Abstract
Objective
Coronary artery disease (CAD) in the general population is characterized by an increased frequency of particular susceptibility single nucleotide polymorphisms (SNPs). Since CAD is increased in rheumatoid arthritis (RA), we sought to determine whether the frequency of these SNPs are increased in RA patients with CAD, hypothesizing that RA could enhance CAD risk by acting through established genetic pathways predisposing to CAD.
Methods
CAD was assessed by coronary artery calcification (CAC) using computed-tomography in 561 patients with RA. 100 SNPs associated with CAD in the general population were genotyped or imputed and their relation to CAC established through multiple regression analysis for individual SNPs and a genetic risk score (GRS) representing their cumulative effect.
Results
91 CAD SNPs were genotyped successfully; 81 exhibited no association with CAC (Agatston units) or different CAC categorizations, either individually or collectively in the GRS. Only rs579459 (ABO) and rs17676451 (HAL) had a consistent positive association between genotype and CAC score, with a significant increase of the effect allele frequency in both homozygous or heterozygous genotype distributions. 5 were variably negatively associated. Furthermore, a positive association was found between DAS28 and CAC and after adjusting for traditional cardiovascular risk factors it was not modified by correcting for the CAD related SNPs GRS.
Conclusion
The increased risk for CAC in RA does not appear to operate primarily through established genetically regulated atherogenic mechanisms that are preponderant in the general population.
Introduction
Individuals with rheumatoid arthritis (RA) have an increased incidence of cardiovascular events, including myocardial infarctions and cardiac deaths (1). This excess in cardiovascular disease cannot be completely explained by traditional atherosclerosis risk factors, and it is thought to be due to an accelerated atherogenic process that appears to be related to one or more elements in the immune response underlying RA (2).
In the general population, 30-60% of the risk of developing coronary artery disease (CAD) is estimated to be genetic (3). Genome-wide association studies (GWAS) have identified and validated over 100 SNPs that are associated with CAD susceptibility. Moreover, the risk of CAD is proportional to the number of these SNPs inherited by an individual (4-21). These SNPs have been associated with increased risk of CAD through diverse mechanisms, including effects on lipid metabolism, plaque stability, and acute myocardial infarction. Still others act through unknown pathways, suggesting varying impacts of different alleles on the biology and pathophysiology of cardiovascular disease. In particular, some SNPs focus attention on novel and intriguing mechanisms contributing to CAD, such as those mapping to the 53 kb region of chromosome 9p21.3, an area without classic genes that encodes an antisense noncoding RNA in the INK4 locus, designated ANRIL, or CDKN2B-AS1 (22). Homozygosity for 9p21.3 risk alleles confers a population-attributable risk of 21% for CAD (23). Similarly, pathways involving glycotransferase activity encoded at the ABO locus are considered to foster development of myocardial infarction in the general population, acting on the stage after development of atherosclerosis (24).
Coronary artery calcification (CAC) detected by computed tomography is a noninvasive validated measure of subclinical CAD in the general population (25). Direct relationships have been established between CAC scores and histologic, intracoronary ultrasonic, and angiographic measures of coronary artery atherosclerosis and plaque burden on a vessel-by-vessel and segment-by-segment basis (26). CAC has been shown to predict incident myocardial infarction even after adjustment for other traditional cardiovascular risk factors (27). Importantly, analogous to CAD, the presence of CAC is also a heritable trait (28) and CAC is significantly higher in individuals with a parental history of CAD (29). In the general population, it is now recognized that many of SNPs predominantly exert their influence at different stages of the biologic events leading to the ultimate development of myocardial infarction, including atherosclerosis, CAC, and CAD (24), and the presence of CAC analogously has been linked to the SNPs associated with CAD across multiple genetic loci (5, 30).
In RA, both the prevalence of positive CAC scores and their magnitude are significantly increased compared with the general population (31). Elevated CAC scores in RA patients have also been shown to independently predict cardiovascular events (32).
Accordingly, we sought to determine whether in the subset of RA patients with CAC, the frequency of the SNPs associated with CAD in the general population would also be significantly increased over their frequency in the subset of RA patients without CAC. If this were found to be the case, it would imply that the effect of RA is to enhance the risk for CAD by acting through established genetic pathways that result in CAD in the general population, resulting in an increase of RA patients having these susceptibility genes in the group with CAC. Alternatively, if the frequencies of these alleles were not found to be significantly increased among RA patients with CAC, it would suggest that increased CAC in RA is a consequence of RA-associated immune mediated mechanisms that act independently of the genetically determined proatherosclerotic pathways operating in the general population.
Material and methods
Study participants and data collection
A total of 561 patients with RA from three different clinic-based cohorts (the ESCAPE-RA cohort from Johns Hopkins University, and two additional cohorts from the University of Pittsburgh and Vanderbilt University) were included in this study. All patients were men or women who were diagnosed by a rheumatologist and who fulfilled the American Rheumatism Association 1987 revised criteria for the classification of RA (33). The ESCAPE-RA (Evaluation of Subclinical Cardiovascular disease And Predictors of Events in Rheumatoid Arthritis) study has been described in detail previously (31) and was initiated to study the prevalence, progression, and risk factors for subclinical cardiovascular disease in RA. A total of 197 patients aged 45 to 80 years old with a disease duration >6 months, were recruited from the Johns Hopkins Arthritis Center. The Pittsburgh cohort consisted of 195 patients recruited from the University of Pittsburgh Medical Center Arthritis Network outpatient practices. For this cohort patients were only women, 16 years old or older, and disease duration was ≥ 2 years. In the Vanderbilt cohort, patients were a minimum age of 18 years old and there were two groups of patients: duration of disease of <5 years (early RA) or >10 years (established RA). In this cohort 169 patients were obtained from a registry of patients with early RA, were referred by local rheumatologists, or were recruited by advertisements. Details of the Pittsburgh and Vanderbilt cohorts have also been previously reported (34, 35). Each study was approved by the Institutional Review Board of the universities and hospitals of the three cohorts with all subjects providing written informed consent prior to enrollment.
Assessments and CAC quantification
Age, gender, race/ethnicity, current and past smoking were assessed from patient-self report. Ninety percent of patients in the three cohorts were of Caucasian ethnicity. Patients filled in a medication usage questionnaire and underwent examination to assess anthropometric measures and blood pressure. Medical records were reviewed to ascertain diagnosis and medication. Current and past use of glucocorticoids, biologic and non-biologic disease modifying antirheumatic drugs use was queried by detailed examiner-administered questionnaires. Disease activity was measured using the Disease Activity Score in 28 joints (DAS28) through erythrocyte sedimentation rate (ESR) or C-reactive protein (CRP), while the degree of disease disability was determined using the Health Assessment Questionnaire (HAQ). Metabolic syndrome was defined using the 2005 National Cholesterol Education Program Adult Treatment Panel III (ATP III) criteria.
Fasting plasma and serum samples were collected and frozen until analysis of circulating lipids, glucose, insulin, high sensitivity C-reactive protein and homocysteine. Rheumatoid factor was assessed by nephelometry with seropositivity defined at or above a level of 40 units. All subjects underwent cardiac computerized tomography using methodology described previously (36). CAC was quantitated using the Agatston method (37). In addition to the Agatston score as a continuous variable, CAC was also categorized as greater than 0 and greater than 10 Agatston units, and 75th and 90th CAC percentiles, adjusted for age, gender and race/ethnicity, were also defined as previously described (38).
Genotyping, imputation, and genetic risk score calculation
We identified SNPs of interest from genome-wide association studies published up to 2014 in which phenotypes studied were myocardial infarction, angiographically defined CAD, and CAC, which we refer to collectively as “CAD”, and where the association between a SNP and CAD exceeded a genome-wide association threshold (p<5×108). We identified 100 CAD related disease SNPs in the literature (4-21). Supplementary Table 1 shows the selected SNPs with details of their gene information, effect and non-effect alleles and published odd ratio (OR), or beta coefficient, association effect. RA patients were genotyped using the Immunochip. This is an Illumina Infinium genotyping chip, containing 196,524 polymorphisms (718 small insertion deletions, 195,806 SNPs) designed both to perform deep replication of major autoimmune and inflammatory diseases, and fine-mapping of established GWAS significant loci (39). Because some CAD SNPs in the GWAS studies were identified using alternative chips, such as the Metabochip (40) with a different selection of SNPs not present on the Immunochip, it was necessary to impute these un-typed SNPs using Impute2 software (41) after being ‘pre-phased’ using SHAPEIT (42). SNPs with low imputation quality, minor allele frequency <1% or Hardy–Weinberg equilibrium <0.001 were excluded. Of the 100 selected SNPs, 9 were neither able to be genotyped nor successfully imputed (rs17114036, rs6783981, rs1878406, rs273909, rs12205331, rs9268402, rs3184504, rs2895811, rs17228213). A multilocus genetic risk score (GRS) was constructed as previously described using Plink 1.07 software (43) for each individual by summing the number of CAD alleles (0/1/2) for each of the 86 SNPs weighted by their estimated effect sizes (OR) (rs6783981, rs2026458, rs10811647, rs17676451, rs3809346, rs8001186 were not include in the genetic risk calculation because OR information was not available). Missing genotype values in the GRS calculation were imputed with the cohort-specific averages of risk allele frequencies. Supplementary Table 1 shows SNP specific weights.
Statistical analysis
The hypothesis to be tested in this study design was that in the subset of RA patients with CAC, the frequency of the SNPs known, a priori from the literature, to be associated with CAD in the general population would be significantly increased in frequency in the subset of RA patients with CAC, versus their frequency in those RA patients without CAC. Accordingly the correction for multiple comparisons required in a study with a posteriori hypotheses is not appropriate in this study design. In terms of study power, on the basis of previously published findings (44) we assumed that smoking, hypertension and diabetes are associated with approximately 50% of the risk of an acute myocardial infarction. In a European cohort (45), a GRS created from three SNPs that have been credibly associated with CAC explained 2.4 % of the variation in CAC, and a 45-SNP GRS constructed from CAD-associated SNPs explained an additional 4 %. Proceeding with these conservative assumptions, in a multivariable regression model with a α level of 0.05 and a β level of 0.10, and assuming a coefficient of determination of 50% and that this would increase to 52% when adding GRS to the model, we estimated that we would need to enroll 425 subjects.
The distributions of all variables were examined. Means and standard deviations were calculated for all normally distributed continuous variables and medians and interquartile ranges were calculated for continuous variables that were not normally distributed. For categorical variables, counts and percentages were calculated. CAC scores were modeled as the natural log of the CAC score plus 1, in order to account for zero scores. In table 2 CAC Agatston units are expressed as median (interquartile range) and U Mann-Whitney test were used to test for the differences in CAC units between allele groups. In this sense, Table 2 describes the comparison of effect allele homozygous vs. heterozygous, and the comparison of the effect allele vs. the non-effect. Similarly, logistic regression analysis adjusted for traditional cardiovascular risk factors (age, sex, diabetes, hypertension, dyslipidemia, and smoking) was used to also study the relation of homozygous against heterozygous, or for the relation of affected allele against the non affected, with CAC expressed as higher than 0 and 10 units or higher than 70th and 90th percentiles. This association analysis in Table 2 was performed using SNPTEST that can account for some imputation uncertainty (46). Linear trends for the association of GRS and CAC were tested using linear regression with orthogonal polynomial contrasts. To assess the relationship of CAC and GRS with DAS28, three models were constructed, one adjusted for age and sex (model 1), model 2 adjusted for model 1 plus traditional cardiovascular risk factors (diabetes, hypertension, smoking and dyslipemia) and model 3 that was constructed by adding GRS to both model 1 and 2. All statistical calculations were performed using SPSS software, version 21 (IBM). In all tests, a two-tailed alpha of 0.05 was defined as the level of statistical significance.
Table 2. Multivariable regression and association analysis of cardiovascular disease related SNPs relation with coronary artery calcification.
