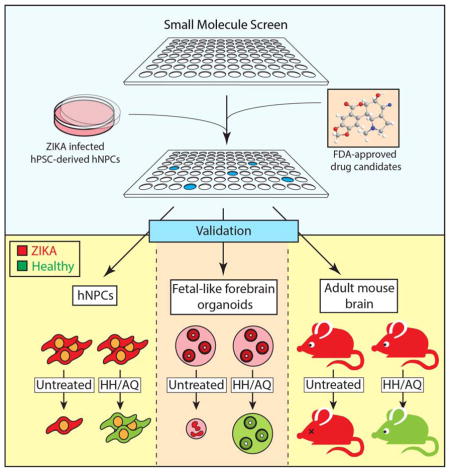
Summary
Zika virus (ZIKV) infects fetal and adult human brain, and is associated with serious neurological complications. To date, no therapeutic treatment is available to treat ZIKV infected patients. We performed a high content chemical screen using human embryonic stem cell derived cortical neuron progenitor cells (hNPCs) and found that hippeastrine hydrobromide (HH) and amodiaquine dihydrochloride dihydrate (AQ), can inhibit ZIKV infection in hNPCs. Further validation showed that HH also rescues ZIKV-induced growth and differentiation defects in hNPCs and human fetal-like forebrain organoids. Finally, HH and AQ inhibit ZIKV infection in adult mouse brain in vivo. Strikingly, HH suppresses viral propagation when administered to adult mice with active ZIKV infection, highlighting its therapeutic potential. Our approach highlights the power of stem cell-based screens and validation in human forebrain organoids and mouse models in identifying drug candidates for treating ZIKV infection and related neurological complications in fetal and adult patients.
Graphical Abstract
Introduction
ZIKV infection is associated with an increased incidence of congenital defects, most prominently microcephaly (Mlakar et al., 2016; Rasmussen et al., 2016). In adults, ZIKV can lead to serious neurological complications, such as Guillain-Barré syndrome, meningoencephalitis and myelitis (Cao-Lormeau et al., 2016; Carteaux et al., 2016; Mecharles et al., 2016; Parra et al., 2016). Recent studies using human pluripotent stem cell (hPSC)-derived human cortical neural progenitor cells (hNPCs) and forebrain organoids demonstrated that ZIKV replication persists in human fetal neural progenitors for weeks, leading to cell death and attenuation of cortical growth (Garcez et al., 2016; Hanners et al., 2016; Qian et al., 2016; Tang et al., 2016). Furthermore, ZIKV can infect neural stem cells in the murine adult brain (Li et al., 2016). These data support a direct link between ZIKV infection and congenital and adult neurologic symptoms.
Two high content chemical screens reported the identification of drug candidates inhibiting ZIKV (Barrows et al., 2016; Xu et al., 2016). Although these studies validated a screening platform to identify ZIKV inhibitors, none of the hit compounds were evaluated for therapeutic use in ZIKV-infected animals. Here, we describe a high content screen using hNPCs identifying two compounds that can inhibit ZIKV infection. One of these drug candidates, hippeastrine hydrobromide (HH) suppresses ZIKV infection in the brain of mice carrying replicating ZIKV, validating the therapeutic potential of this drug candidate.
Results
A high content chemical screen using hNPCs identifies anti-ZIKV compounds
To identify anti-ZIKV compounds, we screened the Prestwick library that includes 1120 FDA-approved drug and drug candidates, selected for known bioavailability and safety in humans. hNPCs were treated with each compound from the library or vehicle control (DMSO) for 1 hour, followed by addition of ZIKV MR766 (MOI=0.125) for 2 hours. After an additional 3 days of culture, cells were stained with antibodies against the ZIKV envelope protein (ZIKV E) and a cell proliferation marker Ki67 (Figure 1A). Quantifications were performed by an automated imaging and analysis system (Figure S1A). Positive hits were defined as compounds that increased total cell number by greater than 100% and suppressed ZIKA infection to less than 20% of control. Based on the fold change of the percentage of ZIKV infection, the signal-to-basal (S/B) ratio is 4.2 and the coefficient of variation (CV) is 6.9%. The Z factor is 0.59 indicating a robust screening system.
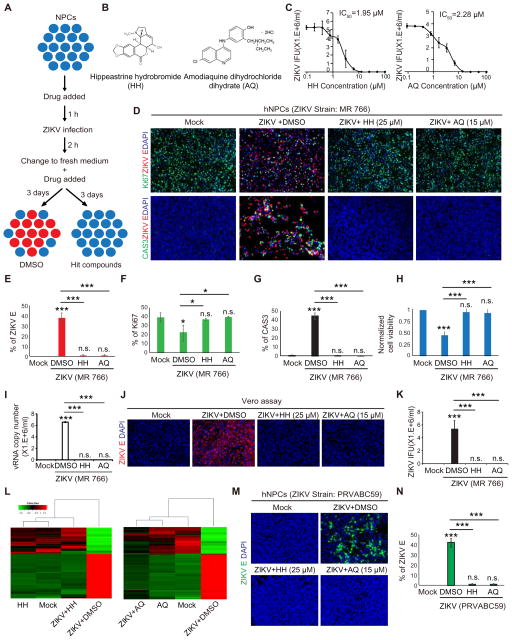
Figure 1. A high content chemical screen identifies anti-ZIKV compounds in hNPCs.
(A) Scheme of high content chemical screen.
(B) Chemical structure of hit compounds.
(C) Inhibitory curve of HH or AQ on hNPCs (n=3) based on infectious particles in supernatant as determined by the Vero reinfection assay.
(D–H) Immunocytochemistry analysis (D), the quantification of the percentage of ZIKV E+ cells (E), proliferation rate (F), cell apoptosis rate (G) and the normalized cell viability (H) of hNPCs at 72 h after ZIKV (MR766) infection. Scale bars, 100 μm. (n=6).
(I) qRT-PCR analysis to monitor the total ZIKV vRNA in the supernatant of hNPC cultures at 72 h after ZIKV infection.
(J and K) Vero cell reinfection assay (J) and the quantification of ZIKV infectious particles produced by ZIKV-infected hNPCs (K). Scale bars, 200 μm. (n=3).
(L) Heatmap of mock-infected hNPCs+DMSO (mock); mock-infected hNPCs+25 μM HH (HH), mock-infected hNPCs+15 μM AQ (AQ), ZIKV infected hNPCs+DMSO (ZIKV+DMSO); ZIKV-infected hNPCs+25 μM HH (ZIKV+HH), and ZIKV-infected hNPCs+15 μM AQ (ZIKV+AQ). Genes were selected from the up/down-regulated genes (fold change >4) in ZIKV-infected hNPCs+DMSO compared to mock-infected hNPCs+DMSO.
