Abstract
Immune checkpoint blockade has shown significant promise as an anti-cancer treatment, yet the determinants of response are not completely understood. Here, we show that somatic mutations in SERPINB3 and SERPINB4 are associated with survival following anti-CTLA4 immunotherapy in two independent cohorts of melanoma patients (n=174). Interestingly, serpins are homologues of the well-known ovalbumin antigen and are associated with autoimmunity. Our findings have implications for the personalization of immunotherapy.
Main Text
Immune checkpoint inhibitors have shown exceptional promise in the treatment of several advanced malignancies. For example, treatment with ipilimumab, an anti-CTLA4 antibody, has increased survival rates for patients with melanoma.1,2 Anti-PD1 blockade has shown therapeutic efficacy in cancers such as melanoma, non-small cell lung cancer, and renal cell cancer, amongst others.3–5 Understanding the genetic determinants of response to immune checkpoint blockade is critical for determining which patients will benefit from immunotherapy and for design more effective treatment options.
We and others have previously shown that the genetic features of cancers can shape the susceptibility of tumors to immune checkpoint blockade therapy.6–9 For example, neoantigen load, mutational load, and tumor clonality can affect the likelihood of response to anti-CTLA4 or anti-PD1.6–10 Prior sequencing studies have suggested that lung cancer patients with elevated smoking-related mutagenesis were more likely to respond to anti-PD1 therapy.7 However, it is unknown whether the presence of mutations in specific genes can influence response rates for immune checkpoint inhibitors in a manner analogous to how EGFR mutations predict response to erlotinib.
To address this issue in a rigorous manner, we analyzed the exomes of matched tumor and normal pairs from 174 melanoma patients treated with anti-CTLA4 therapy. These patients were from two independent cohorts; one from the United States generated by us (n=64, cohort 1) and a second from Germany (n=110 cohort 2).6,9 These data, along with a recently published analysis of mutations in melanoma by the TCGA now enable a gene-centric approach to detect recurrently mutated genes that predict survival.11
A comprehensive analysis of recurrent mutations in these datasets was performed to determine association with overall survival after anti-CTLA4 therapy (see Online Methods; Supplementary Table 1).11 Strikingly, we discovered that SERPINB3 was recurrently mutated in patients deriving clinical benefit from anti-CTLA4. These mutations were strongly associated with overall survival following therapy in both independently collected cohorts (Fig. 1a). SERPINB3 encodes a serine protease inhibitor that functions in apoptosis and auto-immunity.11–14 Interestingly, it is a human homologue of the chicken ovalbumin protein (OVA), which is a classic model antigen. We also found mutations in SERPINB4, a close human homologue of SERPINB3 with which it shares 92% protein sequence identity. Not surprisingly, SERPINB3 and SERPINB4 proteins have overlapping functions and are involved in both oncogenesis and immunity.14–16 We have therefore considered mutations in both genes together.
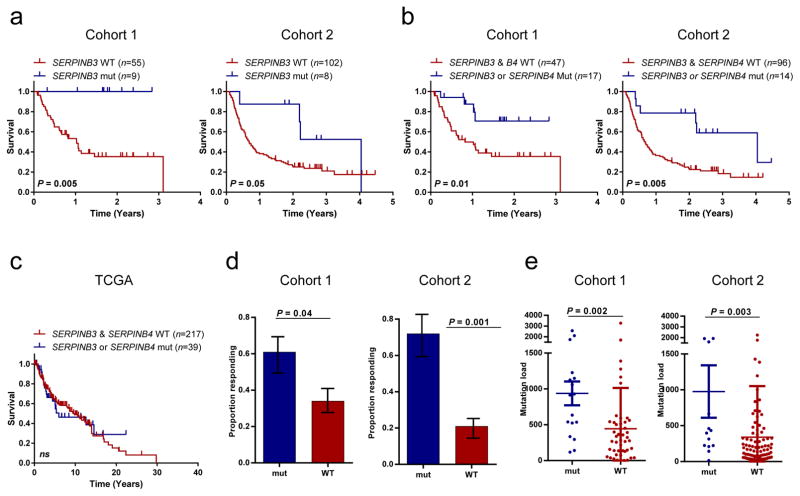
Figure 1. Somatic mutations of SERPINB3 and SERPINB4 predict improved survival from treatment with anti-CTLA4 therapy.