| rs | Chr | Gene/ nearest gene |
Effect/ non effect allele |
Risk allele frequency |
Published OR (95%CI) |
Ref | Effect allele |
n | CAC (units) | OR (95%CI) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||||
| p | CAC > 0 | CAC > 10 | CAC > p75 | CAC > p90 | ||||||||||
| rs6604023 | 1 | CDC7 | C/G | 0.09 | 1.19 (1.13-1.26) | |||||||||
|
| ||||||||||||||
| cell division cycle 7 | 7,8 | CC | 3 | 6 (0-114) | 0.91 | 0.45 (0.21-0.99) | 0.58 (0.26-1.28) | 0.66 (0.31-1.43) | 0.79 (0.34-1.84) | |||||
| intragenic | CG | 24 | 0 (0-14) | |||||||||||
| GG | 131 | 4 (0-123) | ||||||||||||
|
| ||||||||||||||
| C | 27 | 0 (0-14) | 0.05 | 1.08 (0.42-2.79) | 1.11 (0.43-2.88) | 1.13 (0.44-2.89) | 1.21 (0.46-3.13) | |||||||
| G | 131 | 4 (0-123) | ||||||||||||
|
| ||||||||||||||
| rs4845625 | 1 | IL6R | T/C | 0.43 | 1.04 (1.02-1.07) | 6 | ||||||||
|
| ||||||||||||||
| interleukin 6 receptor | TT | 74 | 3 (0-104) | 0.96 | 1.09 (0.66-1.82) | 1.11 (0.66-1.86) | 1.25 (0.76-2.05) | 0.99 (0.57-1.73) | ||||||
| intron | TC | 215 | 1 (0-152) | |||||||||||
| CC | 134 | 2 (0-107) | ||||||||||||
|
| ||||||||||||||
| T | 289 | 1 (0-129) | 0.77 | 1.48 (0.83-2.61) | 1.44 (0.81-2.58) | 1.45 (0.83-2.52) | 1.59 (0.87-2.91) | |||||||
| C | 134 | 2 (0-107) | ||||||||||||
| rs17465637 | 1 | MIA3 | C/A | 0.69 | 1.20 (1.12-1.30) | 8,9,10 | ||||||||
|
| ||||||||||||||
| melanoma inhibitory activity family, member 3 | CC | 196 | 1 (0-116) | 0.68 | 1.14 (0.70-1.84) | 1.33 (0.81-2.17) | 1.01 (0.64-1.62) | 1.24 (0.73-2.10) | ||||||
| intron | AC | 193 | 3 (0-135) | |||||||||||
| AA | 36 | 0 (0-46) | ||||||||||||
|
| ||||||||||||||
| C | 389 | 2 (0-128) | 0.27 | 1.16 (0.56-2.37) | 1.41 (0.67-2.95) | 1.23 (0.60-2.52) | 1.34 (0.61-2.93) | |||||||
| A | 36 | 0 (0-46) | ||||||||||||
| rs11206510 | 1 | PCSK9 | T/C | 0.86 | 1.15 (1.10-1.21) | 6,8,10 | ||||||||
|
| ||||||||||||||
| proprotein convertase subtilisin/kexin type 9 | TT | 76 | 1 (0-123) | 0.97 | - | - | - | - | ||||||
| intragenic | CT | 26 | 1 (0-40) | |||||||||||
| CC | 1 | 202 | ||||||||||||
|
| ||||||||||||||
| T | 102 | 1 (0-102) | 0.37 | - | - | - | - | |||||||
| C | 1 | 202 | ||||||||||||
| rs602633 | 1 | PSRC1 | G/T | 0.72 | 1.13 | 6 | ||||||||
|
| ||||||||||||||
| proline/serine-rich coiled-coil 1 | GG | 113 | 8 (0-116) | 0.03 | 1.14 (0.52-2.50) | 1.17 (0.53-2.61) | 0.97 (0.44-2.11) | 1.04 (0.45-2.39) | ||||||
| intragenic | TG | 77 | 0 (0-26) | |||||||||||
| TT | 21 | 1 (0-335) | ||||||||||||
|
| ||||||||||||||
| G | 190 | 1 (0-114) | 0.85 | 1.69 (0.9-3.2) | 1.75 (0.9-3.38) | 1.60 (0.85-3.02) | 1.46 (0.72-2.93) | |||||||
| T | 21 | 1 (0-335) | ||||||||||||
| rs599839 | 1 | PSRC1 | A/G | 0.69 | 1.29 (1.18-1.40) | 6,9,10 | ||||||||
|
| ||||||||||||||
| proline/serine-rich coiled-coil 1 | AA | 96 | 11 (0-128) | 0.06 | 1.16 (0.53-2.53) | 1.23 (0.55-2.77) | 1.02 (0.47-2.22) | 1.08 (0.47-2.48) | ||||||
| 5′ exon 1 | GA | 78 | 0 (0-117) | |||||||||||
| GG | 21 | 1 (1-335) | ||||||||||||
|
| ||||||||||||||
| A | 174 | 1 (0-128) | 0.96 | 1.64 (0.86-3.13) | 1.85 (0.94-3.63) | 1.58 (0.84-3,00) | 1.30 (0.65-2.61) | |||||||
| G | 21 | 1 (1-335) | ||||||||||||
| rs17464857 | 1 | TAF1A | T/C | 0.84 | 1.02 | 6 | ||||||||
|
| ||||||||||||||
| TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa | TT | 297 | 3 (0-135) | 0.12 | 1.01 (0.41-2.48) | 1.10 (0.44-2.72) | 1.07 (0.44-2.63) | 1.22 (0.49-3.04) | ||||||
| intron 1 | TG | 118 | 0 (0-86) | |||||||||||
| GG | 9 | 5 (0-111) | ||||||||||||
|
| ||||||||||||||
| T | 415 | 2 (0-120) | 0.95 | 1.13 (0.67-1.89) | 1.24 (0.73-2.12) | 1.23 (0.74-2.05) | 1.28 (0.71-2.29) | |||||||
| G | 9 | 5 (0-111) | ||||||||||||
| rs646776 | 1 | CELSR2 | T/C | 0.84 | 1.14 (1-09-1.19) | 11 | ||||||||
|
| ||||||||||||||
| cadherin, EGF LAG seven-pass G-type receptor 2 | TT | 129 | 8 (0-122) | 0.02 | 1.25 (0.64-2.42) | 1.72 (0.85-3.48) | 1.65 (0.82-3.29) | 1.30 (0.63-2.71) | ||||||
| intragenic 3′ | CT | 53 | 0 (0-22) | |||||||||||
| CC | 4 | 0 (0-1) | ||||||||||||
|
| ||||||||||||||
| T | 182 | 1 (0-116) | 0.15 | 1.07 (0.43-2.63) | 1.16 (0.46-2.94) | 0.97 (0.39-2.42) | 1.14 (0.45-2.89) | |||||||
| C | 4 | 0 (0-1) | ||||||||||||
| rs6544713 | 2 | ABCG8 | T/C | 0.31 | 1.06 (1.04-1.09) | 6 | ||||||||
|
| ||||||||||||||
| ATP-binding cassette, sub-family G (WHITE), member 8 | TT | 37 | 0 (0-54) | 0.45 | 1.32 (0.81-2.14) | 1.46 (0.89-2.40) | 1.35 (0.84-2.16) | 1.14 (0.67-1.95) | ||||||
| intron | TC | 185 | 3 (0-119) | |||||||||||
| CC | 202 | 2 (0-128) | ||||||||||||
|
| ||||||||||||||
| T | 222 | 1 (10-116) | 0.70 | 0.98 (0.49-1.97) | 1.05 (0.51-2.15) | 0.88 (0.44-1.75) | 1.08 (0.52-2.23) | |||||||
| C | 202 | 2 (0-128) | ||||||||||||
| rs6725887 | 2 | WDR12 | C/T | 0.12 | 1.17 (1.11-1.23) | 6,8 | ||||||||
|
| ||||||||||||||
| WD repeat domain 12 | CC | 6 | 141 (40-269) | 0.15 | 1.17 (0.46-3.01) | 1.23 (0.48-3.17) | 1.03 (0.41-2.60) | 1.14 (0.45-2.92) | ||||||
| intron | CT | 86 | 0 (0-77) | |||||||||||
| TT | 333 | 2 (0-123) | ||||||||||||
| C | 92 | 0 (0-105) | 0.43 | 1.12 (0.64-1.96) | 1.20 (0.68-2.14) | 1.16 (0.67-2.00) | 1.17 (0.64-2.14) | |||||||
| T | 333 | 2 (0-123) | ||||||||||||
|
| ||||||||||||||
| rs515135 | 2 | APOB | C/T | 0.78 | 1.08 (1.05-1.11) | 6 | ||||||||
|
| ||||||||||||||
| apolipoprotein B | CC | 271 | 3 (0-155) | 0.22 | 0.96 (0.58-1.58) | 1.17 (0.70-1.95) | 1.07 (0.66-1.74) | 1.63 (0.92-2.89) | ||||||
| intragenic | CT | 122 | 0 (0-77) | |||||||||||
| TT | 32 | 2 (0-97) | ||||||||||||
|
| ||||||||||||||
| C | 393 | 1 (0-123) | 0.95 | 0.94 (0.44-1.98) | 1.09 (0.52-2.31) | 0.87 (0.42-1.81) | 1.04 (0.47-2.28) | |||||||
| T | 32 | 2 (0-97) | ||||||||||||
| rs2252641 | 2 | TEX41 | C/T | 0.13 | 1.06 | 6 | ||||||||
|
| ||||||||||||||
| testis expressed 41 | CC | 3 | 3 (0-55) | 0.95 | - | - | - | - | ||||||
| intron | TC | 19 | 1 (0-9) | |||||||||||
| TT | 73 | 4 (0-93) | ||||||||||||
|
| ||||||||||||||
| C | 22 | 1 (0-15) | 0.25 | - | - | - | - | |||||||
| T | 73 | 4 (0-93) | ||||||||||||
| rs1561198 | 2 | VAMP5 | A/G | 0.54 | 1.05 (1.03-1.07) | 6 | ||||||||
|
| ||||||||||||||
| vesicle-associated membrane protein | AA | 30 | 0 (0-77) | 0.31 | - | - | - | - | ||||||
| intragenic | AG | 37 | 8 (0-138) | |||||||||||
| GG | 22 | 28 (0-91) | ||||||||||||
|
| ||||||||||||||
| A | 67 | 3 (0-123) | 0.47 | - | - | - | - | |||||||
| G | 22 | 28 (0-91) | ||||||||||||
| rs2943634 | 2 | IRS1 | C/A | 0.65 | 1.08 (0.90-1.31) | 8,9 | ||||||||
|
| ||||||||||||||
| insulin receptor substrate 1 | CC | 180 | 4 (0-105) | 0.75 | 1.09 (0.67-1.79) | 1.13 (0.68-1.88) | 0.97 (0.60-1.56) | 1.36 (0.79-2.33) | ||||||
| intragenic | AC | 192 | 1 (0-117) | |||||||||||
| AA | 50 | 0 (0-269) | ||||||||||||
|
| ||||||||||||||
| C | 372 | 2 (0-114) | 0.66 | 1.13 (0.59-2.16) | 0.93 (0.48-1.80) | 0.88 (0.47-1.65) | 0.74 (0.38-1.47) | |||||||
| A | 50 | 0 (0-269) | ||||||||||||
| rs2123536 | 2 | WDR35 | T/C | 0.03 | 1.12 (1.08-1.16) | 12 | ||||||||
|
| ||||||||||||||
| WD repeat domain 35 | TT | 1 | 0 | 0.48 | - | - | - | - | ||||||
| intragenic | TC | 8 | 37 (0-81) | |||||||||||
| CC | 144 | 3 (0-129) | ||||||||||||
|
| ||||||||||||||
| T | 9 | 5 (0-68) | 0.86 | - | - | - | - | |||||||
| C | 144 | 3 (0-129) | ||||||||||||
| rs9818870 | 3 | MRAS | T/C | 0.15 | 1.07 | 6 | ||||||||
|
| ||||||||||||||
| muscle RAS oncogene homolog | TT | 13 | 1 (0-510) | 0.73 | 1.07 (0.62-1.84) | 1.20 (0.69-2.08) | 1.17 (0.70-1.98) | 1.19 (0.67-2.13) | ||||||
| 3′UTR | TC | 101 | 2 (0-123) | |||||||||||
| CC | 311 | 1 (0-116) | ||||||||||||
|
| ||||||||||||||
| T | 114 | 2 (0-123) | 0.85 | 0.95 (0.40-2.24) | 0.89 (0.37-2.10) | 1.09 (0.47-2.51) | 1.41 (0.60-3.34) | |||||||
| C | 311 | 1 (0-116) | ||||||||||||
| rs2306374 | 3 | MRAS | C/T | 0.15 | 1.15 (1.11–1.19) | 6,1 | ||||||||
|
| ||||||||||||||
| muscle RAS oncogene homolog | CC | 12 | 0 (0-455) | 0.85 | 0.93 (0.39-2.21) | 0.86 (0.36-2.07) | 0.97 (0.41-2.27) | 1.19 (0.49-2.87) | ||||||
| intron | TC | 103 | 3 (0-126) | |||||||||||
| TT | 310 | 2 (0-116) | ||||||||||||
|
| ||||||||||||||
| C | 115 | 2 (0-129) | 0.85 | 1.03 (0.60-1.76) | 1.24 (0.72-2.16) | 1.22 (0.73-2.05) | 1.17 (0.66-2.09) | |||||||
| T | 310 | 2 (0-116) | ||||||||||||
| rs1199338 | 3 | MRAS | C/A | 0.19 | 1.08 (1-02-1.13) | 11 | ||||||||
|
| ||||||||||||||
| muscle RAS oncogene homolog | CC | 7 | 0 (0-28) | 0.51 | - | - | - | - | ||||||
| intron | CA | 38 | 2 (0-95) | |||||||||||
| AA | 89 | 2 (0-109) | ||||||||||||
|
| ||||||||||||||
| C | 45 | 0 (0-68) | 0.64 | - | - | - | - | |||||||
| A | 89 | 2 (0-109) | ||||||||||||
| rs7692387 | 4 | GUCY1A3 | G/A | 0.89 | 1.06 (1.03-1.09) | 6 | ||||||||
|
| ||||||||||||||
| guanylatecyclase 1, soluble, alpha 3 | GG | 263 | 2 (0-128) | 0.51 | 1.31 (0.71-2.44) | 0.98 (0.52-1.87) | 0.94 (0.51-1.74) | 0.95 (0.48-1.87) | ||||||
| intron | AG | 63 | 0 (0-111) | |||||||||||
| AA | 5 | 8 (6-269) | ||||||||||||
|
| ||||||||||||||
| G | 326 | 1 (0-123) | 0.35 | 0.68 (0.28-1.67) | 0.75(0.30-1.87) | 0.81 (0.33-2.00) | 0.75 (0.30-1.86) | |||||||
| A | 5 | 8 (6-269) | ||||||||||||
| rs1842896 | 4 | GUCY1A3 | T/G | 0.94 | 1.14 (1.10-1.19) | 12 | ||||||||
|
| ||||||||||||||
| guanylatecyclase 1, soluble, alpha 3 | TT | 21 | 0 (0-205) | 0.51 | - | - | - | - | ||||||
| intragenic | GT | 3 | 5 (2-59) | |||||||||||
| GG | 0 | - | ||||||||||||
|
| ||||||||||||||
| T | 24 | 0 (0-90) | - | - | - | - | - | |||||||
| G | 0 | - | ||||||||||||
| rs11748327 | 5 | IRX1 | C/T | 0.79 | 1.25 | 13 | ||||||||
|
| ||||||||||||||
| iroquoishomeobox 1 | CC | 198 | 1 (0-144) | 0.58 | - | - | - | - | ||||||
| intragenic | TC | 100 | 1 (0-68) | |||||||||||
| TT | 17 | 1 (0-110) | ||||||||||||
|
| ||||||||||||||
| C | 298 | 0 (0-90) | 0.98 | - | - | - | - | |||||||
| T | 12 | 1 (0-110) | ||||||||||||
| rs17609940 | 6 | ANKS1A | G/C | 0.82 | 1.07 (1.05-1.10) | 6,1 | ||||||||
|
| ||||||||||||||
| ankyrin repeat and sterile alpha motif domain containing 1A | GG | 285 | 1 (0-121) | 0.50 | 0.96 (0.58-1.57) | 0.76 (0.46-1.27) | 0.83 (0.51-1.34) | 0.68 (0.39-1.17) | ||||||