(M and N) Immunocytochemistry analysis (M) and quantification of the percentage of ZIKV E+ cells (N) of hNPCs at 72 h after ZIKV (PRVABC59, MOI=0.1) infection. Scale bars, 100 μm.
p values were calculated by one-way repeated measures ANOVA with a Bonferroni test. *p<0.05, **p<0.01 and ***p<0.001.
Using these criteria, 9 primary hit compounds were confirmed to significantly inhibit ZIKV infection without inhibiting cell proliferation at 10 μM (Figures S1B–D, Table S1). Two hit compounds that showed the highest efficacy (Figure S1C), HH (IC50=1.95 μM), and Amodiaquine dihydrochloride dihydrate (AQ, IC50=2.28 μM), were chosen for further study (Figure 1B and 1C). Both compounds effectively inhibited ZIKV infection and blocked ZIKV induced growth arrest or apoptosis compared to DMSO controls (Figures 1D–H). HH and AQ also significantly suppressed the production of ZIKV viral RNA (vRNA) (Figure 1I), and production of infectious ZIKV (Figures 1J–K and S1E). Furthermore, transcript profiling suggested that HH or AQ treatment reversed the transcriptional changes induced by ZIKV infection (Figure 1L). Interestingly, gene ontology analysis highlighted neuroactive ligand-receptor interaction pathway in both Mock vs. HH and Mock vs. AQ conditions (Figure S1F). Finally, 25 μM HH or 15 μM AQ effectively inhibited infection of another ZIKV strain, PRVABC59 (Figures 1M and 1N). Merbromin significantly increased hNPC proliferation, but did not affect ZIKV infection (Figures S1G–1K). Therefore, we focused on HH and AQ for further validation as drug candidates.
HH eliminates virus from ZIKV infected hNPCs
HH or AQ was applied 2h post ZIKV infection (hpi, Figure 2A). The compounds eliminated ZIKV infection with IC50 = 3.62 μM (HH, Figure 2B) and IC50 = 2.81 μM (AQ, Figure S2A). In addition, HH successfully rescued ZIKV-induced loss of cell viability with no obvious cytotoxicity (Figure 2C). In contrast, AQ affected cell viability at 25 μM, suggesting that AQ can be toxic to hNPCs (Figure S2B).
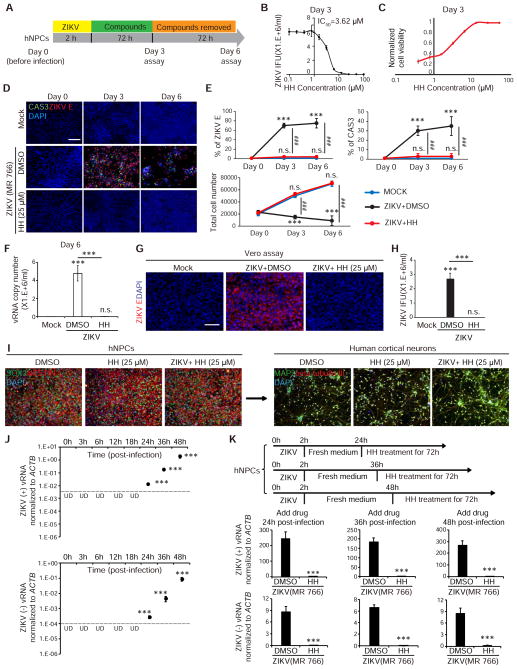
Figure 2. HH eliminates virus in ZIKV-infected hNPCs.
(A) Scheme to evaluate the elimination of ZIKV.
(B) Inhibitory curve of HH calculated based on infectious particles in the supernatant as determined by the Vero reinfection assay (n=3).
(C) Effective curve of HH calculated based on normalized cell viability (n=3).
(D and E) Immunocytochemistry analysis (D) and the quantification of the percentage of ZIKV E+ cells, cell apoptosis rate, and total cell number (E) of hNPCs on day 3 and day 6. Scale bars, 100 μm. (n=3). p values were calculated by one-way repeated measures ANOVA with a Bonferroni test. **p<0.01 and ***p<0.001.
(F) qRT-PCR analysis to monitor the (+) strand ZIKV vRNA in the supernatant of hNPC cultures on day 6. Scale bars, 200 μm.
(G and H) Vero cell reinfection assay (G) and the quantification of ZIKV infectious particles produced by ZIKV-infected hNPCs at day 6. Scale bars, 200 μm. (n=3).
(I) hNPCs with or without ZIKV MR766 infection were treated with 25 μM HH or DMSO for 3 days and then stained for hNPC markers: SOX2 (green) and NESTIN (red), and with DAPI (blue), or were differentiated to cortical neurons. Scale bars, 100 μm.
(J) qRT-PCR analysis to quantify the (+) strand and replicating (−) strand ZIKV vRNAs in the hNPCs at different time points post infection. The dash line shows detection limit. p values were calculated by one-way repeated measures ANOVA with a Bonferroni test. ***p<0.001. UD: undetectable.
(K) qRT-PCR analysis of hNPCs, on which 25 μM HH or DMSO were added at 24, 36 and 48 hpi (MR766) infection and maintained for additional 3 days.
p values were calculated by unpaired two-tailed Student’s t-test. *p<0.05, **p<0.01 and ***p<0.001, if not mentioned specifically.
Related to Figure S2.
ZIKV-infected hNPCs were treated with HH or AQ for 3 days, followed by 3 days culture in drug-free medium. Both compounds suppressed ZIKV infection to undetectable levels by day 3 (Figures 2D and S2C). By day 6, HH-treated hNPCs remained undetectable for ZIKV infection (Figures 2D and 2E). Consistent with these observations, HH treatment also rescued the cell death rate and total cell number at levels comparable to mock infection conditions (Figures 2D and 2E). Neither ZIKV vRNA (Figure 2F), nor production of infectious ZIKV was detected in day 6 supernatant under HH-treated conditions (Figures 2G and 2H). Furthermore, hNPCs with or without ZIKV infection treated with HH for 3 days maintained the expression of neural progenitor markers, SOX2 and NESTIN, and were capable of differentiation into cortical neurons expressing MAP2 and beta-tubulin III (Figure 2I). Therefore, HH can eliminate ZIKV from infected hNPCs without affecting cell identity or differentiation capacity. In contrast, AQ treatment resulted in viral rebound after 3 days of growth in drug-free medium (Figures S2C and S2D), suggesting that AQ only transiently suppresses ZIKV infection.