(a) Overall survival of patients with SERPINB3 mutations in cohort 1 (n=64, p=0.005) and cohort 2 (n=110, p=0.05).
(b) Overall survival of patients with either SERPINB3 or SERPINB4 mutations in cohort 1 (p=0.01) and cohort 2 (p=0.005)
(c) Survival by SERPINB3/4 mutations in the TCGA melanoma cohort. All statistical tests are log-rank. (n=262; p=NS)
(d) Proportion of patients with SERPINB3 or SERPINB4 mutations with clinical benefit or response in cohort 1 (p=0.04) and cohort 2 (p=0.001) (Fisher’s exact test; error bars correspond to standard error).
(e) Mutation load as a function of SERPINB3 or SERPINB4 mutations in cohort 1 (p=0.002) and cohort 2 (p=0.003) (Wilcox rank-sum test).
Mutations in either SERPINB3 or SERPINB4 (SERPINB3/B4) were associated with significantly longer survival following anti-CTLA4 treatment in both cohorts (Fig. 1b). Importantly, mutations in SERPINB3/B4 did not associate with survival in metastatic melanoma patients from the Cancer Genome Atlas Project (TCGA), suggesting that these mutations are predictive of response to immunotherapy and not simply prognostic (Fig. 1c). Patients with SERPINB3/B4 mutations were also significantly more likely to experience clinical benefit from anti-CTLA4 in both cohorts (Fig. 1d). Tumors with SERPINB3/B4 mutations were by no means limited to highly mutated tumors (Fig. 1e), and multivariate analysis revealed that SERPINB3/B4 mutations were associated with overall survival independent of mutation load (cohort 1: p=0.05; cohort 2: p=0.01; Online methods; Supplementary Table 2). Mutations occurred in all 4 subtypes of melanoma and appeared to be mutually exclusive of each other (Fig. 2a).
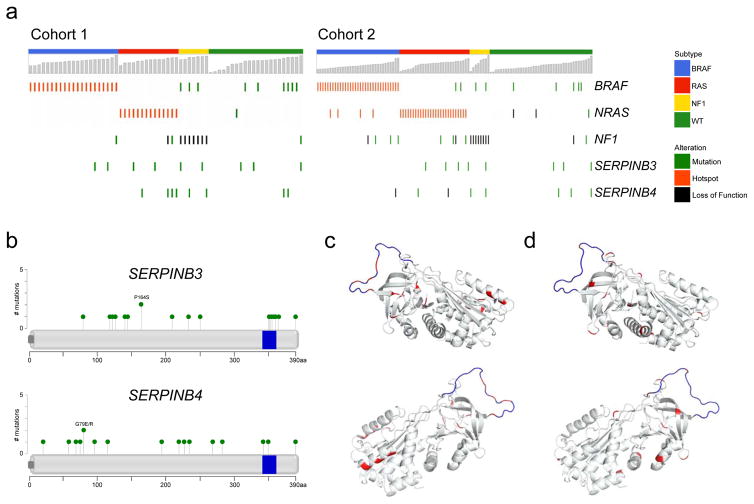
Figure 2. Characteristics of mutations in SERPINB3 and SERPINB4.
(a) Oncoprints of SERPINB3 and SERPINB4 mutations in the two cohorts.
(b) Diagrams showing location of mutations in SERPINB3 and SERPINB4 (data from both cohorts combined). Blue bar represents putative reactive center loop (RCL) domain.
(c) Location of mutations on 3-dimensional protein structure of SERPINB3. Red: mutated amino acid; blue represents putative RCL domain.
(d) Location of mutations on 3-dimensional protein structure of SERPINB4
The characteristics and locations of these mutations are described in Fig. 2b, Supplementary Fig. 1, and Supplementary Table 3. The missense mutations that occur throughout both genes may alter protein activity and in many cases are predicted to produce immunogenic neopeptides (Supplementary Table 4 & 5). Indeed, visualization of mutations on the solved three-dimensional protein structure shows a cluster of mutations near the active site, the reactive center loop (RCL) domain (Fig. 2c,d).