| intron | CG | 123 | 6 (0-123) | |||||||||||
| CC | 17 | 0 (0-63) | ||||||||||||
|
| ||||||||||||||
| G | 408 | 2 (0-122) | 0.50 | 1.09 (0.47-2.53) | 0.94 (0.40-2.20) | 1.15 (0.50-2.65) | 0.88 (0.37-2.09) | |||||||
| C | 17 | 0 (0-63) | ||||||||||||
| rs3869109 | 6 | HCG27 | G/A | 0.59 | 1.14 | 14 | ||||||||
|
| ||||||||||||||
| Human leucocyte antigen C | GG | 145 | 7 (0-138) | 0.12 | 1.45 (0.88-2.38) | 1.59 (0.95-2.64) | 1.50 (0.93-2.42) | 1.35 (0.79-2.32) | ||||||
| intragenic | AG | 213 | 0 (0-116) | |||||||||||
| AA | 66 | 0 (0-111) | ||||||||||||
|
| ||||||||||||||
| G | 358 | 2 (0-123) | 0.49 | 1.40 (0.74-2.66) | 1.62 (0.84-3.15) | 1.27 (0.67-2.38) | 1.81 (0.88-3.76) | |||||||
| A | 66 | 0 (0-111) | ||||||||||||
| rs10947789 | 6 | KCNK5 | T/C | 0.95 | 1.06 (1.03-1.08) | 6 | ||||||||
|
| ||||||||||||||
| potassium channel, subfamily K, member 5 | TT | 96 | 5 (0-81) | 0.91 | - | - | - | - | ||||||
| intron | CT | 11 | 0 (0-320) | |||||||||||
| CC | 0 | - | ||||||||||||
|
| ||||||||||||||
| T | 107 | 5 (0-119) | - | - | - | - | - | |||||||
| C | 0 | - | ||||||||||||
| rs3798220 | 6 | LPA | C/T | 0.02 | 1.92 (1.48-2.49) | 6,10,15 | ||||||||
|
| ||||||||||||||
| lipoprotein A | CC | 0 | - | - | - | - | - | - | ||||||
| exon | TC | 18 | 1 (0-105) | |||||||||||
| TT | 407 | 2 (0-121) | ||||||||||||
|
| ||||||||||||||
| C | 18 | 1 (0-105) | 0.92 | 1.26 (0.58-2.70) | 0.88 (0.40-1.92) | 0.92 (0.43-1.96) | 0.94 (0.42-2.14) | |||||||
| T | 407 | 2 (0-121) | ||||||||||||
| rs10455872 | 6 | LPA | G/A | 0.05 | 1.70 (1.49-1.95) | 15 | ||||||||
|
| ||||||||||||||
| lipoprotein A | GG | 0 | - | - | - | - | - | - | ||||||
| intron | GA | 45 | 7 (0-226) | |||||||||||
| AA | 378 | 1 (0-114) | ||||||||||||
|
| ||||||||||||||
| G | 45 | 7 (0-226) | 0.18 | 1.06 (0.56-2.04) | 0.83 (0.44-1.60) | 0.84 (0.44-1.59) | 1.02 (0.51-2.06) | |||||||
| A | 378 | 1 (0-114) | ||||||||||||
| rs2048327 | 6 | SLC22A3 | G/A | 0.25 | 1.06 | 6 | ||||||||
|
| ||||||||||||||
| solute carrier family 22 (organic cationtransorter), member 3 | GG | 13 | 2 (0-279) | 0.47 | 1.13 (0.64-2.01) | 1.13 (0.63-2.03) | 0.94 (0.54-1.65) | 0.84 (0.44-1.60) | ||||||
| intron | GA | 117 | 3 (0-150) | |||||||||||
| AA | 154 | 0 (0-107) | ||||||||||||
|
| ||||||||||||||
| G | 130 | 0.49 | 1.27 (0.55-2.93) | 0.89 (0.38-2.09) | 0.95 (0.41-2.22) | 1.12 (0.47-2.67) | ||||||||
| A | 154 | 0 (0-107) | ||||||||||||
| rs6922269 | 6 | MTHFD1L | A/G | 0.28 | 1.23 (1.01-1.50) | 8,9 | ||||||||
|
| ||||||||||||||
| methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like | AA | 37 | 0 (0-123) | 0.68 | 1.17 (0.72-1.90) | 1.08 (0.66-1.77) | 0.87 (0.54-1.39) | 0.89 (0.52-1.52) | ||||||
| intron | AG | 167 | 3 (0-114) | |||||||||||
| GG | 221 | 1 (0-128) | ||||||||||||
|
| ||||||||||||||
| A | 204 | 2 (0-116) | 0.78 | 0.92 (0.45-1.88) | 0.86 (0.42-1.77) | 0.64 (0.31-1.31) | 0.88 (0.41-1.88) | |||||||
| G | 221 | 1 (0-128) | ||||||||||||
| rs9369640 | 6 | PHACTR1 | A/C | 0.27 | 1.09 | 6 | ||||||||
|
| ||||||||||||||
| phosphatase and actin regulator 1 | AA | 20 | 18 (0-281) | 0.13 | 0.61 (0.35-1.06) | 0.82 (0.47-1.43) | 0.74 (0.43-1.28) | 0.76 (0.41-1.41) | ||||||
| intron | AC | 117 | 0 (0-32) | |||||||||||
| CC | 159 | 5 (0-150) | ||||||||||||
|
| ||||||||||||||
| A | 137 | 0 (0-38) | 0.01 | 1.00 (0.42-2.35) | 0.81 (0.34-1.93) | 0.68 (0.28-1.62) | 0.78 (0.32-1.90) | |||||||
| C | 159 | 5 (0-150) | ||||||||||||
| rs12526453 | 6 | PHACTR1 | C/G | 0.30 | 1.13 (1.09–1.17) | 6,8,10 | ||||||||
|
| ||||||||||||||
| phosphatase and actin regulator 1 | CC | 39 | 3 (0-218) | 0.64 | 0.71 (0.44-1.15) | 0.96 (0.58-1.57) | 0.87 (0.54-1.39) | 0.79 (0.46-1.35) | ||||||
| intron | CG | 176 | 0 (0-64) | |||||||||||
| GG | 210 | 4 (0-179) | ||||||||||||
|
| ||||||||||||||
| C | 215 | 0 (0-70) | 0.03 | 0.93 (0.43-1.98) | 0.78 (0.36-1.70) | 0.73 (0.34-1.55) | 0.72 (0.32-1.64) | |||||||
| G | 210 | 4 (0-179) | ||||||||||||
| rs9349379 | 6 | PHACTR1 | A/G | 0.91 | 1.26 (1.14-1.14) | 7,16 | ||||||||
|
| ||||||||||||||
| phosphatase and actin regulator 1 | AA | 206 | 5 (0-191) | 0.10 | - | - | - | - | ||||||
| intron | GA | 41 | 0 (0-48) | |||||||||||
| GG | 2 | 109 (0-218) | ||||||||||||
|
| ||||||||||||||
| A | 247 | 4 (0-147) | 0.99 | 1.27 (0.64-2.51) | 0.94 (0.47-1.89) | 1.03 (0.52-2.03) | 1.01 (0.49-2.11) | |||||||
| G | 2 | 109 (0-218) | ||||||||||||
| rs1332844 | 6 | PHACTR1 | C/T | 0.74 | 1.11 (1.07-1.15) | 11 | ||||||||
|
| ||||||||||||||
| phosphatase and actin regulator 1 | CC | 155 | 5 (0-150) | 0.01 | 1.01 (0.43-2.38) | 1.24 (0.52-2.94) | 1.51 (0.64-3.59) | 1.32 (0.54-3.23) | ||||||
| intron | TC | 115 | 0 (0-31) | |||||||||||
| TT | 18 | 24 (0-343) | ||||||||||||
|
| ||||||||||||||
| C | 270 | 1 (0-95) | 0.08 | 1.70 (0.99-2.93) | 1.29 (0.74-2.25) | 1.47 (0.85-2.54) | 1.28 (0.69-2.35) | |||||||
| T | 18 | 24 (0-343) | ||||||||||||
| rs2026458 | 6 | PHACTR1 | T/C | 0.07 | 0.16 (0.03)* | 7 | ||||||||
|
| ||||||||||||||
| phosphatase and actin regulator 1 | TT | 1 | 218 | 0.44 | - | - | - | - | ||||||
| intron | TC | 17 | 29 (0-119) | |||||||||||
| CC | 113 | 3 (0-150) | ||||||||||||
|
| ||||||||||||||
| T | 18 | 30 (0-121) | 0.80 | - | - | - | - | |||||||
| C | 113 | 3 (0-150) | ||||||||||||
| rs4252120 | 6 | PLG | T/C | 0.95 | 1.06 (1.03-1.09) | 6 | ||||||||
|
| ||||||||||||||
| plasminogen | TT | 48 | 4 (0-173) | 0.76 | - | - | - | - | ||||||
| intron | TC | 5 | 3 (0-13) | |||||||||||
| CC | 0 | - | ||||||||||||
|
| ||||||||||||||
| T | 53 | 4 (0-166) | - | - | - | - | - | |||||||
| C | 0 | - | ||||||||||||
| rs12190287 | 6 | TCF21 | C/G | 0.35 | 1.08 (1.06-1.10) | 6, 10 | ||||||||
|
| ||||||||||||||
| transcription factor 21 | CC | 52 | 2 (0-116) | 0.83 | 0.75 (0.46-1.22) | 0.71 (0.43-1.18) | 0.77 (0.47-1.24) | 0.93 (0.54-1.61) | ||||||
| exon | CG | 192 | 1 (0-129) | |||||||||||
| GG | 181 | 2 (0-118) | ||||||||||||
|
| ||||||||||||||
| C | 244 | 1 (0-123) | 0.99 | 0.99 (0.52-1.91) | 0.76 (0.39-1.48) | 1.11 (0.59-2.11) | 0.96 (0.48-1.94) | |||||||
| G | 52 | 2 (0-118) | ||||||||||||
| rs6905288 | 6 | VEGFA | T/C | 0.58 | 1.23 | 14 | ||||||||
|
| ||||||||||||||
| vascular endothelial growth factor A | TT | 146 | 3 (0-158) | 0.33 | 0.83 (0.48-1.44) | 0.76 (0.44-1.34) | 0.89 (0.52-1.52) | 0.89 (0.49-1.62) | ||||||
| intragenic | TC | 199 | 1 (0-125) | |||||||||||
| CC | 80 | 2 (0-61) | ||||||||||||
|
| ||||||||||||||
| T | 345 | 1 (0-129) | 0.43 | 1.17 (0.71-1.94) | 1.08 (0.64-1.80) | 1.26 (0.78-2.05) | 1.08 (0.62-1.86) | |||||||
| C | 80 | 2 (0-61) | ||||||||||||
| rs12539895 | 7 | COG5 | A/C | 0.03 | 1.02 | 6 | ||||||||
|
| ||||||||||||||
| component of oligomericgolgi complex 5 | AA | 0 | - | - | - | - | - | - | ||||||
| intron | AC | 6 | 46 (1-160) | |||||||||||
| CC | 114 | 2 (0-116) | ||||||||||||
|
| ||||||||||||||
| A | 6 | 46 (1-160) | 0.66 | - | - | - | - | |||||||
| C | 120 | 2 (0-116) | ||||||||||||
| rs2023938 | 7 | HDAC9 | G/A | 0.09 | 1.07 (1.04-1.11) | 6 | ||||||||
|
| ||||||||||||||
| histone deacetylase 9 | GG | 7 | 0 (0-202) | 0.72 | 0.88 (0.49-1.59) | 1.11 (0.61-2.04) | 0.97 (0.54-1.72) | 1.15 (0.61-2.16) | ||||||
| exon | GA | 63 | 1 (0-128) | |||||||||||
| AA | 343 | 2 (0-121) | ||||||||||||
|
| ||||||||||||||
| G | 70 | 0 (0-128) | 0.84 | 0.80 (0.34-1.92) | 0.92 (0.38-2.22) | 0.97 (0.41-2.29) | 0.97 (0.40-2.35) | |||||||
| A | 343 | 2 (0-121) | ||||||||||||
| rs11556924 | 7 | ZC3HC1 | C/T | 0.91 | 1.09 (1.07-1.12) | 6, 10 | ||||||||
|
| ||||||||||||||
| zinc finger, C3HC-type containing 1 | CC | 30 | 0 (0-66) | 0.98 | - | - | - | - | ||||||
| exon | CT | 7 | 0 (0-28) | |||||||||||
| TT | 0 | - | ||||||||||||
|
| ||||||||||||||
| C | 37 | 0 (0-47) | - | - | - | - | - | |||||||
| T | 0 | - | ||||||||||||
| rs10953541 | 7 | BCAP29 | C/T | 0.97 | 1.08 (1.05–1.11) | 11 | ||||||||
|
| ||||||||||||||
| B-cell receptor-associated protein 29 | CC | 103 | 2 (0-129) | 0.55 | - | - | - | - | ||||||
| intron | CT | 6 | 1 (0-46) | |||||||||||
| TT | 0 | - | ||||||||||||
|
| ||||||||||||||
| C | 109 | 1 (0-121) | - | - | - | - | - | |||||||
| T | 0 | - | ||||||||||||
| rs264 | 8 | LPL | G/A | 0.95 | 1.05 (1.02-1.08) | 6 | ||||||||
|
| ||||||||||||||
| lipoprotein lipase | GG | 241 | 2 (0-116) | 0.83 | 1.11 (0.51-2.39) | 0.94 (0.44-2.02) | 1.02 (0.48-2.16) | 0.84 (0.38-1.87) | ||||||
| intron | AG | 26 | 1 (0-179) | |||||||||||
| AA | 0 | - | ||||||||||||
|
| ||||||||||||||
| G | 267 | 2 (0-117) | - | - | - | - | - | |||||||
| A | 0 | - | ||||||||||||
| rs2954029 | 8 | (TRIB1)LOC105375745 | A/T | 0.55 | 1.04 (1.02-1.06) | 6 | ||||||||
|
| ||||||||||||||
| (tribbles homolog 1) LOC105375745 | AA | 120 | 5 (0-128) | 0.20 | 0.72 (0.41-1.24) | 0.81 (0.47-1.42) | 0.76 (0.45-1.29) | 0.73 (0.41-1.30) | ||||||
| intron | TA | 223 | 0 (0-116) | |||||||||||
| TT | 78 | 4 (0-137) | ||||||||||||
|
| ||||||||||||||
| A | 343 | 1 (0-121) | 0.33 | 1.10 (0.65-1.86) | 0.88 (0.51-1.51) | 0.91 (0.54-1.52) | 1.02 (0.57-1.82) | |||||||
| T | 78 | 4 (0-137) | ||||||||||||
| rs579459 | 9 | ABO | C/T | 0.23 | 1.10 (1.07-1.13) | 6, 10 | ||||||||
|
| ||||||||||||||
| ABO blood group | CC | 23 | 122 (4-619) | 0.01 | 1.25 (0.59-2.64) | 1.46 (0.70-3.06) | 1.34 (0.66-2.71) | 1.34 (0.63-2.85) | ||||||
| intragenic | CT | 146 | 4 (0-129) | |||||||||||
| TT | 256 | 0 (0-110) | ||||||||||||
|
| ||||||||||||||
| C | 169 | 6 (0-186) | 0.03 | 1.32 (0.81-2.14) | 1.24 (0.75-2.03) | 1.00 (0.63-1.60) | 1.18 (0.70-2.01) | |||||||
| T | 256 | 0 (0-110) | ||||||||||||
| rs3217992 | 9 | CDKN2B-AS1 | A/G | 0.25 | 1.14 | 6 | ||||||||
|
| ||||||||||||||
| CDKN2B antisense RNA 1 | AA | 12 | 34 (0-290) | 0.38 | 1.05 (0.47-2.37) | 1.42 (0.63-3.19) | 1.30 (0.59-2.89) | 1.31 (0.56-3.07) | ||||||
| antisense RNA | AG | 84 | 2 (0-116) | |||||||||||
| GG | 121 | 0 (0-105) | ||||||||||||
|
| ||||||||||||||
| A | 96 | 3 (0-147) | 0.34 | 1.07 (0.58-1.95) | 1.16 (0.62-2.15) | 1.10 (0.6-2.01) | 1.06 (0.53-2.11) | |||||||
| G | 121 | 0 (0-105) | ||||||||||||