We used qRT-PCR analysis to monitor the dynamics of ZIKV vRNA expression (MR766 strain) in hNPCs. Both (+) and replicating (−) vRNA strands could be detected at 24 hpi, and the vRNA level increased progressively at 36 and 48 hpi (Figure 2J). Immunocytochemistry analysis showed that the percentage of ZIKV E+ cells started to plateau at 48 hpi. Moreover, cell apoptosis was detected at 48 hpi with substantial cell death from 48 to 72 hpi indicated by induction of CAS3 expression and decrease of the total cell number (Figure S2E). HH treatment at 24, 36 or 48 hpi successfully suppressed ZIKV vRNA levels (Figure 2K). HH consistently suppressed the production of ZIKV vRNAs in hNPCs derived from different donors (Figure S2F). HH treatment at 48 hpi, 12h after the initial detection of replicating (−) strand vRNA (Figure S2H), completely suppressed the production of vRNAs of another ZIKV strain (FSS13025, Figure S2G). The data suggest that HH can eliminate virus from hNPCs carrying replicating ZIKV.
HH inhibits ZIKV infection in a human fetal-like forebrain organoid model
hPSC-derived forebrain organoids provide a unique platform to study disorders of brain growth including microcephaly (Bershteyn and Kriegstein, 2013). We tested both short- and long-term effects of HH and AQ on day 20 (D20) forebrain organoids following ZIKV infection. D20 forebrain organoids were inoculated with ZIKV (MR766) for 24h, followed by treatment with 15 μM AQ or 25 μM HH for 3 days (D20+4, short-term) or 17 days (D20+18, long-term). Organoids were analyzed for ZIKV infection, cell proliferation, and apoptosis (Figure 3A). Immunohistochemical analysis of SOX2 and ZIKV E showed that D20 organoids consist mainly of SOX2+ NPCs, and more than 80% of the cells were infected by ZIKV at D20+4 (Figures 3B and 3C), HH was effective at suppressing the percentage of ZIKV E+ cells (Figures 3B and 3C) and total ZIKV vRNA level in the supernatant (Figure 3D) to undetectable levels. Infection of D20 organoids with ZIKV reduced the relative number of Ki67+ proliferating cells, and increased the number of CAS3+ apoptotic cells, phenotypes that were completely rescued by HH (Figure 3E). Although 15 μM AQ suppresses ZIKV infection in organoids, it resulted in a substantial reduction in mitotic cells and increase in apoptotic cells (Figure S3A).
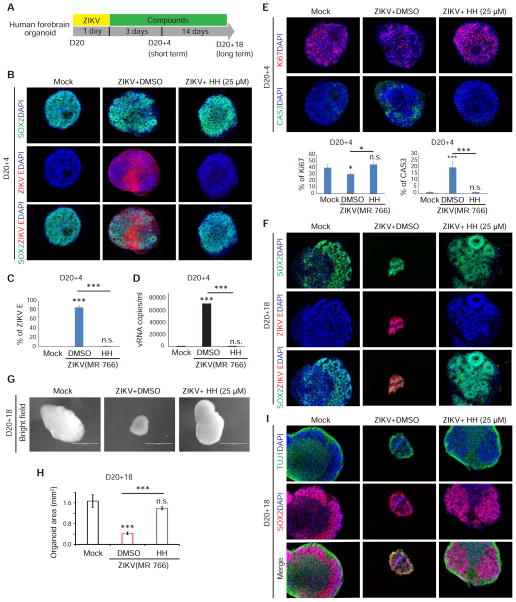
Figure 3. HH inhibits ZIKV infection in short and long term forebrain organoid cultures.
(A) Scheme to evaluate HH using forebrain organoids.
(B and C) Immunocytochemistry analysis (B) and the quantification of the percentage of ZIKV E+ cells (C) in mock, DMSO or 25 μM HH treated ZIKV-infected D20 organoids at D20+4 (n=3). Scale bars, 100 μm.
(D) qRT-PCR analysis of (+) strand ZIKV vRNA in the supernatant of DMSO or 25 μM HH treated ZIKV-infected D20 organoids (n=3).
(E) HH rescues ZIKV-related proliferation and apoptosis defects in D20+4 organoids. (n=3). Scale bars, 100 μm.
(F) D20+18 organoids are highly infiltrated by ZIKV (red), while HH suppressed any detectable ZIKV infection (n=3). Scale bars, 50 μm.
(G and H) Brightfield images of whole organoids (G) and the quantification of average cross sectional area of D20+18 organoids (H). Values represent mean ± SEM (n=3). Scale bars, 1 mm.
(I) HH rescues ZIKV-induced structural defects in forebrain organoids. (n=3). Scale bars, 100 μm.
p values were calculated by unpaired two-tailed Student’s t-test. *p<0.05, **p<0.01 and ***p<0.001 Related to Figure S3.
D20 organoids were infected with ZIKV and after 18 additional days of growth, analyzed for ZIKV infection. While ZIKV infected organoids without drug treatment were strongly infiltrated by virus, HH-treated organoids were devoid of detectable ZIKV (Figure 3F). Organoid size was measured 18 days post infection (dpi). The cross-sectional area of ZIKV infected organoids was significantly decreased compared to mock infected conditions, modeling ZIKV-associated congenital microcephaly (Figures 3G and 3H). In contrast, ZIKV-infected organoids treated with 25 μM HH grew to a similar size as mock-infected organoids (Figures 3G and 3H). Moreover, HH successfully rescued ZIKV-induced structural defects seen in long-term cultures. In the ZIKV-infected organoids treated with HH, there are clearly defined zones of progenitors (SOX2+) and neurons (TUJ1+), similar as mock conditions, while in the ZIKV infected organoids without HH, the progenitor and neuronal zones are intermixed and lack discernible organization (Figure 3I). We also monitored ZIKV vRNA expression dynamics using qRT-PCR. Both (+) strand and replicating (−) ZIKV vRNA strands were detected at 42 and 48 hpi (Figure S3B). Significant cell apoptosis was detected in ZIKV infected cells in organoids at 42 hpi (Figure S3C). HH treatment significantly decreased the ZIKV vRNA levels when added at 42 or 48 hpi (Figure S3D). Therefore, HH successfully inhibits ZIKV activity during long-term forebrain organoid growth and rescues crucial aspects associated with fetal brain development.
HH or AQ suppresses ZIKV infection in vivo
To evaluate the prophylactic activity of drug candidates in vivo, HH or AQ was administrated to 6–8 week old SCID-beige mice 12h before they were infected with ZIKV (MR766). HH was injected at a dose of 100 mg/kg/day subcutaneously (Figure 4A), and AQ was administrated at a dose of 40 mg/kg/day through intraperitoneal injection (Figure S4A). The mice were euthanized at 7 dpi. The levels of (+) strand ZIKV vRNA was low in liver, spleen and kidney, whereas, strong ZIKV infection was detected in brains of infected mice (Figure 4B). HH treatment significantly suppressed levels of ZIKV vRNA in brain (Figure 4C). AQ treatment achieved a similar prophylactic effect (Figure S4B).