Due to the pleiotropic functions of serpin proteins, a mechanistic link between mutations in genes encoding serpins and immunotherapy response is likely multi-faceted and complex. While elucidation of these molecular details will require additional functional work, our genetic data may provide insight regarding which of the several aspects of serpin biology may be involved. SERPINB3 has been identified as a significantly and recurrently mutated gene in melanoma by the TCGA and another group, pinpointing it as a driver of oncogenesis.11,17 Indeed, serpins are known to exhibit anti-apoptotic functions,13 including suppression of ultraviolet-induced apoptosis in human keratinocytes.18 There are also a number of possible mechanisms by which the observed SERPINB3/B4 mutations may influence tumor immunogenicity. Mutations in various serpin family proteins are known to cause misfolding and self-polymerization, leading to the formation of inflammatory aggregates or plaques. These, in turn, function as targets in various autoimmune diseases, including systemic lupus erythematosus and psoriasis.13,14,19,20 Serpin polymers can also induce autophagy, thereby potentially enhancing auto-antigen presentation.13,21 Therefore, mutant SERPINB3/4 may act as both a driver of melanoma tumorigenesis and/or also as an immunodeterminant, similar to mutant IDH1 in glioma.22
Interestingly, SERPINB3 is a human homologue of the chicken egg protein ovalbumin (OVA), a classic model antigen that contributes to egg allergies and atopic dermatitis in humans.23 OVA and SERPINB3 share sequence similarity, including distinct regions functionally validated as epitopes of human OVA-reactive T cells.24 It has not escaped our attention that many of the observed SERPINB3/B4 mutations occur within these regions of homology (Supplementary Fig. 2). However, these epitopes may or may not serve as direct targets for the adaptive immune system throughout the course of metastatic disease, and alternative mechanisms such as cross presentation and epitope spreading may be involved. Expression data from TCGA suggest SERPINB3/B4 are broadly expressed in primary tumors, but are significantly down-regulated in regional lymph nodes and metastatic sites, perhaps suggesting the occurrence of immuno-editing as tumors evolve or that silencing of these genes occur during metastasis (Supplementary Fig. 3). We hypothesize that, in light of these expression, mutation, and survival data, SERPINB3/B4 mutations may exert an early immunogenic effect, thereby helping to initiate a broad immune response that can later be reinvigorated through checkpoint blockade. Additional mechanistic work will be required to clarify the role of SERPINB3/B4 mutations in immunotherapy response. We believe our findings have broad implications for efforts to personalize immunotherapy for cancer patients.
ONLINE METHODS
Mutational analysis and whole-exome sequencing
Details of the anti-CTLA4 treated patient cohorts are previously described.6,9 Whole-exome sequencing for cohort 1 and cohort 2 had been previously completed with mean depths of coverage of 103 and 183.7 respectively.6,9 Analysis was performed as previously described by DePristo et. al.25 Briefly, paired-end reads in FASTQ format were aligned to the reference human genome GRCh37 using Burrows-Wheeler Aligner (BWA v0.7.10).26 Subsequently, local re-alignment was performed using the Genome Analysis Toolkit (GATK) version 3.2.2.27 Duplicate reads were removed using Picard version 1.119. Somatic single nucleotide variants (SNVs) were identified using a combination of four mutation callers, namely Mutect 1.1.4, Varscan 2.3.7, Somatic Sniper 1.0.4, and Strelka 1.0.13.28–31 Sequence data from both cohorts were analyzed in the same manner. SNVs with an allele read count of less than 5 or with a normal coverage of less than 7 were removed. Small indels were called using GATK 3.2.2.
Statistics and survival analysis
Overall survival information and classification of patients into those with durable clinical benefit and those with minimal benefit were obtained from the original publications.6,9 Survival analysis was performed with the Kaplan-Meier method with differences in survival being determined with the log-rank test. Multivariate survival analysis was performed using a Cox proportional hazards model. Differences in the distribution of quantitative variables between groups were determined with the Wilcox rank-sum test, unless otherwise indicated. Difference in proportions between groups were determined with Fisher’s exact test, unless otherwise indicated. All statistical analyses were performed in the R statistical environment (v3.2).
