Abstract
We describe an approach to accelerate the search for competitive inhibitors for carbohydrate-recognition domains (CRDs). Genetically encoded fragment-based discovery (GE-FBD) uses selection of phage-displayed glycopeptides to dock a glycan fragment at the CRD and guide selection of synergistic peptide motifs adjacent to the CRD. Starting from concanavalin A (ConA), a mannose (Man)-binding protein, as a bait, we narrowed a library of 108 glycopeptides to 86 leads that share a consensus motif, Man-WYD. Validation of synthetic leads yielded Man-WYDLF that exhibited 40–50-fold enhancement in affinity over methyl α-D-mannopyranoside (MeMan). Lectin array suggested specificity: Man-WYD derivative bound only to 3 out of 17 proteins—ConA, LcH, and PSA—that bind to Man. An X-ray structure of ConA:Man-WYD proved that the trimannoside core and Man-WYD exhibit identical CRD docking, but their extra-CRD binding modes are significantly different. Still, they have comparable affinity and selectivity for various Man-binding proteins. The intriguing observation provides new insight into functional mimicry of carbohydrates by peptide ligands. GE-FBD may provide an alternative to rapidly search for competitive inhibitors for lectins.
Carbohydrate–protein interactions are central to a variety of normal and pathological processes, including inflammation, cell-to-cell adhesion, metastasis, and recognition of pathogens by the immune system. Although inhibition of some of these interactions would be therapeutically valuable, the development of effective inhibitors for lectins is challenging due to the synthetic complexity and usually low intrinsic affinity of the native oligosaccharides.1 The generally shallow landscape of many carbohydrate-binding sites and the lack of strong hydrophobic interactions with the ligand results in fast off-rates (koff).2 The interaction can be reinforced by using a non-carbohydrate synergistic motif that can fill the remaining space in the carbohydrate-binding site or occupy area on the protein surface adjacent to the principal binding site.
Partial3 or complete4 replacement of glycan with simple structural blocks has been successful in the discovery of lectin ligands for therapeutic targets, such as E-Selectin5 and MAG.6 This replacement often yields ligands with improved metabolic stability and pharmacokinetic properties for applications such as probing lectin function,7 design of therapeutics,2,8 and vaccines.8,9 Generally, these ligands are developed through rational design, requiring multistep synthesis and complex chemical manipulations.1b Genetic encoding of peptide libraries offers a 105-fold increase in the throughput and selection of active peptides that bind lectins.10 However, binding of peptides lacking a carbohydrate fragment to lectins is often non-specific, and these peptides often target non-carbohydrate-binding sites.11 Selections that use peptide solely as a linker, rather than as a recognition element, can yield peptides as a multivalent scaffold for Man9 and yield glyco-oligomers with avidity in the sub-nanomolar range.12 We pursued an approach in which we aim to identify peptides that can serve as a moiety that synergizes with glycans in binding to lectins (Figure 1), rather than acting as a neutral linker or standalone recognition element. Indeed, synthetic conjugates of peptides and carbohydrates are known to yield effective inhibitors of carbohydrate–protein interactions,13 but the throughput of synthesis of these ligands is limited. Here, we employ genetically encoded fragment-based discovery (GE-FBD) to identify peptide fragments (Figure 1) capable of forming a synergistic interaction together with a glycan moiety by taking advantage of the immense diversity of a genetically encoded peptide library.14
Figure 1.
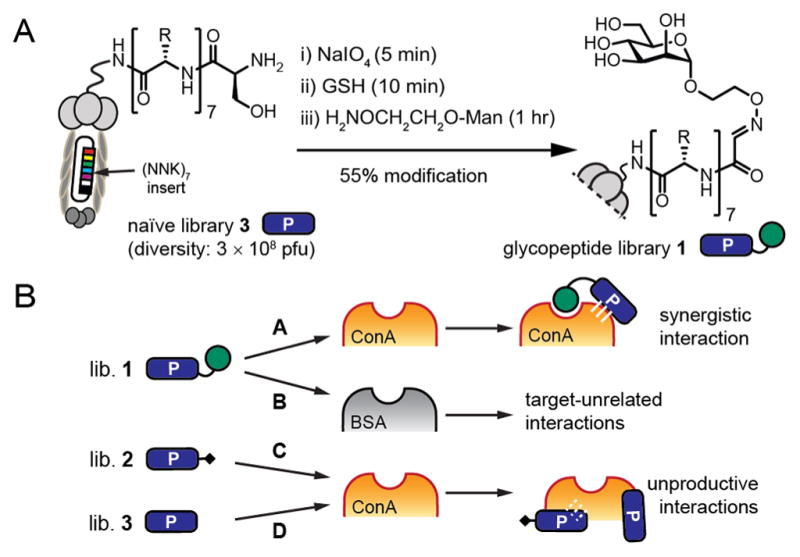
(A) Modification of phage-displayed peptide library (3) yielded Man-X7 (1). (B) Libraries 1 (Man-X7), 2 (methyl-X7), and 3 (Ser-X7) were panned against two targets (ConA or BSA) in parallel.
