Abstract
We analyzed the der(11) and der(4) genomic breakpoint junctions of a t(4;11) in the leukemia of a patient previously administered etoposide and dactinomycin by molecular and biochemical approaches to gain insights about the translocation mechanism and the relevant drug exposure. The genomic breakpoint junctions were amplified by PCR. Cleavage of DNA substrates containing the normal homologues of the MLL and AF-4 translocation breakpoints was examined in vitro upon incubation with human DNA topoisomerase IIα and etoposide, etoposide catechol, etoposide quinone, or dactinomycin. The der(11) and der(4) genomic breakpoint junctions both involved MLL intron 6 and AF-4 intron 3. Recombination was precise at the sequence level except for the overall gain of a single templated nucleotide. The translocation breakpoints in MLL and AF-4 were DNA topoisomerase II cleavage sites. Etoposide and its metabolites, but not dactinomycin, enhanced cleavage at these sites. Assuming that DNA topoisomerase II was the mediator of the breakage, processing of the staggered nicks induced by DNA topoisomerase II, including exonucleolytic deletion and template-directed polymerization, would have been required before ligation of the ends to generate the observed genomic breakpoint junctions. These data are inconsistent with a translocation mechanism involving interchromosomal recombination by simple exchange of DNA topoisomerase II subunits and DNA-strand transfer; however, consistent with reciprocal DNA topoisomerase II cleavage events in MLL and AF-4 in which both breaks became stable, the DNA ends were processed and underwent ligation. Etoposide and/or its metabolites, but not dactinomycin, likely were the relevant exposures in this patient.
Leukemia has become an increasingly common complication of effective chemotherapy (reviewed in ref. 1). Two classes of chemotherapy are associated with leukemia—alkylating agents and DNA topoisomerase II inhibitors (reviewed in refs. 1–3). The hallmarks of leukemias associated with DNA topoisomerase II inhibitors are balanced chromosomal translocations, many of which involve the MLL gene at chromosome band 11q23 (reviewed in refs. 2, 4, and 5). It has been suggested that the MLL translocation mechanism may involve DNA topoisomerase II-mediated chromosomal breakage and formation of the translocations when the breakage is repaired (2, 4, 5).
DNA topoisomerase II catalyzes the relaxation of supercoiled DNA by transiently cleaving and religating both strands of the double helix (6). The DNA topoisomerase II homodimer introduces four-base staggered nicks in DNA as each subunit covalently binds and cleaves one strand (6). In the presence of ATP, the DNA open gate allows passage of a second DNA helix through the cleaved strands (6). Next, the cleaved DNA strands are religated, and after ATP hydrolysis, the enzyme homodimer attains its original conformational state to catalyze another cycle (6). DNA topoisomerase II has been implicated in MLL translocations, because several anticancer drugs interfere with its cleavage–religation equilibrium (6, 7).
Etoposide is the anticancer drug most often associated with leukemia with MLL translocations. Anticancer drugs that target DNA topoisomerase II increase the concentration of enzyme–DNA covalent complexes (7). Many drugs such as etoposide decrease the rate of religation (7). The resultant increased cleavage is associated with chromosomal breakage, which may be repaired by translocation. It has been suggested that the simultaneous trapping of two DNA topoisomerase II covalent complexes on different homologous chromosomal regions might facilitate DNA topoisomerase II subunit exchange and lead to precise interchromosomal DNA recombination (8). We used a combined molecular and biochemical approach to determine the feasibility of DNA topoisomerase II subunit exchange as the translocation mechanism in a case of treatment-related acute lymphoblastic leukemia (ALL). Because different DNA topoisomerase II cleavage sites are enhanced by specific anticancer drugs with varied sequence preferences (9–12), a second objective was to determine which anticancer drug exposure was relevant to the translocation in the leukemia of this patient.