| rs1333049 | 9 | CDKN2B-AS1 | C/G | 0.55 | 1.28 (1.07.1.53) | 7,9 | ||||||||
|
| ||||||||||||||
| CDKN2B antisense RNA 1 | CC | 123 | 1 (0-116) | 0.63 | 0.95 (0.54-1.68) | 0.95 (0.53-1.72) | 1.14 (0.64-2.01) | 0.77 (0.42-1.43) | ||||||
| antisense RNA | CG | 215 | 3 (0-149) | |||||||||||
| GG | 82 | 0 (0-81) | ||||||||||||
|
| ||||||||||||||
| C | 338 | 2 (0-128) | 0.25 | 0.96 (0.57-1.63) | 0.89 (0.52-1.54) | 0.82 (0.49-1.39) | 0.99 (0.56-1.77) | |||||||
| G | 82 | 0 (0-81) | ||||||||||||
| rs10757274 | 9 | CDKN2B-AS1 | G/A | 0.46 | 1.29 (1.06-1.58) | 17 | ||||||||
|
| ||||||||||||||
| CDKN2B antisense RNA 1 | GG | 94 | 0 (0-95) | 0.38 | 0.96 (0.56-1.65) | 1.07 (0.61-1.87) | 1.06 (0.62-1.81) | 1.24 (0.69-2.23) | ||||||
| antisense RNA | GA | 203 | 3 (0-175) | |||||||||||
| AA | 128 | 0 (0-111) | ||||||||||||
|
| ||||||||||||||
| G | 297 | 3 (0-128) | 0.39 | 1.33 (0.79-2.24) | 1.38 (0.81-2.37) | 1.50 (0.89-2.53) | 1.18 (0.66-2.11) | |||||||
| A | 128 | 0 (0-111) | ||||||||||||
| rs1333040 | 9 | CDKN2B-AS1 | C/T | 0.42 | 1.13 | 6 | ||||||||
|
| ||||||||||||||
| CDKN2B antisense RNA 1 | CC | 76 | 0 (0-118) | 0.65 | 0.62 (0.34-1.13) | 0.79 (0.43-1.46) | 0.60 (0.33-1.10) | 0.95 (0.49-1.81) | ||||||
| antisense RNA | CT | 206 | 3 (0-128) | |||||||||||
| TT | 143 | 1 (0-114) | ||||||||||||
|
| ||||||||||||||
| C | 282 | 2 (0-124) | 0.75 | 0.71 (0.43-1.18) | 0.75 (0.45-1.25) | 0.75 (0.46-1.22) | 0.76 (0.44-1.32) | |||||||
| T | 143 | 1 (0-114) | ||||||||||||
| rs10811647 | 9 | CDKN2B-AS1 | C/G | 0.36 | -0.212 (0.031)* | 7 | ||||||||
|
| ||||||||||||||
| CDKN2B antisense RNA 1 | CC | 36 | 0 (0-95) | 0.48 | 1.17 (0.59-2.33) | 0.96 (0.48-1.92) | 0.94 (0.48-1.86) | 0.97 (0.45-2.11) | ||||||
| antisense RNA | CG | 115 | 3 (0-129) | |||||||||||
| GG | 108 | 0 (0-110) | ||||||||||||
|
| ||||||||||||||
| C | 151 | 2 (0-125) | 0.57 | 1.25 (0.54-1.63) | 1.23 (0.51-1.67) | 1.01 (0.41-1.35) | 1.11 (0.43-1.65) | |||||||
| G | 108 | 0 (0-110) | ||||||||||||
| rs2891168 | 9 | CDKN2B-AS1 | G/A | 0.46 | 1.29 (1.20-1.38) | 18 | ||||||||
|
| ||||||||||||||
| CDKN2B antisense RNA 1 | GG | 90 | 0 (0-68) | 0.27 | 0.95 (0.55-1.64) | 1.03 (0.58-1.81) | 1.02 (0.59-1.76) | 1.23 (0.68-2.25) | ||||||
| antisense RNA | GA | 199 | 0 (0-147) | |||||||||||
| AA | 125 | 0 (0-113) | ||||||||||||
|
| ||||||||||||||
| G | 289 | 2 (0-121) | 0.52 | 1.32 (0.77-2.24) | 1.30 (0.76-2.24) | 1.46 (0.86-2.48) | 1.10 (0.61-1.98) | |||||||
| A | 125 | 0 (0-113) | ||||||||||||
| rs4977574 | 9 | CDKN2B-AS1 | G/A | 0.45 | 1.29 (1.23-1.36) | 6,8,10 | ||||||||
|
| ||||||||||||||
| CDKN2B antisense RNA 1 | GG | 90 | 0 (0-68) | 0.24 | 0.94 (0.54-1.63) | 1.02 (0.58-1.80) | 1.01 (0.59-1.74) | 1.23 (0.67-2.23) | ||||||
| antisense RNA | GA | 201 | 3 (0-175) | |||||||||||
| AA | 128 | 0 (0-111) | ||||||||||||
|
| ||||||||||||||
| G | 291 | 2 (0-128) | 0.46 | 1.31 (0.78-2.21) | 1.35 (0.79-2.32) | 1.46 (0.86-2.46) | 1.16 (0.64-2.07) | |||||||
| A | 128 | 0 (0-111) | ||||||||||||
| rs10757278 | 9 | CDKN2B-AS1 | G/A | 0.46 | 1.27 (1.19-1.36) | 17 | ||||||||
|
| ||||||||||||||
| CDKN2B antisense RNA 1 | GG | 87 | 0 (0-68) | 0.22 | 1.11 (0.63-1.95) | 1.12 (0.62-2.02) | 0.96 (0.55-1.68) | 1.24 (0.67-2.27) | ||||||
| antisense RNA | GA | 213 | 3 (0-175) | |||||||||||
| AA | 125 | 1 (0-113) | ||||||||||||
|
| ||||||||||||||
| G | 300 | 2 (0-128) | 0.59 | 1.05 (0.62-1.77) | 1.10 (0.64-1.90) | 1.17 (0.69-1.96) | 1.02 (0.57-1.83) | |||||||
| A | 125 | 1 (0-113) | ||||||||||||
| rs2383207 | 9 | CDKN2B-AS1 | G/A | 0.56 | 1.27 (1.18-1.36) | 17 | ||||||||
|
| ||||||||||||||
| CDKN2B antisense RNA 1 | GG | 140 | 0 (0-68) | 0.13 | 0.97 (0.58-1.60) | 0.81 (0.48-1.37) | 0.85 (0.51-1.39) | 0.97 (0.55-1.70) | ||||||
| antisense RNA | GA | 192 | 3 (0-177) | |||||||||||
| AA | 89 | 1 (0-116) | ||||||||||||
|
| ||||||||||||||
| G | 332 | 2 (0-123) | 0.72 | 1.12 (0.63-1.99) | 0.99 (0.55-1.80) | 1.11 (0.63-1.98) | 1.02 (0.54-1.92) | |||||||
| A | 89 | 1 (0-116) | ||||||||||||
| rs2383206 | 9 | CDKN2B-AS1 | G/A | 0.50 | 1.30 (1.06-1.58) | 17 | ||||||||
|
| ||||||||||||||
| CDKN2B antisense RNA 1 | GG | 98 | 0 (0-68) | 0.19 | 0.99 (0.57-1.70) | 0.98 (0.56-1.72) | 0.96 (0.56-1.65) | 1.14 (0.63-2.06) | ||||||
| antisense RNA | GA | 192 | 3 (0-177) | |||||||||||
| AA | 97 | 1 (0-116) | ||||||||||||
|
| ||||||||||||||
| G | 290 | 2 (0-128) | 0.66 | 0.86 (0.68-2.11) | 0.81 (0.60-1.92) | 1.00 (0.76-2.37) | 0.86 (0.61-2.15) | |||||||
| A | 97 | 1 (0-116) | ||||||||||||
| rs1537373 | 9 | CDKN2B-AS1 | G/T | 0.54 | 1.19 (1.07-1.31) | 16 | ||||||||
|
| ||||||||||||||
| CDKN2B antisense RNA 1 | GG | 131 | 0 (0-95) | 0.27 | 1.27 (0.72-2.22) | 1.06 (0.60-1.88) | 1.23 (0.71-2.14) | 0.99 (0.54-1.81) | ||||||
| antisense RNA | GT | 194 | 3 (0-147) | |||||||||||
| TT | 99 | 0 (0-116) | ||||||||||||
|
| ||||||||||||||
| G | 325 | 2 (0-123) | 0.61 | 1.33 (0.59-1.63) | 1.21 (0.50-1.44) | 1.33 (0.57-1.55) | 1.26 (0.61-1.86) | |||||||
| T | 99 | 0 (0-116) | ||||||||||||
| rs10965219 | 9 | CDKN2B-AS1 | G/A | 0.49 | 1.16 (1.05-1.29) | 16 | ||||||||
|
| ||||||||||||||
| CDKN2B antisense RNA 1 | GG | 66 | 4 (0-147) | 0.50 | 1.08 (0.59-1.96) | 1.19 (0.65-2.20) | 0.98 (0.54-1.77) | 0.95 (0.48-1.84) | ||||||
| antisense RNA | GA | 123 | 0 (0-65) | |||||||||||
| AA | 70 | 2 (0-129) | ||||||||||||
|
| ||||||||||||||
| G | 189 | 0 (0-81) | 0.41 | 0.86 (0.46-1.59) | 0.75 (0.4-1.41) | 0.89 (0.48-1.64) | 0.80 (0.41-1.57) | |||||||
| A | 70 | 2 (0-129) | ||||||||||||
| rs7025486 | 9 | DAB2IP | A/G | 0.15 | 1.10 (1.06-1.14) | 19 | ||||||||
|
| ||||||||||||||
| DAB2 interacting protein | AA | 5 | 0 (0-28) | 0.93 | - | - | - | - | ||||||
| intronic | AG | 16 | 0 (0-11) | |||||||||||
| GG | 65 | 0 (0-111) | ||||||||||||
|
| ||||||||||||||
| A | 21 | 0 (0-17) | 0.40 | - | - | - | - | |||||||
| G | 65 | 0 (0-111) | ||||||||||||
| rs12413409 | 10 | CNNM2 | G/A | 0.92 | 1.12 (1.08-1.16) | 6,10 | ||||||||
|
| ||||||||||||||
| cyclin M2 | GG | 360 | 1 (0-128) | 0.91 | 1.02 (0.56-1.86) | 0.95 (0.51-1.76) | 0.94 (0.52-1.69) | 1.03 (0.54-1.96) | ||||||
| intronic | AG | 60 | 3 (0-119) | |||||||||||
| AA | 5 | 24 (0-30) | ||||||||||||
|
| ||||||||||||||
| G | 420 | 2 (0-121) | 0.97 | 0.88 (0.34-2.26) | 0.84 (0.32-2.18) | 0.87 (0.34-2.24) | 1.07 (0.41-2.77) | |||||||
| A | 5 | 24 (0-30) | ||||||||||||
| rs501120 | 10 | CXCL12 | A/G | 0.85 | 1.33 (1.20-1.48) | 9 | ||||||||
|
| ||||||||||||||
| chemokine (C-X-C motif) ligand 12 | AA | 307 | 3 (0-128) | 0.33 | 0.87 (0.39-1.90) | 1.31 (0.59-2.91) | 0.83 (0.38-1.82) | 1.19 (0.51-2.79) | ||||||
| intragenic | GA | 96 | 0 (0-93) | |||||||||||
| GG | 17 | 4 (0-155) | ||||||||||||
|
| ||||||||||||||
| A | 403 | 1 (0-119) | 0.41 | 1.40 (0.83-2.37) | 1.96 (1.13-3.41) | 1.25 (0.74-2.11) | 1.05 (0.59-1.88) | |||||||
| G | 17 | 4 (0-155) | ||||||||||||
| rs1746048 | 10 | CXCL12 | C/T | 0.84 | 1.33 (1.20-1.48) | 6,8,10 | ||||||||
|
| ||||||||||||||
| chemokine (C-X-C motif) ligand 12 | CC | 306 | 3 (0-128) | 0.34 | 1.45 (0.86-2.44) | 1.97 (1.14-3.40) | 1.33 (0.79-2.23) | 1.05 (0.59-1.88) | ||||||
| intragenic | CT | 100 | 0 (0-105) | |||||||||||
| TT | 19 | 4 (0-155) | ||||||||||||
|
| ||||||||||||||
| C | 406 | 1 (0-120) | 0.53 | 0.84 (0.39-1.84) | 1.23 (0.56-2.71) | 0.90 (0.42-1.95) | 1.23 (0.53-2.86) | |||||||
| T | 19 | 4 (0-155) | ||||||||||||
| rs2047009 | 10 | CXCL12 | C/A | 0.30 | 1.05 | 6 | ||||||||
|
| ||||||||||||||
| chemokine (C-X-C motif) ligand 12 | CC | 13 | 44 (0-167) | 0.58 | - | - | - | - | ||||||
| intragenic | AC | 61 | 0 (0-111) | |||||||||||
| AA | 73 | 13 (0-222) | ||||||||||||
|
| ||||||||||||||
| C | 74 | 0 (0-121) | 0.07 | - | - | - | - | |||||||
| A | 73 | 13 (0-222) | ||||||||||||
| rs2505083 | 10 | KIAA1462 | C/T | 0.23 | 1.07 (1.04-1.09) | 20 | ||||||||
|
| ||||||||||||||
| KIAA1462 | CC | 5 | 0 (0-104) | 0.69 | - | - | - | - | ||||||
| intron | CT | 23 | 1 (0-129) | |||||||||||
| TT | 44 | 9 (0-357) | ||||||||||||
|
| ||||||||||||||
| C | 28 | 1 (0-116) | 0.21 | - | - | - | - | |||||||
| T | 44 | 9 (0-357) | ||||||||||||
| rs2246833 | 10 | LIPA | T/C | 0.16 | 1.04 | 6 | ||||||||
|
| ||||||||||||||
| lipase A, lysosomal acid, cholesterol esterase | TT | 1 | 0 (0-0) | 0.27 | - | - | - | - | ||||||
| intron | TC | 5 | 5 (0-110) | |||||||||||
| CC | 16 | 22 (1-338) | ||||||||||||
|
| ||||||||||||||
| T | 6 | 2 (0-110) | 0.26 | - | - | - | - | |||||||
| C | 16 | 22 (1-338) | ||||||||||||
| rs1412444 | 10 | LIPA | T/C | 0.20 | 1.09 (1.07-1.12) | 11 | ||||||||
|
| ||||||||||||||
| lipase A, lysosomal acid, cholesterol esterase | TT | 2 | 0 (0-0) | 0.10 | - | - | - | - | ||||||
| intron | TC | 5 | 5 (0-110) | |||||||||||
| CC | 16 | 22 (1-338) | ||||||||||||
|
| ||||||||||||||
| T | 7 | 0 (0-110) | 0.15 | - | - | - | - | |||||||
| C | 16 | 22 (1-338) | ||||||||||||
| rs11203042 | 10 | LIPA | T/C | 0.31 | 1.03 | 6 | ||||||||
|
| ||||||||||||||
| lipase A, lysosomal acid, cholesterol esterase | TT | 2 | 254 (0-508) | 0.82 | - | - | - | - | ||||||
| intron | TC | 6 | 7 (0-66) | |||||||||||
| CC | 8 | 63 (2-114) | ||||||||||||
|
| ||||||||||||||
| T | 8 | 7 (0-88) | 0.44 | - | - | - | - | |||||||
| C | 8 | 63 (2-114) | ||||||||||||
| rs1075724 | 10 | MIKI67 | G/A | 0.21 | 1.78 (1.45-2.18) | 21 | ||||||||
|
| ||||||||||||||
| antigen identified by monoclonal antibody Ki-67 | GG | 0 | - | - | - | - | - | - | ||||||
| intergenic | AG | 25 | 5 (0-214) | |||||||||||
| AA | 34 | 63 (2-114) | ||||||||||||
|
| ||||||||||||||
| G | 25 | 5 (0-214) | 0.65 | - | - | - | - | |||||||
| A | 34 | 63 (2-114) | ||||||||||||
| rs974819 | 11 | PDGFD | A/G | 0.31 | 1.07 (1.04-1.09) | 11 | ||||||||
|
| ||||||||||||||
| platelet derived growth factor D | AA | 2 | 0 | 0.38 | - | - | - | - | ||||||
| intergenic | AG | 1 | 34 (0-175) | |||||||||||
| GG | 5 | 0 | ||||||||||||
|
| ||||||||||||||
| A | 3 | 0 (0-175) | - | - | - | - | - | |||||||
| G | 5 | 0 | ||||||||||||
| rs964184 | 11 | ZPR1 | G/C | 0.15 | 1.13 (1.10–1.16) | 6,10 | ||||||||
|
| ||||||||||||||