Association of recurrent mutations with survival
Since our initial report on anti-CTLA4 therapy in melanoma, the TCGA published a comprehensive genomic analysis of melanoma.11 We analyzed the 19 recurrently mutated genes in melanoma identified by InVex and described by the TCGA (Supplemental Table 1).11 All 19 genes were tested for association with overall survival using the chi-square test statistic from the survival package in R and a permutation procedure (Strona et. al.32,33). The overall procedure follows the concept described by Kim et al.34: We created N=10000 permutations of the binary mutation matrix (genes x samples), keeping row- and column sums constant, thereby accounting for potential confounding factors such as mutation load. In each iteration, we recorded the chi-square test statistic for association between permuted mutations and overall survival for all genes. An empiric p-value was derived for each recurrently mutated gene by comparing observed test statistics to the distribution of simulated test statistics. The association of SERPINB3 with OS was significant after Bonferroni correction for multiple testing at P=0.037 (uncorrected p-value = 0.005). No other genes were significant.
We subsequently verified that SERPINB3 was also associated with overall survival in an independently collected group of patients, cohort 2 (from Germany) (p=0.05; Fig. 1a). As SERPINB3/B4 are close homologues, we grouped mutations in these genes together (see main text). Multivariate analysis correcting for M-stage and mutation load demonstrated that SERPINB3/B4 mutations were associated with overall survival in cohort 1 (HR=0.34, 95% CI=0.11—0.98, p=0.05) and cohort 2 (HR=0.32, 95% CI=0.13—0.76, p=0.01) (Supplemental Table 2).
Alignment of Ovalbumin and SERPINB3
Clustal Omega was used to align ovalbumin and SERPINB3. Presented epitopes for SERPINB3 were determined from the literature.35 Immunogenic epitopes from ovalbumin were also determined from the literature.24 The immune epitope database was accessed on 21-March-2016, and was used to identify all relevant epitopes.
Computational Neoantigen Prediction
Class I HLA typing was performed manually for the Cohort I and for Cohort II was computed with polysolver from the exome data by the original authors. Each non-synonymous SNV was translated into a 17-mer peptide sequence, centered on the mutated amino acid. This 17-mer was then used to create 9-mers via a sliding window approach for determination of MHC-Class I binding.36 netMHC version 3.4 was used to determine the binding strength of mutated peptides to patient specific HLA alleles.37 All peptides with a binding score of IC50 < 500 nM were considered as putative class I neoantigens. For class II antigens, a 29-mer peptide sequence was created and a 15-mer sliding window approach was utilized. Class II HLA typing was determined with SOAP-HLA on both cohorts from the exome data. netMHCpan version 3.1 was used to determine the affinity of mutated peptides to patient specific HLA alleles, and those with a rank less than 2 percent, were considered putative Class II neoantigens.38
Supplementary Material
Recurrently mutated genes in melanoma as identified by InVex analysis performed by TCGA.
Multivariate model of overall survival and SERPINB3/B4 mutations.
SERPINB3/B4 mutations in both cohorts of patients.
MHC Class I predicted neoantigens from SERPINB3/B4 mutations.
MHC Class II predicted neoantigens from SERPINB3/B4 mutations.
Acknowledgments
We thank Jedd Wolchok, Taha Merghoub, Jianda Yuan, Phillip Wong, and Alexandra Snyder for collaborative interactions. We thank the Integrated Genomics Operation and the Ludwig Immune Monitoring Facility at MSK for technical assistance. We thank Adriana Heguy and the Genome Technology Center at NYU and Levi Mangarin for assistance with validation sequencing. This work was funded by a Pershing Square Sohn Cancer Research grant (TAC), the Frederick Adler Chair (TAC), Stand Up 2 Cancer (TAC), and the STARR Cancer Consortium (TAC). TAC is a co-founder of Gritstone Oncology.
Footnotes
URLs: R-project available from http://www.r-project.org/. IEDB access available at http://www.iedb.org
Code Availability: Code and ancillary data necessary to reproduce results and figures is available upon request
AUTHOR CONTRIBUTIONS: T.C. and N.R. designed and conceived the study. Analysis of mutations in individual genes with outcome was performed by S.K, N.W., N.R., V.M., and J.H. Neoantigen analysis was performed by J.H., S.K., and V.M. Analysis of expression data was performed by L.W., A.D., and N.R.. N.R., J.H., and T.C. prepared the manuscript. All authors participated in the discussion of the final manuscript and interpretation of results.