Methods
A 9-year 11-month-old girl, previously designated patient 33 (13) and patient 7 (14), presented with primary alveolar rhabdomyosarcoma of the forearm and received cyclophosphamide, dactinomycin, vincristine, ifosphamide, etoposide, and local radiation (13). At 15 months after the primary cancer was diagnosed, the peripheral WBC count was 122.7 × 109 cells per liter with 64% circulating blasts. The marrow aspirate revealed French–American–British L1 ALL. The marrow karyotype was 46,XX,t(4;11)(q21;q23)[4]/46,XX[3] (ref. 13). The patient received ALL induction therapy and an allogeneic bone-marrow transplant from an unrelated donor (14) and is disease-free at 5 years from leukemia diagnosis.
Characterization of der(11) and der(4) Genomic Breakpoint Junctions of MLL Translocation.
The Institutional Review Board at The Children's Hospital of Philadelphia approved this research. MLL gene rearrangement was examined in genomic DNA prepared from cryopreserved leukemic marrow cells by using the B859 fragment of ALL-1 cDNA and has been reported (13). Isolation of the der(11) genomic breakpoint junction in a 2.6-kb panhandle PCR product has been described (GenBank accession no. AF024543) (13). AF-4- and MLL-specific primers were designed from the der(11) sequence to amplify the der(4) genomic breakpoint junction in a predicted 745-bp product. The sense primer from AF-4 intron 3 positions 10,405–10,424 (GenBank accession no. AJ238093) was 5′-TCCGTAAGCTCGACCTCAGT-3′. The antisense primer from MLL exon 7 positions 2,468 to 2,449 (GenBank accession no. U04737) was 5′-ATCCTGTGGACTCCATCTGC-3′. PCRs were performed in triplicate. The products were purified from an agarose gel and directly sequenced.
DNA Topoisomerase II in Vitro Cleavage Assays.
Human DNA topoisomerase IIα was isolated from Saccharomyces cerevisiae containing the human TOP2α gene (15, 16). Preparation of etoposide catechol and etoposide quinone from etoposide and structural confirmation and purification were performed as described (17). DNA fragments composed of MLL breakpoint cluster region positions 2,050–2,587 from intron 6, exon 7, and intron 7 (GenBank accession no. U04737) or AF-4 intron 3 positions 7,318–7,841 (GenBank accession no. A238093) were subcloned into the EcoRI–BamHI sites of pBluescript II SK(+) (Stratagene). Plasmid DNA (20 μg) first was treated with 400 units of T4 DNA ligase, digested with 32 units of BssHII, and dephosphorylated with 20 units of calf intestinal phosphatase (all from New England Biolabs; ref. 17). The DNA was 5′ end-labeled by using [γ-32P]ATP (Amersham Pharmacia) and 10 units of T4 polynucleotide kinase (New England Biolabs; ref. 17). Cold substrate DNA (20 μg) was prepared in similar reactions (17). Singly 5′ end-labeled or cold BssHII–SacI fragments containing the normal homologues of the MLL or AF-4 translocation breakpoints of interest were obtained by digestion with 80 units of SacI (New England Biolabs; ref. 17). A total of 25 ng of substrate DNA containing 30,000 cpm labeled DNA and the remainder cold DNA were incubated at 37°C for 10 min in 50-μl reactions in 147 nM human DNA topoisomerase IIα/1 mM ATP/137.5 mM KCl/12.5 mM Tris, pH 7.6/5 mM MgCl2/105 μM EDTA/25 μM DTT/4.5% (vol/vol) glycerol/8% (vol/vol) DMSO in the absence of drug and in the presence of 20 μM etoposide, 20 μM etoposide catechol, 20 μM etoposide quinone, or 20 μM dactinomycin. Additional reactions were performed without DNA topoisomerase IIα. Covalent complexes were irreversibly trapped by adding 5 μl of 10% (vol/vol) SDS without or with prior incubation for 10 min at 65°C. The latter was done to evaluate the heat stability of the complexes. After addition of 3.75 μl of 250 mM EDTA, cleavage products were deproteinized by incubation with proteinase K (Amersham Pharmacia) at a final concentration of 60 μg/ml for 30 min at 45°C. Yeast tRNA (20 μg) was added, and the cleavage products were ethanol-precipitated twice, resuspended in 6 μl of Sequenase Stop Solution (Amersham Pharmacia), and resolved in an 8% polyacrylamide-7.0 M urea gel in parallel with dideoxy sequencing reactions primed at the same 5′ end. Cleavage products were visualized by autoradiography and quantitated with a PhosphorImager and imagequant software (Molecular Dynamics).