The GE-FBD procedure starts with the synthesis of a glycopeptide-presenting phage library with diversity of ~108, via periodate oxidation of a peptide library with fixed N-terminal serine residues, and subsequent chemical modification of the resulting bioorthogonal aldehyde functionality (Figure 1A).14a Next, we employed deep-sequencing15 and multiple panning controls to identify functional ligands, even after a single round of panning. In the proof-of-concept experiment with the model lectin, concanavalin A (ConA), we panned an α-mannopyranoside-oxime-terminated library (Man-X7, 1) against ConA and a control target (bovine serum albumin, BSA) (screens A and B, Figure 1B). In parallel, we panned control libraries terminated with O-methyl-oxime (methyl-X7, 2) and an unmodified naïve library (3) against ConA (screens C and D, Figure 1B). The four combinations of the target and the library—A–D—were incubated, rinsed, and processed on a 96-well plate, each in 3–6 replicates. After panning, recovery of phage DNA, and ion torrent sequencing, we acquired ~105 sequences per replicate (Figure S1). Student’s t-test identified a set of 231 sequences that were significantly (p ≤ 0.05) enriched by >5-fold in the screen against ConA but not against BSA (designated as set A/B in Figure 2A). In the A/B set, we identified a weak “consensus motif” (Figure 2C). By adding data from the control sets A/C and A/D, and calculating the intersection of (A/B)∩(A/C)∩(A/D), we reduced the number of hits from 231 to 86 (Figure 2B); these 86 hits shared a pronounced consensus motif: Man-[WYF]Y[SDEA] (Figure 2C).
Figure 2.
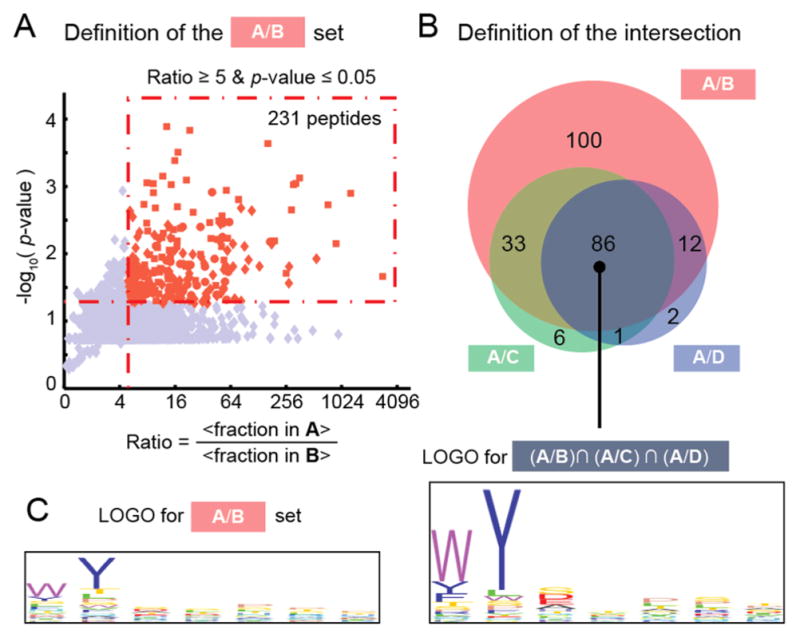
(A) A volcano plot defined sequences from library 1 that were significantly enriched in the ConA screen more than in the BSA screen. We abbreviate the set of these sequences as A/B and defined the sets A/C and A/D analogously. (B) 86 sequences belong to all three sets (“intersection”). Refer to Table S1 for the complete list of the sequences. (C) Consensus motifs defined by LOGO in set A/B and in the intersections with sets A/C and A/D.
