Abstract
Non-functional pituitary adenomas (NFPAs) are among the commonest intracranial neoplasms. While histologically benign, NFPAs sometimes become large enough to limit therapeutic options and reduce quality of life. Investigations of the molecular etiology of NFPAs have failed to identify prevalent genetic changes and, while a role for p53 has been suggested, TP53 gene alterations have yet to be described in NFPAs. We found that the polymorphism rs1042522:C > G in codon 72 of exon 4 of the TP53 gene, whose C variant produces a proline and is more common in most ethnicities, has a G variant producing an arginine in 79.8% of NFPAs (n = 42; p < 1.411 × 10−18 vs. 1000 Genomes database), causing patients to present a decade earlier with symptomatic NFPAs. In cultured NFPA cells, transfection with the rs1042522 G variant versus the C variant reduced expression of cell arrest gene p21 and increased proliferation. These findings suggest that this TP53 polymorphism influences NFPA growth.
Keywords: p53, Polymorphism, Pituitary adenoma
1. Introduction
Non-functional pituitary adenomas (NFPAs) are among the most common primary brain tumors (Trouillas et al., 2013), and range from benign, slow growing tumors to more aggressive versions that can exert a devastating impact on a patient’s quality of life through mass effect on neuroanatomical structures causing hypopituitarism, vision loss, and debilitating headaches. Once an MRI is obtained showing a lesion suspected to be an NFPA, endonasal transsphenoidal surgery remains the most common surgical approach used for lesions that require surgery based on their size and/or associated symptoms. Transsphenoidal surgery offers the benefits of confirming the diagnosis pathologically and cytoreductive treatment, but sometimes fails to provide symptomatic relief or adequate extent of resection for patients whose tumors at presentation have already extended into critical adjacent neuroanatomic structures.
TP53 is a ubiquitously expressed tumor suppressor known to be downregulated or dysfunctional in several human tumors (Lakin and Jackson, 1999), but remains understudied and poorly understood in the pathogenesis of pituitary adenoma. The existing literature includes only limited studies using immunohistochemistry to support a role for p53 loss in promoting growth of an already developed pituitary adenoma into an adenoma of sufficient size to become symptomatic. Recognizing this data, the World Health Organization classifies the 15–20% of adenomas with MIB-1 proliferative index greater than 3%, excessive p53 immunoreactivity (an indirect correlate of mutation), and increased mitotic activity as atypical adenomas (Zada et al., 2011; Zhou et al., 2014). However, no study to date has identified TP53 somatic mutations or polymorphisms associated with pituitary adenomas, leading some to suggest that adenomas are instead driven by alterations in accessory proteins that lead to inactivation of p53 signaling (Zhou et al., 2014). Of note, the only reported search for TP53 mutations or polymorphisms associated with pituitary adenomas was a single study sequencing exons 7 and 8 of the TP53 gene in 15 NFPAs. While the choice of these exons to screen was reasonable due to the fact that they house 98% of the TP53 mutations observed in other cancers (Levy et al., 1994), the coverage of the TP53 gene in this study was still incomplete. To address this knowledge gap, we sequenced the entire TP53 gene in a large cohort of NFPAs and matched patient lymphocytes. In doing so, we sought to determine whether the p53 immunostaining abnormalities felt to drive a more aggressive phenotype in pituitary adenomas are reflective of genetic changes in TP53 and whether the identified genetic changes are biologically and clinically significant. Besides somatic mutations, polymorphisms arising in TP53 may also contribute to alterations of the normal functioning of its gene product, such that single nucleotide polymorphisms can impact susceptibility to cancers. Thus, we initiated a search for TP53 mutations and polymorphisms as part of our comprehensive analysis.
2. Materials and methods
2.1. Obtaining and preparing patient samples for TP53 gene sequencing: Collection protocols, inclusion criteria, and ethics
Screening of TP53 for potential mutations was carried out on both fresh pituitary adenoma tissue, obtained from transsphenoidal surgeries performed at University of California San Francisco on patients with no other cancers, and lymphocytes isolated from paired whole blood obtained from these same patients at the time of surgery through venous needle stick allowing blood to be siphoned into a vacutainer sprayed with EDTA solution to prevent clotting.
Samples were preserved through the services of the Brain Tumor Research Center Tissue Core, which obtained the samples after informed consent from patients. Tissue samples were flash frozen in liquid nitrogen at explantation, while blood was kept refrigerated. In order to be included in our primary cohort, the pathologic diagnosis of NFPA was rendered by the neuropathologist from the surgical specimen, with the nonfunctional status confirmed by lack of hormonal hypersecretion in assays of blood hormone levels run from the clinic. In order to be included in our secondary cohort, the pathologic diagnosis of prolactinoma was rendered by the neuropathologist from the surgical specimen based on immunohistochemistry revealing pituitary adenoma staining for prolactinoma, with the biochemical hypersecretion of prolactin confirmed by assays of blood hormone levels run from the clinic.
2.2. PCR reactions for gene sequencing
Samples for sequencing were retrieved from storage, dissociated through a combination of ATL buffer, a tissue lysis buffer for use in purification of nucleic acids (Qiagen; Valencia, CA), and Proteinase K digestion as well as passaged through a 21-gauge syringe, and then processed to obtain genomic DNA using the DNeasy Blood and Tissue Kit (Qiagen), following standard manufacturer’s protocol. A set volume of paired whole blood from each patient was diluted in a mixture of PBS and Proteinase K, then processed for DNA extraction, again following manufacturer’s protocol of the DNeasy Blood and Tissue Kit.
Primers for PCR of coding regions of TP53 were created utilizing UCSC genome browser data (GRCh38/hg38) for genomic sequences and Primer3 primer design software (http://biotools.umassmed.edu/bioapps/primer3_www.cgi). Primer sequences were subject to Nucleotide BLAST against the human genome to confirm binding specificity. Due to intron size, coding regions were amplified with four independent PCR reactions with the following primer sequences: PCR1_FW5′–CTTGGGTTGTGGTGAAACATTG–3′; PCR1_RV 5′ – GGAATCCCAAAGTTCCAAAC –3′; PCR2_FW 5′–GAGGTGCTT ACGCATGTTTG–3′; PCR2_RV 5′–TGGGGTTATAGGGAGGTCAA–3′; PCR3_FW 5′–TGCCACAGGTCTCCCCAAGGCG–3′; PCR3_RV 5′-CCCCAATTGCAGGTAAAACA–3′; PCR4_FW 5′–CATGTTGCTTTTG TACCGTCAT–3′; PCR4_RV 5′–GCAAGCAAGGGTTCAAAGAC–3′. The primers were designed in intronic regions and required to be > 50 bp from any exon boundary. Amplification was performed using Platinum Taq polymerase (Invitrogen; Grand Island, NY) on a thermal cycler following standard manufacturer’s guidelines for Platinum Taq: 94° C for 2 min, 35 cycles of a) 94° C for 30 s, b) specific Tm for primer set for 35 s and c) 72° C for 1 min/kb, followed by 72° C completion for 10 min and holding at 4° C until collection. Tm values for each primer set are: PCR1 61° C; PCR2 62° C; PCR3 62° C and PCR4 64° C. PCR product sizes were subsequently confirmed using a 1% agarose gel. Exonuclease and shrimp alkaline phosphatase (Exo-SAP-IT; Affymetrix; Santa Clara, CA) was used to clean up unincorporated primers, leftover dNTPs and partial single stranded DNA from the PCR product. The primers used for PCR amplification were also used for sequencing; the 50 + base pair cushion was utilized to ensure that all coding regions were clearly sequenced. Samples were submitted, with both forward and reverse primers, to QuintaraBio, and sequence files were retrieved from the company servers the next day.