Results
Genomic Breakpoint-Junction Sequences in der(11) and der(4) Chromosomes.
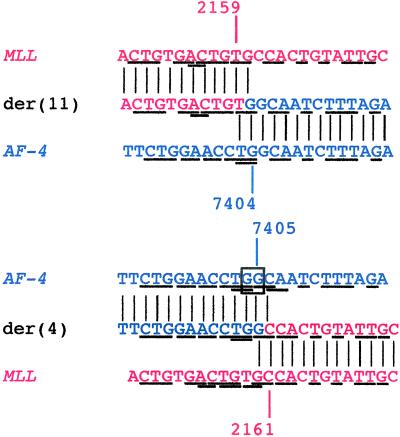
The der(11) genomic breakpoint junction summarized in Fig. 1 has been described (13). Additional information on the sequence of the partner gene, AF-4, has since become available (18). The MLL breakpoint was in intron 6, 5′ in the 8.3-kb breakpoint cluster region. The AF-4 breakpoint was in intron 3. The breakpoint positions in MLL and AF-4 could not be ascertained exactly from the der(11) sequence, because MLL and AF-4 contain homologous (TG) bases at their respective breakpoints (Fig. 1). Additional homologous sequences of a few base pairs in length also were detected in MLL and AF-4 (Fig. 1), similar to those observed at other MLL genomic breakpoint junctions (13, 19–25). If the MLL der(11) breakpoint is defined as the last base of MLL 5′ of the fused sequence from AF-4, possible MLL breakpoint positions are nucleotides 2,158, 2,159, or 2,160. If the AF-4 der(11) breakpoint is defined as the first base of AF-4 3′ of the MLL sequence to which it has fused, possible AF-4 breakpoint positions are nucleotides 7,403, 7,404, or 7,405 (GenBank accession no. AF024543).
Figure 1.
der(11) and der(4) genomic breakpoint-junction sequences in treatment-related ALL. Vertical lines indicate homologies in derivative chromosomes with MLL intron 6 and AF-4 intron 3. Underlines indicate homologies between MLL and AF-4. From the sequences alone, the breakpoint positions cannot be ascertained exactly because MLL and AF-4 contain homologous bases (TG) at the der(11) junction (Upper Middle) and homologous bases (GC) at the der(4) junction (Lower Middle). From the sequences alone, the MLL der(11) breakpoint was position 2,158, 2,159, or 2,160, and the AF-4 der(11) breakpoint was position 7,403, 7,404, or 7,405 (Upper). The AF-4 der(4) breakpoint was position 7,403, 7,404, 7,405, or 7,406, and the MLL der(4) breakpoint was position 2,160, 2,161, or 2,162 (Lower). There was an overall gain of a single G at both genomic breakpoint junctions together relative to the normal MLL and AF-4 sense sequences. DNA topoisomerase II in vitro cleavage assays provided additional information about the possible sites of breakage (compare Fig. 2). Red and blue indicate sequences derived from MLL and AF-4, respectively, with breakpoint positions in each derivative chromosome consistent with DNA topoisomerase II in vitro cleavage sites after processing of ends (compare Fig. 3).