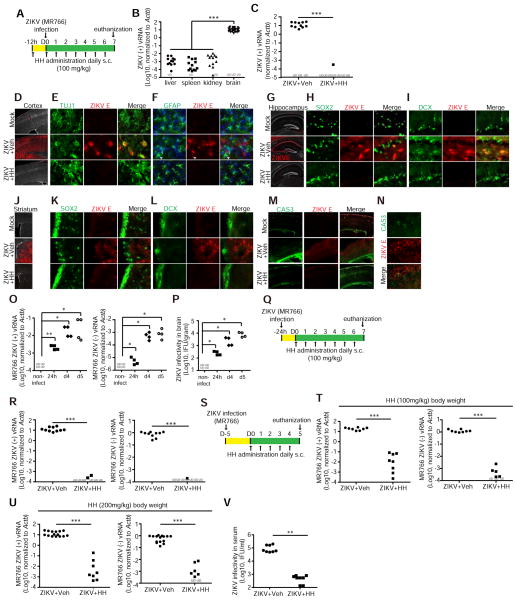
Figure 4. HH suppresses ZIKV infection in vivo.
(A) Scheme of prophylactic treatment of HH in vivo.
(B) qRT-PCR analysis of (+) strand ZIKV vRNA in liver, spleen, kidney and brain of ZIKV infected mice. n=13 for liver, spleen and kidney and n=30 for brain.
(C) qRT-PCR analysis of (+) strand ZIKV vRNA in brains of ZIKV-infected mice treated with vehicle (n=14) or HH (n=8). UD, undetectable.
(D–F) Immunohistochemical analysis of adult cortex using antibodies against ZIKV E (D), βIII-tubulin (E) and GFAP (F). Scale bar, 100 μm in (D) and 10 μm in (E, F).
(G–I) Immunohistochemical analysis of adult hippocampus including the post-mitotic CA regions using antibodies against ZIKV E (G), SOX2 (H) and DCX (I). Scale bar, 100 μm in (G) and 10 μm in (H, I).
(J–L) Immunohistochemical analysis of adult striatum using antibodies against ZIKV E (J), SOX2 (K) and DCX (L). Scale bar, 100 μm in (J) and 10 μm in (K, L).
(M and N) Immunohistochemical analysis of cellular apoptosis in adult cortex antibodies against CAS3. Scale bar, 100 μm in (M) and 10 μm in (N).
(O, P) Kinetics of ZIKV (MR766). Mice were infected with (1 x106 PFU) through intraperitoneal injection. Mice were euthanized at 24 hpi, 4 and 5 dpi. vRNA level was analyzed by qRT-PCR (O). Infectivity of viral particles was analyzed by Vero assay (P). n ≥ 4 mice at each time point.
(Q) Scheme of therapeutic intervention with HH 24 hpi (MR766) in (R).
(R) qRT-PCR analysis of ZIKV vRNA in brains of mice treated with vehicle (n=10) or HH (n=8) for 5 days starting at 24 hpi.
(S) Scheme of therapeutic intervention with HH 5 dpi (MR766) in (T–V).
(T) qRT-PCR analysis of ZIKV vRNA in the brains of mice treated with vehicle (n=8) or 100 mg/kg HH (n=8) for 5 days starting at 5 dpi.
(U) qRT-PCR analysis of ZIKV vRNA in the brains of mice treated with vehicle (n=15) or 200 mg/kg HH (n=8) for 5 days starting at 5 dpi.
(V) Quantification of ZIKV infectious particles in serum of ZIKV-infected mice (MR766) treated with vehicle (n=8) or 200 mg/kg HH (n=8) for 5 days starting at 5 dpi.
p values were calculated by one-way repeated measures ANOVA or two-way repeated measures ANOVA with a Bonferroni test. *p<0.05, **p<0.01, ***p<0.001.
Related with Figure S4
Immunohistochemical analysis revealed that ZIKV broadly infects forebrain structures, with strong infiltration of the adult cortex (Figure 4D). ZIKV was found in many post-mitotic TUJ1+ cortical neurons (Figure 4E), but not GFAP+ cortical astrocytes (Figure 4F). HH fully inhibited infection of adult cortex (Figures 4D–F). ZIKV also strongly infiltrated hippocampal structures, including the cornu ammonis (CA1 and CA3) regions, suggesting infection of post-mitotic neurons (Figure 4G). High magnification images revealed that ZIKV can infect SOX2+ neural progenitors (Figure 4H) and DCX+ immature neurons (Figure 4I). HH completely inhibited infection of the adult hippocampus, including immature cells (Figures 4G–I). ZIKV strongly infiltrated the striatum (Figure 4J), localized outside the neurogenic subventricular zone (SVZ), with few ZIKV-infected SOX2+ neural progenitors (Figure 4K) or DCX+ immature neurons (Figure 4L). HH fully inhibited infection of the adult striatum (Figures 4J–L). Given the strong infiltration of adult brain structures, we analyzed whether ZIKV exerts detrimental cellular effects in the adult nervous system. Immunostaining for CAS3 revealed that ZIKV infection increases the number of apoptotic cells in the cortex (Figure 4M). ZIKV induced apoptosis in both infected and non-infected cells (Figure 4N). Treatment of mice with HH rescued cellular apoptosis associated with ZIKV infection in the cortex (Figure 4M). AQ treatment was also able to inhibit ZIKV infection from all cell-types in the adult (Figures S4C–K) and to rescue ZIKV-induced cellular apoptosis (Figure S4L).
To evaluate the therapeutic potential of HH, we first monitored the kinetics of ZIKV infection in adult mouse organs. SCID-beige mice were inoculated with ZIKV (MR766) and euthanized at 24 hpi, 4 and 5 dpi. Organs were analyzed for vRNA by qRT-PCR (Figures 4O and S4M) and Vero re-infection assays (Figures 4P and S4O). Both (+) (Figure 4O, left) and replicating (−) (Figure 4O, right) vRNAs strands were detected in brains of ZIKV-infected mice at 24 hpi, 4 and 5 dpi. The (+) vRNA strand was occasionally detectable in liver, spleen and kidney (Figure S4M, left). However, the (−) strand was only detectable in spleen and became undetectable after 4 dpi (Figure S4M, right). Consistently, infectious viral particles were detected in brain (Figure 4P), but not liver, spleen or kidney (Figure S4O). Therefore, we evaluated the effect of HH on the infected brain.