2.3. Analyzing archived sequences
Sequences previously deposited into the SRA database (Sequence Read Archive; https://www.ncbi.nlm.nih.gov/sra) were manually searched for pituitary adenoma projects, excluding those that focused solely on hormonally active adenomas. The project initiated by the Wang Lab at University of Southern California (SRA #SRP035646) was identified and consisted of RNA-sequencing of 24 human pituitary adenomas, of which 17 were classified as NFPA. Sequence reads were downloaded, then set as the reference genome in BLAST and compared to the TP53 coding sequence surrounding the P72R variant to obtain genotypes at that locus for all 17 NFPA samples.
TCGA archived sequence data was analyzed using the official NIH data repository for genomic data sharing, the Genomic Data Commons (https://gdc.cancer.gov) and cBioportal, a secondary analysis software that assists in TCGA data visualization (www.cbioportal.org) (Cerami et al., 2012; Gao et al., 2013). TCGA data in all available cancer types was limited to TP53 and then manually searched for P72R alterations. When available, allelic frequencies of this variant were collected and used for comparison to our 42 patient NFPA cohort.
2.4. Culturing and transfecting primary pituitary adenoma cells
To establish SF10052 primary pituitary adenoma cells in culture, an NFPA was provided fresh from the operating room by the UCSF Neurosurgery Tissue Bank. This tumor was confirmed to have the C/ C genotype of the rs1042522 polymorphism through the methods described above. The tumor was mechanically dissociated with a razor blade in 150 µL digestion buffer. Enzymatic digestion buffer consisted of RPMI-1640 (UCSF Cell Culture Facility) with 10% FBS (UCSF Cell Culture Facility; San Francisco, CA), 100 µL/mL DNase I (Roche Diagnostics; Indianapolis, IN; #10104159001), and 200 U/mL Collagenase IV (Life Technologies; San Francisco, CA; #17104019). Once fine, tissue suspensions were transferred to a vial containing 7 mL digestion buffer and incubated at 37 °C for 45 min while rotating. Tumor suspensions were then washed with 5% FBS in DPBS (UCSF Cell Culture Facilities) and passed through a 70 µm cell strainer (Corning; Corning, NY; #352350). Unstrained tumor chunks were transferred to a 100 mm petri dish (Corning, #430167) with DMEM + 10% FBS + 100 U/mL Penicillin and 100 µg/mL Streptomycin (UCSF CCF). Strained cells were washed and pelleted with 5% FBS in DPBS twice before treatment with 1x RBC Lysis Buffer by manufacturer’s instructions (eBioscience; San Diego, CA; #00–4300-54). Cell suspensions were then washed with 5% FBS in DPBS, pelleted, and plated on matrigel coated flasks at 3.5 × 105 viable cells/mL in complete DMEM. To establish matrigel coated flasks, T-75 flasks (Greiner Bio-One; Monroe, NC; #82050-856) were coated with 3 mL Matrigel (Corning, CB40230) at 1:30 dilution in DPBS. Vented flasks were allowed to air dry in a biosafety cabinet for 4 h at room temperature before use. Once established, these SF10052 human pituitary adenoma cells were transfected with the pCMV6-P72 or −R72 plasmids described above, using FuGene transfection reagent according to manufacturer’s protocol (Promega; Madison, WI).
2.5. Plasmid construction
A vector for expressing p53 cDNA was purchased from Origene (Origene #RC200003; Rockville, MD); the construct was a pCMV6-Entry plasmid containing a CMV promoter upstream of the full-length p53 cDNA ORF with the more common C nucleotide variant of the rs1042522 polymorphism, followed by an attached FLAG tag (DYKDDDDK motif) and was dubbed pCMV-P72-FLAG. The G variant of the rs1042522 polymorphism was introduced to replace the C allele using the Agilent Quik-Change II Site Directed Mutagenesis PCR kit (Agilent #200523; Santa Clara, CA), following all manufacturers’ directions for primer design and PCR, resulting in the vector pCMV6-R72-FLAG. The FLAG tag was then removed from both plasmids utilizing the same Site Directed mutagenesis PCR kit and a stop codon introduced to establish a tag-less version of each plasmid type (pCMV6-P72 and pCMV6-R72 respectively). Base pair changes and insertions/deletions were confirmed by sequencing before use.
For the dual luciferase reporter vector, we utilized the pGL4.23 luciferase reporter vector (Promega; Madison, WI) containing a minimal promoter upstream of the luciferase reporter gene luc2 (Photinus pyralis) and a multiple cloning region upstream of the minimal promoter to allow cloning of a regulatory sequence of choice. The reporter vector was then constructed by inserting a known p53 consensus sequence (5′ - AGGCATGTCTTCTGTACGGA −3′) (el-Deiry et al., 1992) upstream of the minimal promoter of the luciferase gene in the pGL4 vector (pGL4-p53-Luc).
2.6. Dual luciferase transfection assay
SF10052 cells cultured from resected pituitary samples were used as a host for expression analysis, as described above. Trans-fection of the pGL4-p52-Luc vector was done simultaneously with 1) either pCMV6-P72 or pCMV6-R72 for dual-luciferase experimental data and 2) hRenilla vector utilized as a transfection control. Transfections were carried out using FuGene reagent (Promega #E2691; Madison, WI) following manufacturer’s guidelines. Promega Dual Luciferase Reporter Assay System (#E1910) was used to provide luciferin substrate for luminescence production, which was measured with a GloMax 96 microplate luminometer. Trans-fection efficiency was normalized with the luminescence of the Renilla luciferase. The DNA ratio of p53 constructs to the hRenilla vectors was set at 50 to 1 after a series of optimization trials. Untransfected cells produced negligible levels of both the firefly and Renilla luciferase activities (data not shown).
2.7. Quantitative PCR
SF10052 cells for quantitative PCR (qPCR) experiments were left as untransfected cells or transfected with pCMV6-P72 or −R72, as described above. Cells were washed and removed with manual scraping 48 h post-transfection. Cellular RNA was then isolated using the RNeasy kit (Qiagen #74104) and reverse-transcribed into cDNA with Superscript III (Invitrogen #18080051). This cDNA was diluted to a constant concentration for all samples to ensure similar nucleic acid loading levels. Quantitative PCR was carried out using Power Syber Green Master Mix (Applied Biosystems) with the following primers: p21 FW 5′- GGATGTCCGTCAGAACCCATG −3′ and RV 5′- CCATTAGCGCATCACAGTCGC −3′; VEGF FW 5′-CTTGCCTTGCTGCTCTACCT –3′ and RV 5′ – CACA-CAGGATGGCTTGAAGA –3′; GAPDH FW 5′- GCTGA-GAACGGGAAGCTTGT-3′ and RV 5′-TCTCCATGGTGGTGAAGA CG-3′. qPCR was carried out on an Applied Biosystems StepOne Real-Time PCR cycler following the recommended amplification guidelines described by Applied Biosystems for Syber: 95° C for 10 min, followed by 40 cycles of 95° C for 15 s and 60° C for 1 min. Ct values were calculated using the StepOne software accompanying the real-time cycler. Samples were prepared with three technical replicates for each primer pair and used GAPDH as a control housekeeping gene. Results obtained from this experiment were normalized to both GAPDH and sample control using the 2^(−ΔΔCt) method.
