ABSTRACT
Follicular regulatory T (TFR) cells are a subset of CD4+ T cells in secondary lymphoid follicles. TFR cells were previously included in the follicular helper T (TFH) cell subset, which consists of cells that are highly permissive to HIV-1. The permissivity of TFR cells to HIV-1 is unknown. We find that TFR cells are more permissive than TFH cells to R5-tropic HIV-1 ex vivo. TFR cells expressed more CCR5 and CD4 and supported higher frequencies of viral fusion. Differences in Ki67 expression correlated with HIV-1 replication. Inhibiting cellular proliferation reduced Ki67 expression and HIV-1 replication. Lymph node cells from untreated HIV-infected individuals revealed that TFR cells harbored the highest concentrations of HIV-1 RNA and highest levels of Ki67 expression. These data demonstrate that TFR cells are highly permissive to R5-tropic HIV-1 both ex vivo and in vivo. This is likely related to elevated CCR5 levels combined with a heightened proliferative state and suggests that TFR cells contribute to persistent R5-tropic HIV-1 replication in vivo.
IMPORTANCE In chronic, untreated HIV-1 infection, viral replication is concentrated in secondary lymphoid follicles. Within secondary lymphoid follicles, follicular helper T (TFH) cells have previously been shown to be highly permissive to HIV-1. Recently, another subset of T cells in secondary lymphoid follicles was described, follicular regulatory T (TFR) cells. These cells share some phenotypic characteristics with TFH cells, and studies that showed that TFH cells are highly permissive to HIV-1 included TFR cells in their definition of TFH cells. The permissivity of TFR cells to HIV-1 has not previously been described. Here, we show that TFR cells are highly permissive to HIV-1 both ex vivo and in vivo. The expression of Ki67, a marker of proliferative capacity, is predictive of expression of viral proteins, and downregulating Ki67 leads to concurrent decreases in expression of viral proteins. Our study provides new insight into HIV-1 replication and a potential new cell type to target for future treatment.
KEYWORDS: HIV pathogenesis, HIV replication, follicular helper T cell, follicular regulatory T cell, secondary lymphoid follicle
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) replication is concentrated in T follicular helper (TFH) cells within B cell follicles of secondary lymphoid tissues during chronic asymptomatic disease (1–5). Heightened permissivity of TFH cells (5), a paucity of virus-specific cytotoxic T lymphocytes (CTL) (4, 6), and the presence of potently infectious virions on follicular dendritic cells (FDC) (7) all likely contribute to heightened virus replication at those sites (8). TFH cells are a distinct CD4 T cell population in which differentiation is driven by expression of the transcription factor Bcl-6 (9). They function as a crucial part of the germinal center (GC) reaction, providing help to B cells undergoing somatic hypermutation (SHM) and affinity maturation via cytokines such as interleukin-21 (IL-21) and IL-4 (9–11). Despite their heightened permissivity to HIV-1, TFH cell populations expand in untreated, chronically HIV-1-infected individuals as a percentage of CD4 T cells found in lymph nodes (LN) (12) and in chronic simian immunodeficiency virus (SIV) infection as a percentage of central memory (CM) CD4 T cells (13).
A novel T cell subset, follicular regulatory T (TFR) cells, was previously included within the phenotypic definition of TFH cells. Originally described in mice (14–16) and more recently in humans (17–19), TFR cells express Foxp3, the canonical regulatory T (Treg) transcription factor, Bcl-6, and Blimp-1 (14, 16). TFR cells derive from extrafollicular Treg cell precursors and localize to the follicle and GC. They have been shown to directly suppress TFH cell function in mice (14), nonhuman primates (19, 20), and humans (19) and have been shown to regulate antibody production in mice (14–16). We recently demonstrated that TFR cells are expanded in chronic HIV-1 and SIV infection and that ex vivo spinoculation of disaggregated, uninfected tonsil cells with X4- or R5-tropic HIV-1 increases TFR cell expression and secretion of regulatory proteins, such as CTLA-4 and IL-10 (19). However, this study did not determine the permissivity of TFR cells to HIV-1 relative to that of other lymphoid CD4 T cell subsets.
Using tonsil cells from children at low risk of HIV-1 infection, we show that TFR cells are more permissive to R5-tropic HIV-1 infection ex vivo than TFH and extrafollicular CD4 T cells. This was true for both an HIV-1 green fluorescent protein (GFP) reporter virus as well as transmitted/founder (T/F) R5-tropic HIV-1. TFR cells are more susceptible to R5 viral fusion than other cells and express the highest levels of CCR5 and CD4. HIV-1 coreceptor expression does not fully account for increased TFR cell permissivity to HIV-1, however, nor does it fully explain increased HIV-1 fusion. We show that increased ex vivo permissivity of TFR cells is related to Ki67 expression. In LN cells from asymptomatic HIV-1-infected humans, we determined that TFR cells harbor the highest concentrations of HIV-1 RNA and, furthermore, express the largest amount of Ki67. These data indicate that TFR cells are a highly proliferative subset of follicular T cells that directly contribute to the follicular concentration of HIV-1 replication in vivo.
RESULTS
Tonsillar TFR cells are highly permissive to R5-tropic HIV-1 ex vivo.
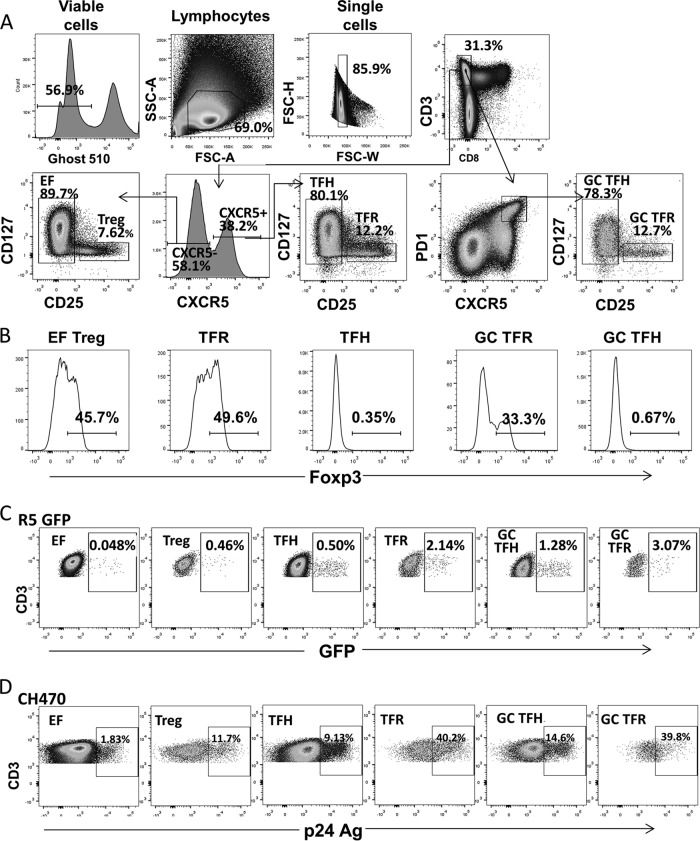
To determine the permissivity of TFR cells to HIV-1 ex vivo, we spinoculated disaggregated tonsil cells from children at low risk of HIV-1 infection with four different R5-tropic HIV-1 isolates, including a laboratory-adapted GFP reporter virus and three transmitted/founder (T/F) viruses. At 2 days postspinoculation, GFP or intracellular p24 expression of T cell subsets was measured by flow cytometry. CD3+ CD8− T cells were analyzed instead of the CD4+ T cells as CD4 is downregulated during HIV-1 replication (21–27). As shown in representative flow cytometry plots (Fig. 1A), CD3+ CD8− T cell subsets were defined as follows: extrafollicular ([EF] CXCR5− CD25− CD127+/−), EF Treg (CXCR5− CD25+ CD127−), TFH (CXCR5+ CD25− CD127−/+), GC TFH (CXCR5+ PD1+ CD25− CD127−/+), TFR (CXCR5+ CD25+ CD127−), and GC TFR (CXCR5+ PD1+ CD25+ CD127−). To define regulatory T cells, the surface phenotype CD25+ CD127− was used (28–30) rather than intranuclear Foxp3 staining as the latter cannot be used for cell sorting experiments that require live, functional cells postsorting. We confirmed that Foxp3 expression was restricted to EF Treg, TFR, and GC TFR cells as defined by these cell surface markers and not expressed in EF, TFH, and GC TFH cells (Fig. 1B). HIV-1 permissivity was assessed by measuring GFP expression for HIV-1 GFP reporter viruses (Fig. 1C), and p24 antigen was measured for expression for T/F viruses (Fig. 1D).
FIG 1.