Accession Numbers
Exome sequencing data for cohort 1 is available in dbGap at accession number phs001041.v1.p1, while cohort 2 is available with accession number phs000452.v2.p1
References
- 1.Hodi FS, et al. N Engl J Med. 2010;363:711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robert C, et al. N Engl J Med. 2011;364:2517–26. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- 3.Brahmer J, et al. N Engl J Med. 2015;373:123–35. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motzer RJ, et al. N Engl J Med. 2015;373:1803–13. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robert C, et al. N Engl J Med. 2015;372:320–30. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 6.Snyder A, et al. N Engl J Med. 2014;371:2189–99. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rizvi NA, et al. Science. 2015;348:124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le DT, et al. N Engl J Med. 2015;372:2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Allen EM, et al. Science. 2015;350:207–11. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGranahan N, et al. Science. 2016 [Google Scholar]
- 11.Cancer Genome Atlas, N. Cell. 2015;161:1681–96. [Google Scholar]
- 12.Vidalino L, et al. Autoimmun Rev. 2009;9:108–12. doi: 10.1016/j.autrev.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 13.Gatto M, et al. Clin Rev Allergy Immunol. 2013;45:267–80. doi: 10.1007/s12016-013-8353-3. [DOI] [PubMed] [Google Scholar]
- 14.Sivaprasad U, et al. J Invest Dermatol. 2015;135:160–9. doi: 10.1038/jid.2014.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider SS, et al. Proc Natl Acad Sci U S A. 1995;92:3147–51. doi: 10.1073/pnas.92.8.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Catanzaro JM, et al. Nat Commun. 2014;5:3729. doi: 10.1038/ncomms4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guan J, Gupta R, Filipp FV. Sci Rep. 2015;5:7857. doi: 10.1038/srep07857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katagiri C, Nakanishi J, Kadoya K, Hibino T. J Cell Biol. 2006;172:983–90. doi: 10.1083/jcb.200508064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Rachkidy RG, Young HS, Griffiths CE, Camp RD. J Invest Dermatol. 2008;128:2219–24. doi: 10.1038/jid.2008.71. [DOI] [PubMed] [Google Scholar]
- 20.Lysvand H, Hagen L, Klubicka L, Slupphaug G, Iversen OJ. Biochim Biophys Acta. 2014;1842:734–8. doi: 10.1016/j.bbadis.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 21.Munz C. Front Immunol. 2012;3:9. doi: 10.3389/fimmu.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schumacher T, et al. Nature. 2014;512:324–7. doi: 10.1038/nature13387. [DOI] [PubMed] [Google Scholar]
- 23.Mine Y, Yang M. J Agric Food Chem. 2008;56:4874–900. doi: 10.1021/jf8001153. [DOI] [PubMed] [Google Scholar]
- 24.Holen E, Elsayed S. Clin Exp Allergy. 1996;26:1080–8. [PubMed] [Google Scholar]
- 25.DePristo MA, et al. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Durbin R. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenna A, et al. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cibulskis K, et al. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koboldt DC, et al. Genome Res. 2012;22:568–76. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larson DE, et al. Bioinformatics. 2012;28:311–7. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saunders CT, et al. Bioinformatics. 2012;28:1811–7. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 32.Strona G, Nappo D, Boccacci F, Fattorini S, San-Miguel-Ayanz J. Nat Commun. 2014;5:4114. doi: 10.1038/ncomms5114. [DOI] [PubMed] [Google Scholar]
- 33.Gotelli NJ, Ellison AM. EcoSimR 1.00 [Google Scholar]
- 34.Kim J, et al. Nat Genet. 2016 [Google Scholar]
- 35.Schellens IM, et al. PLoS One. 2015;10:e0136417. doi: 10.1371/journal.pone.0136417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snyder A, Chan TA. Curr Opin Genet Dev. 2015;30:7–16. doi: 10.1016/j.gde.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen M, et al. Protein Sci. 2003;12:1007–17. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andreatta M, et al. Immunogenetics. 2015;67:641–50. doi: 10.1007/s00251-015-0873-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Recurrently mutated genes in melanoma as identified by InVex analysis performed by TCGA.
Multivariate model of overall survival and SERPINB3/B4 mutations.
SERPINB3/B4 mutations in both cohorts of patients.
MHC Class I predicted neoantigens from SERPINB3/B4 mutations.
MHC Class II predicted neoantigens from SERPINB3/B4 mutations.