Based on the dynamics of viral replication, we tested the anti-ZIKV potential of HH at 24 hpi and 5 dpi, two time points when vRNA is detected. At 24 hpi, the ZIKV-infected mice were randomly separated into two groups and treated with vehicle or HH (100 mg/kg/day subcutaneously) for 7 days (Figure 4Q). The (+) and (−) ZIKV vRNA strands were rarely detectable following HH treatment (Figure 4R). In addition, HH treatment at 5 dpi, 4 days after the emergence of detectable ZIKV, consistently decreased levels of both (+) and replicating (−) vRNA strands when compared to the vehicle treated cohort (Figures 4S and 4T). High dose HH treatment (200 mg/kg/day subcutaneously) applied at 5 dpi also suppressed (+) and (−) ZIKV vRNA strands in the brain (Figure 4U). Importantly, high dose HH treatment starting at 5 dpi also significantly suppressed the infectious capacity of ZIKV in mouse serum (Figure 4V). To test the breadth of HH therapeutic activity, we inoculated mice with a second ZIKV strain (Asian strain, FSS13025). HH also significantly decreased levels of this strain when administered at 7 dpi, 6 days after the initial detection of ZIKV in brain (Figures S4O–R). Therefore, in immuno-compromised adult hosts, ZIKV achieves disseminated infection outside of progenitor zones, where it typically resides. HH has broad anti-ZIKV activity in mouse brain carrying replicating ZIKV, highlighting its therapeutic potential.
Discussion
We carried out a high content chemical screen using an FDA-approved drug library and discovered two drug candidates with anti-ZIKV activity. Both HH and AQ can inhibit ZIKV infection of hNPCs. HH, but not AQ, eliminates ZIKV from hNPCs previously infected with ZIKV. As a result, HH can rescue microcephaly-related defects in human fetal-like forebrain organoid cultures, with little or no evidence of cellular toxicity. Finally, HH exhibits anti-ZIKV activity in vivo in the adult mouse brain. Two other anti-ZIKV compounds were reported in mouse models, NITD008 (Deng et al., 2016) and 7-deaza-2′-C-methyladenosine (Zmurko et al., 2016), but only tested for their prophylactic activity. Here we demonstrated that HH has strong in vivo anti-ZIKV activity, inhibiting virus production and spread in the brains of ZIKV-infected immuno-compromised SCID-beige mice. Additional work needs to be done to further confirm the HH’s anti-ZIKV effect on other tissues that can potentially be infected by ZIKV, including eyes (Miner et al., 2016), testis (Govero et al., 2016), and spinal cord (Lazear et al., 2016).
HH was first isolated from the amaryllidaceae family plant species Lycoris radiata, and reported to exhibit anti-avian influenza H5N1 activity (He et al., 2013). A chemical analog of HH inhibits another flavivirus, Hepatitis C (Chen et al., 2015). HH shows no toxicity even at high doses (100 μM in vitro and 200 mg/kg in vivo). Given the safety and efficacy of HH to suppress ZIKV infection in hNPCs, human forebrain organoids, and adult mice, HH is a highly promising drug candidate for further clinical development to treat ZIKV-infected patients across the lifespan.
We also identified AQ, also called Amodiaquine, as another FDA-approved drug with anti-ZIKV activity. The WHO recommended AQ as an optional treatment of uncomplicated malaria (Olliaro and Mussano, 2003). AQ inhibits replication and infectivity of another flavavirus, Dengue virus type 2 (Boonyasuppayakorn et al., 2014). We found that AQ effectively inhibits ZIKV infection in NPCs in vitro and in a mouse model. However, AQ exhibited high cytotoxicity in brain organoid cultures, which raises safety concerns for using this drug during pregnancy.
Few options are currently available to treat potentially devastating infections from the ZIKV pandemic, but chemical-based drugs may provide a first response option. Our findings suggest a strong drug candidate for the treatment of ZIKV infection, in addition to indicating targets for drug development against other flaviviruses, including West Nile Virus, Dengue Virus, and yellow fever virus, which all cause severe illness.
STAR Methods
Detailed methods are provided in the online version of this paper and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to the Lead Contact, Dr. Shuibing Chen (shc2034@med.cornell.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
3 human iPSC lines were generated from skin biopsy samples of 3 different donors, including a healthy female at 16 years old (3113-3-21 line, donor 1), a healthy female at 18 years old (3182-1-2 line, donor 2) and a healthy female at 12 years old (3183-3-2 line, donor 3). hNPCs were differentiated from the 3 hiPSC lines as previously reported (Topol et al., 2015). The detailed protocol was described in Method details. hNPC 3182-1-2 line and 3183-3-2 line were used in Figure S2F, and the hNPC 3113-3-21 line were used in all the other hNPC experiments in this study. Human forebrain organoids were generated from H9 hESC line (WA-09, WiCell).
Mice
6–8 weeks old immunocompromised SCID-Beige female mice (CB17.Cg-PrkdcscidLystbg-J/Crl) purchased from Charles River Laboratories were used for in vivo compound testing under a protocol approved by the WCMC Institutional Animal Care and Use Committee. All mice were housed in bio-hazardous animal facility and the experiments were performed in BSL2+ hood.
METHOD DETAILS
hNPC generation, culture and cortical neuron differentiation
Human iPSC lines were generated from skin biopsy samples of a healthy female at 16 years old (3113-3-21 line), a healthy female at 18 years old (3182-1-2 line) and a healthy female at 12 years old (3183-3-2 line), had been fully characterized and cultured on a MEF feeder layer in hESC medium consisting of DMEM/F12 (Thermo Fisher Scientific), 20% KO-Serum Replacement (Life Technologies), 1x Glutamax (Thermo Fisher Scientific) 1x NEAA (Thermo Fisher Scientific) 1x 2 mercaptoethanol (Thermo Fisher Scientific20ng/ml FGF2 (Peprotech). The three human iPSCs were differentiated into forebrain-specific hNPCs as previously described (Topol et al., 2015). Briefly, on day 1, hiPSCs colonies were detached from the feeder layer with 1 mg/ml collagenase treatment for 1 hour and suspended in N2/B27 medium consisting of DMEM/F12 (Thermo Fisher Scientific), 1x N2 (Thermo Fisher Scientific) and 1x B27-RA (Thermo Fisher Scientific). The colonies were then transferred to non-adherent 6-well dishes to form EBs overnight. On day 2, feed the EBs with N2/B27 medium supplemented with 0.1 μM LDN193189 (Stem Cell Technologies) and 10 μM SB431542 (Tocris Bioscience). On days 3–7, feed the EBs every second day with N2/B27 medium supplemented with 0.1 μM LDN193189 and 10 μM SB431542. On days 7–14, plating of EBs to Matrigel (BD Biosciences) coated plates. Neural rosettes began to appear within a few days (characterized as round clusters of neuroepithelial cells with apico-basal polarity). On day 14, the rosettes were picked mechanically and dissociated with Accutase (Sigma-Aldrich) at 37° C for 15 minutes, and placed onto matrigel-coated 6-well plates to form monolayer hNPCs in NPC medium consisting of Dulbecco’s Modified Eagle Medium/Ham’s F12 Nutrient Mixture (ThermoFisher Scientific), 1xN2 (ThermoFisher Scientific), 1xB27 minus RA (ThermoFisher Scientific) and 20 ng/ml recombinant human basic FGF (Peprotech). The hNPCs were split at 1:3 ratio every week with Accutase. hNPCs at passage 5–8 were used for ZIKV infection in the study.