Representative flow plots. (A) Representative flow plots showing gating used to define tonsillar CD4 T cell subsets. All populations were first gated as live, lymphocyte, single, and CD3+ CD8− cells. The following populations are defined: EF (CXCR5− CD25− CD127+/−), EF Treg (CXCR5− CD25+ CD127−), TFH (CXCR5+ CD25− CD127−/+), GC TFH (CXCR5+ PD1+ CD25− CD127−/+), TFR (CXCR5+ CD25+ CD127−), and GC TFR (CXCR5+ PD1+ CD25+ CD127−) cells. (B) Representative flow plots of Foxp3 expression on T cell subsets defined as regulatory or nonregulatory based on surface CD25 and CD127 expression. (C) Representative flow plots showing GFP expression following ex vivo infection with an R5-tropic GFP reporter virus. (D) Representative flow plots showing p24 antigen expression in a CH470 spinoculated tonsil.
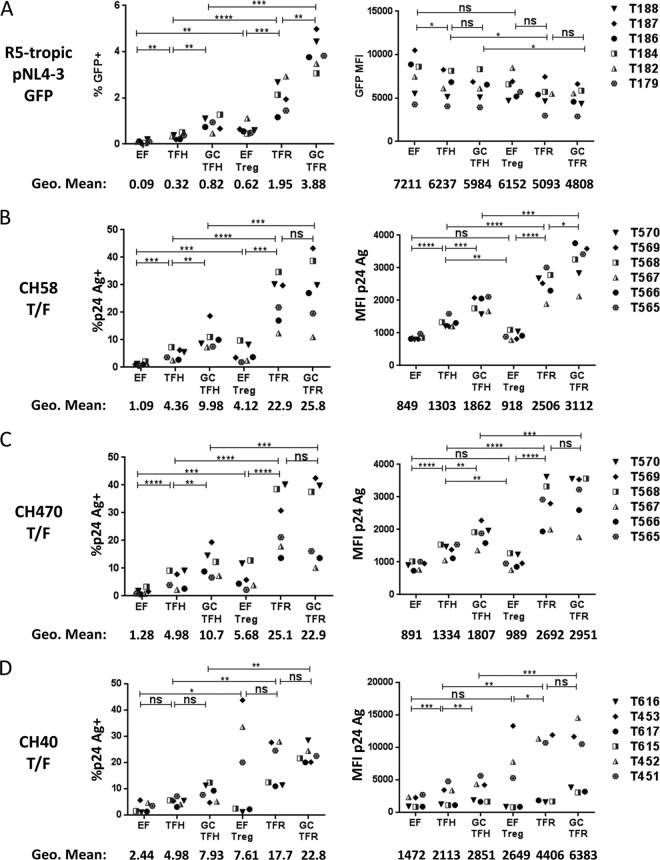
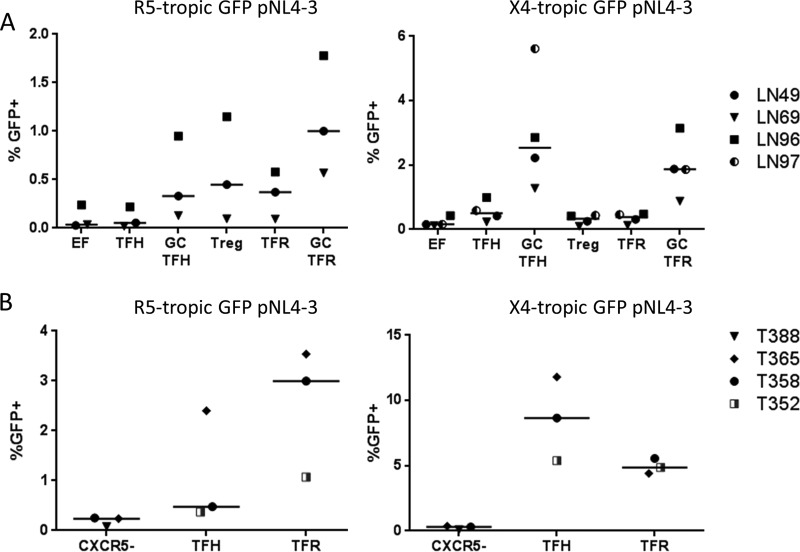
Compared to TFH and GC TFH cells, TFR and GC TFR cells demonstrated high percentages of R5-tropic HIV-1 GFP positive (GFP+) (Fig. 2A) and T/F p24 antigen-positive (Ag+) cells (Fig. 2B to D) following ex vivo HIV-1 infection. EF Treg cells demonstrated a higher percentage of R5-tropic GFP+ or p24 Ag+ cells than EF cells for all four viruses investigated (Fig. 2A to D). Similar results were obtained following infection with the R5-tropic GFP reporter virus when regulatory cells were defined as Foxp3+ instead of CD25+ CD127− (Fig. 3A and B). In this case, permissivity was assessed by measuring p24 Ag instead of GFP expression as some GFP expression was lost when intranuclear permeabilization was performed for Foxp3 staining. While TFR and GC TFR cells demonstrated the highest geometric mean fluorescence intensity (MFI) of p24 Ag when infected with three different T/F viruses (Fig. 2B to D, right panels), they demonstrated the lowest GFP MFI when infected with the lab-adapted R5-tropic HIV-1 GFP reporter virus (Fig. 2A, right panel). TFR cell permissivity to X4-tropic HIV-1 was also investigated using a lab-adapted GFP reporter virus and two X4-tropic infectious molecular clones. TFR and GC TFR cells demonstrated similar or higher percentages of GFP+ or p24 Ag+ cells than TFH and GC TFH cells, respectively (Fig. 4A to C). Differences in CXCR4 expression levels measured in the same cells as the GFP experiments (Fig. 4D) paralleled frequencies of GFP+ T cells in each subset (Fig. 4A). As previously reported (21, 22, 31, 32), the percentages of GFP+ or p24+ cells in each population were consistently higher in the X4-tropic infections than in R5-tropic infections (compare Fig. 3A to D and 4A to C). To determine whether the heightened permissivity of TFR cells extended to other secondary lymphoid tissues, we spinoculated previously cryopreserved, disaggregated cells from LN of HIV-1-seronegative individuals with R5- and X4-tropic GFP reporter viruses. The highest percentage of GFP+ cells was in GC TFR cells in R5-tropic HIV-1 infection but not X4-tropic infection (Fig. 5A), similar to what was observed in tonsil cell infections (Fig. 2A and 4A). To exclude the possibility that productive HIV-1 infection induced cells to acquire a TFR cell phenotype, disaggregated tonsil cells were first sorted into CXCR5−, TFH, and TFR cell populations, then spinoculated with R5- and X4-tropic GFP reporter viruses, and analyzed for GFP expression after 2 days. TFR cells consistently harbored more GFP+ cells than TFH cells in R5-tropic but not X4-tropic HIV-1 infection (Fig. 5B). Taken together, these data demonstrate that TFR cells were the most permissive lymphoid tissue CD4 T cell subset to R5-tropic HIV-1 ex vivo.
FIG 2.
TFR cells are highly permissive to R5-tropic HIV ex vivo. Tonsils were disaggregated and spinoculated with virus at room temperature for 2 h and cultured for 48 h. Productive infection of the following viruses is shown: R5-tropic GFP reporter virus (A), CH58 transmitted/founder virus (B), CH470 transmitted/founder virus (C), and CH40 transmitted/founder virus (D). MFI values and the percentages of GFP+ and p24 Ag+ cells were determined, as indicated. Statistical significance was determined using repeated-measures one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant. Only pairwise comparisons of interest are shown. Overall, the P value was <0.05 (by ANOVA) for all. Tonsil (T) sample numbers are indicated on the right side of the figure.
FIG 3.
Tonsil TFR cells are highly permissive to R5-tropic HIV as defined by Foxp3 expression. Fresh, disaggregated tonsil cells were spinoculated with R5-tropic GFP reporter virus for 2 h at room temperature and analyzed by flow cytometry after 48 h. The following T cell subsets were first defined by surface and intranuclear proteins: EF (CD3+ CD8− CXCR5− Foxp3−); EF Treg (CD3+ CD8− CXCR5− Foxp3+); TFH (CD3+ CD8− CXCR5+ Foxp3−); TFR (CD3+ CD8− CXCR5+ Foxp3+); GC TFH (CD3+ CD8− CXCR5+ PD1+ Foxp3−); GC TFR (CD3+ CD8− CXCR5+ PD1+ Foxp3+) (A) Percent p24 Ag+ cells. (B) MFI of p24 Ag. Statistical significance was determined using repeated-measures one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Only pairwise comparisons of interest are shown. The overall P value was <0.05 (by ANOVA) for all.
FIG 4.
TFR cells are highly permissive to X4-tropic HIV. Tonsils were disaggregated and spinoculated with virus at room temperature for 2 h and cultured for 48 h. Productive infection of the following viruses is shown: X4-tropic GFP reporter virus (A), HIVMN/H9 (B), and SG3.1 (C). MFI values and the percentages of GFP+ and p24 Ag+ cells were determined, as indicated. (D) Frequency of CXCR4 expression at day 0. Statistical significance was determined using repeated-measures one-way ANOVA. *,P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant. Only pairwise comparisons of interest are shown.