| zinc finger protein 259 | GG | 9 | 4 (0-175) | 0.34 | 1.00 (0.59-1.69) | 0.86 (0.50-1.46) | 0.86 (0.52-1.42) | 0.96 (0.54-1.69) | ||||||
| intergenic | CG | 110 | 5 (0-197) | |||||||||||
| CC | 306 | 1 (0-93) | ||||||||||||
|
| ||||||||||||||
| G | 119 | 5 (0-197) | 0.08 | 0.89 (0.37-2.12) | 1.16 (0.48-2.81) | 0.93 (0.39-2.23) | 1.00 (0.41-2.47) | |||||||
| C | 306 | 1 (0-93) | ||||||||||||
| rs9326246 | 11 | ZPR1 | C/G | 0.06 | 1.04 | 6 | ||||||||
|
| ||||||||||||||
| zinc finger protein 259 | CC | 1 | 175 | 0.43 | 1.06 (0.56-1.99) | 1.36 (0.72-2.59) | 1.44 (0.79-2.65) | 1.29 (0.67-2.49) | ||||||
| intergenic | CG | 49 | 28 (0-227) | |||||||||||
| GG | 365 | 1 (0-114) | ||||||||||||
|
| ||||||||||||||
| C | 50 | 31 (0-207) | 0.11 | - | - | - | - | |||||||
| G | 365 | 1 (0-114) | ||||||||||||
| rs17676451 | 12 | HAL | A/G | 0.12 | -0.17 (0.04)* | 7 | ||||||||
|
| ||||||||||||||
| histidine ammonia lyase | AA | 2 | 157 (34-279) | 0.25 | 2.06 (1.05-4.05) | 2.03 (1-03-3.99) | 1.98 (1.02-3.85) | 1.33 (1.57-2.77) | ||||||
| intronic | AG | 41 | 48 (0-175) | |||||||||||
| GG | 141 | 0 (0-54) | ||||||||||||
|
| ||||||||||||||
| A | 43 | 48 (0-185) | 0.00 | 1.28 (0.51-3.25) | 1.36 (0.54-3.47) | 1.35 (0.53-3.43) | 1.15 (0.45-2.97) | |||||||
| G | 141 | 0 (0-54) | ||||||||||||
| rs7136259 | 12 | ATP2B1 | T/C | 0.48 | 1.11 (1-08-1.15) | 12 | ||||||||
|
| ||||||||||||||
| ATPase, Ca++ transporting, plasma membrane 1 | TT | 13 | 150 (5-393) | 0.06 | - | - | - | - | ||||||
| intronic | TC | 30 | 0 (0-198) | |||||||||||
| CC | 15 | 18 (0-166) | ||||||||||||
|
| ||||||||||||||
| T | 43 | 11 (0-257) | 0.77 | - | - | - | - | |||||||
| C | 15 | 18 (0-166) | ||||||||||||
| rs11066015 | 12 | ACAD10 | A/G | 0.01 | 1.41 | 20,22 | ||||||||
|
| ||||||||||||||
| acyl-CoA dehydrogenase family, member 10 | AA | 0 | - | - | 1.05 (0.42-2.63) | 0.91 (0.35-2.34) | 1.08 (0.43-2.71) | 0.91 (0.35-2.34) | ||||||
| intronic | AG | 6 | 1 (0-332) | |||||||||||
| GG | 418 | 2 (0-121) | ||||||||||||
|
| ||||||||||||||
| A | 6 | 1 (0-332) | 0.87 | - | - | - | - | |||||||
| G | 418 | 2 (0-121) | ||||||||||||
| rs4773144 | 13 | COL4A2 | G/A | 0.43 | 1.07 (1.05-1.09) | 6,10 | ||||||||
|
| ||||||||||||||
| collagen, type IV, alpha 2 | GG | 80 | 3 (0-155) | 0.47 | 1.50 (0.85-2.67) | 1.68 (0.94-3.01) | 1.70 (0.99-2.95) | 1.63 (0.90-2.96) | ||||||
| intronic | GA | 201 | 2 (0-108) | |||||||||||
| AA | 141 | 0 (0-123) | ||||||||||||
|
| ||||||||||||||
| G | 281 | 2 (0-116) | 0.63 | 1.44 (0.86-2.39) | 1.28 (0.75-2.17) | 1.42 (0.85-2.36) | 1.26 (0.71-2.24) | |||||||
| A | 141 | 0 (0-123) | ||||||||||||
| rs9515203 | 13 | COL4A2 | T/C | 0.96 | 1.08 | 6,11 | ||||||||
|
| ||||||||||||||
| collagen, type IV, alpha 2 | TT | 68 | 0 (0-91) | 0.04 | - | - | - | - | ||||||
| intronic | TC | 6 | 38 (31-348) | |||||||||||
| CC | 0 | - | ||||||||||||
|
| ||||||||||||||
| T | 74 | 2 (0-98) | - | - | - | - | - | |||||||
| C | 0 | - | ||||||||||||
| rs3809346 | 13 | COL4A2 | A/G | 0.43 | 0.15 (0.03)* | 7 | ||||||||
|
| ||||||||||||||
| collagen, type IV, alpha2 | AA | 80 | 3 (0-155) | 0.48 | 1.43 (0.86-2.37) | 1.27 (0.75-2.15) | 1.40 (0.84-2.32) | 1.28 (0.72-2.28) | ||||||
| intronic | AG | 201 | 2 (0-108) | |||||||||||
| GG | 142 | 0 (0-122) | ||||||||||||
|
| ||||||||||||||
| A | 281 | 2 (0-116) | 0.66 | 1.50 (0.84-2.66) | 1.67 (0.93-3.01) | 1.70 (0.98-2.94) | 1.64 (0.91-2.98) | |||||||
| G | 142 | 0 (0-122) | ||||||||||||
| rs9319428 | 13 | FLT1 | A/G | 0.30 | 1.05 (1.03-1.08) | 6 | ||||||||
|
| ||||||||||||||
| fms-related tyrosine kinase 1 | AA | 37 | 0 (0-104) | 0.45 | 1.07 (0.66-1.73) | 1.29 (0.78-2.12) | 1.15 (0.71-1.84) | 1.23 (0.72-2.10) | ||||||
| intronic | AG | 173 | 4 (0-123) | |||||||||||
| GG | 208 | 1 (0-123) | ||||||||||||
|
| ||||||||||||||
| A | 210 | 3 (0-119) | 0.92 | 0.72 (0.35-1.45) | 0.76 (0.37-1.55) | 0.75 (0.37-1.51) | 0.94 (0.45-1.97) | |||||||
| G | 208 | 1 (0-123) | ||||||||||||
| rs8001186 | 13 | IRS2 | A/G | 0.36 | -0.15 (0.03)* | 7 | ||||||||
|
| ||||||||||||||
| insulin receptor substrate 2 | AA | 8 | 65 (1-169) | 0.64 | - | - | - | - | ||||||
| intronic | AG | 17 | 34 (0-115) | |||||||||||
| GG | 21 | 3 (0-311) | ||||||||||||
|
| ||||||||||||||
| A | 25 | 34 (0-135) | 0.77 | - | - | - | - | |||||||
| G | 21 | 3 (0-311) | ||||||||||||
| rs7173743 | 15 | ADAMTS7 LOC105370915 | T/C | 0.54 | 1.06 | 6 | ||||||||
|
| ||||||||||||||
| (ADAM metallopeptidase with thrombospondin type 1 motif, 7) LOC105370915 | TT | 112 | 1 (0-128) | 0.72 | 1.01 (0.59-1.71) | 1.02 (0.60-1.75) | 1.25 (0.75-2.07) | 1.08 (0.61-1.91) | ||||||
| intragenic | CT | 235 | 1 (0-105) | |||||||||||
| CC | 78 | 5 (0-128) | ||||||||||||
|
| ||||||||||||||
| T | 347 | 1 (0-117) | 0.32 | 1.73 (0.32-1.03) | 0.94 (0.43-1.39) | 0.91 (0.45-1.38) | 0.82 (0.52-1.83) | |||||||
| C | 78 | 5 (0-128) | ||||||||||||
| rs17514846 | 15 | FURIN | A/C | 0.48 | 1.05 (1.03-1.08) | 6 | ||||||||
|
| ||||||||||||||
| furin (paired basic amino acid cleaving enzyme) | AA | 96 | 0 (0-128) | 0.16 | 1.05 (0.61-1.83) | 0.94 (0.54-1.64) | 0.80 (0.47-1.36) | 0.88 (0.48-1.58) | ||||||
| intronic | AC | 156 | 5 (0-105) | |||||||||||
| CC | 107 | 3 (0-129) | ||||||||||||
|
| ||||||||||||||
| A | 252 | 1 (0-111) | 0.36 | 1.15 (0.47-1.41) | 0.86 (0.39-1.19) | 1.30 (0.60-1.75) | 1.16 (0.65-2.15) | |||||||
| C | 107 | 3 (0-129) | ||||||||||||
| rs3825807 | 15 | ADAMTS7 | A/G | 0.60 | 1.08 (1.06–1.10) | 6,10 | ||||||||
|
| ||||||||||||||
| ADAM metallopeptidase with thrombospondin type 1 motif, 7 | AA | 155 | 1 (0-119) | 0.84 | 0.84 (0.47-1.51) | 1.11 (0.61-2.03) | 1.16 (0.64-2.08) | 0.76 (0.40-1.43) | ||||||
| exon | GA | 202 | 2 (0-123) | |||||||||||
| GG | 68 | 2 (0-121) | ||||||||||||
|
| ||||||||||||||
| A | 357 | 2 (0-121) | 0.88 | 1.33 (0.64-1.75) | 1.32 (0.59-1.65) | 1.36 (0.63-1.66) | 0.97 (0.42-1.28) | |||||||
| G | 68 | 2 (0-121) | ||||||||||||
| rs4380028 | 15 | ADAMTS7 | C/T | 0.63 | 1.07 (1.05–1.10) | 11,23 | ||||||||
|
| ||||||||||||||
| ADAM metallopeptidase with thrombospondin type 1 motif, 7 | CC | 162 | 2 (0-129) | 0.61 | 0.79 (0.41-1.51) | 0.96 (0.50-1.86) | 0.90 (0.48-1.70) | 0.82 (0.41-1.62) | ||||||
| exon | TC | 212 | 1 (0-91) | |||||||||||
| TT | 51 | 5 (0-150) | ||||||||||||
|
| ||||||||||||||
| C | 374 | 1 (0-116) | 0.36 | 1.06 (0.65-1.73) | 0.95 (0.58-1.58) | 1.09 (0.68-1.75) | 0.94 (0.55-1.61) | |||||||
| T | 51 | 5 (0-150) | ||||||||||||
| rs12936587 | 17 | RAI1 | G/A | 0.69 | 1.07 (1.05–1.09) | 6,10 | ||||||||
|
| ||||||||||||||
| retinoic acid induced 1 | GG | 97 | 4 (0-81) | 0.32 | 1.24 (0.66-2.32) | 0.88 (0.48-1.63) | 0.69 (0.38-1.28) | 0.91 (0.47-1.77) | ||||||
| intragenic | AG | 92 | 17 (0-250) | |||||||||||
| AA | 17 | 0 (0-64) | ||||||||||||
|
| ||||||||||||||
| G | 189 | 5 (0-129) | 0.16 | 1.08 (0.48-2.44) | 1.39 (0.61-3.16) | 0.92 (0.41-2.08) | 1.51 (0.64-3.6) | |||||||
| A | 17 | 0 (0-64) | ||||||||||||
| rs2281727 | 17 | SMG6 | C/T | 0.32 | 1.04 | 11 | ||||||||
|
| ||||||||||||||
| smg-6 homolog, nonsense mediated mRNA decay factor LOC105371487 | CC | 36 | 0 (0-28) | 0.28 | 0.83 (0.41-1.66) | 0.67 (0.33-1.39) | 0.81 (0.40-1.65) | 0.86 (0.40-1.85) | ||||||
| RNA anitsense | CT | 144 | 0 (0-129) | |||||||||||
| TT | 161 | 3 (0-197) | ||||||||||||
|
| ||||||||||||||
| C | 180 | 0 (0-105) | 0.18 | 0.76 (0.45-1.27) | 0.77 (0.45-1.32) | 0.67 (0.4-1.12) | 0.72 (0.4-1.27) | |||||||
| T | 161 | 3 (0-197) | ||||||||||||
| rs1231206 | 17 | SMG6 | A/G | 0.30 | 1.07 (1.05-1.09) | 6 | ||||||||
|
| ||||||||||||||
| smg-6 homolog, nonsense mediated mRNA decay factor | AA | 30 | 0 (0-23) | 0.30 | 0.81 (0.48-1.37) | 0.80 (0.46-1.38) | 0.75 (0.44-1.26) | 0.77 (0.43-1.37) | ||||||
| RNA anitsense | AG | 134 | 0 (0-129) | |||||||||||
| GG | 161 | 3 (0-197) | ||||||||||||
|
| ||||||||||||||
| A | 164 | 0 (0-114) | 0.22 | 1.14 (0.49-2.08) | 1.01 (0.37-1.63) | 1.12 (0.44-1.92) | 1.09 (0.43-2.04) | |||||||
| G | 161 | 3 (0-197) | ||||||||||||
| rs216172 | 17 | SMG6 | C/G | 0.29 | 1.07 (1.05–1.09) | 10 | ||||||||
|
| ||||||||||||||
| smg-6 homolog, nonsense mediated mRNA decay factor | CC | 30 | 0 (0-18) | 0.24 | 0.85 (0.50-1.43) | 0.81 (0.47-1.39) | 0.76 (0.45-1.28) | 0.81 (0.46-1.44) | ||||||
| RNA anitsense | CG | 132 | 0 (0-129) | |||||||||||
| GG | 164 | 2 (0-189) | ||||||||||||
|
| ||||||||||||||
| C | 162 | 0 (0-114) | 0.25 | 0.91 (0.45-1.86) | 0.71 (0.34-1.48) | 0.85 (0.41-1.76) | 0.91 (0.42-1.99) | |||||||
| G | 164 | 2 (0-189) | ||||||||||||
| rs15563 | 17 | UBE2Z | C/T | 0.53 | 1.01 (0.99-1.03) | 6 | ||||||||
|
| ||||||||||||||
| ubiquitin-conjugating enzyme E2Z | CC | 36 | 0 (0-46) | 0.68 | - | - | - | - | ||||||
| exon | CT | 47 | 0 (0-41) | |||||||||||
| TT | 29 | 1 (0-105) | ||||||||||||
|
| ||||||||||||||
| C | 83 | 0 (0-43) | 0.26 | - | - | - | - | |||||||
| T | 29 | 1 (0-105) | ||||||||||||
| rs46522 | 17 | UBE2Z | T/C | 0.53 | 1.06 (1.04–1.08) | 6,10 | ||||||||
|
| ||||||||||||||
| ubiquitin-conjugating enzyme E2Z | TT | 36 | 0 (0-46) | 0.64 | - | - | - | - | ||||||
| intron | CT | 46 | 0 (0-43) | |||||||||||
| CC | 29 | 1(0-105) | ||||||||||||
|
| ||||||||||||||
| T | 82 | 0 (0-43) | 0.28 | - | - | - | - | |||||||
| C | 29 | 1 (0-105) | ||||||||||||
| rs1122608 | 19 | SMARCA4 | G/T | 0.78 | 1.14 (1.09–1.18) | 6,8,10 | ||||||||
|
| ||||||||||||||
| SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | GG | 260 | 1 (0-121) | 0.44 | 0.71 (0.43-1.16) | 0.89 (0.54-1.47) | 0.71 (0.44-1.15) | 0.83 (0.48-1.42) | ||||||
| intron | GT | 139 | 3 (0-147) | |||||||||||
| TT | 24 | 1(0-66) | ||||||||||||
|
| ||||||||||||||
| G | 399 | 2 (0-129) | 0.65 | 0.71 (0.35-1.46) | 0.77 (0.37-1.59) | 0.66 (0.33-1.33) | 1.05 (0.48-2.28) | |||||||
| T | 24 | 1(0-66) | ||||||||||||
| rs2228671 | 19 | LDLR | C/T | 0.94 | 1.09 (1.02-1.15) | 11 | ||||||||
|
| ||||||||||||||
| low density lipoprotein receptor | CC | 307 | 0 (0-121) | 0.03 | - | - | - | - | ||||||
| exon | TC | 42 | 35 (0-129) | |||||||||||
| TT | 1 | 0 | ||||||||||||
|
| ||||||||||||||
| C | 349 | 2 (0-128) | - | 0.55 (0.28-1.07) | 0.63 (0.32-1.23) | 0.54 (0.29-1.03) | 0.48 (0.25-0.94) | |||||||
