, Ding S, Alsahafi N, Tolbert WD, Prevost J, Chapleau JP, Coutu M, Jia M, Brassard N, Park J, Courter JR, Melillo B, Martin L, Tremblay C, Hahn BH, Kaufmann DE, Wu X, Smith AB III, Sodroski J, Pazgier M, Finzi A. 2016. Co-receptor binding site antibodies enable CD4-mimetics to expose conserved anti-cluster A ADCC epitopes on HIV-1 envelope glycoproteins. EBioMedicine 12:208–218. doi: 10.1016/j.ebiom.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tolbert WD, Gohain N, Veillette M, Chapleau JP, Orlandi C, Visciano ML, Ebadi M, DeVico AL, Fouts TR, Finzi A, Lewis GK, Pazgier M. 2016. Paring down HIV Env: design and crystal structure of a stabilized inner domain of HIV-1 gp120 displaying a major ADCC target of the A32 region. Structure 24:697–709. doi: 10.1016/j.str.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H, Cohen AA, Galimidi RP, Gristick HB, Jensen GJ, Bjorkman PJ. 2016. Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop. Proc Natl Acad Sci U S A 113:E7151–E7158. doi: 10.1073/pnas.1615939113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. 2010. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog 6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O'Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, NISC Comparative Sequencing Program, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR. 2014. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Israel ZR, Gorny MK, Palmer C, McKeating JA, Zolla-Pazner S. 1997. Prevalence of a V2 epitope in clade B primary isolates and its recognition by sera from HIV-1-infected individuals. AIDS 11:128–130. [PubMed] [Google Scholar]
- 51.Kayman SC, Wu Z, Revesz K, Chen H, Kopelman R, Pinter A. 1994. Presentation of native epitopes in the V1/V2 and V3 regions of human immunodeficiency virus type 1 gp120 by fusion glycoproteins containing isolated gp120 domains. J Virol 68:400–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKeating JA, Shotton C, Jeffs S, Palmer C, Hammond A, Lewis J, Oliver K, May J, Balfe P. 1996. Immunogenicity of full length and truncated forms of the human immunodeficiency virus type I envelope glycoprotein. Immunol Lett 51:101–105. doi: 10.1016/0165-2478(96)02562-X. [DOI] [PubMed] [Google Scholar]
- 53.Mayr LM, Cohen S, Spurrier B, Kong XP, Zolla-Pazner S. 2013. Epitope mapping of conformational V2-specific anti-HIV human monoclonal antibodies reveals an immunodominant site in V2. PLoS One 8:e70859. doi: 10.1371/journal.pone.0070859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klein F, Nogueira L, Nishimura Y, Phad G, West AP Jr, Halper-Stromberg A, Horwitz JA, Gazumyan A, Liu C, Eisenreich TR, Lehmann C, Fatkenheuer G, Williams C, Shingai M, Martin MA, Bjorkman PJ, Seaman MS, Zolla-Pazner S, Karlsson Hedestam GB, Nussenzweig MC. 2014. Enhanced HIV-1 immunotherapy by commonly arising antibodies that target virus escape variants. J Exp Med 211:2361–2372. doi: 10.1084/jem.20141050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pegu P, Vaccari M, Gordon S, Keele BF, Doster M, Guan Y, Ferrari G, Pal R, Ferrari MG, Whitney S, Hudacik L, Billings E, Rao M, Montefiori D, Tomaras G, Alam SM, Fenizia C, Lifson JD, Stablein D, Tartaglia J, Michael N, Kim J, Venzon D, Franchini G. 2013. Antibodies with high avidity to the gp120 envelope protein in protection from simian immunodeficiency virus SIV(mac251) acquisition in an immunization regimen that mimics the RV-144 Thai trial. J Virol 87:1708–1719. doi: 10.1128/JVI.02544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaccari M, Gordon SN, Fourati S, Schifanella L, Liyanage NP, Cameron M, Keele BF, Shen X, Tomaras GD, Billings E, Rao M, Chung AW, Dowell KG, Bailey-Kellogg C, Brown EP, Ackerman ME, Vargas-Inchaustegui DA, Whitney S, Doster MN, Binello N, Pegu P, Montefiori DC, Foulds K, Quinn DS, Donaldson M, Liang F, Lore K, Roederer M, Koup RA, McDermott A, Ma ZM, Miller CJ, Phan TB, Forthal DN, Blackburn M, Caccuri F, Bissa M, Ferrari G, Kalyanaraman V, Ferrari MG, Thompson D, Robert-Guroff M, Ratto-Kim S, Kim JH, Michael NL, Phogat S, Barnett SW, Tartaglia J, Venzon D, Stablein DM, Alter G, Sekaly RP, Franchini G. 2016. Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition. Nat Med 22:762–770. doi: 10.1038/nm.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barouch DH, Liu J, Li H, Maxfield LF, Abbink P, Lynch DM, Iampietro MJ, SanMiguel A, Seaman MS, Ferrari G, Forthal DN, Ourmanov I, Hirsch VM, Carville A, Mansfield KG, Stablein D, Pau MG, Schuitemaker H, Sadoff JC, Billings EA, Rao M, Robb ML, Kim JH, Marovich MA, Goudsmit J, Michael NL. 2012. