ABSTRACT
The UL112-113 gene is one of the few alternatively spliced genes of human cytomegalovirus (HCMV). It codes for four phosphoproteins, p34, p43, p50, and p84, all of which are expressed with early kinetics and accumulate at sites of viral DNA replication within the host cell nucleus. Although these proteins are known to play important, possibly essential, roles in the viral replication cycle, little is known about the contribution of individual UL112-113 protein products. Here we used splice site mutagenesis, intron deletion and substitution, and nonsense mutagenesis to prevent the individual expression of each UL112-113 protein isoform and to investigate the importance of each isoform for viral replication. We show that HCMV mutants lacking p34 or p50 expression replicated to high titers in human fibroblasts and endothelial cells, indicating that these proteins are nonessential for viral replication, while mutant viruses carrying a stop mutation within the p84 coding sequence were severely growth impaired. Viral replication could not be detected upon the inactivation of p43 expression, indicating that this UL112-113 protein is essential for viral replication. We also analyzed the ability of UL112-113 proteins to recruit other viral proteins to intranuclear prereplication compartments. While UL112-113 expression was sufficient to recruit the UL44-encoded viral DNA polymerase processivity factor, it was not sufficient for the recruitment of the viral UL84 and UL117 proteins. Remarkably, both the p43 and p84 isoforms were required for the efficient recruitment of pUL44, which is consistent with their critical role in the viral life cycle.
IMPORTANCE Human cytomegalovirus requires gene products from 11 genetic loci for the lytic replication of its genome. One of these loci, UL112-113, encodes four proteins with common N termini by alternative splicing. In this study, we inactivated the expression of each of the four UL112-113 proteins individually and determined their requirement for HCMV replication. We found that two of the UL112-113 gene products were dispensable for viral replication in human fibroblasts and endothelial cells. In contrast, viral replication was severely reduced or absent when one of the other two gene products was inactivated, indicating that they are of crucial importance for the viral replication cycle. We further showed that the latter two gene products are involved in the recruitment of pUL44, an essential cofactor of the viral DNA polymerase, to specific sites within the cell nucleus that are thought to serve as starting points for viral DNA replication.
KEYWORDS: human cytomegalovirus, RNA splicing, UL112, UL113, mutagenesis, replication factor
INTRODUCTION
Human cytomegalovirus (HCMV) (human herpesvirus 5) is an opportunistic pathogen that causes serious disease in immunocompromised individuals. HCMV infection can also be transmitted to the fetus during pregnancy and is the leading cause of congenital neurological damage (1).
HCMV has a large double-stranded DNA genome of 235 kb and codes for more than 200 proteins (1). The viral genome is linear and circularizes after entering the nucleus during infection. During lytic infection, DNA replication initiates from the replication origin (oriLyt) (2). oriLyt-dependent DNA replication requires six core replication proteins that are highly conserved among the Herpesviridae (3): DNA polymerase and its processivity factor (UL54 and UL44, respectively), single-stranded DNA-binding protein (UL57), and the heterotrimeric helicase-primase complex (UL70, UL105, and UL102). Moreover, factors expressed from five additional genetic loci were required for the replication of an oriLyt-containing plasmid in transient-transfection assays: immediate early 2 (IE2) (UL122), TRS1/IRS1, UL36-38, UL84, and UL112-113 (4, 5). These auxiliary replication proteins are not conserved in the subfamilies Alphaherpesvirinae and Gammaherpesvirinae, and their contribution to viral DNA replication remains incompletely defined. Some of these auxiliary factors, such as the TRS1/IRS1 and UL36-38 proteins, might promote viral DNA replication indirectly by maintaining essential cellular functions (6–10).
The UL112-113 region codes for four phosphoproteins (p34, p43, p50, and p84) from alternatively spliced transcripts, all of which share an N terminus encoded by exon 1 (11–13). These proteins can bind to DNA (14) and enhance the IE2-mediated transactivation of viral early promoters (15–17). Moreover, the UL112-113 proteins accumulate in viral prereplication foci and replication compartments within the nucleus (18–20) and interact with IE2 and the viral DNA polymerase processivity factor pUL44 (21–23). The crucial role of UL112-113 in viral DNA replication was confirmed by genetic studies demonstrating that their expression is required for efficient viral replication (19, 24, 25).
The alternatively spliced UL112-113 gene is conserved in primate and rodent cytomegaloviruses, and its four protein products are thought to have nonredundant functions. However, the specific functions of individual isoforms remain poorly understood, and their importance in the viral life cycle has not been determined. Mutagenesis procedures commonly used for the inactivation of herpesvirus genes, such as open reading frame (ORF) deletion or the introduction of a stop codon, are of limited use for the genetic dissection of an alternatively spliced gene; for example, a deletion within UL112-113 exon 1 affected all four protein products (23). In this study, we made use of a strategy recently developed for the genetic dissection of an alternatively spliced murine cytomegalovirus (MCMV) gene (26). We inactivated the expression of each of the four UL112-113 protein isoforms individually by a combination of splice site mutagenesis, intron deletion and substitution, and nonsense mutagenesis. We analyzed the necessity of each isoform for HCMV replication and found that the UL112-113 p34 and p50 isoforms are dispensable for viral replication in fibroblasts and endothelial cells, while in contrast, HCMV mutants lacking the p87 and p43 isoforms were severely growth defective or did not replicate at all. We further show that the p43 and p87 isoforms are also required for the efficient recruitment of the viral DNA polymerase processivity factor pUL44 to intranuclear prereplication foci, suggesting that the recruitment of pUL44 is of crucial importance for viral replication.
RESULTS
HCMV UL112-113 gene products can be inactivated individually.
The four UL112-113 protein isoforms share an identical N-terminal segment composed of 252 amino acids encoded by exon 1 (12, 13). The p34 isoform results from readthrough into intron 1. The p43 isoform is encoded by a transcript from which introns 1 and 2 have been removed by splicing, and the p50 isoform results from the removal of introns 1 and 2′. The p84-coding transcript lacks intron 1 but retains the second intron (Fig. 1A). Thus, the p43, p50, and p84 proteins share the amino acids encoded by exons 1 and 2 but have different C termini encoded in different reading frames.