2.8. Western blot
SF10052 cells were left as untransfected cells or transfected with pCMV6-P72 or –R72, as described above. At 24 h post-transfection, cells were then washed twice in cold PBS, then removed from the culture plate surface by addition of RIPA lysis buffer (Cell Signaling Technology; Danvers, MA; #9806S), followed by manual scraping of the plate bottom utilizing a sterilized plastic scraper. Lysate was collected in a 1.5 mL Eppendorf tube and allowed to chill on ice for 30 min, then centrifuged at 4 °C for 20 min at 14,000 rpm. The supernatant was removed and saved in a fresh tube on ice. Protein concentrations were determined against a BSA standard curve, using BCA Protein Assay kit (Thermo Fisher #23225; Waltham, MA) and following standard manufacturer’s protocol. Loading samples were prepared with the same amount of protein per well and run on an SDS gel. Completed gels were transferred to a PVDF membrane and blocked for 1 h in TBS-T with 5% milk powder. Blocked membranes were then incubated with primary antibody (p21: Cell Signaling #2947S, 1:1000; VEGF: Abcam #183100, 1:500; MMP2: Cell Signaling #4022S, 1:1000; GAPDH Millipore #MAB374, 1:50,000) overnight. Membranes were then washed in TBS-T, incubated with secondary antibody (Cell Signaling #7076S at 1:8000 for GAPDH, Cell Signaling #7074S, 1:1000 for all other antibodies) for 1 h, rewashed and then exposed using Clarity Western ECL substrate (Bio-Rad #1705060; Hercules, CA). Films were scanned to the computer and imaged, followed by band densitometry quantification, if necessary, using manual boxing of protein bands followed by densitometry measurements in ImageJ.
2.9. Transwell migration
Experiments to compare migration rates of pituitary cell lines were conducted using a Boyden chamber transwell system (8 µm PET membrane, Becton Dickinson; San Diego, CA; #354578), which separated plates into upper and lower chambers. SF10052 pituitary adenoma cells homozygous for the C allele of rs1042522 were either left untransfected or transfected with pCMV6-P72 or pCMV6-R72, as described above. After 24 h to allow vector uptake and expression, cells were seeded at a density of 5 × 105 cells/well in each upper chamber and given media in both chambers. At migration time points, inserts were removed and washed of media before drying for 2 h. The PET membrane was cut off the insert and placed on a slide for imaging with DAPI reagent. Imaging was carried out using a Zeiss Axio Observer inverted microscope (Zeiss; Gottingen, Germany) outfitted with an Axi.Observer.Z1m stand and AxioVision software. Cells in each 40x frame were counted using ImageJ software (Version 1.5, National Institutes of Health; Bethesda, MD) (Schneider et al., 2012). Files were first converted to 16-bit grayscale images, then the threshold was manually adjusted to highlight cells while reducing background noise. The resultant image was then examined and noticeably overlapping cells were separated using a white line divider. Finally, cells were counted using ImageJ’s automated “Analyze Particles” feature.
2.10. CyQuant assay
SF10052 cells that were either untransfected or transfected with pCMV6-P72 or pCMV6-R72, as described above, were plated at a density of 105 cells/well in a 96 well culture plate, then incubated at 37 °C for multiple independent time points. At the specified time point, plate was removed from 37 °C incubation, centrifuged for 3 min at 3000 rpm and each well was aspirated of media. Plates were then frozen at −80 °C for at least 2 h (longer times permitted). Once all time points were frozen, plates were brought to room temperature and proliferation assays were carried out using the CyQUANT Cell Proliferation Assay Kit (ThermoFisher #C7026), following standard manufacturer’s protocol.
2.11. Metabolism measurement
SF10052 cells were seeded at 5 × 105 cells per well in a 6 well plate, then specified wells were transfected with either pCMV6-P72 or pCMV6-R72. Cells were cultured in either normoxia (5% CO2, 20% O2, 74% N2) or hypoxia (5% CO2, 1.5% O2, 94% N2) in a humidified O2 Control incubator (Sanyo; Burlingame, CA) that contained only the cells being studied and was not opened during incubation periods. These cells were then analyzed via a colorometric Glycolysis Cell-Based Assay Kit (Cayman #600450; Ann Arbor, MI).
2.12. Apoptosis measurement
SF10052 cells were seeded at 5 × 105 cells per well in a 6 well plate, then specified wells were transfected with either pCMV6-P72 or pCMV6-R72. Cells were left for 24 h in reduced serum media, then trypsinized off the plate. Cells were washed twice in cold PBS, counted and resuspended in 1X Binding Buffer from the FITC Annexin V Apoptosis Detection kit (BD Biosciences #556547; Franklin Lakes, NJ). Cell concentration was adjusted and Annexin V/PI added according to manufacturer’s protocol. Cells were then flow sorted on a Sony SH800S Cell Sorter, including an unstained population to correct for background signal.
2.13. Measuring p53 degradation
2.13.1. Pulse-chase assay
SF10052 cells were plated at a density of 5.0 × 105 per 60 mm dish two days prior to the experiment. Transient transfections with either pCMV6-P72-FLAG or pCMV6-R72-FLAG were carried out the following day using Fugene reagent according to standard manufacturer’s protocol. After allowing 24 h for transfection and vector expression, pulse-chase experiment was started by replacing media with DMEM media deficient in cysteine and methionine DMEM (ThermoFisher #21013024) for 20 min for depletion. Cells were then aspirated, washed once with phosphate-buffered saline (PBS) and replaced with additional deficient media spiked with 20 µCi of [35S]methionine/cysteine (Perkin Elmer # NEG072014MC; SanJose, CA) per well. Pulse was allowed to continue for 1 h uninterrupted before beginning the chase. For each time point, the cells were washed with PBS and chased with complete DMEM containing 10% calf serum at 37 °C for the indicated amount of time. At end of time point, cells were washed and lysed using 1X RIPA lysis buffer.
2.13.2. Immunoprecipitation and imaging
Cell lysates that underwent the pulse-chase assay described above were cleared by centrifugation at 15,000 revolutions per minute (rpm) in a cold room followed by removal of the supernatant. Immunoprecipitation of the p53 protein tagged with FLAG was extracted from 20 µg of total protein using Protein-A sepharose magnetic beads (VWR international #89129-078) with anti-FLAG antibody (Cell Signaling Technology #2386), following manufacturer’s protocol. The immunoprecipitated protein was run on an SDS-PAGE gel and transferred to a PVDF membrane, then left in darkroom conditions to develop for 7 days. Band intensities were measured by scanning densitometry (ImageJ) of autoradiographs.
2.14. Statistics
Control allele frequencies were obtained from large-scale genomic control databases (HapMap Rel #28 and the SNP500Cancer database from the National Cancer Institute). Allele frequencies for various ethnic groups were weighted in proportion to match the frequency of self-reported ethnicity found in our adenoma cohort. Adjusted allelic frequency was then subjected to statistical analysis against our adenoma (NFPA or prolactinoma) cohorts to determine odds ratio and p-value. Equations calculating the lifetime risk of developing surgical pituitary adenoma was adapted from previous work by Yang et al. (Yang et al., 2009). The 42 patient NFPA cohort and 11 patient prolactinoma cohort used for data analyses in this manuscript were subdivided by age and subjects with a G allele were counted in each bracket for comparison with the general population. The student’s t-test was used to test the difference in means for continuous variables. Fisher’s exact test was used to compare proportions. P values are two-tailed and P < 0.05 was considered statistically significant.