To induce cortical neuronal differentiation, NPCs were dissociated with Accutase and plated at 2.0 × 105 cells per cm−2 in NPC media on growth factor reduced Matrigel-coated plates. After overnight culture, medium was changed to neural differentiation medium, including DMEM/F12, 1xN2, 1xB27 minus RA, 20 ng/ml BDNF (Peprotech), 20 ng/ml GDNF (Peprotech), 20 ng/ml NT-3 (Peprotech), 1 mM dibutyryl-cyclic AMP (Sigma), 200 nM ascorbic acid (Sigma) and 1 μg/ml laminin (ThermoFisher Scientific). NPC-derived neurons were differentiated for 1~2 months.
Generation and culture of human forebrain organoids
To induce forebrain organoid differentiation, H9 hESC line (WA-09, WiCell) hPSCs were dissociated to single cells using EDTA, and 9000 cells were reaggregated using low-adhesion V-bottom 96 well plates (Wako) in Essential8 Medium (Fisher Scientific) with 10 μM Y-27632 (Tocris Biosciences). After 24 h (day 0), medium was changed to Essential6 (Fischer Scientific) supplemented with 10 μM SB431542 (Tocris Biosciences), 500 nM LDN193189 (Stem Cell Technologies), and 2 μM XAV939 (Tocris Biosciences) until day 4. From day 4 to day 18, XAV939 was removed. Medium was changed every other day. From Day 18, organoids were maintained in organoid differentiation medium (50% DMEM F-12 (Fisher Scientific), 50% mL Neurobasal (Fisher Scientific), 0.5xN2 supplement (Stem Cell Technologies), 0.025% insulin (Sigma), 5 mM L-Glutamine (1x, Fischer Scientific), 0.7 mM MEM-NEAA (1x, Fischer Scientific), 50 U/mL Penicillin-Streptomycin (1x, Fischer Scientific), 55 μM 2-mercaptoethanol (1x, Fischer Scientific), 1xB27 supplement without Vitamin A (Fischer Scientific).
ZIKV infection of hNPCs and forebrain organoids
Vero cell line was maintained in EMEM medium (ATCC) plus 10% heat-inactivated FBS with penicillin/streptomycin at 37°C. MR766 strain and P RVABC59 strain of ZIKV were obtained commercially (ZeptoMetrix) and then titrated by plaque assay in Vero cells. FSS13025 strain was propagated using a mosquito cell line C6/36 cells (ATCC® CRL-1660™). For virus propagation, C6/36 cells were maintained in DMEM medium plus 10% FBS at 28°C and infected with FSS13025 ZIKV (MOI=0.01) for 2 h. Half of the culture medium was changed to fresh medium at 1 and 3 dpi. Virus was harvested at 5 dpi and stored at −80°C.
hNPCs were plated on 96-well plates at 8.5×104/cm2. After overnight incubation, hNPCs were infected with ZIKV (MR766 strain, MOI=0.125) for 2 h, and changed to virus-free medium. The cells were maintained in NPC medium with daily medium change. Three days later, the supernatant and cells were collected and used for assays. hESC-derived forebrain organoids were infected with ZIKV MR766 (5 × 105 PFU/ml) for 24 h. After removal of virus containing medium, forebrain organoids were maintained in forebrain organoid medium for an additional 3 or 17 days and then used for assays.
To monitor ZIKV dynamics in hNPCs, hNPCs were infected with ZIKV (MR766 strain, MOI=0.125, FSS13025 strain MOI=0.1) for 2 h, and changed to virus-free medium. The cells were maintained in NPC medium with daily medium change. The cells were lysed at the different time points and analyzed using qRT-PCR. The MOI was calculated by dividing the number of virus particles by the cell number during infection. The cell number was counted using hemocytometer when plating. The number of virus particles was determined with focus-forming assay on Vero cells. In brief, ZIKV stocks were titrated to infect Vero cells for 2 h. Vero cells were then fixed in 4% paraformaldehyde 2 dpi and stained with antibodies against ZIKV envelop protein (ZIKV E) to detect the infected foci. Number of infected foci was counted under fluorescent microscope.
High-content Screen
hNPCs were dissociated and plated on Matrigel-coated 384-well plates for the chemical screen. After overnight incubation, the cells were treated with compounds from an FDA-approved drug library for 1 h with final concentration at 10 μM. DMSO was used as a negative control. ZIKV (MR766 strain) was added at MOI=0.125. After 2 h infection, the supernatant was replaced with fresh medium and compounds from the same chemical library were added to the same wells. After an additional 3 days incubation, cells were fixed, stained with antibodies against ZIKV envelop protein (ZIKV E) and proliferation marker Ki67, and analyzed with the ImageXpress Micro Widefield High-Content Analysis System. A two dimensional analysis was performed to pick the primary hit compounds (Figure S1A). X-axis represents the fold change of total cell number, which was calculated by dividing the total cell number of the chemical treated well by the average of total cell number of DMSO treated wells. Y-axis represents the fold change of the percentage of ZIKV infection, which was calculated by dividing the percentage of ZIKV infection of the chemical treated well by the average of the percentage of ZIKV infection of DMSO treated wells. The compounds in which the fold change of total cell number >1 and the fold change of the percentage of ZIKV infection <20% were further evaluated.
Generation of inhibitory curves
hNPCs were plated on 96-well plates with Accutase digestion at 8.5×104/cm2 density and incubated overnight. For generating inhibitory curves of compound for inhibiting ZIKV infection, cells were first treated with DMSO or different doses of hit compounds for 1 h before ZIKV infection. The cells were then infected with ZIKV (MR766 strain) at a MOI=0.125. After 2 h of infection, the virus-containing medium was replaced by fresh virus-free medium and compounds were added again to the same wells. For generating inhibitory curves of compound for eliminating ZIKV, compounds were instead added after ZIKV infection. After 3 days incubation, the culture medium was harvested and applied to the Vero re-infection assay. Vero cells were plated to 96-well plates one night before re-infection. ZIKV containing medium was serial diluted 10, 102, 103, 104, 105 and106 fold for Vero infection.