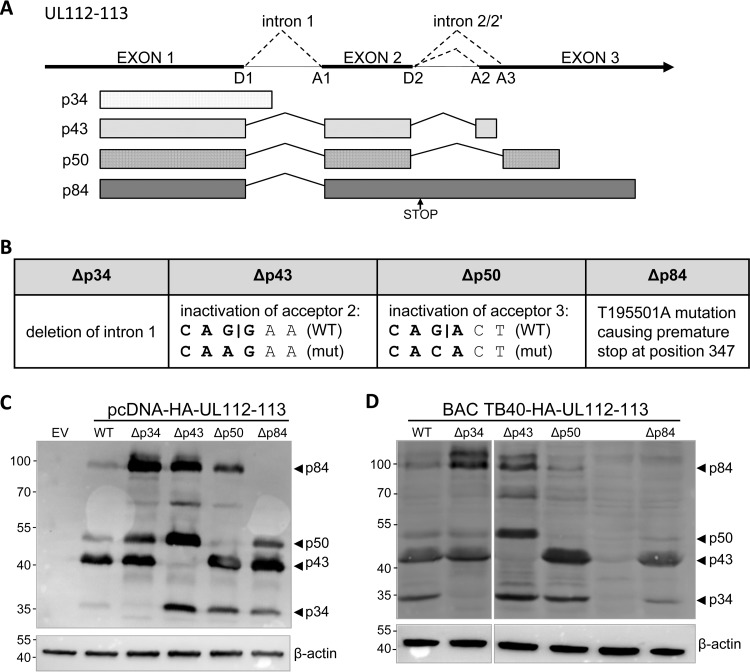
FIG 1.
Mutagenesis of the UL112-113 gene. (A) Schematic view of the HCMV UL112-113 locus and the four proteins expressed from this locus by alternative splicing. Splice donor and acceptor sites are marked with D and A, respectively. The stop codon introduced for the Δp84 mutant is marked with an arrow. (B) Mutations introduced to delete individual isoforms. (C) HEK 293A cells were transfected with the empty pcDNA3 vector (EV) or WT or mutant plasmids expressing HA-tagged UL112-113 proteins. The proteins were detected by using an anti-HA antibody; β-actin served as a loading control. (D) RPE-1 cells were transfected with BAC DNA of WT or mutant HCMV genomes. Protein expression was detected as described above.
We wanted to test whether we could mutate the UL112-113 coding region in a way so that individual protein isoforms would no longer be expressed while maintaining the expression of the other isoforms. As a first step, we PCR amplified and cloned the entire UL112-113 coding region from HCMV strain TB40/E in the plasmid expression vector pcDNA3. Because we were unable to obtain a UL112-113-specific antibody, we attached a hemagglutinin (HA) epitope tag sequence at the 5′ end of exon 1. The p34 isoform was then deleted by removing intron 1, resulting in a fusion of exons 1 and 2, while the Δp43 and Δp50 mutants were generated by splice site mutagenesis, where single nucleotide changes were introduced into splice acceptor sites A2 and A3, respectively (Fig. 1B). These changes destroyed the minimal splice acceptor consensus sequence but did not alter the amino acid sequence of the p84 protein. Since it was impossible to completely eliminate the large p84 isoform without affecting other isoforms, we introduced a single nucleotide change resulting in a stop codon shortly after the shared D2 splice donor site. This stop mutation should lead to a truncated p84 protein lacking the last 335 amino acids with a calculated molecular weight similar to that of p43.
Expression plasmids carrying the wild-type (WT) or mutant UL112-113 sequence were transfected into HEK 293A cells, and the expression of the UL112-113 protein isoforms was analyzed by immunoblotting using an anti-HA antibody (Fig. 1C). As expected, the mutations introduced led to a loss (or at least a massive reduction) of the respective isoforms. Unexpectedly, weak bands of sizes similar to those of the p43 and p50 isoforms were detected in cells transfected with the Δp43 and the Δp50 plasmids, respectively. However, these bands were not detected in all transfection experiments and appeared to migrate slightly faster than the expected bands, suggesting that they might represent degradation products of larger UL112-113 isoforms. In most cases, the inactivation of one isoform resulted in the stronger expression of the other isoforms. Moreover, an additional band of approximately 65 kDa was detected in cells expressing higher levels of p84 (Fig. 1C). This band probably represents a proteolytic degradation product of p84 (27).
As our mutagenesis strategy appeared to work with plasmid-expressed UL112-113, we decided to mutate UL112-113 within the HCMV genome. First, an HA tag followed by a 7-amino-acid tobacco etch virus (tev) protease cleavage site was introduced by site-directed mutagenesis at the 5′ end of UL112-113 into TB40/Ewt-GFP, a bacterial artificial chromosome (BAC) clone of HCMV strain TB40/E. The same isoform mutations described above were then introduced by site-directed mutagenesis. In order to ensure that the gene was mutated as intended, the mutant BACs were analyzed by restriction digestion and gel electrophoresis, and the entire UL112-113 region was sequenced (data not shown). To test whether the mutant BACs were capable of expressing the UL112-113 proteins, we transfected retinal pigment epithelial (RPE-1) cells with WT and mutant BACs using polyethylenimine 2000. RPE-1 cells can be transfected with high efficiency and express immediate early and early proteins upon transfection with HCMV BACs, even if the viral genome is replication incompetent (28). As shown in Fig. 1D, the transfected RPE-1 cells expressed the expected sets of UL112-113 protein isoforms. We noticed differences in the relative abundances of the different UL112-113 protein isoforms in plasmid-transfected (Fig. 1C) versus BAC-transfected (Fig. 1D) cells. The reasons for these differences are unknown but might be related to cell type (HEK 293A versus RPE-1 cells), expression level, and the influence of other viral proteins in BAC-transfected cells.
Differential requirement of individual UL112-113 isoforms for HCMV replication.
Next, we investigated the importance of individual UL112-113 proteins for viral replication. Mutant BACs were transfected into MRC-5 human embryonic lung fibroblasts to reconstitute mutant viruses. The Δp50 mutant replicated in MRC-5 cells and attained titers equivalent to those of the parental WT virus in multistep growth curves (Fig. 2A). In contrast, we could not reconstitute infectious virus from the Δp43 mutant BAC in several attempts. Individual green fluorescent protein (GFP)-expressing MRC-5 cells were observed after transfection; however, the mutant virus did not replicate and spread to neighboring cells, suggesting that the UL112-113 p43 isoform is essential for viral replication. To verify this, we constructed a revertant (Δp43Rev) by repairing the splice site mutation in the mutant BAC. The infectious virus reconstituted from the Δp43Rev BAC replicated with WT kinetics in fibroblasts (Fig. 2B). To further confirm the necessity of the isoform, we expressed p43 in MRC-5 cells by retroviral transduction (Fig. 2C). Transfection of these complementing cells with the Δp43 BAC resulted in the formation of foci and plaques (Fig. 2D). However, viral replication and spread were slow, and titers of cell-free virus were low, suggesting that complementation was not optimal. Cell-free virus harvested from the complementing cells infected WT MRC-5 cells but did not spread to neighboring cells (data not shown). Collectively, these results demonstrated that p43 is essential for HCMV replication and that the splice site mutation, which leads to the loss of p43 expression, was responsible for the inability of the Δp43 mutant to replicate.