3. Results
3.1. Sequencing
Sequencing of genomic DNA coding regions of the TP53 gene was performed in 42 NFPAs and the circulating lymphocytes from these patients. While no mutations were found in NFPAs compared to genomic DNA obtained from paired blood, alignment analysis of the combined sequencing data revealed a previously described polymorphism (rs1042522:C > G) (Supplementary Table 1), which exhibited a much higher frequency of the G allele leading to an arginine residue (R72) (G = 0.798) than the expected values determined using published ethnic frequencies from the 1000 Genomes (Table 1; G = 0.304; P < 1.411 × 10−17) and HapMap databases (Table 2; G = 0.312; P < 2.364 × 10−17) weighted to match the distribution of ethnicities in our adenoma cohort. As an additional control, we obtained DNA from blood cells of neurologically normal cancer-free control individuals provided to us by the Coriell Institute for Medical Research in a manner that did not include analysis of ethnicity and performed a similar screen for variants in the TP53 coding region. Once again, our NFPA cohort exhibited a distinct allelic frequency for rs1042522 that differed significantly from what we observed in the Coriell cohort (Table 3; G = 0.467; P < 2.728 × 10−8). Thus, using three distinct control groups for comparisons, the G allele leading to an arginine residue (R72) consistently had a much greater frequency in our NFPA cohort than in control groups, in which the C allele leading to a proline residue (P72) was more common. Among our 42 patient NFPA cohort, four patients retained the C/C genotype that predominated control databases across most ethnicities, nine displayed a heterozygous C/G genotype, and the remaining 29 members of the cohort all had the G/G genotype in both pituitary tissue and paired lymphocytes. As with the allelic analysis, these genotypic frequencies deviated significantly from those of the adjusted control databases. The G/G genotypic frequency was 0.690 in NFPAs, compared to 0.090 in weighted controls derived from the 1000 Genomes database (Table 1; P < 2.211 × 10−18), 0.14 in the HapMap database (Table 2; P < 2.355 × 10−14), and 0.332 in the Coriell database (Table 3; P < 2.455 × 10−5). In order to corroborate this finding, we obtained a validation cohort from genome-wide sequencing data on a cohort of 17 NFPAs deposited in the NIH Sequence Read Archive (SRA #SRP035646). Although the SRA validation sequences did not come with paired blood to verify that the tumor sequence at this position was a polymorphism not a mutation, because our cohorts of NFPA and prolactinomas contained only a polymorphism and no mutation at this location and a full search of the TCGA database revealed no mutation at this loci in any other tumors, we felt that the SRA sequences could be used for validation. Reads were screened for variation in TP53 and the resultant genotypes were compared to our two control cohorts, which showed similar trends and odds ratios as our initial cohort (Supplementary Tables 2 and 3).
Table 1.
Adjusted Odds Ratio of rs1042522 in pituitary adenoma samples with weighted control populations derived from 1000 Genomes database.
| Large Scale 1000G
|
Pituitary Adenoma
|
Odds ratio
|
|||||
|---|---|---|---|---|---|---|---|
| n | Freq | N | Freq | Odds Ratio vs database (95% CI) | p | ||
| Genotype | C/C | 1902 | 0.482 | 4 | 0.095 | 22.635 (11.661–40.501)a | p < 2.211 × 1018 |
| C/G | 1690 | 0.428 | 9 | 0.214 | |||
| G/G | 354 | 0.090 | 29 | 0.690 | |||
| Allele | C | 5494 | 0.696 | 17 | 0.202 | 9.029 (5.291–15.409) | p < 1.411 × 10−17 |
| G | 2398 | 0.304 | 67 | 0.798 | |||
Genotypic odds ratio and subsequent p-value were calculated for G/G genotype as compared to both C/C and C/G.
Table 2.
Adjusted Odds Ratio of rs1042522 in pituitary adenoma samples with weighted control populations calculated from HapMap and SNP500 databases.
| Database Controls
|
Pituitary Adenoma
|
Odds ratio
|
|||||
|---|---|---|---|---|---|---|---|
| n | Freq | n | Freq | Odds Ratio vs database (95% CI) | p | ||
| Genotype | C/C | 1165 | 0.516 | 4 | 0.095 | 13.702 (7.047–26.642)a | p < 2.355 × 10−14 |
| C/G | 776 | 0.344 | 9 | 0.214 | |||
| G/G | 316 | 0.140 | 29 | 0.690 | |||
| Allele | C | 3106 | 0.688 | 17 | 0.202 | 8.694 (5.087–14.859) | p < 2.364 × 10−17 |
| G | 1408 | 0.312 | 67 | 0.798 | |||
Genotypic odds ratio and subsequent p-value were calculated for G/G genotype as compared to both C/C and C/G.
Table 3.
Adjusted Odds Ratio of rs1042522 in pituitary adenoma samples compared to non-weighted control frequencies calculated from in-house screening of patient controls from Coriell database.
| Coriell Controls
|
Pituitary Adenoma
|
Odds ratio
|
|||||
|---|---|---|---|---|---|---|---|
| n | Freq | n | Freq | Odds Ratio vs database (95% CI) | p | ||
| Genotype | C/C | 73 | 0.397 | 4 | 0.095 | 4.498 (2.184–9.265)a | p < 2.455 × 10−5 |
| C/G | 50 | 0.272 | 9 | 0.214 | |||
| G/G | 61 | 0.332 | 29 | 0.690 | |||
| Allele | C | 196 | 0.533 | 17 | 0.202 | 4.491 (2.539–7.944) | p < 2.728 × 10−8 |
| G | 172 | 0.467 | 67 | 0.798 | |||
Genotypic odds ratio and subsequent p-value were calculated for G/G genotype as compared to both C/C and C/G.
The dramatically elevated incidence of the G allele of the rs1042522 polymorphism we noted in NFPAs also occurred in a cohort of 11 prolactinomas (Supplementary Table 4–7), suggesting the applicability of our findings to both nonfunctional and functional pituitary adenomas. To lend additional support to these associations, we analyzed allelic frequencies for the P72R variant in all cancer types available through the TCGA database. The resultant allelic frequencies showed significant difference from our NFPA cohort (P < 1.0 × 10−10 Supplementary Table 8). While the G allele predominated in our NFPA cohort, TCGA patients with bladder (n = 131), pancreatic (n = 109), or stomach (n = 295) cancer resembled our 3 control cohorts in exhibiting C allele predominance.
3.2. Population analysis
The striking incidence of this polymorphism in our adenoma cohort suggests that the R72 may contribute to pituitary adenoma development relative to P72, which is more common in the general population. We quantified this possibility at a general population level by integrating our finding with epidemiologic data on the age-related incidence of NFPAs derived form the Surveillance, Epidemiology, and End Results (SEER) database (McDowell et al., 2011) to calculate the lifetime risk of developing an NFPA requiring surgery in patients with the G allele versus the population in general. We found a significantly higher risk in patients with the G allele that grew more rapidly as patients aged such that, once they reached age 75, patients with the G allele had a 0.22% risk of having developed an NFPA requiring surgery (Fig. 1a). Similarly, we then assessed the impact of the G allele on NFPA development in our NFPA cohort. While patients with the C/C or C/G genotypes had comparable NFPA size as patients with the G/G genotype (mean diameter 2.8 vs. 2.5 cm; p = 0.4; Supplementary Table 9), NFPA patients with the C/C or C/G genotypes were older than patients with the G/G genotype at diagnosis (67 vs. 55 years; p = 0.002; Supplementary Table 9; Fig. 1b), consistent with the population-based data. Similarly, prolactinoma patients with the C/C or C/G genotypes also had comparable adenoma size as patients with the G/G genotype (mean diameter 2 vs. 1.5 cm; p = 0.6; Supplementary Table 10), while prolactinoma patients with the C/C or C/G genotypes were older than patients with the G/G genotype at diagnosis (43 vs. 28 years; p = 0.03; Supplementary Table 10).