Several ligands chosen from the pool of 86 hits, based on their correlation to the consensus motif (Figure 2C) and their rank of abundance in screen A (Table S1), were chemically synthesized and their activities evaluated by several methods. All ligands were able to inhibit the binding of ConA to the dextran-coated surface in a surface plasmon resonance (SPR) assay and exhibited lower IC50 values (11–34 μM) than methyl α-D-mannopyranoside (MeMan) (144 ± 7 μM, Figure S2). The monosaccharide was essential to the activity; the corresponding unmodified peptides did not inhibit the binding of ConA to dextran (Figure S3). Specific peptide sequences were required for synergistic binding, as the affinities of glycopeptides with randomly chosen sequences were indistinguishable from those of MeMan (Figure S4).
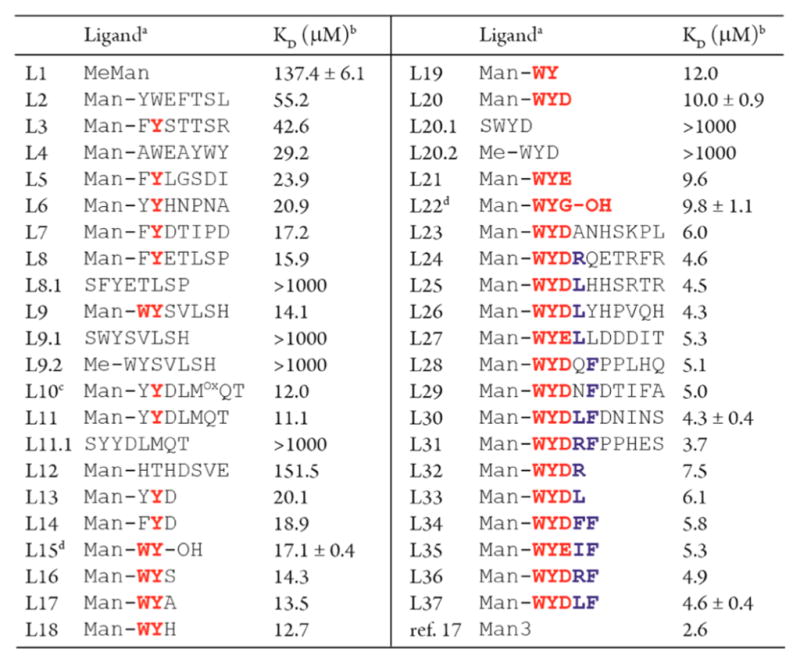
We validated the SPR findings using isothermal titration calorimetry (ITC) and found that all 10 selected Man-X7 conjugates (L2–L11), including the six SPR-validated hits, could bind to ConA with higher affinity than MeMan (Table 1). ITC also confirmed the lack of activity in false positive hits. For example, L12 ligand present in the A/B set but not the intersect (Figure 2B) showed no improvement in affinity over MeMan. By including C and D controls, we eliminate these false positives. The dissociation constants (KD) of the corresponding peptides lacking Man fragment were found to be >1 mM (L8.1, L9.1, L9.2, L11.1; Table 1 and Figure S5). Therefore, the synergistic sequences discovered in our screen would not be identified by screening unmodified peptide libraries.
Table 1.
KD Values of Synthetic Ligands with ConA
|
In general, red residues are essential for the strongest enhancement of affinity, while blue residues provide additional benefit.
KD was derived from ITC data (Figure S6). The errors are one standard deviation of the mean value measured by 2–5 independent experiments.
Mox is methionine sulfoxide.
With the exception of L15 and L22, all peptides were C-terminal amides (CONH2).