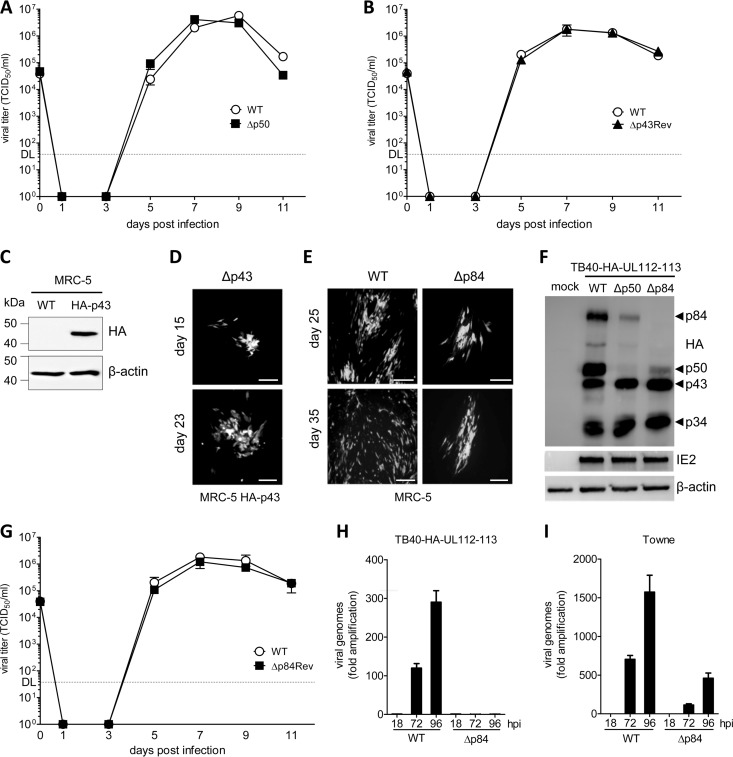
FIG 2.
Replication of HCMV UL112-113 mutants and revertants in fibroblasts. (A) Replication kinetics of TB40-HA-UL112-113 (WT) and the Δp50 mutant virus in MRC-5 cells infected at an MOI of 0.2 TCID50/cell. Means ± standard errors of the means of data from triplicates are shown. DL, detection limit. (B) Replication kinetics of the WT and the Δp43Rev mutant viruses. (C) MRC-5 cells were transduced with a retroviral vector encoding HA-tagged p43. The expression of HA-p43 in MRC-5 cells was analyzed by immunoblotting. (D) Fluorescence micrographs of p43-expressing MRC-5 cells transfected with the Δp43 BAC. Cells were not passaged between days 15 and 23 posttransfection. Small foci of Δp43-infected cells slowly expanded to larger foci and plaques. Bar, 100 μm. (E) Fluorescence micrographs of MRC-5 cells transfected with WT or Δp84 BACs. Cells were not passaged between days 25 and 35 posttransfection. On day 35, the WT virus had infected the entire monolayer, whereas the Δp84 mutants had produced only small plaques. Bar, 100 μm. (F) MRC-5 cells completely infected with the WT or mutant were lysed and analyzed by immunoblotting. UL112-113 proteins were detected by using an anti-HA antibody. IE2 and β-actin were detected as infection and loading controls, respectively. (G) Replication kinetics of TB40-HA-UL112-113 (WT) and the Δp43Rev mutant. (H) Viral genome amplification of TB40-HA-UL112-113 (WT) and Δp84 mutant viruses. MRC-5 cells were infected at an MOI of 0.02 TCID50/cell, and total DNA was extracted at different times postinfection. Viral genome copy numbers were quantified by qPCR and normalized to the cellular genome copy numbers. Data are shown as means ± standard errors of the means of data from triplicates. (I) Viral genome amplification of WT Towne and Δp84 mutant viruses was analyzed as described above.
Transfection of the Δp84 BAC yielded very small and slow-growing foci of GFP-expressing cells (Fig. 2E). These foci slowly expanded to plaques, and eventually, the entire cell monolayer was infected. This took several weeks, suggesting that the Δp84 mutant is replication competent but severely growth impaired. A lysate of a completely infected cell monolayer was used to analyze UL112-113 expression by immunoblotting. Lysates of MRC-5 cells infected with the parental WT virus or the Δp50 mutant were used for comparison. As expected, the Δp50 mutant did not express p50, and the Δp84 mutant did not express p84 (Fig. 2F). We could not detect a distinct truncated p84 protein species resulting from the introduced stop mutation. The predicted molecular mass of the truncated p84 protein differs from the predicted molecular mass of p43 by only 1 kDa; thus, the truncated p84 protein might comigrate with p43 in polyacrylamide gels. A revertant virus (Δp84Rev) constructed by site-directed mutagenesis replicated with WT kinetics (Fig. 2G), indicating that the stop mutation was responsible for the massively impaired replication of the Δp84 mutant. Since the authors of a previous publication reported that a p84 stop mutant of the HCMV Towne strain did not replicate in human fibroblasts (21, 22), we tested a second independent TB40/E Δp84 BAC clone as well as two Δp84 BAC clones constructed on the basis of the HCMV Towne strain. All Δp84 mutants showed a growth-attenuated phenotype compared to their parental virus. The Towne Δp84 mutants appeared to be somewhat less growth attenuated than the TB40/E Δp84 mutants. However, it should be noted that the laboratory-adapted Towne strain generally replicates faster than the low-passage-number clinical isolate TB40/E. These observations were confirmed when we quantified viral genome replication by quantitative real-time PCR (qPCR) (Fig. 2H and I). Thus, we concluded that the UL112-113 p84 isoform is not strictly essential but is required for the efficient replication of the viral genome.
UL112/113 intron 1 is essential for HCMV replication.
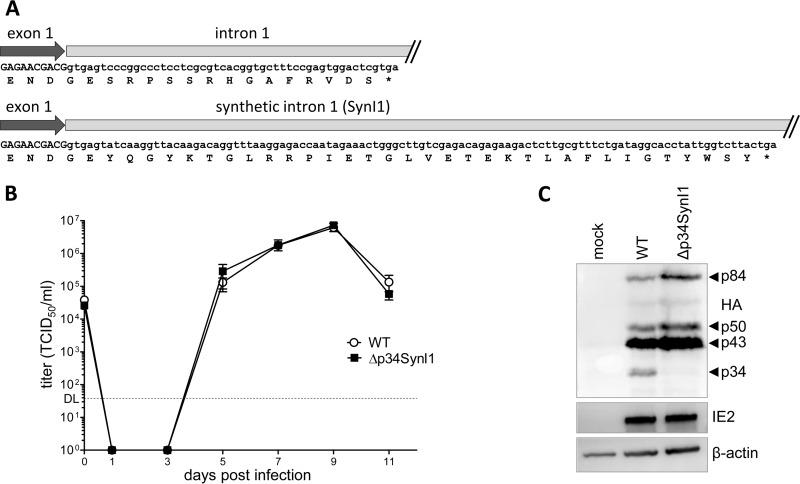
Interestingly, the TB40/E Δp34 mutant, which lacks intron 1, could not be reconstituted, even after several attempts with two independent clones of Δp34 were made. Two explanations seemed possible: either the p34 protein or the presence of intron 1 is essential for viral replication. To differentiate between these two possibilities, we introduced an unrelated synthetic intron (SynI1) into the Δp34 BAC at the position of the authentic intron 1. The protein encoded by exon 1 and SynI1 would differ from p34 in amino acid composition (Fig. 3A) and predicted molecular mass (approximately +2.5 kDa). Transfection of the Δp34SynI1 BAC mutant resulted in a virus that replicated with WT kinetics in MRC-5 fibroblasts (Fig. 3B). The Δp34SynI1 mutant expressed the UL112-113 p43, p50, and p84 isoforms but not p34 (Fig. 3C). We also did not detect a protein encoded by exon 1 and SynI1 with the predicted molecular weight, suggesting that this protein is unstable or synthesized in quantities below the detection threshold due to highly efficient splicing. Thus, the successful rescue of the Δp34 mutant by the insertion of an unrelated synthetic intron without restoring p34 expression suggests that p34 is nonessential, whereas the presence of intron 1 is essential.