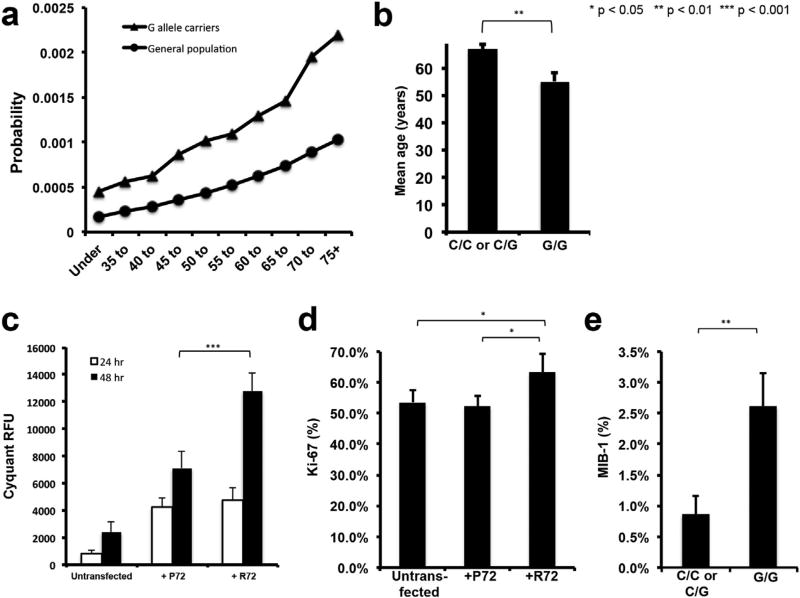
Fig. 1. The G variant of p53 polymorphism rs1042522 drives proliferation in pituitary adenoma cells.
(a) Statistical analysis of the lifetime risk of developing an NFPA needing surgery was calculated using data from adenoma patient cohort and neurologically normal samples sequenced as controls. (b) Patients with C/C or C/G genotypes were found to be older than patients with G/G genotype (P = 0.002); error bars represent standard error of the mean. (c) CyQuant assay of transfected and untransfected SF10052 pituitary adenoma cells at specified time points showed an increase in RFU, reflective of increased cell numbers, in R72-transfected cells at 48 h (n = 6/group; P < 10−7). (d) Ki-67 labeling of transfected and untransfected SF10052 pituitary adenoma cells at 24 h revealed increased percentage of cells labeled with Ki-67 in R72-vs. P72-transfected cells (P = 0.02) and R72-transfected cells vs. untransfected (P = 0.04), indicating that the R72 variant led to increased proliferation. (e) MIB-1 labeling of adenoma specimens from patients revealed increased MIB-1-labeling of adenomas from patients with the G/G genotype than patients with C/C or C/G genotypes (P = 0.008).
3.3. Impact of TP53 polymorphism on adenoma proliferation
Although a number of previous studies have shown an association between the R72 variant and disorders such as endometriosis (Li et al., 2015), squamous cell carcinoma (Brooks et al., 2000), and lung cancer (Papadakis et al., 2002), among others, the functional effect of this variant remains uncertain and may be cell type-specific. To this end, we performed functional assays to better elucidate the downstream effects of the arginine versus proline residues associated with the rs1042522 polymorphism.
We transiently transfected SF10052, primary human pituitary adenoma cells cultured from a patient NFPA specimen, whose genotype was homozygous C/C for rs1042522 (resulting in proline residues for both alleles), to comparably overexpress both variants (Supplementary Fig. 1). We found that over-expressing the R72 variant through transient transfection with a plasmid in SF10052 cells led to increased cell numbers over time compared to comparable over-expression of the P72 variant (p < 10−7; Fig. 1c). To determine if these increased cell numbers represented increased proliferation associated with the arginine variant, we performed Ki-67 labeling of cultured transfected cells in chamber slides and found greater Ki-67 labeling of cells transfected with R72 than P72 (P < 0.05; Fig. 1d; Supplementary Fig. 2). Consistent with this finding, the 29 adenomas with the G/G genotype in our 42 patient nonfunctional adenoma cohort had an average MIB-1 labeling index of 3%, more than 3-fold higher than the 0.9% seen in the 13 patients with C/C or C/G genotypes (P = 0.008; Fig. 1e; Supplementary Table 9). A similar finding could not be identified in prolactinomas due to the smaller sample size of the surgical prolactinoma cohort (Supplementary Table 10).
3.4. Impact of TP53 polymorphism on adenoma gene transcription
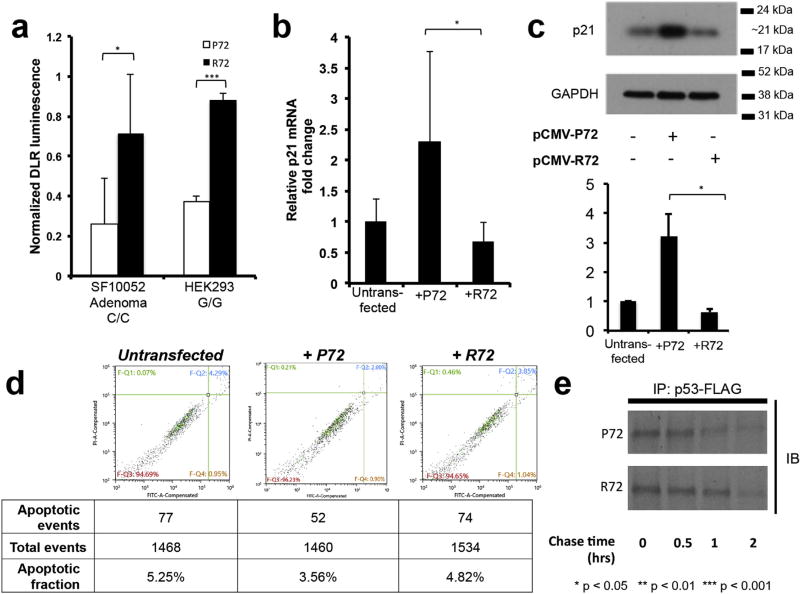
We then attempted to determine the downstream effects of the R72 variant as compared to the P72 by utilizing a modified dual luciferase assay to test the effects of the various residues on the ability of p53 to bind its consensus DNA binding sequence. To control for host genotypes, we transfected our reporter and expression vectors, as well as an hRenilla transfection control, into SF10052, the primary human pituitary adenoma cells described above with the C/C genotype of the rs1042522 polymorphism, as well as HEK293 human embryonic kidney cells which carry the G/G genotype of rs1042522. While we hypothesized that R72 would drive less transcription from the p53 consensus binding sequence, which regulates various cell cycle arrest genes, we instead found that the R72 variant in SF10052 led to 2.76-fold increase in reporter gene expression compared to adding P72 in SF10052 (P = 0.04) and a 2.35-fold increased in transfected HEK293 (p < 0.0001) respectively (Fig. 2a). While the dual-luciferase assay suggested increased ability of the R72 variant to drive a reporter gene from the p53 consensus DNA binding sequence, we decided to further investigate a specific mechanism by which this variant could promote adenoma growth. We examined the effect of R72 on transcription of p21, a crucial mediator of p53-mediated cell cycle arrest (Macleod et al., 1995), including in pituitary adenomas (Chesnokova et al., 2008). To do this, we again transiently transfected SF10052 pituitary adenoma cells, with a C/C homozygous genotype for rs1042522, to comparably overexpress both variants. We found that overexpression through transient transfection of the R72 variant, was significantly associated with reduced p21 transcription (P = 0.02; Fig. 2b) and translation (P = 0.03; Fig. 2c; Fig. 3) as compared to overexpression of the P72 variant in SF10052. Transient transfection of the R72 variant did not significantly alter apoptosis compared to transient transfection of the P72 variant (P = 0.06; Fig. 2d)
Fig. 2. Defining mechanisms allowing the G variant of p53 polymorphism rs1042522 to drive proliferation in pituitary adenoma cells.