FIG 5.
R5-tropic permissivity of seronegative LN cells and sorted tonsil cells. (A) Cryopreserved seronegative LN cells were thawed and spinoculated with R5-tropic or X4-tropic GFP reporter virus for 2 h at room temperature; cells were cultured in R10 medium for 48 h and then analyzed by flow cytometry. LN sample numbers are shown at right. (B) Fresh disaggregated tonsil cells were stained, sorted, and then spinoculated with R5- or X4-tropic GFP reporter virus for 2 h at room temperature. Cell were cultured in R10 medium for 48 h and then analyzed by flow cytometry. The percentages of GFP+ cells are shown. Horizontal lines indicate medians.
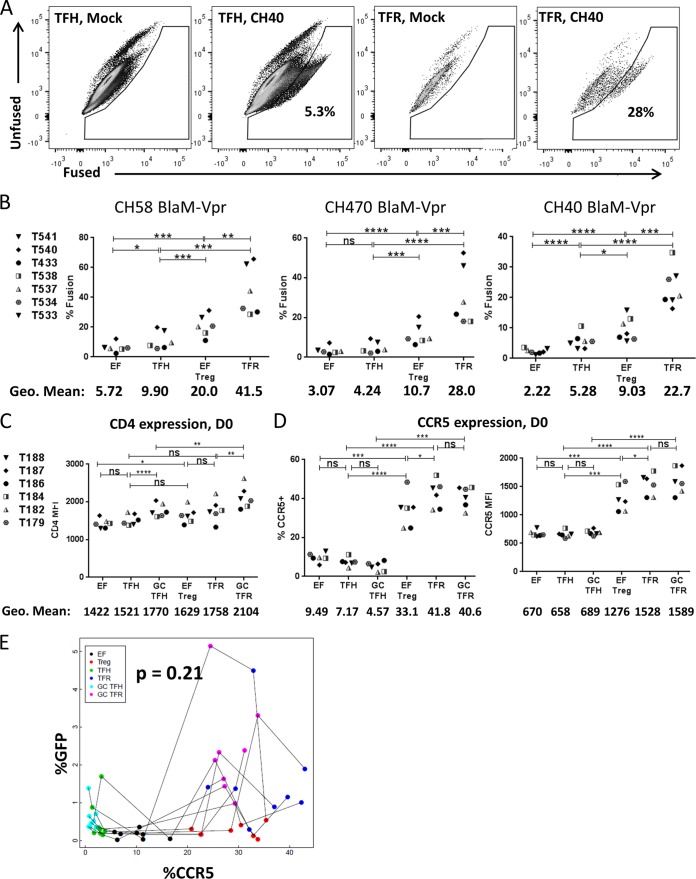
TFR cells express high levels of CCR5 and CD4 and exhibit heightened R5-tropic HIV-1 fusion.
We next investigated potential mechanisms to explain the heightened permissivity of TFR cells to HIV-1. Given that differences in GFP expression and p24 Ag were more pronounced for R5-tropic than for X4-tropic HIV-1 strains, we hypothesized that R5-tropic HIV-1 entry may be a critical step. To investigate HIV-1 entry, we used a flow cytometry-based assay to quantify viral fusion (33). Briefly, T/F HIV-1 viruses containing β-lactamase-Vpr (BlaM-Vpr) were constructed. For this assay, target cells are loaded with coumarin cephalosporin fluorescein acetoxymethyl ester (CCF2-AM), a fluorescent BlaM substrate; if fusion occurs in a cell, BlaM cleaves CCF2-AM, changing the fluorescence emission spectrum from 520 nm to 447 nm, which can be measured by a flow cytometer (Fig. 6A). As shown in Fig. 6B, fusion of CH58, CH470, and CH40 occurred in a greater percentage of TFR cells than in EF Treg, TFH, and EF CD4 T cells. Interestingly, fusion occurs in a greater percentage of EF Treg cells than TFH cells in all three T/F viruses (Fig. 6B); however, the percentage of productively infected EF Treg cells was not different from the percentage of TFH cells that were productively infected when spinoculated with CH58 (Fig. 3B) or CH470 (Fig. 3C). Similar to what was observed when productive infection was measured, fusion occurred in a greater percentage of EF Treg cells than EF cells in all three T/F viruses (Fig. 6B). The percentage of cells in which viral fusion occurred was consistently higher than the percentage of cells that were productively infected in a particular T cell subset for each T/F virus. These data suggest that increased fusion likely was at least partially responsible for the increased R5-tropic HIV permissivity of TFR cells.
FIG 6.
TFR cells are more susceptible to HIV fusion than TFH cells. (A and B) Isolated tonsil CD4+ T cells were spinoculated with BlaM-Vpr transmitted/founder viruses. HIV fusion was evaluated at 2 h postspinoculation by flow cytometry. (A) Representative flow plots showing mock-infected and CH40-infected TFH and TFR cells. (B) Percentage of fusion-positive cells for the indicated virus populations. (C and D) Disaggregated tonsil cells were evaluated for CCR5 and CD4 expression at day 0 (D0). Statistical significance was determined using repeated-measures one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant. Only pairwise comparisons of interest are shown. The overall P value was <0.05 (by ANOVA) for all data in panels B to D. (E) Tonsil cells were disaggregated and immediately stained for CCR5 expression and evaluated by flow cytometry or spinoculated with R5-tropic GFP reporter virus and analyzed for GFP expression after 2 days. A mixed-effects model was used to determine if the percentage of CCR5+ cells predicts the percentage of GFP+ cells for all CD4 T cell subsets investigated. For every log10 increase in the percentage of CCR5, there was a 0.17 (95% CI, −0.01, 0.44) log10 increase in the percentage of GFP (P = 0.21).
Expression of both CD4 and CCR5 is necessary for R5 HIV-1 binding and entry into cells. At day 0, small, statistically significant, differences were observed between EF and EF Treg CD4 cell expression levels as well as between GC TFH and GC TFR cell levels, but not between TFH and TFR cells (Fig. 6C). CCR5 was expressed on a substantially higher proportion of all regulatory subsets, including EF Treg, TFR, and GC TFR cells than the nonregulatory subsets of EF, TFH, and GC TFH cells (Fig. 6D, left). Expression of CCR5 based on the geometric mean fluorescent intensity (MFI) was also higher on EF Treg, TFR, and GC TFR cells than on EF, TFH, and GC TFH cells (Fig. 6D, right). However, CCR5 expression did not differ among regulatory subsets, whereas there were marked differences in permissivity levels to R5-tropic HIV-1 among these subsets (Fig. 3A to D). Furthermore, the frequency of CCR5 expression was not a significant predictor of the frequency of R5-tropic HIV-1 GFP expression (Fig. 6E).
Tonsil CD4 T cell subset permissivity is linked to Ki67 expression.
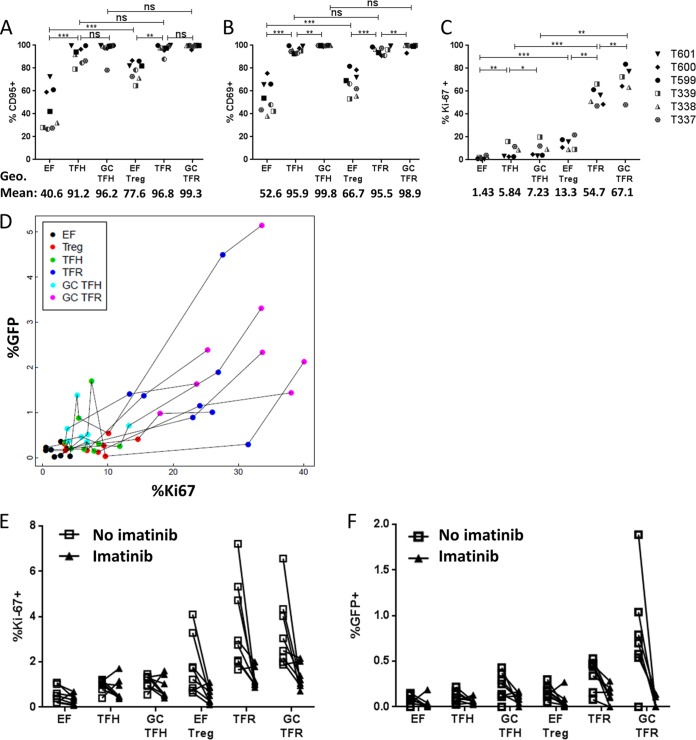
As coreceptor expression did not predict R5-tropic HIV-1 GFP expression, we hypothesized that differences in memory or activation status of the different tonsillar CD4 T cell subsets may be linked to differences in levels of permissivity. Memory CD4 T cells, as well as activated cells, are known to be more permissive to HIV-1 than naive, resting CD4 T cells. We examined markers of memory, cellular activation, and proliferation at day 0 to determine whether any of these parameters were associated with tonsil cell subset permissivity as assessed by GFP expression at day 2. Nearly all cells of the follicular and GC CD4 T cell subsets expressed the memory marker CD95, and there were no significant differences in CD95 expression levels between these subsets (Fig. 7A). The majority of EF Treg cells were CD95+ although significantly fewer than were seen in follicular subsets, and a minority of EF cells were memory cells (Fig. 7A). Assessing T cell activation based on CD69, an early T cell activation marker (Fig. 7B), yielded results very similar to those for CD95 expression (Fig. 7A); nearly all cells of the follicular and GC CD4 T cell subsets express CD69, while the frequency of CD69 expression was significantly reduced in EF CD4 T cells. Thus, differences in expression of CD95 and CD69 did not parallel differences in GFP or p24 Ag expression shown in Fig. 2. To determine the proliferative status of the different tonsillar T cell subsets, Ki67 expression was investigated (Fig. 7C). GC TFR cells harbored the highest proportion of Ki67+ cells, followed by TFR cells, while significantly fewer EF Treg cells expressed Ki67 (Fig. 7C). Nonregulatory T cells (EF, TFH, and GC TFH) expressed Ki67 at a lower frequency than regulatory T cells (Fig. 7C). This pattern most closely paralleled the pattern of GFP expression seen in Fig. 2. To further evaluate whether Ki67 was related to the permissivity of tonsil cell subsets to R5 HIV-1, we analyzed Ki67 expression with respect to GFP expression. All data were log transformed prior to performing the analysis, and a mixed-effects model was used to accommodate repeated-measures data. Using this model, Ki67 expression was predictive of GFP expression; for every log10 increase in the percentage of Ki67, there was a 0.83 log10 increase in the percentage of GFP (P < 0.0001) (Fig. 7D).