FIG 3.
Substitution mutagenesis of UL112-113 intron 1. (A) Schematic of the transition from exon 1 to intron 1 or synthetic intron 1 (SynI1). Nucleotide and amino acid sequences of p34 and the predicted mutant protein are shown. (B) Replication kinetics of TB40-HA-UL112-113 (WT) and Δp34SynI1 mutant viruses in MRC-5 cells infected at an MOI of 0.2 TCID50/cell. Means ± standard errors of the means of data from triplicates are shown. DL, detection limit. (C) MRC-5 cells infected with the WT or mutant virus at an MOI of 3 TCID50/ml were lysed and analyzed by immunoblotting. UL112-113 proteins were detected by using an anti-HA antibody. IE2 and β-actin were detected as infection and loading controls, respectively.
UL112-113 p34 and p50 are dispensable for HCMV replication in endothelial cells.
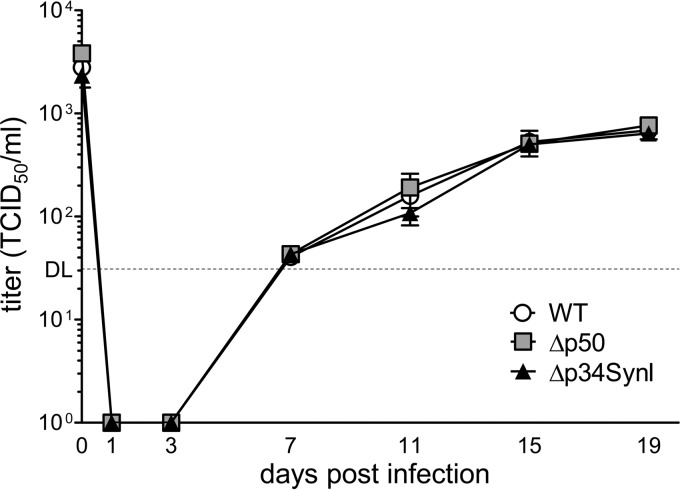
The HCMV Δp50 and Δp34SynI1 mutants replicated with WT kinetics in MRC-5 cells (Fig. 2A and 3B), suggesting that the UL112-113 p34 and p50 proteins are dispensable for viral replication in fibroblasts. To test whether these two proteins are required for efficient replication in another cell type, we analyzed viral replication kinetics in human umbilical vein endothelial cells (HUVECs). As shown in Fig. 4, the Δp34SynI1 and the Δp50 mutants replicated with the same kinetics as the parental WT virus, indicating that HCMV replicates efficiently in the absence of p34 or p50 in both fibroblasts and endothelial cells.
FIG 4.
Replication kinetics of HCMV UL112-113 mutants in endothelial cells. HUVECs were infected with TB40-HA-UL112-113 (WT), Δp50, and Δp34SynI1 viruses at an MOI of 0.03 TCID50/cell. Virus release into the supernatant was determined by titration. Means ± standard errors of the means of data from of triplicates are shown. DL, detection limit.
UL112-113 p43 and p84 are required to recruit pUL44 to intranuclear foci.
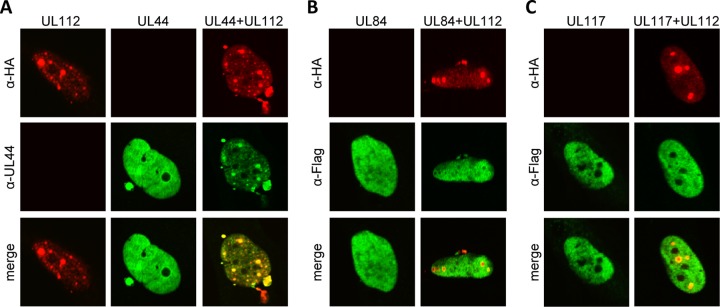
It is well established that the UL112-113 proteins are required and sufficient to recruit the DNA polymerase accessory factor pUL44 to intranuclear prereplication foci (21, 22). We wanted to test the requirement of individual UL112-113 protein isoforms for the recruitment of other viral proteins to prereplication compartments. In addition to pUL44, a known interactor of UL112-113 proteins, we also included pUL84, an auxiliary replication protein (5), and pUL117, a viral protein that inhibits cellular DNA synthesis (29), both of which are known to localize to nuclear replication compartments during viral infection (5, 30). We expressed pUL44, pUL84, and pUL117 in MRC-5 cells by transient transfection either alone or in combination with UL112-113 proteins. In the absence of UL112-113 proteins, the pUL44, pUL84, and pUL117 proteins were detected throughout the nucleus by immunofluorescence analysis. However, when UL112-113 was coexpressed, pUL44 accumulated in intranuclear foci (Fig. 5A). In contrast, pUL84 and pUL117 did not accumulate in UL112-113-positive foci but rather were excluded from the center of these foci (Fig. 5B and C). These results indicated that UL112-113 expression is sufficient to recruit pUL44, but not pUL84 or pUL117, to prereplication compartments.
FIG 5.
Intranuclear localization of HCMV UL44, UL84, and UL117 in the presence of UL112-113 proteins. MRC-5 cells were transfected with expression plasmids encoding the HA-tagged WT UL112-13 proteins and UL44 (A), FLAG-UL84 (B), or FLAG-UL117 (C). Cells were fixed 24 h after transfection and stained with antibodies against HA, FLAG, and UL44 for immunofluorescence analysis. Nuclei were counterstained with Draq5 (not shown).