It was possible to amplify the der(4) genomic breakpoint junction of the t(4;11) translocation predicted by the der(11) sequence by using AF-4- and MLL-specific primers. The AF-4 breakpoint was in intron 3, and the MLL breakpoint was in intron 6 (Fig. 1). Homologous (GC) bases in AF-4 and MLL precluded exact assignment of the der(4) AF-4 and MLL breakpoint positions. Other short homologous sequences in AF-4 and MLL flanked the der(4) genomic breakpoint junction (Fig. 1). Comparison of the der(11) and der(4) sequences indicated that the t(4;11) translocation had resulted from near-precise interchromosomal DNA recombination between MLL and AF-4, except for an overall gain of a single G relative to the normal sense sequences of these genes. If the AF-4 der(4) breakpoint is defined as the last base of AF-4 5′ of the fused sequence from MLL, possible AF-4 breakpoint positions are nucleotides 7,403, 7,404, 7,405, or 7,406. If the MLL der(4) breakpoint is defined as the first base of MLL 3′ of the AF-4 sequence to which it has fused, possible MLL breakpoint positions are nucleotides 2,160, 2,161, or 2,162 (GenBank accession no. AF397907).
DNA topoisomerase II cleavage assays provided additional information about the possible sites of breakage. Fig. 1 indicates breakpoint positions in both derivative chromosomes that are consistent with functional DNA topoisomerase II cleavage sites. The genomic sequences and DNA topoisomerase II cleavage assays together suggest formation of the der(11) by fusion of MLL position 2,159 to AF-4 position 7,404 and formation of the der(4) by fusion of AF-4 position 7,405 to MLL position 2,161 (compare Fig. 3).
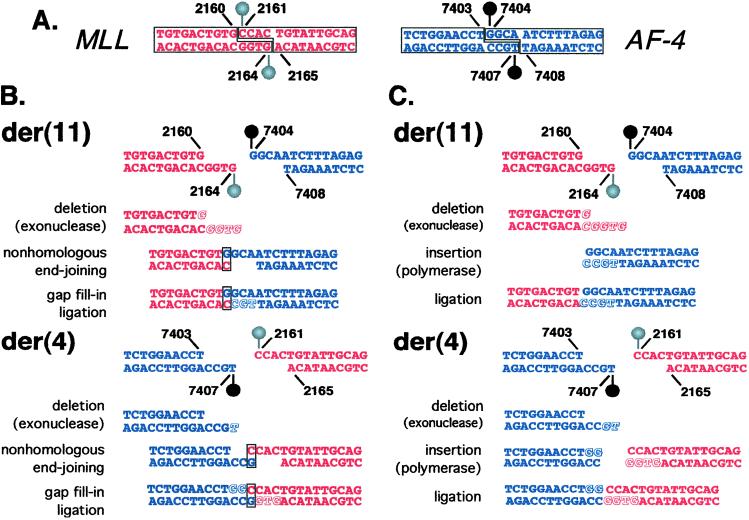
Figure 3.