| T | 0 | 0 | ||||||||||||
| rs2075650 | 19 | TOMM40 | G/A | 0.14 | 1.11 | 6 | ||||||||
|
| ||||||||||||||
| translocase of outer mitochondrial membrane 40 homolog | GG | 8 | 8 (1-262) | 0.43 | 1.49 (0.63-3.50) | 1.18 (0.50-2.80) | 1.20 (0.51-2.84) | 1.49 (0.62-3.60) | ||||||
| intron | GA | 99 | 4 (0-148) | |||||||||||
| AA | 317 | 1 (0-116) | ||||||||||||
|
| ||||||||||||||
| G | 107 | 4 (0-153) | 0.18 | 1.66 (0.97-2.83) | 1.20 (0.70-2.06) | 1.03 (0.62-1.73) | 1.07 (0.60-1.89) | |||||||
| A | 317 | 1 (0-116) | ||||||||||||
| rs9982601 | 21 | KCNE2 | T/C | 0.03 | 1.19 (1.13–1.27) | 6,8,10 | ||||||||
|
| ||||||||||||||
| potassium channel, voltage gated subfamily E regulatory beta subunit 2 | TT | 0 | - | - | - | - | - | - | ||||||
| intergenic | TC | 14 | 66 (0-129) | |||||||||||
| CC | 206 | 1 (0-115) | ||||||||||||
|
| ||||||||||||||
| T | 14 | 66 (0-129) | 0.15 | 1.43 (0.63-3.26) | 1.60 (0.70-3.64) | 1.46 (0.65-3.29) | 1.19 (0.52-2.74) | |||||||
| C | 206 | 1 (0-115) | ||||||||||||
| rs7278204 | 21 | KCNE2 | G/A | 0.02 | 1.09 (1.03-1.15) | 11 | ||||||||
|
| ||||||||||||||
| potassium channel, voltage gated subfamily E regulatory beta subunit 2 | GG | 0 | - | - | - | - | - | - | ||||||
| intergenic | GA | 9 | 79 (0-171) | |||||||||||
| AA | 215 | 1 (0-115) | ||||||||||||
|
| ||||||||||||||
| G | 9 | 79 (0-171) | 0.36 | 1.35 (0.57-3.21) | 1.51(0.64-3.56) | 1.28 (0.54-3.02) | 1.39 (0.58-3.32) | |||||||
| A | 215 | 1 (0-115) | ||||||||||||
CAC is expressed as median (25p-75p). ‘p’ values represent U-Mann Whitney test for the comparison of homozygous for the effect allele versus heterozygous (upper row) and the comparison of the effect allele vs. the non-effect.
Logistic regression analysis (odds ratios, OR -95% confidence interval-) adjusted for traditional cardiovascular risk factors (age, sex, diabetes, hypertension, dyslipidemia, and smoking) express the relation of homozygous against heterozygous, or for the relation of affected allele against the non affected, with CAC expressed as higher than 0 and 10 units or higher than 70th and 90th percentiles.
Effect allele and homozygous forms, and significant p values, are depicted in bold.
Refers to beta coefficient (standard error).
Results
Demographic, laboratory, and clinical characteristics
The clinical characteristics of the patients in the three cohorts are presented in Table 1. Average age across the three cohorts was 57.6 ± 10.5 years old, 77% of the patients were women and 89% of the patients were Caucasian. Cardiovascular comorbidity was not different between groups except for a higher prevalence of hypertension, current smoking, and aspirin intake in the Vanderbilt cohort, where patients with previously known CVD were allowed to enroll. There were no differences between cohorts regarding CAC Agatston score. Median CAC score was 3.12 (interquartile range 0-134.35) in the three cohorts, and 21% and 35% of the patients had a CAC score greater than the 90th and 75th percentile, respectively. RA disease duration was 10 (interquartile range 3-20) years, 44% of the patients were receiving corticosteroid treatment, 32% were receiving anti-TNF therapy, and 65% were seropositive for rheumatoid factor.
Table 1. Demographic and clinical data of rheumatoid arthritis patients in the three cohorts.
| Total (n=561) | Cohorts | |||
|---|---|---|---|---|
|
|
|
|||
| ESCAPE (n=197) | Pittsburgh (n=195) | Vanderbilt (n=169) | ||
|
|
||||
| Demographics | ||||
|
| ||||
| Age, years | 57.6 ± 10.5 | 59.4 ± 8.7 | 58.8 ± 10.2 | 54.2 ± 11.8 |
| Sex, female(%) | 430 (77) | 118 (60) | 195 (100) | 117 (69) |
| BMI, kg/m2 | 28.5 ± 6.0 | 28.3 ± 5.3 | 27.9 ± 6.0 | 29.2 ± 6.8 |
| Hip circumference, cm | ||||
| Female | 106.7 ± 14.8 | 105.5 ± 14.6 | 105.9 ± 14.3 | 109.4 ± 15.6 |
| Male | 103.0 ± 12.3 | 100.6 ± 10.7 | - | 106.7 ± 13.6 |
| Waist circumference, cm | ||||
| Female | 91.9 ± 16.4 | 91.7 ± 15.6 | 91.2 ± 16.7 | 93.1 ± 16.8 |
| Male | 101.4 ± 14.9 | 101.6 ± 13.3 | - | 101.1 ± 17.2 |
| Race | ||||
| White | 502 (89) | 169 (86) | 184 (94) | 149 (88) |
| Others | 57 (10) | 28 (14) | 10 (5) | 19 (11) |
| Agatston score for CAC | ||||
|
| ||||
| CAC Agatston units | 3.12 (0-134.35) | 4.69 (0-175.00) | 2.75 (0-93.37) | 1.85 (0-150.35) |
| CAC greater than 0 units, n(%) | 307 (55) | 107 (55) | 117 (60) | 83 (51) |
| CAC greater than 10 units, n(%) | 248 (49) | 90 (46) | 86 (44) | 72 (44) |
| CAC greater than percentile 75, n(%) | 195 (35) | 63 (32) | 75 (39) | 57 (35) |
| CAC greater than percentile 90, n(%) | 113 (21) | 36 (18) | 39 (20) | 38 (23) |
| Previous cardiovascular disease | ||||
|
| ||||
| Cardiovascular events, n(%) | 61 (11) | 0 (0) | 21 (11) | 40 (24) |
| Comorbidity | ||||
|
| ||||
| Hypertension, n(%) | 235 (42) | 76 (38) | 71 (36) | 88 (52) |
| Systolic blood pressure, mmHg | 129 ± 20 | 128 ± 19 | 125 ± 19 | 133 ± 20 |
| Diastolic blood pressure, mmHg | 76 ± 10 | 76 ± 9 | 76 ± 10 | 75 ± 11 |
| Use of antihypertensives, n(%) | 210 (37) | 79 (40) | 67 (34) | 64 (38) |
| Dyslipemia | 119 (21) | 62 (31) | 57 (29) | 81 (48) |
| On lipid lowering drugs | 60 (11) | 35 (18) | 25 (13) | 19 (12) |
| Diabetes | 22 (4) | 12 (6) | 10 (5) | 17 (10) |
| Previous smoking | 291 (52) | 115 (58) | 96 (49) | 80 (47) |
| Current smoking | 81 (14) | 23 (12) | 17 (9) | 41 (24) |
| Packs/years | 0 (0-22) | 7 (0-30) | 0 (0-14) | 0 (0-22) |
| Postmenopause | 249 (44) | 92 (47) | 157 (81) | 74 (47) |
| Hormone replacement use | 85 (15) | 16 (8) | 69 (35) | - |
| Metabolic syndrome | 90 (16) | 44 (22) | 46 (24) | 55 (36) |
| Aspirin | 102 (18) | 34 (17) | 14 (7) | 54 (32) |
| Analytical parameters | ||||
|
| ||||
| Total cholesterol, mg/dL | 197 ± 39 | 195 ± 38 | 208 ± 37 | 186 ± 39 |
| Triglycerides, mg/dL | 1116 (86-157) | - | 120 (90-156) | 111 (80-158) |
| HDL cholesterol, mg/dL | 54 ± 16 | 49 (41-67) | 61 ± 15 | 43 (37-54) |
| LDL cholesterol, mg/dL | 116 ± 33 | 116 ± 31 | 120 ± 35 | 112 ± 33 |
| Glucose, mg/dL | 91 ± 19 | 89 (83-98) | 88 (82-94) | 87 (83-94) |
| Insulin, μU/mL | 10 ± 7 | 6 (4-10) | 12 (9-14) | 10 (5-19) |
| Homocysteine, μmol/L | 10.7 ± 3.6 | 9.1 (7.5-10.6) | 11.1 (9.6-13.7) | 10.5 ± 3.4 |
| Creatinine, mg/dL | 0.8 ± 0.3 | 0.8 ± 0.2 | 0.9 ± 0.3 | 0.8 ± 0.2 |
| CRP, mg/L | 4.65 (1.75-12.50) | - | 5.62 (1.97-13.35) | 4.00 (1.22-13.02) |
| Rheumatoid arthritis | ||||
|
| ||||
| Disease duration, years | 10 (3-20) | 9 (4-17) | 13 (7-23) | 3 (2-18) |
| Prednisone | ||||
| Ever prednisone treatment | 458 (82) | 147 (75) | 172 (88) | 139 (82) |
| Current with prednisone | 246 (44) | 76 (39) | 78 (40) | 92 (54) |
| Current pred. dosage, mg/day | 0 (0-5) | 0 (0-5) | 0 (0-5) | 2 (0-5) |
| Hydroxycloroquine | 125 (22) | 47 (24) | 36 (18) | 42 (25) |
| Methotrexate | 360 (64) | 125 (63) | 115 (59) | 120 (71) |
| TNF inhibitors | 179 (32) | 85 (43) | 59 (30) | 35 (21) |
| Anakinra | 2 (0) | 1 (1) | 1 (1) | 0 (0) |
| NSAIDs | 313 (73) | 127 (64) | 135 (69) | 51 (30) |
| COX2 inhibitors | 154 (72) | 47 (24) | 56 (29) | 51 (30) |
| Minutes of morning stiffness | 30 (5-60) | 15 (5-30) | 30 (0-60) | 30 (10-90) |
| Current biologic DMARD use | 149 (27) | 89 (45) | 60 (31) | 32 (19) |
| Any current use of non-biologic DMARDs | 487 (87) | 165 (84) | 175 (90) | 147 (87) |
| Joint surgery | 156 (28) | 55 (28) | 101 (52) | - |
| Rheumatoid nodules | 197 (35) | 89 (45) | 108 (55) | - |
| Global ass. of disease activity | 24 (9-47) | 21 (5-47) | 19 (6-34) | 30 (16-55) |
| MModified HAQ (VU cohort only) | 0.500 (0.000-0.875) | 0.500 (0.000-0.875) | ||
| DAS28 (ESCAPE and VU cohorts) | 3.72 ± 1.35 | 3.66 ± 1.08 | - | 3.79 ± 1.61 |
| Full HAQ (ESCAPE cohort only) | 0.625 (0.125-1.250) | 0.625 (0.125-1.250) | - | - |
| RF (>40 units) | 363 (65) | 129 (65) | 117 (60) | 117 (69) |
Data expressed as mean (± standard deviation) or median (interquartile range). Dichotomous variables are expressed as number (percentage).
CRP=C reactive protein; HAQ=Health Assessment Questionnaire; DAS28=Disease Activity Score; HDL=High density lipoprotein; LDL=Low density lipoprotein.
DMARD=Disease-modifying Antirheumatic Drug; TNF=tumor necrosis factor; COX-2=cyclooxygenase-2
Modified HAQ is only available for Vanderbilt cohort. DAS28 only for ESCAPE and Vanderbilt coh ort. Full HAQ only in ESCAPE series.
CAC -coronary artery calcification- percentiles were adjusted for age and race as previously described
Coronary artery disease related SNPs in relation to RA CAC
Table 2 shows the relation of the 91 SNPS related to CAD in the general population to the presence of CAC in the RA cohort. 86 of these SNPs did not exhibit any positive association with CAC in the RA cohort, either considering the affected versus the non-affected allele in homozygous or heterozygous inheritance with the Agatston score as a continuous variable, or in the association analysis using CAC categorizations. Only rs646776 (CELSR2), rs602633 (PSCR1), rs579459 (ABO), rs501120 (CXCL12) and rs17676451 (HAL) expressed any positive relation with CAC in one or more of the analyses. rs579459 (ABO) and rs17676451 (HAL) genotype exhibited the most consistent association between genotype and CAC Agatston units, with a significant increase in the frequency of the effect alleles in both homozygous or heterozygous genotype distributions, however, in the case of ABO, CAC categorizations, although demonstrating trends to significance, were not significantly associated. The positive associations for the other three SNPs were much less consistent, e.g. for rs501120 (CXCL12) there was no association of homozygous or heterozygous genotype distribution with CAC Agatston units, however in heterozygotes there was a positive association with the effect allele A in higher than 10 units CAC categorization (odds ratio 1.96 [1.13-3.41)]), although there was no association of any CAC categorizations with the homozygous AA genotype, which would be expected to be more sensitive (Table 2).