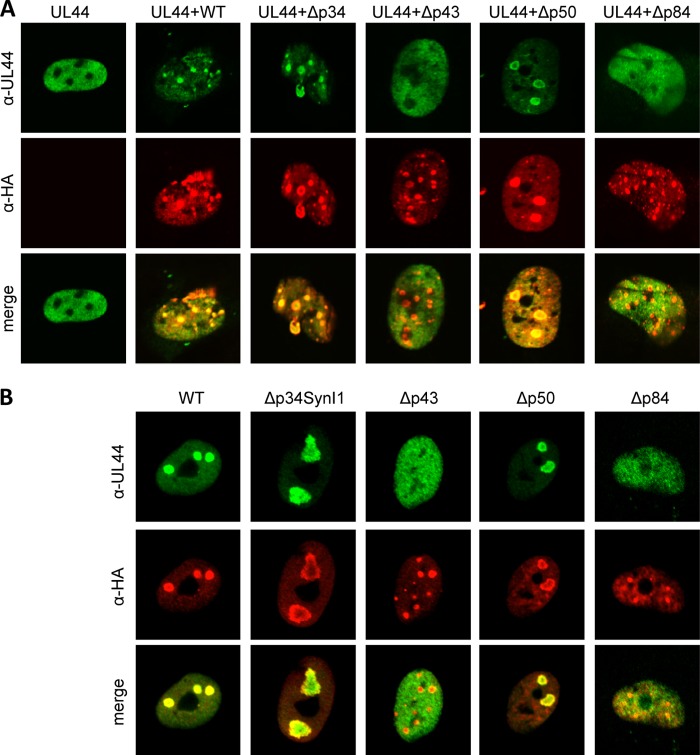
Next, we investigated which UL112-113 protein isoforms were required for pUL44 recruitment. To do this, we cotransfected MRC-5 cells with a UL44 expression plasmid and the mutant UL112-113 plasmids described above. As shown in Fig. 6A, pUL44 accumulated in UL112-113-positive foci even in the absence of UL112-113 p34 or p50 expression. However, in the absence of p43 or p84, pUL44 remained diffusely distributed throughout the nucleus (Fig. 6A). We also investigated pUL44 recruitment to intranuclear replication compartments in HCMV-infected cells. To this end, MRC-5 fibroblasts were infected with WT or mutant TB40-HA-UL112-113, and the intranuclear distribution of pUL44 and pUL112-113 was analyzed at 48 h postinfection (p.i.) by immunofluorescence (Fig. 6B). As expected, pUL44 accumulated in UL112-113-positive replication compartments in cells infected with the WT virus or the Δp34SynI1 or Δp50 mutant. In cells infected with the Δp43 mutant (harvested from complementing cells), pUL44 recruitment to replication compartments was not observed. pUL44 also remained diffusely distributed throughout the nucleus in the majority (>70%) of Δp84-infected cells (Fig. 6B), but pUL44 accumulation in UL112-113-positive foci was observed in the remaining cells (not shown), suggesting that pUL44 accumulation in replication compartments is less efficient or delayed in the absence of p84. Thus, the UL112-113 protein isoforms required for the efficient recruitment of the viral DNA polymerase processivity factor pUL44 are the same as those that are required for the efficient replication of the virus, suggesting that the recruitment of pUL44 to prereplication foci is a crucial function of the UL112-113 p43 and p84 proteins.
FIG 6.
Requirement of individual UL112-113 isoforms for UL44 recruitment to intranuclear foci. (A) MRC-5 cells were transfected with expression plasmids encoding UL44 and WT or mutant UL112-113 (HA tagged). Cells were fixed 24 h after transfection and stained with antibodies against UL44 and HA for immunofluorescence analysis. (B) MRC-5 cells were infected with TB40-HA-UL112-113 (WT) or UL112-113 mutant viruses. Cells were fixed at 48 h postinfection and stained as described above.
DISCUSSION
Recently, we have shown that individual products of an alternatively spliced herpesvirus gene can be inactivated by splice site mutagenesis, intron deletion and substitution, and the introduction of nonsense mutations (26). In that proof-of-concept study, we mutated the M112-113 gene of MCMV. Here we used a similar approach to selectively inactivate the expression of individual products of the HCMV UL112-113 gene and investigate their importance for viral replication. The UL112-113 gene is conserved among primate and rodent cytomegaloviruses; however, only its MCMV homolog has been characterized. The smallest products of the M112-113 and UL112-113 genes are 30% identical at the amino acid level, although stretches of higher similarity exist (31). Both genes give rise to four protein products encoded on alternatively spliced transcripts (12, 13, 31, 32). Although the arrangements of introns and exons are similar, there is one notable difference: UL112-113 has a common splice donor and two alternative splice acceptors for introns 2 and 2′, respectively, whereas M112-113 has two alternative splice donors and a shared acceptor (26). In each of the two viruses, the smallest and one of the medium-sized isoforms are dispensable for virus replication in fibroblasts. Only one protein isoform is strictly essential for replication in each virus; however, in HCMV, this is the other medium-sized isoform, p43 (Fig. 2), whereas in MCMV, it is the large p87 isoform (26). In contrast, the deletion of the M112-113 p38 isoform resulted in a moderately growth-attenuated virus, whereas the deletion of the large UL112-113 p84 isoform resulted in a severely growth-retarded virus (summarized in Table 1). The reason for this difference is not known, but it is tempting to speculate that the two viruses have distributed the various gene functions in a different fashion among the four protein isoforms. It is also noteworthy that in spite of their homology, the M112-113 and UL112-113 genes cannot substitute for one another: MCMV and HCMV substitution mutants carrying the homologous gene of the other virus were not viable (33). Interestingly, adaptive mutations in M112-113 were sufficient to allow MCMV replication in human RPE-1 cells, a cell type not permissive for the replication of WT MCMV (34).
TABLE 1.
Replication of HCMV UL112-113 and MCMV M112-113 mutants in fibroblasts
This study.
See reference 26.
−, no replication; +++, replication like the WT; ++, moderately attenuated replication; +, severely attenuated replication.
At present, it is unclear why introns 1 of UL112-113 and M112-113 are required for the replication of HCMV and MCMV, respectively. The fusion of exons 1 and 2 by the deletion of intron 1 abolished the expression of the smallest isoform in transfected cells (Fig. 1C and D), but infectious virus could not be reconstituted from mutant BACs, which could be interpreted as an indication of an essential role of the smallest isoform. However, the replacement of intron 1 by an unrelated synthetic intron completely restored HCMV and MCMV replication in fibroblasts, even though the synthetic intron did not restore the expression of p34 and p33, respectively. Moreover, predicted proteins derived from a transcript retaining the synthetic intron could not be detected in cells infected with either virus (26; this study). Nevertheless, the remote possibility that both HCMV and MCMV need a small isoform containing exon 1- but not exon 2-encoded sequences cannot be excluded. We favor the alternative hypothesis that intron 1 is required to ensure the balanced expression of the remaining isoforms.