(a) Dual luciferase assay was performed by co-transfection of the expression vector containing p53-P72 or p53-R72 and reporter luciferase vector and revealed increased luminescence with the R variant in both SF10052 adenoma cells which carry the C/C genotype for rs1042522 (P = 0.04) and HEK293 cells which carry the G/G genotype for rs1042522 (P < 0.0001) (n = 6/group). (b) Quantitative PCR and (c) western blot were used to analyze p21 expression in SF10052 cells transfected with P72 or R72. Transfected vectors were allowed 24 h for expression before lysis and RNA and protein extraction. PCR results normalized to GAPDH showed a notable decrease in p21 expression in cells transfected with R72 (P = 0.02), and western blot confirmed this finding at the protein level (P = 0.03). (d) Flow cytometry of SF10052 cells transfected either P72 or R72. Gating shows viable non-apoptotic cells (FITC- PI−) and apoptotic cells, including both early (FITC + PI−) and late (FITC + PI+) apoptotic events. Early apoptotic events were noticeably rare, so all apoptotic events types were counted against the total events screened to determine the apoptotic fraction, which was comparable between P72 versus R72-transfected cells (P = 0.06). (e) Pulse-chase assay of P72 and R72-transfected cells shows the R72 variant has a longer-half life than the P72 variant.
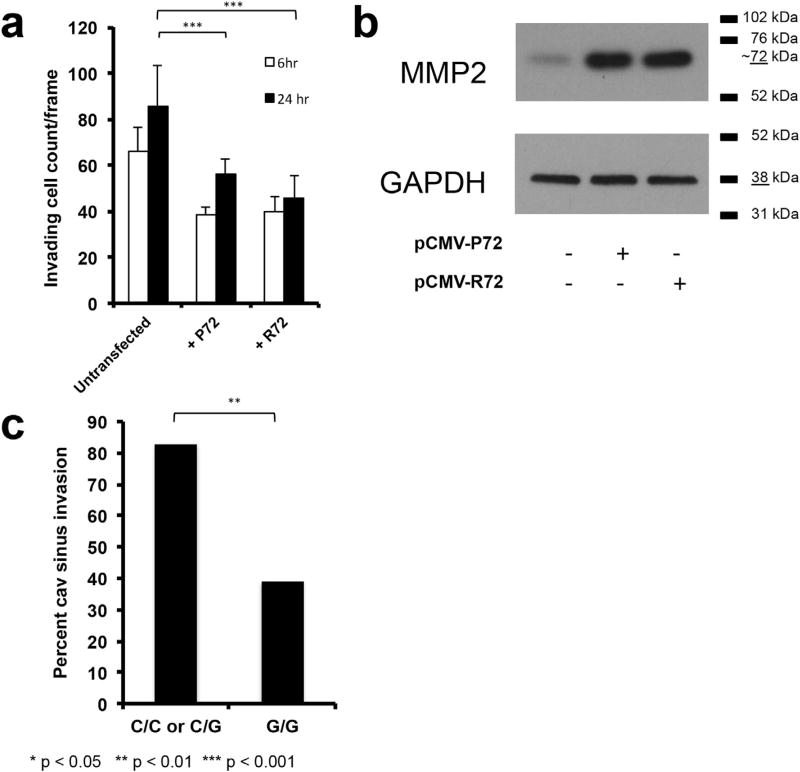
Fig. 3. Impact of the G variant of p53 polymorphism rs1042522 on invasion in pituitary adenoma cells.
(a) SF10052 cells (C/C homozygous for the rs1042522 TP53 polymorphism) were transfected with plasmids carrying the P72 or R72 variant and analyzed for invasion in transwell assays. Transwell invasion assays were imaged and cell count/ frame averaged over 10 frames. The P72 and R72 variants did not show a significant difference in their migrating cell counts (P = 0.8 at 6 h, P = 0.1 at 24 h). (b) MMP2 expression did not vary between SF10052 cells overexpressing the P72 and R72 variants. (c) Cavernous sinus invasion was increased in adenomas in patients with the C/C genotype compared to those with the G/G genotype (P = 0.009).
Thus, despite the increased ability of the R72 variant to drive reporter gene expression, the reduced expression of p21 associated with the R72 variant suggest that the R72 variant may drive proliferation of pituitary adenoma cells by reducing the amount of p21 regulating cell cycle checkpoints.
3.5. Impact of polymorphism on p53 protein turnover
In order to determine the impact of the rs1042522 polymorphism on p53 turnover, we performed a pulse-chase assay performed on SF10052 cells transfected with pCMV6-p53-P72 or −R72. After these cells were pulsed with [35S]methionine/cysteine, then chased at various time points with complete media, lysates were immunoprecipitated for the attached FLAG tag and the precipitates were run on an SDS-PAGE gel and blotted over a week to expose radioactive FLAG tagged p53 and monitor its decay over time. We found that the R72 variant has a longer half-life than the P72 (Fig. 2e), an effect that would augment the short-term functional differences noted in cultured cells.
3.6. Impact of TP53 polymorphism on adenoma invasion
In order to evaluate the effects of the R72 variant on cell invasiveness, we employed a transwell matrigel assay. In this assay, SF10052 cells overexpressing the P72 and R72 variants had comparable levels of invasion in matrigel chamber assays (P = 0.8 at 6 h and P = 0.1 at 24 h; Fig. 3a; Supplementary Fig. 4) and comparable expression of MMP2 (Fig. 3b), a principal mediator of invasiveness in pituitary adenoma cells (Liu et al., 2005). In contrast to this lack of invasiveness differences at a cellular level between cells with the P72 and R72 variants of p53, at a radiographic level, review of preoperative MRIs from NFPA patients with the C/C genotype revealed more frequent cavernous sinus invasion of the adenomas (P = 0.009), which has been shown to correlate with MMP2 expression (Liu et al., 2005), than patients with the G/G genotype (Fig. 3c).