LOGO analysis16 suggested that residues 4–7 contribute minimally to the binding. We therefore truncated the heptapeptide to a tripeptide and, indeed, observed similar KD values for the corresponding Man-peptides (compare L7 to L14, L9 to L16, and L11 to L13 in Table 1). We then performed two more rounds of panning using a focused library of Man-WY[D/E]X7 and a panning–sequencing–analysis routine as described above. All of the hits bind to ConA at single-digit μM affinity (L23–L31, Table 1) and showed on average 5-fold lower KD than ligands from round one (L2–L11, Table 1). Truncation of the hits revealed that again the residues proximal to the glycan dominate the interactions. Man-WYDLFDNINS (L30) exhibited 33-fold lower KD over MeMan (4.3 vs 140 μM), and retained most of its potency when truncated to Man-WYDLF (L37). The KD of L37 (4.6 μM) was close to that of α-Man-(1→3)-[α-Man-(1→6)]-α-Man (Man3, KD = 2.6 μM).17
To determine whether Man-WYD is mimicking Man3, we obtained the crystal structure of its complex with ConA at 1.73 Å resolution (Figure 3A). The mannopyranosyl moiety of the synthetic ligand occupies the same binding site as the (1→6)-linked Man residue of the Man3 (Figure 3B) and displays the same H-bonding pattern (Figure S7). However, the peptide moiety, WYD, does not occupy the same shallow area as the remaining disaccharide portion of the Man3 but instead resides in a somewhat deeper cavity located next to Tyr12, which forms a wall between the two cavities on the protein surface at the tip of each arm of the ConA tetramer. Bound Man-WYD has more extensive van der Waals contact with the protein (contact area 662 Å, Figure S7) when compared to the Man3 (contact area 204 Å). Additionally, the complex revealed a ligand-induced fit: the conformation of the His205 residue has been changed to open a latent hydrophobic site, which is masked by this residue in both native ConA and Man3-ConA structures (Figure 3, asterisks). This site is now occupied by the Tyr side chain of Man-WYD. Hence, our screening identified a novel class of ligands that would be difficult to discover by structure-based design, which commonly employs docking of proposed ligands into a rigid protein.
Figure 3.
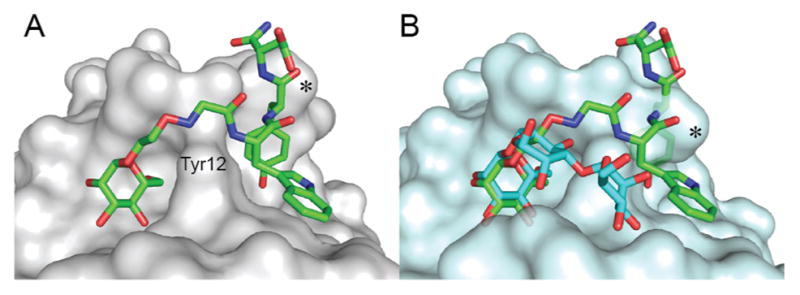
(A) Structure of Man-WYD (green) co-crystallized with ConA (PDB: 4CZS). (B) Superimposition of Man-WYD with the Man3-ConA complex (cyan, PDB: 1CVN), generated by aligning the protein chain A backbone atoms (RMSD = 0.32 Å). * indicates His205 residue.
Molecular dynamics (MD) simulations employing the GLYCAM/AMBER force field18 confirmed the stability of the complex (see MD analysis in Supporting Information) and permitted estimation of the contributions to affinity made by each residue in the protein and the ligand (Figure S8 and Table S2).19 Nearly 60% of the binding free energy is provided by H-bond/electrostatic interactions between the protein and the glycan, while hydrophobic interactions with the Trp and Tyr residues contribute the remainder. This observation is consistent with the view that H-bonds to the sugar provide specificity, whereas hydrophobic interactions enhance affinity.20
A linker of some kind is essential in the GE-FBD approach. Minor changes, such as replacing the oxime fragment in L20 with a hydrolytically stable linker of similar length (L20.5, Figure 4), results in only a 3-fold loss in binding affinity, possibly due to loss of the H-bond between the N atom of the oxime and the hydroxyl group of Tyr12, while shortening of the linker (L20.3) or incorporation of an allyl group (L20.4) completely abolishes the synergistic effect and returns the affinities to the level of the monosaccharide itself. Saturation transfer difference NMR spectroscopy (Figure S9) clearly demonstrates a substantial decrease of the interactions between WYD fragment and ConA in the ligand with a shorter linker, L20.3.
Figure 4.