HCMV oriLyt-dependent DNA replication requires six conserved core replication proteins and the gene products of five additional loci: IE2, TRS1/IRS1, UL36-38, UL84, and UL112-113 (4). How these auxiliary factors contribute to viral DNA replication is not well understood. IE2 is the major transactivator protein of HCMV and is required for the expression of HCMV early and late proteins. This argues for an indirect or regulatory role of IE2, although IE2 may have additional functions more directly involved in DNA replication. Regulatory functions have also been attributed to TRS1/IRS1 and proteins expressed form the UL36-38 locus. Proteins encoded by these two loci do not accumulate in viral replication compartments and are nonessential for HCMV replication, at least when deleted individually from the viral genome (24, 25). In contrast, the auxiliary replication factor UL84 is essential for the replication of several HCMV strains, such as AD169, Towne, and Merlin, but is surprisingly dispensable for the replication of HCMV strains TB40/E and TR (35–37). UL84 of strain AD169 was reported previously to be required for the formation of replication compartments in cotransfection assays (5), and pUL84 localized to the periphery of viral replication compartments in cells infected with HCMV AD169 (38). Its role in viral DNA replication remains incompletely understood but is thought to involve interactions with oriLyt, UL44, and IE2 (39, 40).
It is well established that the UL112-113 proteins localize to intranuclear replication compartments during HCMV infection. Moreover, they form similar intranuclear foci when expressed by plasmid transfection (20), suggesting that they might play a crucial role in the formation of replication compartments and the recruitment of other replication factors. Indeed, UL112-113 proteins interacted with pUL44 in yeast two-hybrid and glutathione S-transferase (GST) pulldown assays, and the coexpression of UL112-113 and UL44 was sufficient to recruit pUL44 to intranuclear prereplication foci (22). We show in this study that the UL112-113 p43 and p84 isoforms are required for the efficient recruitment of pUL44, whereas the p34 and p50 isoforms are dispensable (Fig. 6). The requirement for p43 and p84 correlates with the crucial role that the two proteins play in viral replication, suggesting that the recruitment of UL44 to prereplication compartments is a key function. However, this does not exclude additional important functions of p43 and p84. Interestingly, only the last 13 amino acid residues of p43 are not present in p84, suggesting that the unique C-terminal part of p43 is involved in its essential function.
UL112-113 expression was not sufficient for the recruitment of pUL84 to intranuclear foci (Fig. 5), even though UL112-113 interacts with pUL84 and IE2 in HCMV-infected cells (22). Since pUL84 interacts with pUL44 and IE2 (39), it seems likely that the recruitment of pUL84 to UL112-113-positive foci requires pUL44, IE2, or other factors such as the viral oriLyt. UL112-113 expression was also not sufficient to recruit pUL117 to intranuclear foci (Fig. 5). During infection, pUL117 accumulates in viral replication compartments but functions as an inhibitor of cellular DNA synthesis (29, 30) rather than a promoter of viral DNA replication. Thus, the reason for its association with viral replication compartments remains to be determined.
In summary, the results of this study show that the four UL112-113 protein isoforms have nonredundant functions. One of the isoforms, p43, is essential for viral replication, and another one, p84, is required for efficient replication. The same two isoforms are also required for the recruitment of pUL44 to intranuclear prereplication foci, suggesting that pUL44 recruitment is of crucial importance for viral DNA replication. The remaining two isoforms, p34 and p50, appear to be of less importance, as viral replication in fibroblasts and endothelial cells was not impaired when one of them was inactivated. However, the fact that alternative splicing and the expression of four proteins from this locus are conserved among cytomegaloviruses indicates that p34 and p50 are probably not entirely redundant. They might have more subtle roles as regulators of the other isoforms or fulfill relevant functions only under specific circumstances.
MATERIALS AND METHODS
Cells and viruses.
MRC-5 human embryo lung fibroblasts (ATCC CCL-171) and hTERT-RPE-1 human retinal pigment epithelial cells (ATCC CRL-4000) were obtained from the ATCC. HEK 293A human embryonic kidney cells (catalog number R70507) and primary HUVECs (catalog number C2519A) were purchased from Invitrogen and Lonza, respectively. All cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. HCMV strain TB40/Ewt-GFP (41), a GFP-expressing derivative of TB40-BAC4 (42), was kindly provided by Eain Murphy (Cleveland Clinic, OH). Towne-BAC, a GFP-expressing derivative of the HCMV Towne strain cloned as a BAC, was described previously (43). Viruses were propagated on MRC-5 cells. Titers were determined by the median tissue culture infective dose (TCID50) method. For replication kinetics, cells were infected in six-well dishes. After 4 h, cells were washed twice with phosphate-buffered saline (PBS), and fresh medium was added. Supernatants were harvested at different times postinfection for virus titration.
BAC mutagenesis.
An HA-tev tag sequence was inserted into the TB40/Ewt-GFP BAC at the 5′ end of UL112-113 by en passant mutagenesis (44). The TB40 HA-UL112-113 BAC was used as the parental strain for subsequent mutations introduced into the UL112-113 locus by en passant mutagenesis. Revertants were constructed by using the same method. Independent clones were picked from separate plates after the first step of en passant mutagenesis. The second step was done separately for each clone. For the construction of the HCMV SynI1 mutant, a synthetic intron of the pCI-Neo plasmid (Promega) was inserted into the Δp34 BAC at the position of the deleted intron 1 essentially as described previously (26). In order to verify the integrity of the UL112-113 locus within the mutant BACs, it was PCR amplified from either BAC or viral DNA and analyzed by DyeDeoxy sequencing (Microsynth). RPE-1 cells were transfected with purified HCMV BAC DNA using polyethylenimine 2000 (Sigma) according to a transfection protocol described previously (28) but without the use of adenoviral particles. To reconstitute infectious virus, 107 MRC-5 cells were suspended in Opti-MEM-I (Invitrogen) and combined with 5 μg BAC DNA and 2 μg pCGN71 (45). Transfection in a total volume of 500 μl was done by using a GenePulser Xcell electroporation device (Bio-Rad).
Plasmids and transfection.
The HA-tagged UL112-113 coding sequence was PCR amplified from the TB40 HA-UL112-113 BAC and inserted between the HindIII and EcoRV restriction sites of pcDNA3 (Invitrogen). Point mutations were introduced according to the GeneArt site-directed mutagenesis protocol (ThermoFisher). Deletion of the first intron was performed by PCR-driven overlap extension (46). The UL44 and UL84 ORFs were PCR amplified from TB40-BAC4 and inserted into pcDNA3 via the NotI and XhoI restriction sites. For UL84, an N-terminal FLAG tag was introduced through the PCR primer. UL117 with a 5′ 3×FLAG tag sequence was PCR amplified and inserted between the HindIII and EcoRI sites of pcDNA3. All plasmids were verified by sequencing, and protein expression was checked by immunoblotting using lysates of transiently transfected HEK 293A cells. HEK 293A cells were transfected with expression plasmids by using polyethylenimine 10000 (Sigma). MRC-5 cells were transfected by using Lipofectamine 2000 (ThermoFisher).