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature 482:89–93. doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pegu A, Yang ZY, Boyington JC, Wu L, Ko SY, Schmidt SD, McKee K, Kong WP, Shi W, Chen X, Todd JP, Letvin NL, Huang J, Nason MC, Hoxie JA, Kwong PD, Connors M, Rao SS, Mascola JR, Nabel GJ. 2014. Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor. Sci Transl Med 6:243ra288. doi: 10.1126/scitranslmed.3008992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yates NL, Liao HX, Fong Y, deCamp A, Vandergrift NA, Williams WT, Alam SM, Ferrari G, Yang ZY, Seaton KE, Berman PW, Alpert MD, Evans DT, O'Connell RJ, Francis D, Sinangil F, Lee C, Nitayaphan S, Rerks-Ngarm S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Pinter A, Zolla-Pazner S, Gilbert PB, Nabel GJ, Michael NL, Kim JH, Montefiori DC, Haynes BF, Tomaras GD. 2014. Vaccine-induced Env V1-V2 IgG3 correlates with lower HIV-1 infection risk and declines soon after vaccination. Sci Transl Med 6:228ra239. doi: 10.1126/scitranslmed.3007730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barouch DH, Stephenson KE, Borducchi EN, Smith K, Stanley K, McNally AG, Liu J, Abbink P, Maxfield LF, Seaman MS, Dugast AS, Alter G, Ferguson M, Li W, Earl PL, Moss B, Giorgi EE, Szinger JJ, Eller LA, Billings EA, Rao M, Tovanabutra S, Sanders-Buell E, Weijtens M, Pau MG, Schuitemaker H, Robb ML, Kim JH, Korber BT, Michael NL. 2013. Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell 155:531–539. doi: 10.1016/j.cell.2013.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV, Nussenzweig MC. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 63.Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu JY, Emini EA, Koenig S, Zolla-Pazner S. 1992. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J Virol 66:7538–7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nyambi PN, Gorny MK, Bastiani L, van der Groen G, Williams C, Zolla-Pazner S. 1998. Mapping of epitopes exposed on intact human immunodeficiency virus type 1 (HIV-1) virions: a new strategy for studying the immunologic relatedness of HIV-1. J Virol 72:9384–9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liao HX, Bonsignori M, Alam SM, McLellan JS, Tomaras GD, Moody MA, Kozink DM, Hwang KK, Chen X, Tsao CY, Liu P, Lu X, Parks RJ, Montefiori DC, Ferrari G, Pollara J, Rao M, Peachman KK, Santra S, Letvin NL, Karasavvas N, Yang ZY, Dai K, Pancera M, Gorman J, Wiehe K, Nicely NI, Rerks-Ngarm S, Nitayaphan S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Sinangil F, Kim JH, Michael NL, Kepler TB, Kwong PD, Mascola JR, Nabel GJ, Pinter A, Zolla-Pazner S, Haynes BF. 2013. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity 38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Protocol G Principal Investigators, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Protocol G Principal Investigators, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karwowska S, Gorny MK, Buchbinder A, Gianakakos V, Williams C, Fuerst T, Zolla-Pazner S. 1992. Production of human monoclonal antibodies specific for conformational and linear non-V3 epitopes of gp120. AIDS Res Hum Retroviruses 8:1099–1106. doi: 10.1089/aid.1992.8.1099. [DOI] [PubMed] [Google Scholar]
- 69.Jeffs SA, Gorny MK, Williams C, Revesz K, Volsky B, Burda S, Wang XH, Bandres J, Zolla-Pazner S, Holmes H. 2001. Characterization of human monoclonal antibodies selected with a hypervariable loop-deleted recombinant HIV-1(IIIB) gp120. Immunol Lett 79:209–213. doi: 10.1016/S0165-2478(01)00289-9. [DOI] [PubMed] [Google Scholar]
- 70.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Banerjee K, Andjelic S, Klasse PJ, Kang Y, Sanders RW, Michael E, Durso RJ, Ketas TJ, Olson WC, Moore JP. 2009. Enzymatic removal of mannose moieties can increase the immune response to HIV-1 gp120 in vivo. Virology 389:108–121. doi: 10.1016/j.virol.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gigler A, Dorsch S, Hemauer A, Williams C, Kim S, Young NS, Zolla-Pazner S, Wolf H, Gorny MK, Modrow S. 1999. Generation of neutralizing human monoclonal antibodies against parvovirus B19 proteins. J Virol 73:1974–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee B, Richards FM. 1971. The interpretation of protein structures: estimation of static accessibility. J Mol Biol 55:379–400. doi: 10.1016/0022-2836(71)90324-X. [DOI] [PubMed] [Google Scholar]
- 74.Shrake A, Rupley JA. 1973. Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J Mol Biol 79:351–371. doi: 10.1016/0022-2836(73)90011-9. [DOI] [PubMed] [Google Scholar]