(A) DNA topoisomerase II cleavage sites identified in vitro. The DNA topoisomerase II homodimer creates four-base staggered nicks in duplex DNA. Cleavage assays indicated that MLL position 2,160 is at the 5′ side (−1 position) of a cleavage site on the sense strand. The corresponding cleavage site on the antisense strand staggered by four bases from MLL position 2,160 is position 2,164 (Left, red). Cleavage assays indicated that AF-4 position 7,403 is at the 5′ side (−1 position) of a cleavage site on the sense strand. The corresponding cleavage site on the antisense strand staggered by four bases from position AF-4 position 7,403 is position 7,407 (Right, blue). (B and C) Models for processing of four-base 5′ overhangs of DNA topoisomerase II cleavage sites in MLL and AF-4 to generate observed der(11) and der(4) genomic breakpoint junctions. (B) To form the der(11), one possibility on the MLL side is that the entire four-base 5′ antisense-strand overhang (positions 2,161–2,164) and G at position 2,160 on the sense strand are removed by exonucleolytic nibbling (Red, italic, shadowed). On the AF-4 side, the entire four-base 5′ sense-strand overhang is preserved. The exonucleolytic nibbling of the MLL overhang would create a single-base microhomology, which facilitates base-pairing between the C at MLL antisense position 2,160 and the G at position 7,404 in the AF-4 sense-strand overhang (box). Template-directed polymerization of the antisense strand fills in the gap at AF-4 positions 7,107–7,105 (Blue, shadowed, nonitalic). The der(11) forms by fusion of position 2,159 on the MLL sense strand to position 7,404 on the AF-4 sense strand. To form the der(4) junction, exonucleolytic nibbling on the AF-4 side removes position 7,407 from the four-base 5′ antisense-strand overhang (Blue, italic, shadowed), whereas the entire four-base MLL 5′ sense-strand overhang remains intact. The exonucleolytic nibbling of the AF-4 overhang would create a single-base microhomology, and there is base-pairing between the G at AF-4 antisense position 7,406 and the C at position 2,161 of the MLL sense-strand overhang (box). Template-directed polymerization fills in gaps at AF-4 sense positions 7,404–7,405 (Blue, shadowed, nonitalic) and MLL antisense positions 2,164–2,162 (Red, shadowed, nonitalic). The der(4) forms by fusion of position 7,405 on the AF-4 sense strand to position 2,161 on the MLL sense strand. There are three G residues in the normal MLL and AF-4 sense sequences that form the breakpoint junctions at MLL position 2,160 and at AF-4 positions 7,404 and 7,405. During the complex processing, the G at MLL position 2,160 is removed; both G residues at AF-4 positions 7,404 and 7,505 are preserved at the der(11) junction; and both G residues at AF-4 positions 7,404 and 7,505 are added by gap fill-in at the der(4) junction (compare box, Fig. 1), resulting in an overall gain of a single G relative to the normal MLL and AF-4 sense sequences. (C) Compared with B, there is more exonucleolytic nibbling in this model. The entire four-base 5′ antisense-strand overhang, the G at position 2,160 on the sense strand, and the complementary C are deleted from the MLL end (Red, italic, shadowed) that forms the der(11). Positions 7,406–7,407 of the 5′ antisense-strand overhang are deleted from the AF-4 end (Blue, italic, shadowed) forming the der(4). After template-directed polymerization (Blue, Red, shadowed, nonitalic), ligations at both junctions are blunt-ended. The der(11) forms by fusion of MLL position 2,159 to AF-4 position 7,404. The der(4) forms by fusion of AF-4 position 7,405 to MLL position 2,161. This model is consistent with the deletion of a single G from MLL to form the der(11) and the gain of two templated Gs in AF-4 to form the der(4), resulting in an overall gain of a single G relative to the normal MLL and AF-4 sense sequences. Because of the required processing, simple reciprocal exchange of DNA topoisomerase II subunits alone could not produce the observed der(11) and der(4) breakpoint-junction sequences.
MLL and AF-4 Translocation Breakpoints in t(4;11) Are DNA Topoisomerase II Cleavage Sites.
Cleavage assays of substrates containing normal MLL and AF-4 sequences indicated that the translocation breakpoints in MLL and AF-4 in this leukemia coincided with DNA topoisomerase II cleavage sites. Because prior anticancer therapy in the patient included etoposide and dactinomycin, in vitro cleavage of MLL and AF-4 DNA substrates was analyzed after incubation with human DNA topoisomerase IIα in the absence of drug and in the presence of etoposide, its catechol or quinone metabolites, or dactinomycin. In the absence of drug, little cleavage was detectable in either substrate (Fig. 2). Fragments of various sizes and intensities indicated that multiple DNA topoisomerase II cleavage sites were induced in both substrates by etoposide, etoposide catechol, and etoposide quinone, but there was minimal cleavage in the presence of dactinomycin [Fig. 2, No Heat, (+)Topo].