Notably, none of the group of the 12 SNPs mapping in the 53 kb region of chromosome 9p21.3, encoding ANRIL that are among the strongest predictors of CAD in the general population exhibited any association with CAD. Moreover, in the instance of PHACTR1, also strongly positively associated with CAD risk in the general population, three SNPs (rs9369640, rs12526453, and rs1332844) consistently showed a limited and incomplete protective relation with CAC. E.g., in the case of rs1332844, the p value for a homozygous or heterozygous mode of inheritance was respectively 0.01 and 0.08. Additionally COL4A2 (rs9515203) and LDLR (rs2228671) were negatively associated with CAC in the homozygous mode of inheritance.
Number of homozygous SNPs in relation to RA CAC
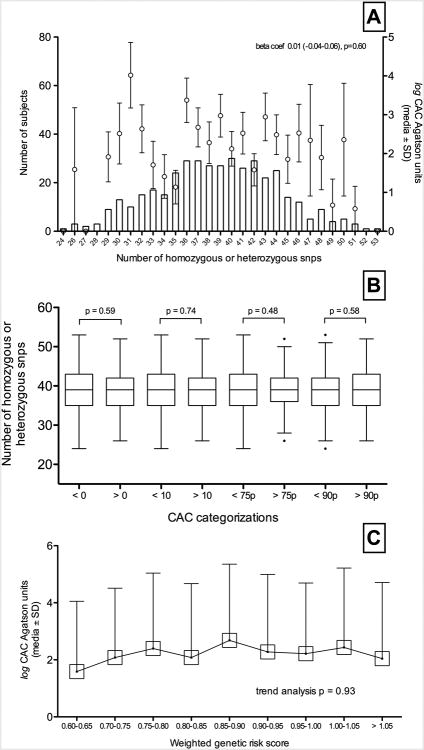
When the association of the CAD related SNPS with CAC was studied through the non weighted and weighted set of SNPs, no significant association was found. In particular, an increase in the number of both homozygous or heterozygous cardiovascular disease related SNPs was not significantly associated with an increase in log CAC Agatston units (Figure 1, panel A). Correspondingly, CAC categorizations in CAC higher than 0 and 10 units, and higher than 75th and 90 percentiles did not exhibit an increased frequency of homozygous or heterozygous SNPs (Figure 1, panel B).
Figure 1.
Panel A shows log coronary artery calcification (CAC) (mean and standard deviation -SD- Agatston units, right axis) relation with number of coronary artery disease related SNPs (left axis). Panel B shows number of CAD related SNPs (Tukey's bloxplots) in every CAC categorization - CAC higher than 0 and 10 Agatston units and higher than percentile 75th and 90th; ‘p’ values of the difference in every categorization is showed. Panel C depicts a trend analysis of log CAC (mean and SD) and its relation with the weighted genetic risk score.
Additionally, when CAD related SNPs were weighted by their effect sizes, the resulting GRS was not significantly associated with CAC. Furthermore, trend analysis showed no relation between GRS and log CAC Agatson units (p=0.93) (Figure 1, panel C).
Genetic risk score, disease activity, and CAC
The relationship between CAC and GRS or disease activity is illustrated in Table 3 through the value of the beta coefficient, which reflects the change in GRS as a function of the change in CAC categorizations. All the categorizations of CAC were associated with higher disease activity scores (DAS 28) after adjusting for age and sex. When this analysis was performed adjusting for traditional cardiovascular risk factors like diabetes, dyslipidemia, hypertension and smoking this relation was maintained in higher than CAC 90th percentile patients (beta coefficient 0.43 [0.04-0.81], p=0.03) and was nearly significant in both patients with CAC higher than 10 Agatston units (beta coef. 0.34 [-0.01-0.68], p=0.06) or patients within the CAC 75th percentile (beta coef. 0.28 [-0.05-0.60], p=0.09).
Table 3. Coronary artery calcification relation with disease activity score and genetic risk score.
| DAS28 (beta coef. 95% CI, p) | ||||
|---|---|---|---|---|
|
| ||||
| Unadjusted | Model #1 | Model #2 | Model #3 | |
| Univariate model | Adjusted for age and sex | Adjusted for Model 1 plus diabetes, hypertension, smoking and dyslipidemia | Adjusted for Model 1 and 2 plus GRS | |
|
|
||||
| CAC > 0 | 0.15 (-0.13-0.43), 0.30 | 0.37 (0.04-0.70), 0.03 | 0.09 (-0.27-0.45), 0.62 | 0.08 (-0.28-0.44), 0.65 |
| CAC > 10 | 0.24 (-0.04-0.52), 0.09 | 0.48 (0.16-0.80), 0.00 | 0.34 (-0.01-0.68), 0.06 | 0.32 (-0.02-0.68), 0.07 |
| CAC > 75th | 0.39 (0.10-0.68), 0.01 | 0.46 (0.16-0.76), 0.00 | 0.28 (-0.05-0.60), 0.09 | 0.27 (-0.06-0.59), 0.10 |
| CAC > 90th | 0.52 (0.18-0.86), 0.00 | 0.55 (0.21-0.89), 0.00 | 0.43 (0.04-0.81), 0.03 | 0.42 (0.04-0.81), 0.03 |
|
| ||||
| Genetic risk score (beta coef. 95% CI, p) | ||||
| Unadjusted | Model #1 | Model #2 | ||
|
| ||||
| Univariate model | Adjusted for age and sex | Adjusted for Model 1 plus diabetes, hypertension, smoking and dyslipidemia | ||
|
| ||||
| CAC > 0 | - 0.03 (-0.06--0.00), 0.03 | -0.01 (-0.04-0.02), 0.51 | -0.01 (-0.05-0.03), 0.66 | |
| CAC > 10 | - 0.04 (-0.06--0.01), 0.04 | -0.01 (-0.04-0.03), 0.68 | -0.01 (-0.04-0.04), 0.95 | |
| CAC > 75th | - 0.01 (-0.04-0.02), 0.59 | 0.00 (-0.03-0.03), 0.91 | 0.01 (-0.03-0.05), 0.72 | |
| CAC > 90th | 0.01 (-0.03-0.04), 0.69 | 0.01 (-0.03-0.05), 0.57 | 0.02 (-0.03-0.06), 0.50 | |
CAC = coronary artery calcification; CI = confidence interval
Conversely, GRS was inversely associated with CAC higher than 0 (- 0.03 [-0.06--0.00], 0.03) and higher than 10 Agatston units (- 0.04 [-0.06--0.01], 0.04) (Table 3). However, in the multivariate analysis, after adjusting for age and sex and for traditional cardiovascular factors, GRS was not related with CAC. In this sense, patients within CAC 90th percentile did not exhibit higher levels of GRS even after multivariate analysis (beta coef. 0.02 [-0.03-0.06], p=0.50).
Similarly, the relation between CAC and DAS28 was not modified when the analysis was adjusted for GRS (Model 3). In this regard, when we evaluated the association between CAC 90th percentile patients and DAS28 for the contribution of GRS, the statistically significant association (beta coef. 0.42 [0.04-0.81], p=0.03) between CAC and DAS28, was maintained in this set of patients, and was not modified by GRS.
Discussion
The central finding of our study is that RA patients with CAC, compared to those without CAC do not exhibit the elevated frequency of the multiple CAD specific SNPs found at increased frequency among patients in the general population with CAD. This finding suggests that CAC in RA does not develop through the concerted action of the same genes and their related pathways implicated with the development of CAD in the general population. The lack of contribution of these proatherosclerotic genes implies that RA itself should be considered an independent cardiovascular risk factor. Accordingly, we suggest that some factor in the RA immune response primarily contributes to the increased prevalence and incidence of cardiovascular disease in RA through pathways distinct from the proatherosclerotic pathways that operate in the general population to cause CAD.
Cardiovascular disease has an estimated heritability in the general population of 30–60% and genome-wide association studies have identified SNPs at several genomic regions associated with atherosclerosis, CAD and the progression to myocardial infarction (4-24, 47). However, the role of these CAD SNPs known to be related with cardiovascular disease in general population has not been previously explored in RA patients. In this study we used CAC as a complex cardiovascular trait related with CAD. The use of CAC as a validated CAD equivalent is supported by its ability to predict incident coronary heart disease even after adjustment for traditional cardiovascular risk factors (27).
Because each individual genetic variant specified by a SNP explains a small fraction of the variation in a complex trait and thus has limited predictive capacity for disease risk (48), we also constructed a weighted GRS with all the CAD related SNPs. GRSs are one of the most promising ways to aggregate multiple sets of SNP results into a single genetic predictor, and have been previously applied to different cardiovascular traits in the literature (49). Although still under development (49), they have been successfully used to predict cardiovascular outcomes and intermediate traits or subclinical phenotypes, including coronary heart/artery disease (50), myocardial infarction (51), ischemic stroke (52), or hypertension (53).
We note that we had the power to detect an association with an effect size of 0.1 at an alpha of 0.05. However, the observed effect sizes were much lower if not negligible. For example, if we sought to detect an association of 0.5% (that could be considered clearly insignificant) we would need a sample size of 1731. Moreover, the positive association with the ABO SNP supports the notion that this study was sufficiently powered to detect associations of CAD SNPs to confer susceptibility if they were present. Therefore, it is appropriate to infer that the CAD SNPs are unlikely to be a significant source of variability for CAC in RA patients. The ABO locus encodes glyco-transferases that when absent in blood group O individuals protect against myocardial infarction (24), although their relation to CAC in RA patients suggests they may foster CAC development in this setting by mechanisms that differ from those in individuals without RA.
The interpretation that factors intrinsic to RA independently contribute to CAC is supported by the finding that CAC is clearly related to disease activity through the DAS28 score. Indeed the increased DAS28 scores in those with higher CAC remained significant even after adjusting for traditional cardiovascular risk factors, as well as by the GRS for alleles influencing CAD in the general population.
Similarly, one immunological mechanism in RA unrelated to CAD SNPs that could potentially contribute to the association is that CAC in RA patients is significantly and independently associated with an elevated percentage of circulating CD28-CD57+CD56+ memory effector CD4 T cells, and with an increased proportion of the CD14hiCD16+ monocyte subset bearing an intermediate or ‘inflammatory’ phenotype (54). These elevated numbers of memory effector T cell and inflammatory monocyte subsets, which are considered an intrinsic element in the immune response mediating RA, may also provision the nascent and clinically unimportant subintimal lipid deposits with effector T cells and inflammatory monocytes that could accelerate the development of advanced atherosclerotic vascular disease.
Additionally, several studies (55) support the association of polymorphic genes involved in influencing RA susceptibility or the severity of effector pathways of RA with cardiovascular disease. These include the association between alleles encoding the HLA-DR shared epitope with both subclinical and clinically evident CV disease (56), and variants belonging to genes implicated in inflammatory and metabolic pathways, such as rs1800629 (TNFA), rs909253 (LTA), rs1800795 (IL6), rs333 (CCR5), rs2812378 (CCL21), and rs1800470 (TGFB1), which have been associated with a higher risk of endothelial dysfunction, different types of cardiovascular events, and cardiovascular mortality in RA (55, 57-60).
There are some limitations in this study. First, this study should be considered exploratory because the experimental design does not include a validation cohort. Second, although genotyping was facilitated in our study because a considerable proportion of CAD SNPs were fortuitously present on the Immunochip, the studies exploring susceptibility for CAD in the general population utilized different chips, most notably the Metabolic chip, each of which has a different spectrum of SNPs. Therefore we had to use imputation to identify some important polymorphisms, which is still an evolving approach (41). Third, CAC appears at an intermediate stage in the development of atherosclerosis and the assessment of earlier stages of atherosclerosis may prove more sensitive, as would adding the later stage of a myocardial event. Fourth, the association observed between DAS28 and CAC could also be influenced by non-inflammatory comorbidities, such as depression, fibromyalgia, osteoarthritis, etc. that can also lead to higher DAS28 scores.
The role of genetic factors in accelerated cardiovascular disease in RA requires further study to better identify the mechanisms and pathways responsible for this association. To our knowledge, this is the first investigation in which general population CAD related SNPs has been assessed in a large set of RA patients with a reproducible, sensitive, quantitative measure of CAD. Our findings of an absence of association of these CAD related SNPs with CAC in RA is novel and could shed light on the complicated relationship between inflammation and cardiovascular risk in RA patients. In conclusion, we have shown that in RA, even when using all known genetic susceptibility variants associated with CAD in the general population, gene dosage of these susceptible CAD SNPs was not associated with the increased CAC observed in patients. Our findings would support the role of one or more mechanistic features in RA itself as the fundamental responsible factor for the increase of cardiovascular diseases in RA.
Supplementary Material
Acknowledgments
We thank Angela Christiano, PhD, and her laboratory for the extraction of DNA in many of the samples.
Financial support: This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases grant AR-050026 (Dr. Bathon), AR056116 (Dr. Stein), a Pfizer ASPIRE Investigator Initiated Rheumatology Award WI195003 (Dr. Winchester), the NIH National Center for Advancing Translational Sciences grant UL1-TR-000040 [formerly the National Center for Research Resources, grant UL1-RR-024156], and CTSA award No. UL1TR000445 from the National Center for Advancing Translational Sciences; K01AR060848 (Dr. Reynolds); National Institute of Arthritis and Musculoskeletal and Skin Diseases (K23AR064768) and the Rheumatology Research Foundation Career Development K Supplement (Chung); and by the Fundación Canaria Dr. Manuel Morales, Fundación Española de Reumatología and Complejo Hospitalario Universitario de Canarias (Dr. Ferraz-Amaro).
Footnotes
Author contributions: All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Ferraz-Amaro had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design: Bathon, Winchester, Giles, Ferraz-Amaro, Reynolds, Gregersen.
Acquisition of data: Bathon, Giles, Stein, Wasko, Gregersen, Chung, Oeser.
Analysis and interpretation of data: Ferraz-Amaro, Winchester, Reynolds, Gregersen, Wasko, Stein, Giles, Bathon, Chung, Oeser.