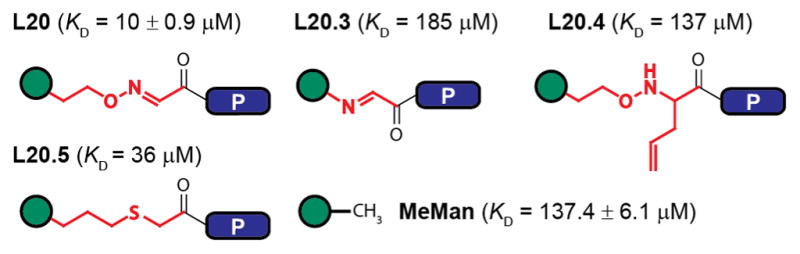
Changes in linker disrupt WYD–ConA interactions. Green circle is mannose, P = WYD.
To assess the selectivity of peptide binding to ConA over other lectins, we used a lectin microarray containing 85 lectins of varied specificities (Table S3).21 We synthesized a Man-WYK-OH probe (L38), which contained a fluorescent probe Cy3 attached to the side chain of lysine. Of the 85 lectins, 17 lectins exhibit selectivity for Man, yet only three show statistically significant binding to L38. Specifically, at 1.85 μM concentration, L38 bound strongly to ConA but only weakly to two other Man-binding lectins (Figure S10), Lens culinaris (LcH) and Pisum sativum (PSA), both of which have a high degree of sequence similarity in their binding site and a folded structure similar to that of ConA (Figure S11). ITC confirmed that the peptide fragment (WYDLF) provides a mere 2-fold enhancement of the binding of monosaccharide moiety to LcH and PSA, but a 35-fold benefit for ConA (Figure 5A, Figure S12).
Figure 5.
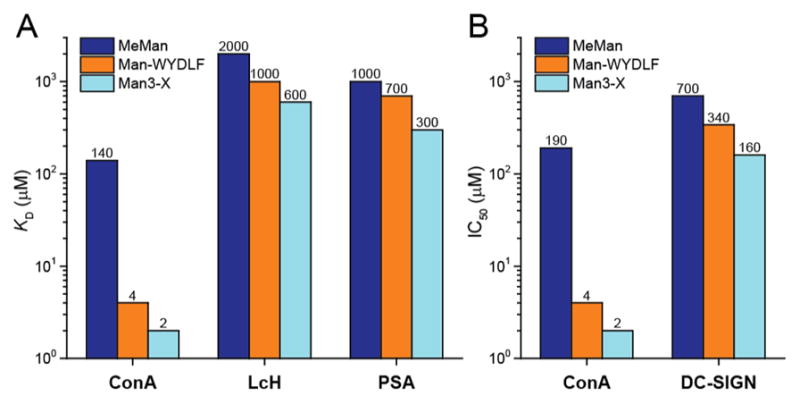
Binding of Man-WYDLF is specific to ConA, providing significantly less affinity for related Man-binding proteins as measured by (A) ITC and (B) inhibition studies.
We also tested binding of Man-peptides to DC-SIGN, an immune system lectin that recognizes the same Man3 as ConA and mediates the transmission of HIV during viral infection.22 ESI-MS assay23 measured an enhanced affinity for binding of DC-SIGN to L20 but not to the control ligand L20.3 (Figure S13). The measured KD of L20 (600 ± 7 μM) closely resembles the KD of Man3 (590 μM) binding to DC-SIGN.24 We demonstrated that Man-WYDLF is able to inhibit the binding of ConA to Man3-containing glycoprotein (horseradish peroxidase) with IC50 = 4 μM (Figure 5B, Figure S14). The inhibitory effect of Man-WYDLF is comparable to that of Man3-X (IC50 = 2 μM, X = 6-azidohexyl). Affinities measured by inhibition are consistent with the ITC experiments (Figure 5A). The binding specificity of Man-WYDLF to ConA, LcH, PSA, and DC-SIGN closely mimics that of Man3. The affinity increases in the order LcH < PSA < DC-SIGN < ConA (Figure 5). While the hydrophobic interactions added by WYDLF in glycopeptide are structurally distinct from the interactions provided by the dimannoside unit in Man3, both interactions add shape complementarity of ligand with the receptor and yield similar affinity and specificity for Man3 and Man-WYDLF.