The cDNA sequence of HA-tagged p43 was PCR amplified from pDEST-SG5-HA-p43 (kindly provided by Jin-Hyun Ahn, Sungkyunkwan University, Suwon, South Korea) and inserted between the BamHI and EcoRI restriction sites of pRetro. The plasmid was transfected into the Phoenix-Ampho packaging cell line (47) to produce p43-expressing retroviral particles, which were used to transduce MRC-5 cells.
Relative quantification of HCMV genomes.
MRC-5 cells were infected with WT or mutant TB40/Ewt-GFP at a multiplicity of infection (MOI) of 0.02. After 2 h, cells were washed with PBS, and fresh medium was added. Genomic DNA was extracted from cells by using an InnuPREP DNA minikit (Analytik Jena). The obtained samples were subjected to qPCR to quantify viral genome replication. Primer set ACGCAAAGAGTTCCTCGTAC and TGAACATAACCACGTCCTCG and primer set ACGTCTTGGTGGACACTTTC and GTGTCCCTTCTTTCCCATGA were used to PCR amplify a piece of the viral UL36 gene for TB40/E and Towne, respectively. Primers GCTGAGGCCCAGTTCTAAAT and TTCAAGTCCCATCCCAGAAAG were used to amplify a piece of the cellular β-actin gene. qPCR was performed with a 7900HT Fast real-time PCR system (ThermoFisher), and numbers of genome copies per cell were calculated by using the ΔΔCT method (48).
Immunoblotting and immunofluorescence.
Monoclonal antibodies against HA (16B12 [Covance] or 3F10 [Roche]), FLAG (M2; Sigma), β-actin (AC-74; Sigma), and UL44 (CA006; Virusys) were purchased from commercial sources. The anti-HCMV IE2 antibody (3H9) was a generous gift from Thomas Shenk (Princeton University). For immunoblotting, transfected or infected cells were lysed in radioimmunoprecipitation assay (RIPA) buffer. Samples were separated by SDS-PAGE and transferred onto positively charged nitrocellulose membranes (Amersham). Proteins of interest were detected with protein-specific primary and horseradish peroxidase-coupled secondary antibodies (Dako) by enhanced chemiluminescence (Amersham). For immunofluorescence analysis, MRC-5 cells were grown on coverslips coated with 0.4% porcine skin gelatin (Sigma), transfected with expression plasmids or infected with HCMV, and fixed for 20 min with 4% paraformaldehyde in PBS. Cells were briefly incubated with 50 mM ammonium chloride, permeabilized with 0.3% Triton X-100, and blocked with 0.2% porcine skin gelatin. Cells were incubated with primary antibodies for 1 h at room temperature (RT), washed three times with PBS, and incubated with secondary antibodies coupled to Alexa Fluor 488, 555, or 647 (Life Technologies) for 1 h at RT. Nuclear DNA was stained with Draq5 (BioStatus) or 4′,6-diamidino-2-phenylindole (DAPI) (Sigma). Samples were washed, mounted onto slides with Aqua-Poly/Mount (Polysciences), and analyzed by confocal laser scanning microscopy using a Zeiss LSM510 Meta/FCS or Nikon C2+ microscope.
ACKNOWLEDGMENTS
We thank Eain Murphy for TB40/Ewt-GFP, Thomas Shenk for the anti-IE2 antibody, Jin-Hyun Ahn for the pDEST-SG5-HA-p43 plasmid, and Theo Potgieter for critical readings of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (BR1730/4-1 to W.B.). J.T. was supported by a scholarship from the China Scholarship Council. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Mocarski ES, Shenk T, Griffiths PD, Pass RF. 2013. Cytomegaloviruses, p 1960–2014. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Anders DG, Punturieri SM. 1991. Multicomponent origin of cytomegalovirus lytic-phase DNA replication. J Virol 65:931–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boehmer PE, Nimonkar AV. 2003. Herpes virus replication. IUBMB Life 55:13–22. doi: 10.1080/1521654031000070645. [DOI] [PubMed] [Google Scholar]
- 4.Pari GS, Anders DG. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J Virol 67:6979–6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarisky RT, Hayward GS. 1996. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J Virol 70:7398–7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall EE, Bierle CJ, Brune W, Geballe AP. 2009. Essential role for either TRS1 or IRS1 in human cytomegalovirus replication. J Virol 83:4112–4120. doi: 10.1128/JVI.02489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skaletskaya A, Bartle LM, Chittenden T, McCormick AL, Mocarski ES, Goldmacher VS. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc Natl Acad Sci U S A 98:7829–7834. doi: 10.1073/pnas.141108798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldmacher VS, Bartle LM, Skaletskaya A, Dionne CA, Kedersha NL, Vater CA, Han J, Lutz RJ, Watanabe S, McFarland ED, Kieff ED, Mocarski ES, Chittenden T. 1999. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc Natl Acad Sci U S A 96:12536–12541. doi: 10.1073/pnas.96.22.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terhune S, Torigoi E, Moorman N, Silva M, Qian Z, Shenk T, Yu D. 2007. Human cytomegalovirus UL38 protein blocks apoptosis. J Virol 81:3109–3123. doi: 10.1128/JVI.02124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moorman NJ, Cristea IM, Terhune SS, Rout MP, Chait BT, Shenk T. 2008. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3:253–262. doi: 10.1016/j.chom.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staprans SI, Spector DH. 1986. 2.2-kilobase class of early transcripts encoded by cell-related sequences in human cytomegalovirus strain AD169. J Virol 57:591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright DA, Staprans SI, Spector DH. 1988. Four phosphoproteins with common amino termini are encoded by human cytomegalovirus AD169. J Virol 62:331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright DA, Spector DH. 1989. Posttranscriptional regulation of a class of human cytomegalovirus phosphoproteins encoded by an early transcription unit. J Virol 63:3117–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwayama S, Yamamoto T, Furuya T, Kobayashi R, Ikuta K, Hirai K. 1994. Intracellular localization and DNA-binding activity of a class of viral early phosphoproteins in human fibroblasts infected with human cytomegalovirus (Towne strain). J Gen Virol 75(Part 12):3309–3318. doi: 10.1099/0022-1317-75-12-3309. [DOI] [PubMed] [Google Scholar]
- 15.Iskenderian AC, Huang L, Reilly A, Stenberg RM, Anders DG. 1996. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J Virol 70:383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerry JA, Priddy MA, Jervey TY, Kohler CP, Staley TL, Vanson CD, Jones TR, Iskenderian AC, Anders DG, Stenberg RM. 1996. Multiple regulatory events influence human cytomegalovirus DNA polymerase (UL54) expression during viral infection. J Virol 70:373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Yamamoto T, Ohtsubo K, Shirakata M, Hirai K. 1999. Major product pp43 of human cytomegalovirus U(L)112-113 gene is a transcriptional coactivator with two functionally distinct domains. Virology 260:89–97. doi: 10.1006/viro.1999.9800. [DOI] [PubMed] [Google Scholar]