FIG 7.
Ki67 expression predicts GFP expression. (A to C) Disaggregated tonsil cells were stained for activation, memory, and proliferation markers at day 0 and immediately analyzed by flow cytometry, as indicated. Statistical significance was determined using repeated-measures one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant. Only pairwise comparisons of interest are shown. The overall P value was <0.05 (by ANOVA) for all. (D) A mixed-effects model was used to determine that the percentage of Ki67+ cells predicts the percentage of GFP+ cells for all T cell subsets investigated (n = 6; P < 0.0001). (E and F) Disaggregated tonsil cells were rested for 2 days with or without imatinib. At day 2, cells were split such that half were analyzed for Ki67 expression, and half were spinoculated with R5-tropic GFP reporter virus and cultured for another 2 days with or without imatinib (n = 6). The percentages of Ki-67+ and GFP+ cells were determined.
To further evaluate whether the relationship between Ki67 expression and virus replication was causative, we treated cells with imatinib to arrest T cell proliferation and decrease Ki67 expression. Imatinib is a tyrosine kinase inhibitor that arrests T cell proliferation in vitro without affecting cell viability (34). Disaggregated tonsil cells were cultured for 2 days with or without imatinib, and Ki67 expression was then measured; cells were then spinoculated with the R5-tropic GFP reporter virus and cultured for 2 more days, when they were analyzed for GFP expression. All CD4 T cell subsets cultured with imatinib expressed significantly lower levels of Ki67 (33.5% reduction; lower confidence interval [CI], 5.7%; upper CI, 53.1%) than those that did not receive imatinib (P = 0.023) (Fig. 7E). Two days after spinoculation, imatinib-treated cells also demonstrated lower R5-tropic HIV-1 GFP expression (77.1%; lower CI, 60.5%; upper CI, 86.8%) than untreated cells (P < 0.0001) (Fig. 7F). These data suggest that the proliferative capacity of tonsillar CD4 T cells, as measured by Ki67 expression, drove differences in R5-tropic HIV GFP expression between tonsillar CD4 T cell subsets ex vivo.
TFR cells harbor high concentrations of HIV-1 and express high levels of Ki67 in vivo.
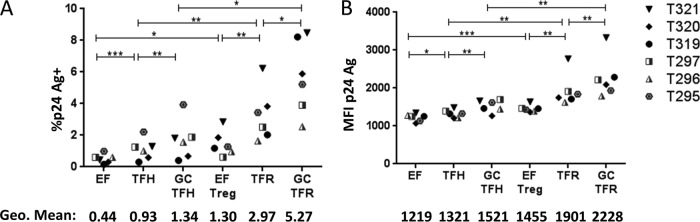
These data, using an ex vivo infection model, revealed that TFR cells were the most permissive tonsillar CD4 T cell subset to R5-tropic HIV-1. However, ex vivo experiments do not always recapitulate events in vivo. To determine if TFR cells were highly permissive in vivo in chronic HIV-1 infection, we measured HIV-1 RNA in LN cells from six asymptomatic, chronically HIV-1-infected, untreated individuals. Five of the six subjects were known to harbor R5-tropic HIV-1, and the tropism of the sixth subject's virus was not determined. Previously cryopreserved, disaggregated LN cells were sorted into EF, TFH, EF Treg, and TFR cell subsets, and HIV-1 RNA was measured by reverse transcription-quantitative PCR (RT-qPCR). TFR cells harbored the highest concentrations of HIV-1 RNA per nanogram of RNA, TFH and EF Treg cells harbored intermediate amounts that did not differ significantly from each other, and EF cells harbored the lowest concentrations (Fig. 8A). Overall, EF cells constituted the largest subset of CD3+ CD8− cells, with a geometric mean (GM) of 40%, and TFH cells were the second largest group, with a geometric mean of 24% (Fig. 8B). EF Treg and TFR cells were distinct minority populations, with geometric means of 4.1% and 3.6%, respectively (Fig. 8B). Because of the higher concentrations of viral RNA in TFR cells, they contributed a disproportionately large amount of HIV-1 RNA than would be expected based on the size of the TFR population: approximately 14.4% of the HIV-1 RNA in the LN CD4 T cell subsets overall (Fig. 8C) and 19.7% of the HIV-1 RNA found in the follicular CD4 T cell subsets. Ki67 expression was elevated in both TFR and EF Treg cells and did not differ significantly between these populations (Fig. 8D). Both regulatory subsets harbored significantly more Ki67+ cells than either TFH or EF cells. These data demonstrate that, similarly to the ex vivo model, TFR cells supported high levels of virus replication in vivo.
FIG 8.
TFR cells are highly permissive to HIV in vivo. Cryopreserved, disaggregated LN cells from chronically HIV-infected, untreated individuals were thawed, stained, and sorted into EF, TFH, EF Treg, and TFR cell populations. RNA was isolated immediately and stored at −80°C. One-step RT-qPCR was performed to measure viral RNA. (A) HIV copy number per nanogram of total RNA in each LN for all sorted T cell populations (overall, P < 0.05; ANOVA). (B) T cell populations as a percentage of total CD3+ CD8− cells (overall, P < 0.0001; ANOVA). (C) Calculated HIV RNA contribution from each T cell population as a percentage of the total population (overall, P = 0.12; ANOVA). (D) Percent Ki67+ cells in each LN T cell population investigated (overall, P = 0.01; ANOVA). Statistical significance was determined using repeated-measures one-way ANOVA. *, P < 0.05; ***, P < 0.001; ns, not significant. Only pairwise comparisons of interest are shown.
DISCUSSION
To our knowledge, this is the first study to investigate the permissivity of TFR cells to HIV-1 ex vivo and in vivo. Using four different R5-tropic viruses, we found that uninfected tonsillar TFR cells are highly permissive ex vivo to R5-tropic HIV-1, the most common virus found in HIV-1-infected individuals in early and mid-stages of disease. TFR cells express the highest levels of CCR5 and CD4, and a greater percentage of tonsillar TFR cells than EF Treg, TFH, and EF cells undergoes viral fusion, indicating that increased permissivity of TFR cells begins with viral entry. Patterns of CCR5 expression parallel HIV-1 fusion but not differences in virus permissivity ex vivo, suggesting that other factors are involved. The cellular proliferation marker Ki67 predicts GFP expression following ex vivo infection with an HIV-1 GFP reporter virus, and reductions in Ki67 expression through addition of imatinib reduce GFP expression. In vivo, TFR cells harbor more HIV-1 RNA per nanogram of total RNA than all other subsets and also exhibit high levels of Ki67 expression. Collectively, these data demonstrate that TFR cells are highly permissive to R5 HIV-1 ex vivo and in vivo and that this elevated permissivity is related to elevated levels of HIV-1 coreceptor expression as well as a heightened proliferative state.