In conclusion, we describe the discovery of ligands for challenging targets (lectins) from chemically modified phage libraries. This report expands the use of GE-FBD, which is known to work effectively with anchor fragments that have nanomolar affinity.25 Here we show that, despite their weak affinity, glycans can also serve as anchors in fragment-based discovery and yield synthetically accessible and potent glycopeptide ligands. These ligands bind competitively to the carbohydrate-recognition domain (CRD) with specificity akin to that of complex oligosaccharides and exhibit novel interactions with the CRD pocket. Therefore, GE-FBD may provide a general route to rapid enhancement of specific binding ligands for any therapeutic target starting from any known ligand or its essential binding fragment.
Supplementary Material
Acknowledgments
We acknowledge financial support from Alberta Glycomics Centre, Canada Foundation for Innovation (CFI). R.J.W. thanks the National Institutes of Health (R01 GM094919 (EUREKA) and P41 GM103390) and the Science Foundation of Ireland (08/IN.1/B2070) for support. S.N., E.L., and K.F.T. thank Alberta Innovates for the fellowship support. We thank Mark Miskolzie, Randy Whittal, Bela Reiz, and Wadim L. Matochko for help with analysis and characterization, and Prof. Todd Lowary for critical review of the paper. X-ray results were derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy (DOE), Office of Biological and Environmental Research (OBER), under contract DE-AC02-06CH11357. The OBER supported research at Oak Ridge National Laboratory’s Center for Structural Molecular Biology, using facilities supported by the Scientific User Facilities Division, Office of Basic Energy Sciences, U.S. DOE.
Footnotes
Notes
The authors declare no competing financial interest.
Figures S1–S18, Tables S1–S3, experimental details, and characterization data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Sears P, Wong CH. Angew Chem, Int Ed. 1999;38:2300. [PubMed] [Google Scholar]; (b) Bernardi A, Cheshev P. Chem—Eur J. 2008;14:7434. doi: 10.1002/chem.200800597. [DOI] [PubMed] [Google Scholar]
- 2.Ernst B, Magnani JL. Nat Rev Drug Discovery. 2009;8:661. doi: 10.1038/nrd2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Andreini M, Doknic D, Sutkeviciute I, Reina JJ, Duan J, Chabrol E, Thepaut M, Moroni E, Doro F, Belvisi L, Weiser J, Rojo J, Fieschi F, Bernardi A. Org Biomol Chem. 2011;9:5778. doi: 10.1039/c1ob05573a. [DOI] [PubMed] [Google Scholar]; (b) Hauck D, Joachim I, Frommeyer B, Varrot A, Philipp B, Möller HM, Imberty A, Exner TE, Titz A. ACS Chem Biol. 2013;8:1775. doi: 10.1021/cb400371r. [DOI] [PubMed] [Google Scholar]; (c) Kadam RU, Bergmann M, Garg D, Gabrieli G, Stocker A, Darbre T, Reymond JL. Chem—Eur J. 2013;19:17054. doi: 10.1002/chem.201302587. [DOI] [PubMed] [Google Scholar]
- 4.(a) Bächle D, Loers G, Guthöhrlein EW, Schachner M, Sewald N. Angew Chem, Int Ed. 2006;45:6582. doi: 10.1002/anie.200601579. [DOI] [PubMed] [Google Scholar]; (b) Borrok MJ, Kiessling LL. J Am Chem Soc. 2007;129:12780. doi: 10.1021/ja072944v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egger J, Weckerle C, Cutting B, Schwardt O, Rabbani S, Lemme K, Ernst B. J Am Chem Soc. 2013;135:9820. doi: 10.1021/ja4029582. [DOI] [PubMed] [Google Scholar]
- 6.Shelke SV, Cutting B, Jiang X, Koliwer-Brandl H, Strasser DS, Schwardt O, Kelm S, Ernst B. Angew Chem, Int Ed. 2010;49:5721. doi: 10.1002/anie.200907254. [DOI] [PubMed] [Google Scholar]