RNA-seq analysis
hNPCs at 72 h post-inoculation were collected for RNA-seq analysis, including mock-infected hNPCs treated with DMSO, mock-infected hNPCs treated with 25 μM HH, mock-infected hNPCs treated with 15 μM AQ, ZIKV (MR766 strain, MOI=0.125) infected hNPCs treated with DMSO, ZIKV-infected hNPCs treated with 25 μM HH, and ZIKV-infected hNPCs treated with 15 μM AQ. The RNA from hNPCs in each condition was extracted with Absolutely RNA Nanoprep kit (Agilent Technologies, 400753). The RNA quality was validated with a bioanalyzer (Agilent). The cDNA libraries were synthesized using the TruSeq RNA Sample Preparation kit (Illumina) and sequenced in single-read with the HiSeq4000 sequencer (Illumina) at Weill Cornell Genomics Resources Core Facility. The reads were aligned to the human hg19 reference genome with Tophat2 (Kim et al., 2013). Gene expression data were analyzed with Cufflinks (Trapnell et al., 2010). To generate heat maps displaying the differential gene expression patterns of different samples, the expression values (RPKM) were normalized per gene over all samples, to be specific, for each gene we calculated the mean and standard deviation (stdev) of expression over all samples, and linearly transformed the expression value using the formula (RPKM-mean)/stdev. The heat maps were then generated by Heatmap.2 in the R plots package. Gene ontology pathway analysis was performed using the DAVID function annotation tool.
Immunofluorescence Staining
Cells were fixed with 4% paraformaldehyde for 20 min at room temperature (RT) and blocked in a solution of Mg2+ and Ca2+ free PBS containing 5% horse serum and 0.3% Triton-X for 1 h at room temperature and followed by incubation with primary antibody at 4°C overnight. The following primary antibodies and dilutions have been used in this study: mouse anti-Flavivirus group antigen antibody (ZIKV E) (1:2000; Millipore, clone D1-4G2-4-15), mouse anti-Ki67 (1:200, DAKO, MIB-1), rabbit anti-Ki67 (1:500, Thermofisher, SP-6), rabbit anti-cleaved caspase-3 (1:1000, Cell Signaling Technology, Asp15), goat anti-SOX2 (1:100, Santa Cruz, sc-17320), rabbit anti-SOX2 (1:200, Biolegend, N-term), mouse anti-NESTIN (1:1000, Neuromics, MO15012), rabbit anti-TUJ1 antibody (1:500, Covance, MRB-435P), and chicken anti-MAP2 (1:1000, Abcam, Ab5392). The secondary antibodies include donkey anti-mouse, goat, rabbit or chicken secondary antibodies conjugated with Alexa-Fluor-488, Alexa-Fluor-594 or Alexa-Fluor-647 fluorophore (1:500, Life Technologies). Nuclei were counterstained by DAPI.
The immunohistochemical analysis of mouse tissues was performed on cryosections. In brief, the tissues were fixed overnight with 4% PFA at 4°C, sunk in 20% sucrose, embedded in OCT, and then sectioned at 30 μm for immunostaining. The primary and secondary antibodies were used as described above.
RNA Extraction and qRT-PCR
Total RNA of cultured cells and organs was extracted with RNeasy plus mini kit (Qiagen). vRNA from cell culture supernatant was extracted with QIAampe viral RNA mini kit (Qiagen). Reverse transcription was carried out with strand specific tagged primers of ZIKV plus ACTB gene specific primer of human or mouse using High Capacity cDNA Reverse Transcription kit (Thermo Fisher). qPCR reactions were performed with PrimeTime Gene Expression 2X Master Mix (IDT DNA) and probes for ZIKV and human or mouse ACTB. The PCR products were analyzed with Sanger sequencing to validate the specificity of “+” and “−” strand primers. ZIKV expression level was then normalized to human ACTB or mouse Actb correspondingly. Primer sets used in RT and qPCR and probes used in qPCR were listed in Table S2.
Adult mouse infection and in vivo drug testing
6–8 week old female SCID-Beige mice were infected with ZIKV (2.5 x105 PFU in 0.5 ml culture fluid) through intraperitoneal injection. For pre-infection treatment, drug candidates were administrated to mice 12 h before ZIKV inoculation and followed by treatment once per day. HH was administrated subcutaneously at the dose of 100 mg/kg body weight. AQ was administrated with intraperitoneal injection at the dose of 40 mg/kg body weight. PBS was used as the vehicle control. For ZIKV kinetics analysis, 6–8 weeks old female SCID-beige mice were inoculated with (1 x106 PFU for MR766 strain or 1 x107 PFU for FSS13025 strain in 0.5 ml culture fluid) through intraperitoneal injection. The mice were euthanized at the indicated time points and analyzed with qRT-PCR to validate the ZIKV level in vivo. At 24 hpi or 5 dpi, HH was administrated subcutaneously at the dose of 100 or 200 mg/kg body weight per day for 5 days (MR766 strain). At 7 dpi, HH was administrated subcutaneously at the dose of 100 mg/kg body weight per day for 7 days (FSS13025 strain). After euthanasia, brain, liver, spleen and kidney were collected and analyzed using qRT-PCR. Blood samples from mice treated with HH with dose of 200 mg/kg body weight after 5 days treatment were analyzed with Vero infection assay.
Vero assay to monitor the infectious particular in vivo
Whole blood samples were collected from tail vein and serum was isolated with centrifuge. Solid samples (brain, liver, spleen and kidney) were harvested from infected mice after euthanasia. Samples were freshly homogenized with cold culture medium (DMEM + 10% fetal bovine serum) and then centrifuged (2000g for 10 minutes at 4 °C). Supernatant and serum were then diluted serially to infect Vero cells in 96-well plates.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification data are presented as mean ± SEM. N=3 independent biological replicates were used if not otherwise specifically indicated. n.s. indicates non-significant difference. For in vitro experiments, p values were calculated by unpaired two-tailed Student’s t-test if not otherwise specifically indicated. For in vivo experiments, p values were calculated by one-way repeated measures ANOVA or two-way repeated measures ANOVA with a Bonferroni test for multiple comparisons using Prism 6. *p<0.05, **p<0.01 and ***p<0.001.
DATA AND SOFTWARE AVAILABILITY
The accession number for the RNA-seq data reported in this paper is GEO: GSE89334.
Supplementary Material
Acknowledgments
S.C. is funded by NIH (DP2 DK098093-01, DP3DK111907-01). S.C. and K.J.B are New York Stem Cell Foundation-Robertson Investigators. T.Z. is funded by a FFPI program at Weill Cornell. G.Y.C. is supported by an F30 Predoctoral fellowship from the NIMH (F30 MH113343-01A1) and a training grant from the NIGMS (T32GM007739) to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program. K.J.B is funded by the Brain and Behavior Research Foundation, Brain Research Foundation, NIH (R01MH101454 and R01MH106056), and the New York Stem Cell Foundation. R.E.S is funded by NIH (1R21AI117213 and 1K08DK101754). This study was also supported by a Shared Facility contract to T.E. and S.C. from NYSTEM (C029156), and a Shared Facility contract to M.T. from NYSTEM (C029153). We thank Drs. Charles M. Rice and Margaret R. MacDonald for providing the ZIKV FSS13025 strain. We are also very grateful for technical support provided by Harold S. Ralph in the Cell Screening Core Facility and Tuo Zhang and Jenny Xiang in the Genomic Core Facility at Weill Cornell Medical College, NY.