Previous studies that investigated the ex vivo permissivity of blood Treg cells to HIV-1 produced mixed results. In one study, ex vivo infection with an R5-tropic GFP reporter virus showed that Treg cells are more permissive than memory T cells (35). Another study found that blood Treg cells are not more permissive than effector T cells to R5-tropic HIV-1 (36). Less than 1% of peripheral blood CD4+ T cells have a TFR cell-like phenotype. It is unknown how their permissivity compares to that of other peripheral blood or LN subsets, and this could be investigated in future studies. A limitation of peripheral blood studies is that peripheral blood mononuclear cells (PBMC) must be stimulated to enable HIV-1 replication, which enhances cellular proliferation and permissivity, thereby clouding results. An advantage of the tonsil cell model over PBMC is that it does not require any exogenous stimulation and therefore more closely mimics the cell state in vivo. Using this model, we observed that TFR cells were more permissive than TFH, EF Treg, and EF non-Treg cells. Additionally, we found that the proliferative status of the CD4 T cell subset is predictive of R5-tropic HIV-1 GFP expression and that changing the proliferative status of the CD4 T cell subsets altered their permissivity. Any exogenous stimulation will likely alter the proliferative status of cells; this provides a possible explanation of differing results. Our results demonstrate that careful attention must be paid to cell type and ex vivo culture conditions in investigations of HIV-1 permissivity as both variables can drastically alter experimental results.
In contrast to the consistent pattern of preferential R5-tropic HIV-1 replication in TFR cells across all viruses investigated, replication of X4-tropic HIV-1 was more variable. Frequencies of GFP+ and p24 Ag+ TFR and GC TFR cells are high, though not always higher than those of TFH and GC TFH cells. Inconsistencies in GFP/p24 expression patterns in X4-tropic viruses may be due to the fact that SG3.1 and HIV-1MN/H9 were previously passaged in cell lines. The R5-tropic viruses are all molecular clones. The HIV-1MN/H9 virus, on the other hand, is not maintained as a molecular clone and is propagated via infecting CD4- and CXCR4-expressing cells. SG3.1 is now maintained as a molecular clone but was originally passaged in a cell line. It is possible that these viruses acquired mutations that were advantageous to productive infection under the specific laboratory conditions by which they were propagated that has changed the way they infect and replicate within primary cells. As it is still unknown why the majority of transmitted viruses are R5 tropic, further investigation into T cell permissivity to X4-tropic compared to R5-tropic HIV-1 could help answer this question.
Interestingly, while patterns of percent p24 Ag+ cells and the p24 Ag MFI parallel each other in all R5 viruses, the percent GFP and the GFP MFI do not. p24 is a viral capsid protein; presence of this protein indicates later stages of productive infection of a cell. The p24 Ag MFI therefore measures the amount of virus produced in each T cell population. GFP, however, is expressed via an internal ribosome entry site (IRES) inserted into the HIV-1 genome, so it is possible that translation of GFP does not necessarily reflect translation of native viral proteins. Additionally, the GFP reporter viruses are lab-adapted strains while the T/F virus sequences are those that have been transmitted from one individual to another. The envelope proteins of the lab-adapted viruses (especially the R5-tropic GFP reporter virus) have been altered from their native configuration. This may result in suboptimal viral entry compared to that of T/F viruses in cells expressing CCR5 and CD4 and thus decreased GFP production in different T cell populations compared to levels in T/F viruses. Further, it is also possible that the GFP reporter virus may be better adapted to replicating in nonregulatory subsets than in Treg cells, whereas T/F viruses may not have evolved such an adaptation in vivo.
TFR cells demonstrate more fusion with three different R5-tropic HIV-1 transmitted/founder viruses than EF Treg, TFH, and EF T cells. Unsurprisingly, the patterns of HIV-1 fusion parallel the differences in coreceptor expression when CCR5 expression and CD4 expression are taken together. However, fusion and coreceptor expression do not directly parallel the frequency of p24 Ag+ cells found in each T cell subset; there is a much higher frequency of CCR5 expression in EF Treg cells than in TFH cells, but the frequency of p24 antigen-positive or GFP+ cells is not higher in EF Treg cells than in TFH cells. While none of the follicular T cell subsets examined differ in their expression levels of CD95 or CD69, TFR cells are more proliferative than EF Treg, TFH, and EF cells, and EF Treg cells are more proliferative than TFH and EF cells, based on Ki67 expression. Proliferating, activated cells are known to support higher levels of HIV-1 integration and replication (37, 38), and here we show that Ki67 expression predicts GFP expression. Further experiments showed that reducing Ki67 expression while leaving CCR5 expression unchanged causes a concurrent reduction in GFP expression. While expression of CCR5 is undoubtedly necessary for viral fusion and entry, this study suggests that heightened proliferation of TFR cells is a key determinant of their heightened permissivity to R5-tropic HIV-1 ex vivo.
Similar to ex vivo HIV-1 infection, TFR cells isolated from LN of chronically HIV-1-infected, untreated individuals contain more HIV-1 RNA than TFH (including GC TFH), EF Treg, and EF LN T cell subsets. Indeed, TFR cells account for only a median of 3.6% of LN CD3+ CD8− T cells but a median of 14.4% of the calculated total HIV-1 RNA from CD3+ CD8− T cell populations. Because TFH cells are more frequent in the LN population, however, constituting a median of 23.7% of CD3+ CD8− T cells, they harbor the largest amount of HIV-1 RNA, a median of 48.1% of the calculated total HIV-1 RNA in LN cells. Similar to findings in human tonsil cells, chronically HIV-1-infected LN TFR cells exhibit a higher frequency of Ki67 expression than TFH and EF cells, which suggests that heightened proliferation promotes HIV-1 replication in these cells. Interestingly, however, the approximately 2-fold difference in the magnitudes of productive infection between TFH and TFR cells as measured by RNA in vivo (Fig. 5) is not as great as the approximately 5-fold difference in productive infections between TFH and TFR cells as measured by GFP or p24 antigen in ex vivo infections (Fig. 2). One possible explanation for this is that levels of Ki67 expression in TFR cells in HIV-1-infected human LN were not as elevated as they were in tonsils; overall a geometric mean of 20% of TFR cells in LN expressed Ki67, versus 55% of tonsil TFR cells. On the other hand, levels of Ki67 in TFH cells were similar in LN TFH cells (geometric mean 5.8%) and tonsil TFR cells (geometric mean, 5.8%). Why TFR cells express less Ki67 in HIV-1-infected LN than in seronegative tonsils in this study is unclear but could be related to intrinsic differences between these tissues, age, or chronic HIV-1 infection as tonsils were obtained from children at low risk of HIV-1 infection whereas LN were obtained from chronically HIV-1-infected adults. Further, this and our imatinib data suggest that it is possible to lower TFR cell permissivity to HIV by reducing Ki67 expression and the proliferative capacity of the cells. Whether this may have any clinical implications remains to be explored.
Levels of Ki67 expression in LN EF Treg cells (GM, 19%) were similar to those in TFR cells (GM, 20%) and higher than those found in TFH cells (GM, 10%). Intriguingly, however, HIV-1 RNA was significantly lower in EF Treg cells than in TFR or TFH cells. We have previously demonstrated that CTL fail to accumulate in B cell follicles and that this is related to high levels of virus replication in B cell follicles in secondary lymphoid tissues in HIV-1-infected humans (4) and SIV-infected rhesus macaques (6). We have also demonstrated in the SIV-infected rhesus macaque model that depletion of CD8 T cells results in substantial increases in HIV-1-producing cells in extrafollicular regions and much smaller increases in follicular regions (39), suggesting that CTL exert much more antiviral activity in extrafollicular than in follicular regions. It seems likely that increased CTL surveillance in the EF region compared to that in the follicles resulted in less viral RNA in EF Treg cells than in TFR and TFH cells despite similar or higher levels of Ki67 expression.
Emerging data suggest that T regulatory cells do not harbor a major fraction of viral DNA in untreated HIV or SIV infection but that cells with a follicular and/or regulatory phenotype increasingly harbor the majority of viral DNA in treated disease (40, 41). The findings of the present study as well as our previous work provide important insight into the mechanisms that may account for this. We previously demonstrated that TFR cells incorporate significantly more bromodeoxyuridine (BrdU) than other lymphoid tissue cells (19), and in the present work we demonstrated that Treg cells, and particularly TFR cells, express heightened levels of the proliferation marker Ki67, suggesting that T regulatory cells are exceptionally proliferative. Proliferation of latently infected T cells has been shown to contribute significantly to the viral reservoir in antiretroviral therapy (ART)-treated patients (42). We previously demonstrated as well that TFR cells are more resistant to apoptosis than TFH cells in ex vivo HIV-1 infection (19) and that CXCR5+ HIV-1-producing cells have more Bcl-2 expression than CXCR5− HIV-1-producing cells in R5-tropic HIV-1 infection (22). These data, when taken together with data in the current study showing that TFR and Treg cells express elevated levels of CCR5, suggest that TFR and Treg cells may be more resistant than other cells to cell death and able to survive despite HIV infection. It seems likely that as these cells proliferate over time, they increasingly contribute to the latent viral reservoir in ART-treated patients. Thus, a better understanding of the establishment and maintenance of latent infection in TFR and Treg cells is essential and could lead to novel strategies to treat and cure HIV-1 infection.
MATERIALS AND METHODS
Ethics statement.