- 18.Penfold ME, Mocarski ES. 1997. Formation of cytomegalovirus DNA replication compartments defined by localization of viral proteins and DNA synthesis. Virology 239:46–61. doi: 10.1006/viro.1997.8848. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto T, Suzuki S, Radsak K, Hirai K. 1998. The UL112/113 gene products of human cytomegalovirus which colocalize with viral DNA in infected cell nuclei are related to efficient viral DNA replication. Virus Res 56:107–114. doi: 10.1016/S0168-1702(98)00032-X. [DOI] [PubMed] [Google Scholar]
- 20.Ahn JH, Jang WJ, Hayward GS. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J Virol 73:10458–10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park MY, Kim YE, Seo MR, Lee JR, Lee CH, Ahn JH. 2006. Interactions among four proteins encoded by the human cytomegalovirus UL112-113 region regulate their intranuclear targeting and the recruitment of UL44 to prereplication foci. J Virol 80:2718–2727. doi: 10.1128/JVI.80.6.2718-2727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim YE, Ahn JH. 2010. Role of the specific interaction of UL112-113 p84 with UL44 DNA polymerase processivity factor in promoting DNA replication of human cytomegalovirus. J Virol 84:8409–8421. doi: 10.1128/JVI.00189-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YE, Park MY, Kang KJ, Han TH, Lee CH, Ahn JH. 2015. Requirement of the N-terminal residues of human cytomegalovirus UL112-113 proteins for viral growth and oriLyt-dependent DNA replication. J Microbiol 53:561–569. doi: 10.1007/s12275-015-5301-3. [DOI] [PubMed] [Google Scholar]
- 24.Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, Liu F. 2003. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A 100:14223–14228. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc Natl Acad Sci U S A 100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schommartz T, Loroch S, Alawi M, Grundhoff A, Sickmann A, Brune W. 2016. Functional dissection of an alternatively spliced herpesvirus gene by splice site mutagenesis. J Virol 90:4626–4636. doi: 10.1128/JVI.02987-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang SK, Jiang MJ, Lin SR, Chen MY, Wang HH, Duh CY. 2015. Calpains mediate the proteolytic modification of human cytomegalovirus UL112-113 proteins. J Gen Virol 96:1115–1126. doi: 10.1099/vir.0.000040. [DOI] [PubMed] [Google Scholar]
- 28.Elbasani E, Gabaev I, Steinbruck L, Messerle M, Borst EM. 2014. Analysis of essential viral gene functions after highly efficient adenofection of cells with cloned human cytomegalovirus genomes. Viruses 6:354–370. doi: 10.3390/v6010354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian Z, Leung-Pineda V, Xuan B, Piwnica-Worms H, Yu D. 2010. Human cytomegalovirus protein pUL117 targets the mini-chromosome maintenance complex and suppresses cellular DNA synthesis. PLoS Pathog 6:e1000814. doi: 10.1371/journal.ppat.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian Z, Xuan B, Hong TT, Yu D. 2008. The full-length protein encoded by human cytomegalovirus gene UL117 is required for the proper maturation of viral replication compartments. J Virol 82:3452–3465. doi: 10.1128/JVI.01964-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bühler B, Keil GM, Weiland F, Koszinowski UH. 1990. Characterization of the murine cytomegalovirus early transcription unit e1 that is induced by immediate-early proteins. J Virol 64:1907–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciocco-Schmitt GM, Karabekian Z, Godfrey EW, Stenberg RM, Campbell AE, Kerry JA. 2002. Identification and characterization of novel murine cytomegalovirus M112-113 (e1) gene products. Virology 294:199–208. doi: 10.1006/viro.2001.1311. [DOI] [PubMed] [Google Scholar]
- 33.Brost R. 2013. Characterization of early 1 proteins of human cytomegalovirus. MSc thesis. University of Hamburg, Hamburg, Germany. [Google Scholar]
- 34.Schumacher U, Handke W, Jurak I, Brune W. 2010. Mutations in the M112/M113-coding region facilitate murine cytomegalovirus replication in human cells. J Virol 84:7994–8006. doi: 10.1128/JVI.02624-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Y, Cei SA, Huete AR, Pari GS. 2004. Human cytomegalovirus UL84 insertion mutant defective for viral DNA synthesis and growth. J Virol 78:10360–10369. doi: 10.1128/JVI.78.19.10360-10369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spector DJ, Yetming K. 2010. UL84-independent replication of human cytomegalovirus strain TB40/E. Virology 407:171–177. doi: 10.1016/j.virol.2010.08.029. [DOI] [PubMed] [Google Scholar]
- 37.Spector DJ. 2015. UL84-independent replication of human cytomegalovirus strains conferred by a single codon change in UL122. Virology 476:345–354. doi: 10.1016/j.virol.2014.12.031. [DOI] [PubMed] [Google Scholar]
- 38.Bender BJ, Coen DM, Strang BL. 2014. Dynamic and nucleolin-dependent localization of human cytomegalovirus UL84 to the periphery of viral replication compartments and nucleoli. J Virol 88:11738–11747. doi: 10.1128/JVI.01889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Y, Colletti K, Pari GS. 2008. Identification of human cytomegalovirus UL84 virus- and cell-encoded binding partners by using proteomics analysis. J Virol 82:96–104. doi: 10.1128/JVI.01559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colletti KS, Smallenburg KE, Xu Y, Pari GS. 2007. Human cytomegalovirus UL84 interacts with an RNA stem-loop sequence found within the RNA/DNA hybrid region of oriLyt. J Virol 81:7077–7085. doi: 10.1128/JVI.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Connor CM, Murphy EA. 2012. A myeloid progenitor cell line capable of supporting human cytomegalovirus latency and reactivation, resulting in infectious progeny. J Virol 86:9854–9865. doi: 10.1128/JVI.01278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinzger C, Hahn G, Digel M, Katona R, Sampaio KL, Messerle M, Hengel H, Koszinowski U, Brune W, Adler B. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J Gen Virol 89:359–368. doi: 10.1099/vir.0.83286-0. [DOI] [PubMed] [Google Scholar]
- 43.Marchini A, Liu H, Zhu H. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J Virol 75:1870–1878. doi: 10.1128/JVI.75.4.1870-1878.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol Biol 634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 45.Baldick CJ Jr, Marchini A, Patterson CE, Shenk T. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J Virol 71:4400–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heckman KL, Pease LR. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 47.Kinsella TM, Nolan GP. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum Gene Ther 7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 48.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]