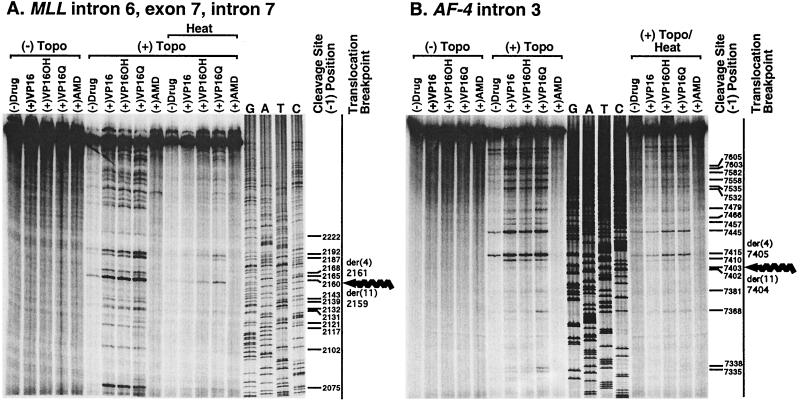
Figure 2.
Etoposide-, etoposide metabolite-, and dactinomycin-induced DNA topoisomerase II cleavage of MLL intron 6, exon 7, and intron 7 coordinates 2,050 to 2,587 (A) and AF-4 intron 3 coordinates 7,318 to 7,841 (B). Shown are autoradiographs of cleavage products after 10-min incubation of 25 ng (30,000 cpm) of singly 5′ end-labeled DNA with 147 nM human DNA topoisomerase IIα, 1 mM ATP, and, where indicated, 20 μM etoposide (VP16), etoposide catechol (VP16-OH), etoposide quinone (VP16-Q), or dactinomycin (AMD). “Heat” indicates that reactions were incubated for 10 min at 65°C before trapping of covalent complexes by adding SDS. The indicated nucleotide is the 5′ side or −1 position of the cleavage site; the cleaved phosphodiester bond is 3′ to the indicated nucleotide. Corkscrew arrows at far right of A and B indicate positions of MLL or AF-4 translocation breakpoints contained within the substrates.
For the substrate containing positions 2,050 to 2,587 in MLL intron 6, exon 7, and intron 7, the sequence encompassing the der(11) and der(4) breakpoint was studied in detail. In the analysis of this MLL sequence in the gel in Fig. 2A, if the coordinate indicates the base at the 5′ side of the cleavage site, also called the −1 position, the strongest cleavage site in the absence of drug was position 2,160. This site was used as a reference for phosphorimage quantitation [Fig. 2A, No Heat, (+)Topo]. Position 2,160 was the second strongest cleavage site in the substrate in assays performed with etoposide and its catechol metabolite but was the strongest cleavage site in the substrate in assays performed with etoposide quinone. Cleavage at position 2,160 in the presence of etoposide was enhanced 3.1-fold in comparison with the reference site but 3.8-fold and 5.0-fold, respectively, in the presence of etoposide catechol and etoposide quinone. To evaluate the relative stability of the enzyme–DNA covalent complexes, cleavage was quantitated upon incubation for 10 min at 65°C after the reactions were complete. Detectable cleavage at many sites decreased significantly or was eliminated after heating [Fig. 2A, Heat, (+)Topo]. However, even in the absence of drug, cleavage at position 2,160 was detectable after heating, indicating that the enzyme–DNA covalent complexes formed at this site were particularly stable. Relative to the other cleavage sites shown in the gel in Fig. 2A, position 2,160 was the most heat-stable cleavage site in this substrate in the absence of drug and in the presence of etoposide and both etoposide metabolites. These results indicate that the MLL genomic breakpoint in the t(4;11) translocation was a preferred and stable DNA topoisomerase II cleavage site in the absence of drug and in the presence of etoposide and both etoposide metabolites but not dactinomycin.