3.7. Impact of TP53 polymorphism on adenoma angiogenesis and metabolism
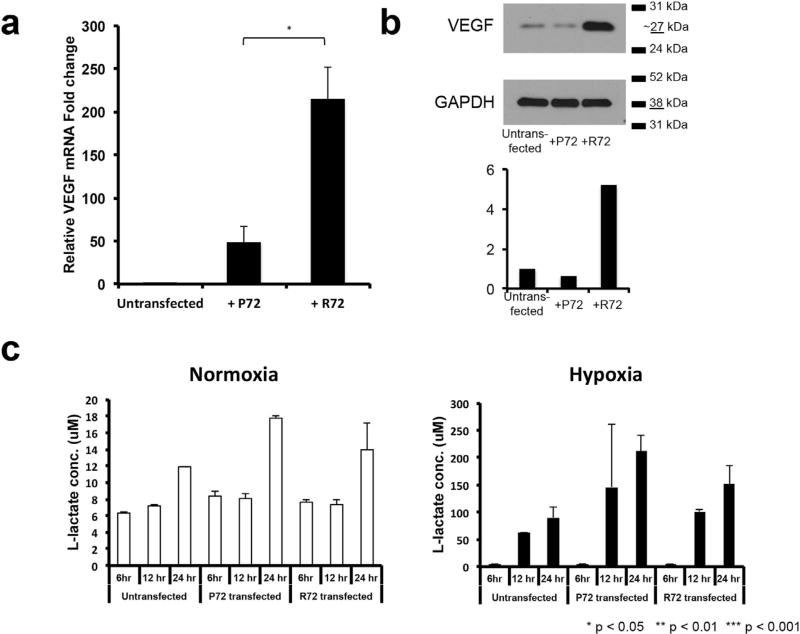
We then assessed the impact of the R72 variant on the ability of pituitary adenoma cells to adapt to the hypoxia that arises as an adenoma reaches critical size (Xiao et al., 2011). We investigated two biological mechanisms that can allow cells from tumors like pituitary adenomas to adapt to hypoxia: increased VEGF, which drives angiogenesis, and increased glycolysis, the anaerobic metabolism upregulated by tumor cells to adapt to hypoxia as part of the Warburg effect.
SF10052 cells overexpressing the R72 variant had greater VEGF expression by both qPCR (P = 0.02; Fig. 4a) and western (Fig. 4b) blot than those overexpressing the P72 variant. A metabolic assay measuring lactate production as a measure of anaerobic glycolytic metabolism revealed no significant difference between SF10052 cells overexpressing the R72 versus P72 variant in either normoxia or hypoxia (P = 0.08–0.43; Fig. 4c).
Fig. 4. Impact of the G variant of p53 polymorphism rs1042522 on ability of pituitary adenoma cells to adapt to hypoxia.
(a–b) SF10052 cells (C/C homozygous for the rs1042522 TP53 polymorphism) exhibited increased VEGF expression by (a) qPCR (P = 0.02) and (b) western blot after transfection with a plasmid carrying the R72 variant compared to the P72 variant. (c) A colormetric assay to measure L-lactate production as a measure of anaerobic glycolysis revealed no differences (P = 0.08–0.43) in glycolysis in normoxic or hypoxic SF10052 cells overexpressing the R72 variant compared to those overexpressing the P72 variant.
4. Discussion
In summary, we identified dramatic prevalence of the R72 variant of the rs1042522 polymorphism in nonfunctional pituitary adenoma patients, as well as in a parallel smaller cohort of prolactinomas. While control data from three different databases (1000Genomes, HapMap, and the Coriell Institute for Medical Research) show that the allele encoding arginine is less common in all ethnic groups studied, our findings demonstrate that this allele was dramatically more prevalent in our pituitary adenoma patients. Interestingly, the proline and arginine residues possible at this location have shown distinct associations with a variety of cancers -the proline residue demonstrates association with increased breast (Proestling et al., 2012), colorectal (Cao et al., 2009), and chronic lymphocytic leukemia cancer risk (Bilous et al., 2014), while the arginine residue is associated with increased risk of lung cancer (Papadakis et al., 2002) and squamous cell carcinoma (Brooks et al., 2000). These conflicting cell-type specific influences of rs1042522 variants are consistent with studies on the effects of these two alleles revealing each to have distinct potential oncogenic functions. In some cell types, R72 has demonstrated increased apoptotic potential, partially through enhanced MDM2 binding and ubiquitination leading to increased cellular export and mitochondrial localization (Dumont et al., 2003), as well as increased susceptibility to human papillomavirus (HPV) E6-induced degradation (Storey et al., 1998). In contrast, P72 has shown weaker apoptotic potential (Bergamaschi et al., 2006), but greater induction of cell cycle arrest in G1 phase, through p21/Waf1 activation (Pim and Banks, 2004). Of note, the degree to which the arginine variant was prevalent in nonfunctional adenoma patients far exceeded the more subtle prevalences of rs1042522 polymorphisms in other tumor types. Indeed, in some tumors like colorectal cancer, initial reports suggesting this polymorphism to be subtly associated with the cancer did not prove verifiable upon meta-analysis (Tang et al., 2010). In contrast, we found that the G allele of TP53 polymorphism rs1042522 is nearly ubiquitous in NFPA patients and a smaller cohort of prolactinoma patients despite being present in a minority of the general population, suggesting that this variant may meaningfully influence adenoma formation or growth in the pituitary gland.
Specifically, we found that, compared to P72, the R72 variant led to reduced expression of p21 and increased VEGF expression. Thus, while p53 has been shown to promote p21 (Macleod et al., 1995) and VEGF (Farhang Ghahremani et al., 2013) expression, differences in the degree to which these changes are induced by the polymorphism variants in adenoma cells lead to the G variant lowering expression of cell arrest gene p21 and increased expression of angiogenesis-mediating VEGF, promoting adenoma growth through increased cell proliferation and vascularity in tumors arising in patients with the G variant.
Furthermore, while It has been suggested that in addition to its canonical role as an inhibitor of cell proliferation, p21 can inhibit apoptosis in a number of systems, and this may counteract its tumor-suppressive functions as a growth inhibitor (Gartel and Tyner, 2002), we found that in adenoma cells, the proliferative increase seen with G variant-associated p21 loss was significant but the increased apoptosis was not. In addition, our finding that the R72 variant survives longer would potentiate these biologic differences between the variants over time. The increased proliferation in pituitary cells may be responsible for the growth of the adenomas to surgical proportions at an earlier age in patients with the R72 variant of the rs1042522 polymorphism.
Our findings will need to be validated in a different cohort of NFPA patients, ideally in a multi-institutional manner, as well as functional adenoma patients such as patients with prolactinomas to validate our findings in prolactinoma patients, as well as patients with Cushing’s disease and acromegaly. Once validated, our findings could be expanded beyond revealing the biological differences in these p53 variants that arise specifically in the context of adenoma cells in order to have practical implications. For example, patients with small adenomas undergoing observation through serial imaging could undergo evaluation of their TP53 polymorphism in their blood and resulting growth of the adenomas over time could be correlated with their rs1042522 genotype. If such a study proved consistent with our findings, younger patients with the G variant of rs1042522 may warrant consideration for elective resection of an asymptomatic smaller adenoma while those with the C variant may be readily observed through serial imaging.
5. Conclusion
We have demonstrated that the polymorphism rs1042522:C > G in codon 72 of exon 4 of the TP53 gene, whose C variant produces a proline and is more common in most ethnic groups, has a G variant producing an arginine that is fairly ubiquitous in nonfunctional adenomas and prolactinomas, which causes patients to present a decade earlier with symptomatic adenomas, reduces expression of cell arrest gene p21, and increases adenoma cell proliferation. These findings suggest that that this variant in the TP53 gene influences adenoma development in the pituitary gland.