- 7.Kiessling LL, Splain RA. Annu Rev Biochem. 2010;79:619. doi: 10.1146/annurev.biochem.77.070606.100917. [DOI] [PubMed] [Google Scholar]
- 8.Johnson MA, Pinto BM. Aust J Chem. 2002;55:13. [Google Scholar]
- 9.Vyas NK, Vyas MN, Chervenak MC, Bundle DR, Pinto BM, Quiocho FA. Proc Natl Acad Sci USA. 2003;100:15023. doi: 10.1073/pnas.2431286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Oldenburg KR, Loganathan D, Goldstein IJ, Schultz PG, Gallop MA. Proc Natl Acad Sci USA. 1992;89:5393. doi: 10.1073/pnas.89.12.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Scott JK, Loganathan D, Easley RB, Gong X, Goldstein IJ. Proc Natl Acad Sci USA. 1992;89:5398. doi: 10.1073/pnas.89.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Yu L, Yu PS, Yee Yen Mui E, McKelvie JC, Pham TPT, Yap YW, Wong WQ, Wu J, Deng W, Orner BP. Biorg Med Chem. 2009;17:4825. doi: 10.1016/j.bmc.2009.03.054. [DOI] [PubMed] [Google Scholar]; (d) Matsubara T. J Nucleic Acids. 2012;2012:15. doi: 10.1155/2012/740982. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Fukuda MN. Glycobiology. 2012;22:318. doi: 10.1093/glycob/cwr140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jain D, Kaur K, Sundaravadivel B, Salunke DM. J Biol Chem. 2000;275:16098. doi: 10.1074/jbc.275.21.16098. [DOI] [PubMed] [Google Scholar]
- 12.Horiya S, Bailey JK, Temme JS, Guillen Schlippe YV, Krauss IJ. J Am Chem Soc. 2014;136:5407. doi: 10.1021/ja500678v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) St Hilaire PM, Lowary TL, Meldal M, Bock K. J Am Chem Soc. 1998;120:13312. [Google Scholar]; (b) Ying L, Liu R, Zhang J, Lam K, Lebrilla CB, Gervay-Hague J. J Comb Chem. 2005;7:372. doi: 10.1021/cc049836e. [DOI] [PubMed] [Google Scholar]
- 14.(a) Ng S, Jafari MR, Matochko WL, Derda R. ACS Chem Biol. 2012;7:1482. doi: 10.1021/cb300187t. [DOI] [PubMed] [Google Scholar]; (b) Arai K, Tsutsumi H, Mihara H. Bioorg Med Chem Lett. 2013;23:4940. doi: 10.1016/j.bmcl.2013.06.059. [DOI] [PubMed] [Google Scholar]
- 15.Matochko WL, Chu K, Jin B, Lee SW, Whitesides GM, Derda R. Methods. 2012;58:47. doi: 10.1016/j.ymeth.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Schneider TD, Stephens RM. Nucleic Acids Res. 1990;18:6097. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dam TK, Oscarson S, Brewer CF. J Biol Chem. 1998;273:32812. doi: 10.1074/jbc.273.49.32812. [DOI] [PubMed] [Google Scholar]
- 18.Kirschner KN, Yongye AB, Tschampel SM, González-Outeiriño J, Daniels CR, Foley BL, Woods RJ. J Comput Chem. 2008;29:622. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadirvelraj R, Grant OC, Goldstein IJ, Winter HC, Tateno H, Fadda E, Woods RJ. Glycobiology. 2011;21:973. doi: 10.1093/glycob/cwr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubinyi H, editor. Hydrogen bonding: The last mystery in drug design. Verlag Helvetica Chimica Acta; Zürich: 2001. [Google Scholar]
- 21.Pilobello KT, Agrawal P, Rouse R, Mahal LK. Curr Protoc Chem Biol. 2013;5:1. doi: 10.1002/9780470559277.ch120035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Kooyk Y, Geijtenbeek TBH. Nat Rev Immunol. 2003;3:697. doi: 10.1038/nri1182. [DOI] [PubMed] [Google Scholar]
- 23.El-Hawiet A, Kitova EN, Klassen JS. Biochemistry. 2012;51:4244. doi: 10.1021/bi300436x. [DOI] [PubMed] [Google Scholar]
- 24.Feinberg H, Mitchell DA, Drickamer K, Weis WI. Science. 2001;294:2163. doi: 10.1126/science.1066371. [DOI] [PubMed] [Google Scholar]
- 25.Meyer SC, Shomin CD, Gaj T, Ghosh I. J Am Chem Soc. 2007;129:13812. doi: 10.1021/ja076197d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