Footnotes
Author Contributions. S.C., L.S., T.Z., M.T., Q.Z., and L.T. designed the project, T.Z., L.T., G.Y.C., B.J.H., K.J.B., Y.F., and S.M. performed additional necessary experiments. T.Z., L.T., G.C., and S.C. analyzed data, T.Z., L.T., G.C., Q.Z., R.E.S., T.E., and S.C. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barrows NJ, Campos RK, Powell ST, Prasanth KR, Schott-Lerner G, Soto-Acosta R, Galarza-Munoz G, McGrath EL, Urrabaz-Garza R, Gao J, et al. A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection. Cell host & microbe. 2016;20:259–270. doi: 10.1016/j.chom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershteyn M, Kriegstein AR. Cerebral organoids in a dish: progress and prospects. Cell. 2013;155:19–20. doi: 10.1016/j.cell.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyasuppayakorn S, Reichert ED, Manzano M, Nagarajan K, Padmanabhan R. Amodiaquine, an antimalarial drug, inhibits dengue virus type 2 replication and infectivity. Antiviral Res. 2014;106:125–134. doi: 10.1016/j.antiviral.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao-Lormeau VM, Blake A, Mons S, Lastere S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, et al. Guillain-Barre Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet. 2016;387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carteaux G, Maquart M, Bedet A, Contou D, Brugieres P, Fourati S, Cleret de Langavant L, de Broucker T, Brun-Buisson C, Leparc-Goffart I, et al. Zika Virus Associated with Meningoencephalitis. N Engl J Med. 2016;374:1595–1596. doi: 10.1056/NEJMc1602964. [DOI] [PubMed] [Google Scholar]
- Chen D, Cai J, Cheng J, Jing C, Yin J, Jiang J, Peng Z, Hao X. Design, Synthesis and Structure-Activity Relationship Optimization of Lycorine Derivatives for HCV Inhibition. Scientific reports. 2015;5:14972. doi: 10.1038/srep14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng YQ, Zhang NN, Li CF, Tian M, Hao JN, Xie XP, Shi PY, Qin CF. Adenosine Analog NITD008 Is a Potent Inhibitor of Zika Virus. Open Forum Infect Dis. 2016;3:ofw175. doi: 10.1093/ofid/ofw175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352:816–818. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- Govero J, Esakky P, Scheaffer SM, Fernandez E, Drury A, Platt DJ, Gorman MJ, Richner JM, Caine EA, Salazar V, et al. Zika virus infection damages the testes in mice. Nature. 2016;540:438–442. doi: 10.1038/nature20556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanners NW, Eitson JL, Usui N, Richardson RB, Wexler EM, Konopka G, Schoggins JW. Western Zika Virus in Human Fetal Neural Progenitors Persists Long Term with Partial Cytopathic and Limited Immunogenic Effects. Cell Rep. 2016;15:2315–2322. doi: 10.1016/j.celrep.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Qi WB, Wang L, Tian J, Jiao PR, Liu GQ, Ye WC, Liao M. Amaryllidaceae alkaloids inhibit nuclear-to-cytoplasmic export of ribonucleoprotein (RNP) complex of highly pathogenic avian influenza virus H5N1. Influenza Other Respir Viruses. 2013;7:922–931. doi: 10.1111/irv.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, Diamond MS. A Mouse Model of Zika Virus Pathogenesis. Cell host & microbe. 2016;19:720–730. doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Saucedo-Cuevas L, Regla-Nava JA, Chai G, Sheets N, Tang W, Terskikh AV, Shresta S, Gleeson JG. Zika Virus Infects Neural Progenitors in the Adult Mouse Brain and Alters Proliferation. Cell Stem Cell. 2016;19:593–598. doi: 10.1016/j.stem.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecharles S, Herrmann C, Poullain P, Tran TH, Deschamps N, Mathon G, Landais A, Breurec S, Lannuzel A. Acute myelitis due to Zika virus infection. Lancet. 2016;387:1481. doi: 10.1016/S0140-6736(16)00644-9. [DOI] [PubMed] [Google Scholar]
- Miner JJ, Sene A, Richner JM, Smith AM, Santeford A, Ban N, Weger-Lucarelli J, Manzella F, Ruckert C, Govero J, et al. Zika Virus Infection in Mice Causes Panuveitis with Shedding of Virus in Tears. Cell reports. 2016;16:3208–3218. doi: 10.1016/j.celrep.2016.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlakar J, Korva M, Tul N, Popovic M, Poljsak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodusek V, et al. Zika Virus Associated with Microcephaly. The New England journal of medicine. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- Olliaro P, Mussano P. Amodiaquine for treating malaria. Cochrane Database Syst Rev. 2003:CD000016. doi: 10.1002/14651858.CD000016. [DOI] [PubMed] [Google Scholar]
- Parra B, Lizarazo J, Jimenez-Arango JA, Zea-Vera AF, Gonzalez-Manrique G, Vargas J, Angarita JA, Zuniga G, Lopez-Gonzalez R, Beltran CL, et al. Guillain-Barre Syndrome Associated with Zika Virus Infection in Colombia. N Engl J Med. 2016 doi: 10.1056/NEJMoa1605564. [DOI] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. Zika Virus and Birth Defects--Reviewing the Evidence for Causality. The New England journal of medicine. 2016;374:1981–1987. doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, Yao B, Shin J, Zhang F, Lee EM, et al. Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell. 2016;18:587–590. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol A, Tran NN, Brennand KJ. A guide to generating and using hiPSC derived NPCs for the study of neurological diseases. J Vis Exp. 2015:e52495. doi: 10.3791/52495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Lee EM, Wen Z, Cheng Y, Huang WK, Qian X, Tcw J, Kouznetsova J, Ogden SC, Hammack C, et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med. 2016 doi: 10.1038/nm.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmurko J, Marques RE, Schols D, Verbeken E, Kaptein SJ, Neyts J. The Viral Polymerase Inhibitor 7-Deaza-2′-C-Methyladenosine Is a Potent Inhibitor of In Vitro Zika Virus Replication and Delays Disease Progression in a Robust Mouse Infection Model. PLoS Negl Trop Dis. 2016;10:e0004695. doi: 10.1371/journal.pntd.0004695. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.