References
- 1.Avina-Zubieta JA, Thomas J, Sadatsafavi M, Lehman AJ, Lacaille D. Risk of incident cardiovascular events in patients with rheumatoid arthritis: a meta-analysis of observational studies. Annals of the rheumatic diseases. 2012;71(9):1524–9. doi: 10.1136/annrheumdis-2011-200726. [DOI] [PubMed] [Google Scholar]
- 2.del Rincon ID, Williams K, Stern MP, Freeman GL, Escalante A. High incidence of cardiovascular events in a rheumatoid arthritis cohort not explained by traditional cardiac risk factors. Arthritis and rheumatism. 2001;44(12):2737–45. doi: 10.1002/1529-0131(200112)44:12<2737::AID-ART460>3.0.CO;2-%23. [DOI] [PubMed] [Google Scholar]
- 3.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. The New England journal of medicine. 1994;330(15):1041–6. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 4.Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nature genetics. 2013;45(1):25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Donnell CJ, Kavousi M, Smith AV, Kardia SL, Feitosa MF, Hwang SJ, et al. Genome-wide association study for coronary artery calcification with follow-up in myocardial infarction. Circulation. 2011;124(25):2855–64. doi: 10.1161/CIRCULATIONAHA.110.974899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Myocardial Infarction Genetics C. Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nature genetics. 2009;41(3):334–41. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, et al. Genomewide association analysis of coronary artery disease. The New England journal of medicine. 2007;357(5):443–53. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nature genetics. 2011;43(4):333–8. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coronary Artery Disease Genetics C. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nature genetics. 2011;43(4):339–44. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 10.Lu X, Wang L, Chen S, He L, Yang X, Shi Y, et al. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nature genetics. 2012;44(8):890–4. doi: 10.1038/ng.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aoki A, Ozaki K, Sato H, Takahashi A, Kubo M, Sakata Y, et al. SNPs on chromosome 5p15.3 associated with myocardial infarction in Japanese population. J Hum Genet. 2011;56(1):47–51. doi: 10.1038/jhg.2010.141. [DOI] [PubMed] [Google Scholar]
- 12.Davies RW, Wells GA, Stewart AF, Erdmann J, Shah SH, Ferguson JF, et al. A genome-wide association study for coronary artery disease identifies a novel susceptibility locus in the major histocompatibility complex. Circulation Cardiovascular genetics. 2012;5(2):217–25. doi: 10.1161/CIRCGENETICS.111.961243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke R, Peden JF, Hopewell JC, Kyriakou T, Goel A, Heath SC, et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. The New England journal of medicine. 2009;361(26):2518–28. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 14.Pechlivanis S, Muhleisen TW, Mohlenkamp S, Schadendorf D, Erbel R, Jockel KH, et al. Risk loci for coronary artery calcification replicated at 9p21 and 6q24 in the Heinz Nixdorf Recall Study. BMC Med Genet. 2013;14:23. doi: 10.1186/1471-2350-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen GQ, Li L, Rao S, Abdullah KG, Ban JM, Lee BS, et al. Four SNPs on chromosome 9p21 in a South Korean population implicate a genetic locus that confers high cross-race risk for development of coronary artery disease. Arterioscler Thromb Vasc Biol. 2008;28(2):360–5. doi: 10.1161/ATVBAHA.107.157248. [DOI] [PubMed] [Google Scholar]
- 16.Broadbent HM, Peden JF, Lorkowski S, Goel A, Ongen H, Green F, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17(6):806–14. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 17.Harrison SC, Cooper JA, Li K, Talmud PJ, Sofat R, Stephens JW, et al. Association of a sequence variant in DAB2IP with coronary heart disease. European heart journal. 2012;33(7):881–8. doi: 10.1093/eurheartj/ehr075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prins BP, Lagou V, Asselbergs FW, Snieder H, Fu J. Genetics of coronary artery disease: genome-wide association studies and beyond. Atherosclerosis. 2012;225(1):1–10. doi: 10.1016/j.atherosclerosis.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 19.Abdullah KG, Li L, Shen GQ, Hu Y, Yang Y, MacKinlay KG, et al. Four SNPS on chromosome 9p21 confer risk to premature, familial CAD and MI in an American Caucasian population (GeneQuest) Ann Hum Genet. 2008;72(Pt 5):654–7. doi: 10.1111/j.1469-1809.2008.00454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JY, Lee BS, Shin DJ, Woo Park K, Shin YA, Joong Kim K, et al. A genome-wide association study of a coronary artery disease risk variant. J Hum Genet. 2013;58(3):120–6. doi: 10.1038/jhg.2012.124. [DOI] [PubMed] [Google Scholar]
- 21.Dechamethakun S, Ikeda S, Arai T, Sato N, Sawabe M, Muramatsu M. Associations between the CDKN2A/B, ADTRP and PDGFD polymorphisms and the development of coronary atherosclerosis in Japanese patients. J Atheroscler Thromb. 2014;21(7):680–90. doi: 10.5551/jat.22640. [DOI] [PubMed] [Google Scholar]
- 22.Pasmant E, Sabbagh A, Vidaud M, Bieche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25(2):444–8. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 23.Chen HH, Almontashiri NA, Antoine D, Stewart AF. Functional genomics of the 9p21.3 locus for atherosclerosis: clarity or confusion? Curr Cardiol Rep. 2014;16(7):502. doi: 10.1007/s11886-014-0502-7. [DOI] [PubMed] [Google Scholar]
- 24.Reilly MP, Li M, He J, Ferguson JF, Stylianou IM, Mehta NN, et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet. 2011;377(9763):383–92. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenland P, Bonow RO, Brundage BH, Budoff MJ, Eisenberg MJ, Grundy SM, et al. ACCF/AHA 2007 clinical expert consensus document on coronary artery calcium scoring by computed tomography in global cardiovascular risk assessment and in evaluation of patients with chest pain: a report of the American College of Cardiology Foundation Clinical Expert Consensus Task Force (ACCF/AHA Writing Committee to Update the 2000 Expert Consensus Document on Electron Beam Computed Tomography) Circulation. 2007;115(3):402–26. doi: 10.1161/CIRCULATIONAHA..107.181425. [DOI] [PubMed] [Google Scholar]
- 26.O'Rourke RA, Brundage BH, Froelicher VF, Greenland P, Grundy SM, Hachamovitch R, et al. American College of Cardiology/American Heart Association Expert Consensus document on electron-beam computed tomography for the diagnosis and prognosis of coronary artery disease. Circulation. 2000;102(1):126–40. doi: 10.1161/01.cir.102.1.126. [DOI] [PubMed] [Google Scholar]
- 27.Keelan PC, Bielak LF, Ashai K, Jamjoum LS, Denktas AE, Rumberger JA, et al. Long-term prognostic value of coronary calcification detected by electron-beam computed tomography in patients undergoing coronary angiography. Circulation. 2001;104(4):412–7. doi: 10.1161/hc2901.093112. [DOI] [PubMed] [Google Scholar]
- 28.Peyser PA, Bielak LF, Chu JS, Turner ST, Ellsworth DL, Boerwinkle E, et al. Heritability of coronary artery calcium quantity measured by electron beam computed tomography in asymptomatic adults. Circulation. 2002;106(3):304–8. doi: 10.1161/01.cir.0000022664.21832.5d. [DOI] [PubMed] [Google Scholar]
- 29.Parikh NI, Hwang SJ, Larson MG, Cupples LA, Fox CS, Manders ES, et al. Parental occurrence of premature cardiovascular disease predicts increased coronary artery and abdominal aortic calcification in the Framingham Offspring and Third Generation cohorts. Circulation. 2007;116(13):1473–81. doi: 10.1161/CIRCULATIONAHA.107.705202. [DOI] [PubMed] [Google Scholar]
- 30.Wojczynski MK, Li M, Bielak LF, Kerr KF, Reiner AP, Wong ND, et al. Genetics of coronary artery calcification among African Americans, a meta-analysis. BMC Med Genet. 2013;14:75. doi: 10.1186/1471-2350-14-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giles JT, Szklo M, Post W, Petri M, Blumenthal RS, Lam G, et al. Coronary arterial calcification in rheumatoid arthritis: comparison with the Multi-Ethnic Study of Atherosclerosis. Arthritis research & therapy. 2009;11(2):R36. doi: 10.1186/ar2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yiu KH, Mok MY, Wang S, Ooi GC, Khong PL, Lau CS, et al. Prognostic role of coronary calcification in patients with rheumatoid arthritis and systemic lupus erythematosus. Clinical and experimental rheumatology. 2012;30(3):345–50. [PubMed] [Google Scholar]
- 33.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis and rheumatism. 1988;31(3):315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 34.Kao AH, Krishnaswami S, Cunningham A, Edmundowicz D, Morel PA, Kuller LH, et al. Subclinical coronary artery calcification and relationship to disease duration in women with rheumatoid arthritis. The Journal of rheumatology. 2008;35(1):61–9. [PubMed] [Google Scholar]
- 35.Chung CP, Oeser A, Raggi P, Gebretsadik T, Shintani AK, Sokka T, et al. Increased coronary-artery atherosclerosis in rheumatoid arthritis: relationship to disease duration and cardiovascular risk factors. Arthritis and rheumatism. 2005;52(10):3045–53. doi: 10.1002/art.21288. [DOI] [PubMed] [Google Scholar]
- 36.Carr JJ, Nelson JC, Wong ND, McNitt-Gray M, Arad Y, Jacobs DR, Jr, et al. Calcified coronary artery plaque measurement with cardiac CT in population-based studies: standardized protocol of Multi-Ethnic Study of Atherosclerosis (MESA) and Coronary Artery Risk Development in Young Adults (CARDIA) study. Radiology. 2005;234(1):35–43. doi: 10.1148/radiol.2341040439. [DOI] [PubMed] [Google Scholar]
- 37.Nelson JC, Kronmal RA, Carr JJ, McNitt-Gray MF, Wong ND, Loria CM, et al. Measuring coronary calcium on CT images adjusted for attenuation differences. Radiology. 2005;235(2):403–14. doi: 10.1148/radiol.2352040515. [DOI] [PubMed] [Google Scholar]
- 38.McClelland RL, Chung H, Detrano R, Post W, Kronmal RA. Distribution of coronary artery calcium by race, gender, and age: results from the Multi-Ethnic Study of Atherosclerosis (MESA) Circulation. 2006;113(1):30–7. doi: 10.1161/CIRCULATIONAHA.105.580696. [DOI] [PubMed] [Google Scholar]
- 39.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis research & therapy. 2011;13(1):101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8(8):e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1(6):457–70. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Connell J, Gurdasani D, Delaneau O, Pirastu N, Ulivi S, Cocca M, et al. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 2014;10(4):e1004234. doi: 10.1371/journal.pgen.1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, Lanas F, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 2004;364(9438):937–52. doi: 10.1016/S0140-6736(04)17018-9. [DOI] [PubMed] [Google Scholar]
- 45.van Setten J, Isgum I, Pechlivanis S, Tragante V, de Jong PA, Smolonska J, et al. Serum lipid levels, body mass index, and their role in coronary artery calcification: a polygenic analysis. Circulation Cardiovascular genetics. 2015;8(2):327–33. doi: 10.1161/CIRCGENETICS.114.000496. [DOI] [PubMed] [Google Scholar]
- 46.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature genetics. 2007;39(7):906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 47.Fan M, Dandona S, McPherson R, Allayee H, Hazen SL, Wells GA, et al. Two chromosome 9p21 haplotype blocks distinguish between coronary artery disease and myocardial infarction risk. Circulation Cardiovascular genetics. 2013;6(4):372–80. doi: 10.1161/CIRCGENETICS.113.000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17(10):1520–8. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith JA, Ware EB, Middha P, Beacher L, Kardia SL. Current Applications of Genetic Risk Scores to Cardiovascular Outcomes and Subclinical Phenotypes. Curr Epidemiol Rep. 2015;2(3):180–90. doi: 10.1007/s40471-015-0046-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cox AJ, Hsu FC, Ng MC, Langefeld CD, Freedman BI, Carr JJ, et al. Genetic risk score associations with cardiovascular disease and mortality in the Diabetes Heart Study. Diabetes Care. 2014;37(4):1157–64. doi: 10.2337/dc13-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaarhorst AA, Lu Y, Heijmans BT, Dolle ME, Bohringer S, Putter H, et al. Literature-based genetic risk scores for coronary heart disease: the Cardiovascular Registry Maastricht (CAREMA) prospective cohort study. Circulation Cardiovascular genetics. 2012;5(2):202–9. doi: 10.1161/CIRCGENETICS.111.960708. [DOI] [PubMed] [Google Scholar]
- 52.Ibrahim-Verbaas CA, Fornage M, Bis JC, Choi SH, Psaty BM, Meigs JB, et al. Predicting stroke through genetic risk functions: the CHARGE Risk Score Project. Stroke. 2014;45(2):403–12. doi: 10.1161/STROKEAHA.113.003044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.International Consortium for Blood Pressure Genome-Wide Association S. Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478(7367):103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winchester R, Giles JT, Nativ S, Downer K, Zhang HZ, Bag-Ozbek A, et al. Association of Elevations of Specific T Cell and Monocyte Subpopulations in Rheumatoid Arthritis With Subclinical Coronary Artery Atherosclerosis. Arthritis Rheumatol. 2016;68(1):92–102. doi: 10.1002/art.39419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodriguez-Rodriguez L, Lopez-Mejias R, Fernandez-Gutierrez B, Balsa A, Gonzalez-Gay MA, Martin J. Rheumatoid arthritis: genetic variants as biomarkers of cardiovascular disease. Curr Pharm Des. 2015;21(2):182–201. doi: 10.2174/1381612820666140825123407. [DOI] [PubMed] [Google Scholar]
- 56.Gonzalez-Gay MA, Gonzalez-Juanatey C, Lopez-Diaz MJ, Pineiro A, Garcia-Porrua C, Miranda-Filloy JA, et al. HLA-DRB1 and persistent chronic inflammation contribute to cardiovascular events and cardiovascular mortality in patients with rheumatoid arthritis. Arthritis and rheumatism. 2007;57(1):125–32. doi: 10.1002/art.22482. [DOI] [PubMed] [Google Scholar]
- 57.Panoulas VF, Nikas SN, Smith JP, Douglas KM, Nightingale P, Milionis HJ, et al. Lymphotoxin 252A>G polymorphism is common and associates with myocardial infarction in patients with rheumatoid arthritis. Annals of the rheumatic diseases. 2008;67(11):1550–6. doi: 10.1136/ard.2007.082594. [DOI] [PubMed] [Google Scholar]
- 58.Panoulas VF, Stavropoulos-Kalinoglou A, Metsios GS, Smith JP, Milionis HJ, Douglas KM, et al. Association of interleukin-6 (IL-6)-174G/C gene polymorphism with cardiovascular disease in patients with rheumatoid arthritis: the role of obesity and smoking. Atherosclerosis. 2009;204(1):178–83. doi: 10.1016/j.atherosclerosis.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 59.Palomino-Morales R, Gonzalez-Juanatey C, Vazquez-Rodriguez TR, Rodriguez L, Miranda-Filloy JA, Fernandez-Gutierrez B, et al. A1298C polymorphism in the MTHFR gene predisposes to cardiovascular risk in rheumatoid arthritis. Arthritis research & therapy. 2010;12(2):R71. doi: 10.1186/ar2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lopez-Mejias R, Genre F, Remuzgo-Martinez S, Robustillo-Villarino M, Garcia-Bermudez M, Llorca J, et al. Protective Role of the Interleukin 33 rs3939286 Gene Polymorphism in the Development of Subclinical Atherosclerosis in Rheumatoid Arthritis Patients. PLoS One. 2015;10(11):e0143153. doi: 10.1371/journal.pone.0143153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.