All research involving human subjects conformed to the principles set forth in the Declaration of Helsinki and was approved by the Colorado Multiple Institutional Review Board (COMIRB). Human tonsils were obtained from the Colorado Children's Hospital (Aurora, CO, USA) following routine tonsillectomies from children at low risk of HIV infection. Use of these tonsils for these studies was reviewed by COMIRB and was determined not to constitute human subject research in accordance with guidelines issued by the Office of Human Research Protections (http://www.hhs.gov/ohrp/policy/checklists/decisioncharts.html). Informed consent was therefore not required. Informed consent was obtained from all subjects who donated inguinal LN, and the study was approved by COMIRB.
Human LN.
Inguinal LN were obtained as previously described (43, 44). In this study, four HIV-seronegative LN were used. One (25%) of the seronegative subjects was female, ages ranged from 35 to 74 years (median, 60.5 years), three subjects (75%) were white, and the race of one subject (25%) was unknown. Six HIV-1-seropositive LN were used. Seropositive individuals had documented HIV-1 infection for at least 6 months, were not receiving ART, and had CD4 T-cell counts of ≥300 per mm3. None of the subjects had an opportunistic infection, malignancy, or acute illness at the time of excision. CD4 T-cell counts ranged from 300 to 1,117 cells per mm3 (median, 542.5 cells per mm3), and plasma viral load ranged from 3.84 to 5.24 log10 copies per ml (median, 4.53 log10 copies per ml). Virus tropism was evaluated in all six subjects as previously described (45). In five subjects virus was determined to be R5 tropic, and in the sixth subject tropism could not be determined. A total of four (66.7%) subjects were females, ages ranged from 23 to 42 years (median, 29.5 years), two subjects (33.3%) were white, including one Hispanic, three subjects were black (50%), and one subject (16.7%) was Native American. At the time of excision, LN were minced in phosphate-buffered saline (PBS) to obtain a single-cell suspension and cryopreserved in liquid nitrogen.
Viruses: GFP reporter viruses, T/F viruses, and fusion viruses.
The R5-tropic GFP reporter virus NLYUV3-GFP (21) and X4-tropic GFP reporter virus NLENG1-IRES (46) were prepared by transfecting 293T cells (ATCC) with NLYUV3-GFP or NLENG1-IRES plasmid constructs using Effectene transfection reagent (Qiagen 301425) according to the manufacturer's protocol, and transfected cells were cultured in complete Dulbecco's modified Eagle's medium (DMEM; supplemented with 10% fetal bovine serum [FBS], penicillin-streptomycin [Pen-Strep], and nonessential amino acids) for 48 h. Supernatants were collected, spun at 800 × g to remove cellular debris, and stored at −80°C prior to use. SG3.1 (NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH; pSG3.1 was from Sajal Ghosh, Beatrice Hahn, and George Shaw; [47]) was prepared by transfecting 293T cells (ATCC) with pSG3.1 plasmid constructs using Mirus TransIT-LT1 transfection reagent (MIR 2300) according to the manufacturer's protocol, and transfected cells were cultured in complete DMEM (DMEM supplemented with 10% FBS, Pen-Strep, and nonessential amino acids) for 48 h. Supernatant was collected and spun at 80,000 × g for 2 h over a 20% sucrose cushion to concentrate virus. HIVMN/H9 virus stocks (previously referred to as HTLV-IIIMN/H9; obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH; HTLV-IIIMN/H9 was from Robert Gallo [48, 49]) were prepared by infection of MOLT4-CCR5 (ARRP 510) cells and passage for 9 days. Supernatants were collected and spun at 80,000 × g for 2 h over a 20% sucrose cushion to concentrate virus. CH58, CH470, and CH40 (obtained from Beatrice Hahn, University of Pennsylvania [50]) BLaM-Vpr viruses were constructed by cotransfecting pAdVantage, pBlaM-Vpr, and the appropriate virus plasmid into 293T cells with 2 M CaCl2. Transfected 293T cells were cultured for 48 h in complete DMEM, and supernatants were collected and ultracentrifuged at 80,000 × g for 2 h over a 20% sucrose cushion to concentrate virus. Titers of concentrated virus stocks were determined via a p24 enzyme-linked immunosorbent assay (ELISA) (ABL 5421).
Tonsil cultures.
Fresh tonsils were minced and either cells were stained for day 0 flow cytometry immediately, or 5 × 106 cells were spinoculated with HIV-1 or mock spinoculated in an equal volume of complete DMEM for 2 h at 1,200 × g at room temperature. Infected or mock-spinoculated cells were washed to remove unbound virus and medium and then cultured for 2 days at 37°C with 5% CO2 in RPMI medium with 10% FBS, l-glutamine, and Pen-Strep (R10) in 3.5 ml in a six-well, flat-bottom plate. After 48 h, cells were collected and stained with antibodies for flow cytometry. In select experiments, disaggregated tonsil cells were rested in R10 medium for 48 h before flow cytometry analysis or spinoculation with CH40. Cells spinoculated with CH40 were subsequently analyzed by flow cytometry after 48 h.
Cell sorting.
Human tonsils and LN were sorted using a MoFlo Astios EQ instrument. Disaggregated cells from tonsils or seronegative LN were sorted into EF (CD3+ CD8− CXCR5− CD25− CD127+/−), EF Treg (CD3+ CD8− CXCR5− CD25+ CD127−), TFH (CD3+ CD8− CXCR5+ CD25− CD127+/−), and TFR (CD3+ CD8− CXCR5+ CD25+ CD127−) cell populations. After cell sorting, tonsils were spinoculated with NLYUV3-GFP and analyzed by flow cytometry after 48 h. Sorted LN cells were immediately processed for RNA isolation.
RNA isolation and RT-qPCR.
RNA was isolated from sorted seropositive LN cells using a PicoPure RNA isolation kit (KIT0204; ThermoFisher) according to the manufacturer's protocol. RNA concentration and purity were measured on an Agilent 2200 TapeStation. All RNA samples used had an RNA integrity number (RIN) of 7 or greater. One-step real-time reverse transcription-quantitative PCR (RT-qPCR) was then performed on a Bio-Rad CFX real-time system using a QuantiTect Probe RT-PCR kit (204443; Qiagen) in a final volume of 25 μl using 500 nM primers (forward primer, 5′-TTTGGAAAGGACCAGCAAA-3′; reverse primer, 5′-CCTGCCATCTGTTTTCAA-3′) and 250 nm dually labeled probe (probe, 5′-6FAM-AAAGGTGAAGGGGCAGTAGTAATACA-TAMRA-3′, where FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine). These primers target a 127-bp, highly conserved region of HIV-1 integrase (51). Absolute quantification was achieved using a 519-bp DNA standard amplified from pSG3.1 (forward, 5′-CAATACATACAGACAATGGCAGC-3′; reverse, 5′-GTCTACTTGCCACACAATCATC-3′) which was subsequently gel purified and quantified. The following cycle conditions were used for the RT-qPCRs: 50°C for 30 min (reverse transcription), 95°C for 15 min (hot start transcriptase), and 40 cycles of 94°C for 15 s (denaturation) and 57°C for 40 s (annealing/extension). Efficiency for all runs was between 90 and 100%. To calculate the percentage of total HIV RNA in each CD4 T cell population, HIV RNA copy numbers in each population were calculated using the RNA copy number and number of cells in each CD4 T cell population. The percentage of HIV RNA of total HIV RNA from each population was then calculated using the total number of HIV copies from all CD4 T cell populations divided by the number of HIV RNA copies in each CD4 T cell population.
Fluorescence-activated cell sorting (FACS) antibodies and staining.
Disaggregated tonsil cells or LN cells were washed in PBS and blocked for 20 min with 2% bovine serum albumin (BSA) in PBS at 4°C and then stained for 30 min at 4°C in the dark. The following anti-human fluorescently conjugated antibodies were used: CD3-UCHT1 (25-0038; Tonbo), CD8-RPA-T8 (83-0088; eBioscience), CXCR5-MU5UBEE (18-9185; eBioscience), PD-1-EH12.2H7 (329908; BioLegend), CD25-BC96 (60-0259; Tonbo), CD127-A1095 (351306; BioLegend), CD4-RPA-T4 (560158; BD), CD69-FN50 (562989; BD), CD95-DX2 (556640; BD), p24-KC57 (41116015; Beckman Coulter), Foxp3-PCH101 (17-4776; eBioscience), Ki67-B56 (561284; BD), CCR5-2D7 (555992; BD), and CXCR4-12G5 (555974; BD). Intracellular staining (Foxp3 and p24) was performed using an eBioscience Toxp3/transcription factor permeabilization kit (72-5776) according to the manufacturer's protocol. Live cells were determined using Tonbo Ghost viability dyes (13-0870).
Fusion assay.
Fusion assays were performed according to a previously published protocol (33) with the following modifications: isolated tonsil CD4 cells (EasySep human CD4+ T cell enrichment kit [19052; StemCell]) were substituted for peripheral blood lymphocytes; cells were spinoculated for 2 h at room temperature instead of being incubated at 37°C for 2 h with virus.