Supplementary Material
Acknowledgments
The authors wish to thank all patients who donated biological specimens used in this study. We also acknowledge the members of the Brain Tumor Research Center for sample collection, inventory and documentation. This research was supported by funding from the UCSF Resource Allocation Program (RAP) (MNS7504293). The authors have no conflict of interest to disclose.
Footnotes
Disclosure statement
The authors have nothing to disclose.
Author contributions
G.Y. received pituitary adenoma tissue and blood for nucleic acid extraction and performed all subsequent sequencing, cloning and functional experiments, with M.K.A. providing support and critical suggestions. R.C., A.J., and J.W. collected viable pituitary specimens and adapted cell culture protocol to allow development of cultured pituitary adenoma lines for experimental work and transfections. M.K.A. performed transsphenoidal surgeries to collect limited samples and facilitated collection of samples from other surgeries to the lab via BTRC collection. The manuscript was written equally by G.Y. and M.K.A.
Conflict of interest
The authors have no conflict of interest to disclose.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.mce.2017.02.017.
References
- Bergamaschi D, Samuels Y, Sullivan A, Zvelebil M, Breyssens H, Bisso A, Del Sal G, Syed N, Smith P, Gasco M, et al. iASPP preferentially binds p53 proline-rich region and modulates apoptotic function of codon 72-polymorphic p53. Nat. Genet. 2006;38(10):1133–1141. doi: 10.1038/ng1879. [DOI] [PubMed] [Google Scholar]
- Bilous NI, Abramenko IV, Chumak AA, Dyagil IS, Martsmall iUZV. TP53 codon 72 single nucleotide polymorphism in chronic lymphocytic leukemia. Exp. Oncol. 2014;36(4):258–261. [PubMed] [Google Scholar]
- Brooks LA, Tidy JA, Gusterson B, Hiller L, O’Nions J, Gasco M, Marin MC, Farrell PJ, Kaelin WG, Jr, Crook T. Preferential retention of codon 72 arginine p53 in squamous cell carcinomas of the vulva occurs in cancers positive and negative for human papillomavirus. Cancer Res. 2000;60(24):6875–6877. [PubMed] [Google Scholar]
- Cao Z, Song JH, Park YK, Maeng EJ, Nam SW, Lee JY, Park WS. The p53 codon 72 polymorphism and susceptibility to colorectal cancer in Korean patients. Neoplasma. 2009;56(2):114–118. doi: 10.4149/neo_2009_02_114. [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokova V, Zonis S, Kovacs K, Ben-Shlomo A, Wawrowsky K, Bannykh S, Melmed S. p21(Cip1) restrains pituitary tumor growth. Proc. Natl. Acad. Sci. U. S. A. 2008;105(45):17498–17503. doi: 10.1073/pnas.0804810105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont P, Leu JI, Della Pietra AC, 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 2003;33(3):357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1(1):45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- Farhang Ghahremani M, Goossens S, Nittner D, Bisteau X, Bartunkova S, Zwolinska A, Hulpiau P, Haigh K, Haenebalcke L, Drogat B, et al. p53 promotes VEGF expression and angiogenesis in the absence of an intact p21-Rb pathway. Cell Death Differ. 2013;20(7):888–897. doi: 10.1038/cdd.2013.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;1(8):1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartel AL, Tyner AL. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol. Cancer Ther. 2002;1(8):639–649. [PubMed] [Google Scholar]
- Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18(53):7644–7655. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- Levy A, Hall L, Yeudall WA, Lightman SL. p53 gene mutations in pituitary adenomas: rare events. Clin. Endocrinol. (Oxf) 1994;41(6):809–814. doi: 10.1111/j.1365-2265.1994.tb02797.x. [DOI] [PubMed] [Google Scholar]
- Li J, Chen Y, Mo Z, Li L. TP53 Arg72Pro polymorphism (rs1042522) and risk of endometriosis among Asian and Caucasian populations. Eur. J. Obstet. Gynecol. Reprod. Biol. 2015;189:73–78. doi: 10.1016/j.ejogrb.2015.03.026. [DOI] [PubMed] [Google Scholar]
- Liu W, Kunishio K, Matsumoto Y, Okada M, Nagao S. Matrix metalloproteinase-2 expression correlates with cavernous sinus invasion in pituitary adenomas. J. Clin. Neurosci. 2005;12(7):791–794. doi: 10.1016/j.jocn.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Macleod KF, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, Vogelstein B, Jacks T. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev. 1995;9(8):935–944. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- McDowell BD, Wallace RB, Carnahan RM, Chrischilles EA, Lynch CF, Schlechte JA. Demographic differences in incidence for pituitary adenoma. Pituitary. 2011;14(1):23–30. doi: 10.1007/s11102-010-0253-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadakis ED, Soulitzis N, Spandidos DA. Association of p53 codon 72 polymorphism with advanced lung cancer: the Arg allele is preferentially retained in tumours arising in Arg/Pro germline heterozygotes. Br. J. Cancer. 2002;87(9):1013–1018. doi: 10.1038/sj.bjc.6600595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pim D, Banks L. p53 polymorphic variants at codon 72 exert different effects on cell cycle progression. Int. J. Cancer. 2004;108(2):196–199. doi: 10.1002/ijc.11548. [DOI] [PubMed] [Google Scholar]
- Proestling K, Hebar A, Pruckner N, Marton E, Vinatzer U, Schreiber M. The Pro allele of the p53 codon 72 polymorphism is associated with decreased intratumoral expression of BAX and p21, and increased breast cancer risk. PLoS One. 2012;7(10):e47325. doi: 10.1371/journal.pone.0047325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F, Breuer J, Leigh IM, Matlashewski G, Banks L. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature. 1998;393(6682):229–234. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- Tang NP, Wu YM, Wang B, Ma J. Systematic review and meta-analysis of the association between P53 codon 72 polymorphism and colorectal cancer. Eur. J. Surg. Oncol. 2010;36(5):431–438. doi: 10.1016/j.ejso.2010.03.010. [DOI] [PubMed] [Google Scholar]
- Trouillas J, Roy P, Sturm N, Dantony E, Cortet-Rudelli C, Viennet G, Bonneville JF, Assaker R, Auger C, Brue T, et al. A new prognostic clinicopathological classification of pituitary adenomas: a multicentric case-control study of 410 patients with 8 years post-operative follow-up. Acta Neuropathol. 2013;126(1):123–135. doi: 10.1007/s00401-013-1084-y. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Liu Q, Zhao B, Wu J, Lei T. Hypoxia induces hemorrhagic transformation in pituitary adenomas via the HIF-1alpha signaling pathway. Oncol. Rep. 2011;26(6):1457–1464. doi: 10.3892/or.2011.1416. [DOI] [PubMed] [Google Scholar]
- Yang Q, Flanders WD, Moonesinghe R, Ioannidis JP, Guessous I, Khoury MJ. Using lifetime risk estimates in personal genomic profiles: estimation of uncertainty. Am. J. Hum. Genet. 2009;85(6):786–800. doi: 10.1016/j.ajhg.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zada G, Woodmansee WW, Ramkissoon S, Amadio J, Nose V, Laws ER., Jr Atypical pituitary adenomas: incidence, clinical characteristics, and implications. J. Neurosurg. 2011;114(2):336–344. doi: 10.3171/2010.8.JNS10290. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhang X, Klibanski A. Genetic and epigenetic mutations of tumor suppressive genes in sporadic pituitary adenoma. Mol. Cell Endocrinol. 2014;386(1–2):16–33. doi: 10.1016/j.mce.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.