Statistical analyses.
Statistical significance was determined using repeated-measures one-way analysis of variance (ANOVA) with pairwise comparisons if the overall P value was less than 0.05. Only pairwise comparisons of interest are shown in the figures. No further adjustment was made for multiple comparisons. For the percentage of Ki67 and percentage of CCR5 predicting percentage of GFP, mixed-effects models were used to accommodate within-subject correlations; cell type was not considered a predictor. Data were log transformed as appropriate. Repeated-measures one-way ANOVA tests were performed using GraphPad Prism, version 6.
ACKNOWLEDGMENTS
This work was funded by NIH/NIAID grants R01 AI096966 to E.C., T32 AI007405 to S.M.M., T32AI007447-22 T32 to B.M., and the University of Colorado Denver Department of Medicine Early Career Scholar Program (M.L.S.). Cell sorting was performed by the University of Colorado Cancer Flow Cytometry Shared Resource, which is supported by Cancer Center Support Grant P30CA046934 and the Skin Diseases Research Cores Grant P30AR057212.
The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: pSG3.1 from Sajal Ghosh, Beatrice Hahn, and George Shaw and HTLV-IIIMN/H9 from Robert Gallo.
We declare that we have no conflicts of interest and that funding sources had no role in the experimental design, data collection, interpretation of data, or decision to submit the manuscript for publication.
S.M.M., B.M., K.G., J.F., M.L.S., A.L.M., M.D.M., D.N.L., and E.C. designed experiments; S.M.M., B.M., and K.G. performed experiments; A.L.M., M.D.M., D.N.L., and M.L.S. contributed reagents and samples; S.M.M., S.M., B.M., K.G., J.F., M.L.S., A.L.M., M.D.M., D.N.L., and E.C. interpreted and analyzed data; S.M.M. and E.C. wrote the manuscript.
REFERENCES
- 1.Biberfeld P, Chayt KJ, Marselle LM, Biberfeld G, Gallo RC, Harper ME. 1986. HTLV-III expression in infected lymph nodes and relevance to pathogenesis of lymphadenopathy. Am J Pathol 125:436–442. [PMC free article] [PubMed] [Google Scholar]
- 2.Tenner-Racz K, Stellbrink H-J, Van Lunzen J, Schneider C, Jacobs J-P, Raschdorff B, Großchupff G, Steinman RM, Racz P. 1998. The unenlarged lymph nodes of HIV-1-infected, asymptomatic patients with high CD4 T cell counts are sites for virus replication and CD4 T cell proliferation. The impact of highly active antiretroviral therapy. J Exp Med 187:949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Folkvord JM, Armon C, Connick E. 2005. Lymphoid follicles are sites of heightened human immunodeficiency virus type 1 (HIV-1) replication and reduced antiretroviral effector mechanisms. AIDS Res Hum Retroviruses 21:363–370. doi: 10.1089/aid.2005.21.363. [DOI] [PubMed] [Google Scholar]
- 4.Connick E, Mattila T, Folkvord JM, Schlichtemeier R, Meditz AL, Ray MG, McCarter MD, MaWhinney S, Hage A, White C, Skinner PJ. 2007. CTL fail to accumulate at sites of HIV-1 replication in lymphoid tissue. J Immunol 178:6975–6983. doi: 10.4049/jimmunol.178.11.6975. [DOI] [PubMed] [Google Scholar]
- 5.Kohler SL, Pham MN, Folkvord JM, Arends T, Miller SM, Miles B, Meditz AL, McCarter M, Levy DN, Connick E. 2016. Germinal center T follicular helper cells are highly permissive to HIV-1 and alter their phenotype during virus replication. J Immunol 196:2711–2722. doi: 10.4049/jimmunol.1502174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connick E, Folkvord JM, Lind KT, Rakasz EG, Miles B, Wilson NA, Santiago ML, Schmitt K, Stephens EB, Kim HO, Wagstaff R, Li S, Abdelaal HM, Kemp N, Watkins DI, MaWhinney S, Skinner PJ. 2014. Compartmentalization of simian immunodeficiency virus replication within secondary lymphoid tissues of rhesus macaques is linked to disease stage and inversely related to localization of virus-specific CTL. J Immunol 193:5613–5625. doi: 10.4049/jimmunol.1401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heath SL, Tew JG, Tew JG, Szakal AK, Burton GF. 1995. Follicular dendritic cells and human immunodeficiency virus infectivity. Nature 377:740–744. doi: 10.1038/377740a0. [DOI] [PubMed] [Google Scholar]
- 8.Miles B, Connick E. 2016. TFH in HIV latency and as sources of replication-competent virus. Trends Microbiol 24:338–344. doi: 10.1016/j.tim.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, Rolph MS, Mackay CR. 2004. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J Immunol 173:68–78. doi: 10.4049/jimmunol.173.1.68. [DOI] [PubMed] [Google Scholar]
- 10.Eto D, Lao C, DiToro D, Barnett B, Escobar TC, Kageyama R, Yusuf I, Crotty S. 2011. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLoS One 6:e17739. doi: 10.1371/journal.pone.0017739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crotty S. 2011. Follicular helper CD4 T cells (Tfh). Annu Rev Immunol 29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 12.Lindqvist M, J van Lunzen Soghoian DZ, Kuhl BD, Ranasinghe S, Kranias G, Flanders MD, Cutler S, Yudanin N, Muller MI, Davis I, Farber D, Hartjen P, Haag F, Alter G, Schulze zur Wiesch J, Streeck H. 2012. Expansion of HIV-specific T follicular helper cells in chronic HIV infection. J Clin Invest 122:3271. doi: 10.1172/JCI64314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrovas C, Yamamoto T, Gerner MY, Boswell KL, Wloka K, Smith EC, Ambrozak DR, Sandler NG, Timmer KJ, Sun X, Pan L, Poholek A, Rao SS, Brenchley JM, Alam SM, Tomaras GD, Roederer M, Douek DC, Seder RA, Germain RN, Haddad EK, Doup RA. 2012. CD4 T follicular helper cell dynamics during SIV infection. J Clin Invest 122:3281–3294. doi: 10.1172/JCI63039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, Srivastava M, Divekar DP, Beaton L, Hogan JJ, Fagarasan S, Liston A, Smith KG, Vinuesa CG. 2011. Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med 17:975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wollenberg I, Agua-Doce A, Hernández A, Almeida C, Oliveira VG, Faro J, Graca L. 2011. Regulation of the germinal center reaction by Foxp3+ follicular regulatory T cells. J Immunol 187:4553–4560. doi: 10.4049/jimmunol.1101328. [DOI] [PubMed] [Google Scholar]
- 16.Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang Y-H, Lim H, Reynolds JM, Zhou XH, Fan HM, Liu ZM, Neelapu SS, Dong C. 2011. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med 17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colineau L, Rouers A, Yamamoto T, Xu Y, Urrutia A, Pham H-P, Cardinaud S, Samri A, Dorgham K, Coulon P-G, Cheynier R, Hosmalin A, Oksenhendler E, Six A, Kelleher AD, Aaunders J, Koup RA, Autran B, Moris A, Graff-Dubois S. 2015. HIV-infected spleens present altered follicular helper T cell (Tfh) subsets and skewed B cell maturation. PLoS One 10:e0140978. doi: 10.1371/journal.pone.0140978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wallin EF, Jolly EC, Suchánek O, Bradley JA, Espéli M, Jayne DR, Linterman MA, Smith KG. 2014. Human T-follicular helper and T-follicular regulatory cell maintenance is independent of germinal centers. Blood 124:2666–2674. doi: 10.1182/blood-2014-07-585976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miles B, Miller SM, Folkvord JM, Kimball A, Chamanian M, Meditz AL, Arends T, McCarter MD, Levy DN, Rakasz EG, Skinner PJ, Connick E. 2015. Follicular regulatory T cells impair follicular T helper cells in HIV and SIV infection. Nat Commun 6:8608. doi: 10.1038/ncomms9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blackburn MJ, Zhong-Min M, Caccuri F, McKinnon K, Schifanella L, Guan Y, Gorini G, Venzon D, Fenizia C, Binello N, Gordon SN, Miller CJ, Franchini G, Vaccari M. 2015. Regulatory and helper follicular T cells and antibody avidity to simian immunodeficiency virus glycoprotein 120. J Immunol 195:3227–3236. doi: 10.4049/jimmunol.1402699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meditz AL, Haas MK, Folkvord JM, Melander K, Young R, McCarter M, MaWhinney S, Campbell TB, Lie Y, Coakley E, Levy DN, Connick E. 2011. HLA-DR+ CD38+ CD4+ T lymphocytes have elevated CCR5 expression and produce the majority of R5-tropic HIV-1 RNA in vivo. J Virol 85:10189–10200. doi: 10.1128/JVI.02529-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haas MK, Levy DN, Folkvord JM, Connick E. 2015. Distinct patterns of Bcl-2 expression occur in R5-and X4-tropic HIV-1-producing lymphoid tissue cells infected ex vivo. AIDS Res Hum Retroviruses 31:298–304. doi: 10.1089/aid.2014.0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDougal J, Mawle A, Cort S, Nicholson J, Cross G, Scheppler-Campbell J, Hicks D, Sligh J. 1985. Cellular tropism of the human retrovirus HTLV-III/LAV. I. Role of T cell activation and expression of the T4 antigen. J Immunol 135:3151–3162. [PubMed] [Google Scholar]
- 24.Kaminsky LS, McHugh T, Stites D, Volberding P, Henle G, Henle W, Levy JA. 1985. High prevalence of antibodies to acquired immune deficiency syndrome (AIDS)-associated retrovirus (ARV) in AIDS and related conditions but not in other disease states. Proc Natl Acad Sci U S A 82:5535–5539. doi: 10.1073/pnas.82.16.5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoxie JA, Alpers JD, Rackowski JL, Huebner K, Haggarty BS, Cedarbaum AJ, Reed JC. 1986. Alterations in T4 (CD4) protein and mRNA synthesis in cells infected with HIV. Science 234:1123–1127. doi: 10.1126/science.3095925. [DOI] [PubMed] [Google Scholar]
- 26.Dalgleish A, Beverley P, Clapham P, Crawford D, Greaves M, Weiss R. 1984. The T4 (CD4) molecule is an essential component of the HTLV-III/LAV-1 receptor. Nature 312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 27.Popovic M, Gallo R, Mann D. 1985. OKT-4 antigen bearing molecule is a receptor for the human retrovirus HTLV-III. Clin Res 33:560A. [Google Scholar]
- 28.Hartigan-O'Connor DJ, Poon C, Sinclair E, McCune JM. 2007. Human CD4+ regulatory T cells express lower levels of the IL-7 receptor alpha chain (CD127), allowing consistent identification and sorting of live cells. J Immunol Methods 319:41–52. doi: 10.1016/j.jim.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 29.Shen L-S, Wang J, Shen D-F, Yuan X-L, Dong P, Li M-X, Xue J, Zhang F-M, Ge H-L, Xu D. 2009. CD4+ CD25+ CD127low/− regulatory T cells express Foxp3 and suppress effector T cell proliferation and contribute to gastric cancers progression. Clin Immunol 131:109–118. doi: 10.1016/j.clim.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 30.Seddiki N, Santner-Nanan B, Martinson J, Zaunders J, Sasson S, Landay A, Solomon M, Selby W, Alexander SI, Nanan R, Kelleher A, Fazekas de St Groth B. 2006 Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J Exp Med 203:1693–1700. doi: 10.1084/jem.20060468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grivel J-C, Elliott J, Lisco A, Biancotto A, Condack C, Shattock RJ, McGowan I, Margolis L, Anton P. 2007. HIV-1 pathogenesis differs in rectosigmoid and tonsillar tissues infected ex vivo with CCR5- and CXCR4-tropic HIV-1. AIDS 21:1263–1272. doi: 10.1097/QAD.0b013e3281864667. [DOI] [PubMed] [Google Scholar]
- 32.Grivel J-C, Margolis LB. 1999. CCR5- and CXCR4-tropic HIV-1 are equally cytopathic for their T-cell targets in human lymphoid tissue. Nat Med 5:344–346. doi: 10.1038/6565. [DOI] [PubMed] [Google Scholar]
- 33.Cavrois M, Neidleman J, Bigos M, Greene WC. 2004. Fluorescence resonance energy transfer-based HIV-1 virion fusion assay. Methods Mol Biol 263:333–344. [DOI] [PubMed] [Google Scholar]
- 34.Dietz AB, Souan L, Knutson GJ, Bulur PA, Litzow MR, Vuk-Pavlovic S. 2004. Imatinib mesylate inhibits T-cell proliferation in vitro and delayed-type hypersensitivity in vivo. Blood 104:1094–1099. doi: 10.1182/blood-2003-12-4266. [DOI] [PubMed] [Google Scholar]
- 35.Oswald-Richter K, Grill SM, Shariat N, Leelawong M, Sundrud MS, Haas DW, Unutmaz D. 2004. HIV infection of naturally occurring and genetically reprogrammed human regulatory T-cells. PLoS Biol 2:e198. doi: 10.1371/journal.pbio.0020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno-Fernandez ME, Zapata W, Blackard JT, Franchini G, Chougnet CA. 2009. Human regulatory T cells are targets for human immunodeficiency virus (HIV) infection, and their susceptibility differs depending on the HIV type 1 strain. J Virol 83:12925–12933. doi: 10.1128/JVI.01352-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevenson M, Stanwick TL, Dempsey MP, Lamonica CA. 1990. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J 9:1551–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213–222. doi: 10.1016/0092-8674(90)90802-L. [DOI] [PubMed] [Google Scholar]
- 39.Li S, Folkvord JM, Rakasz EG, Abdelaal HM, Wagstaff RK, Kovacs KJ, Kim HO, Sawahata R, MaWhinney S, Masopust D, Connick E, Skinner PJ. 2016. Simian immunodeficiency virus-producing cells in follicles are partially suppressed by CD8+ cells in vivo. J Virol 90:11168–11180. doi: 10.1128/JVI.01332-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paiardini M. 2017. Immune-based interventions targeting inflammation and viral persistence, abstr 48. Abstr 24th Conf Retrovir Oppor Infect, Seattle, WA, 13 to 16 February 2017. [Google Scholar]
- 41.Banga R, Procopio FA, Noto A, Pollakis G, Cavassini M, Ohmiti K, Corpataux J-M, de Leval L, Pantaleo G, Perreau M. 2016. PD-1+ and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat Med 22:754–761. doi: 10.1038/nm.4113. [DOI] [PubMed] [Google Scholar]
- 42.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel M-R, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Folkvord JM, Anderson DM, Arya J, MaWhinney S, Connick E. 2003. Microanatomic relationships between CD8+ cells and HIV-1-producing cells in human lymphoid tissue in vivo. J Acquir Immune Defic Syndr 32:469–476. doi: 10.1097/00126334-200304150-00001. [DOI] [PubMed] [Google Scholar]
- 44.Meditz AL, Connick E, McCarter M. 2014. Safety of excisional inguinal lymph node biopsies performed for research purposes in HIV-1-infected women and men. Surg Infect (Larchmt) 15:399–403. doi: 10.1089/sur.2013.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meditz AL, Folkvord JM, Lyle NH, Searls K, Lie Y, Coakley E, McCarter M, MaWhinney S, Connick E. 2014. CCR5 expression is reduced in lymph nodes of HIV type 1-infected women, compared with men, but does not mediate sex-based differences in viral loads. J Infect Dis 209:922–930. doi: 10.1093/infdis/jit575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levy DN, Aldrovandi GM, Kutsch O, Shaw GM. 2004. Dynamics of HIV-1 recombination in its natural target cells. Proc Natl Acad Sci U S A 101:4204–4209. doi: 10.1073/pnas.0306764101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghosh SK, Fultz PN, Keddie E, Saag MS, Sharp PM, Hahn BH, Shaw GM. 1993. A molecular clone of HIV-1 tropic and cytopathic for human and chimpanzee lymphocytes. Virology 194:858–864. doi: 10.1006/viro.1993.1331. [DOI] [PubMed] [Google Scholar]
- 48.Gallo RC, Salahuddin SZ, Popovic M, Shearer GM, Kaplan M, BF Haynes, Palker TJ, Redfield R, Oleske J, Safai B, White G, Foster P, Markham PD. 1984. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science 224:500–503. doi: 10.1126/science.6200936. [DOI] [PubMed] [Google Scholar]
- 49.Shaw GM, Hahn BH, Arya SK, Groopman JE, Gallo RC, Wong-Stall F. 1984. Molecular characterization of human T-cell leukemia (lymphotropic) virus type III in the acquired immune deficiency syndrome. Science 226:1165–1171. doi: 10.1126/science.6095449. [DOI] [PubMed] [Google Scholar]
- 50.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, Hora B, Berg A, Cai F, Hopper J, Denny TN, Ding H, Ochsenbauer C, Kappes JC, Galimidi RP, West AP Jr, Bjorkman PJ, Wilen CB, Doms RW, O'Brien M, Bhardwaj N, Borrow P, Haynes BF, Muldoon M, Theiler JP, Korber B, Shaw GM, Hahn BH. 2013. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci U S A 110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cillo AR, Vagratian D, Bedison MA, Anderson EM, Kearney MF, Fyne E, Koontz D, Coffin JM, Piatak M, Mellors JW. 2014. Improved single-copy assays for quantification of persistent HIV-1 viremia in patients on suppressive antiretroviral therapy. J Clin Microbiol 52:3944–3951. doi: 10.1128/JCM.02060-14. [DOI] [PMC free article] [PubMed] [Google Scholar]