Similarly, for the substrate that spanned AF-4 intron 3 positions 7,318 to 7,841, the sequence encompassing the der(11) and der(4) AF-4 breakpoint was studied in detail. In the analysis of this AF-4 sequence in the gel in Fig. 2B, the strongest cleavage site in the absence of drug was position 7,415. This site was used as a reference for phosphorimage quantitation [Fig. 2B, No Heat, (+)Topo]. There was detectable but weaker cleavage at the site with nucleotide 7,403 at the −1 position, even without drug. Relative to the reference site [Fig. 2B, position 7,415 (−)Drug, No Heat, (+)Topo], there was 0.38-, 0.39- and 0.39-fold cleavage, respectively, at position 7403 in the presence of etoposide, etoposide catechol, and etoposide quinone. Whereas position 7403 did not seem to be a highly preferred cleavage site in the AF-4 substrate, evaluation after heating indicated that the DNA topoisomerase II covalent complexes formed at position 7,403 were heat-stable. The most heat-stable complexes in this substrate were formed at position 7415 in the absence of drug and at position 7,445 in the presence of etoposide or either etoposide metabolite, but there was detectable cleavage after heating at position 7,403 in the absence of drug and in the presence of etoposide or either etoposide metabolite [Fig. 2B, Heat, (+)Topo]. Again, little cleavage was detectable in the presence of dactinomycin even without heating. These results indicate that the AF-4 genomic breakpoint in the t(4;11) translocation was a stable DNA topoisomerase II cleavage site.
Assays of both substrates were repeated with consistent results. Because of the reciprocal nature of the translocation at the sequence level, and because the translocation breakpoints in MLL and AF-4 seemed to coincide with preferred and/or stable cleavage sites, we asked whether, based on the genomic breakpoint-junction sequences and the cleavage assays, interchromosomal recombination by DNA topoisomerase II subunit exchange was feasible as the mechanism. The genomic-sequence findings and in vitro cleavage sites together suggest that, if DNA topoisomerase II subunit exchange was involved in the mechanism, nick processing would have been required to generate the der(11) and der(4) sequences observed in the leukemia. DNA topoisomerase II introduces four-base staggered nicks in duplex DNA and creates four-base 5′ overhangs. Fig. 3 details two possible models for the processing of the overhangs created by DNA topoisomerase II cleavage at MLL position 2,160 and AF-4 position 7,403 that would create the der(11) and der(4) genomic breakpoint junctions. In both models, exonucleolytic nibbling and template-directed polymerization of staggered nicks occur, but the AF-4 overhang at the der(11) junction and the MLL overhang at the der(4) junction are preserved completely. In the model in Fig. 3B, the exonucleolytic nibbling creates single nucleotide homologies at the ends, and base-pairing may promote nonhomologous end-joining (Fig. 3B, boxed G:C and C:G; ref. 23).
Discussion
We have demonstrated that the der(11) and der(4) genomic breakpoint junctions of a t(4;11) in a case of treatment-related ALL were the result of near-precise interchromosomal DNA recombination at the sequence level, and that there was an overall gain of a single templated nucleotide relative to the MLL intron 6 and AF-4 intron 3 sequences that formed this translocation. As is true of MLL genomic breakpoint junctions previously reported (13, 20–25), there was evidence from the genomic sequences of short-sequence homologies after processing at the der(11) and der(4) junctions of this t(4;11) (Fig. 1). We performed DNA topoisomerase II cleavage assays on the involved genomic regions of MLL and AF-4 to explore the mechanism. It was of interest to determine whether the MLL and AF-4 genomic translocation breakpoints were cleaved by DNA topoisomerase II because of the reciprocal nature of the breakpoint junctions. Because prior anticancer therapy in this patient included the DNA topoisomerase II inhibitor etoposide and the DNA topoisomerase I/II inhibitor dactinomycin, separate cleavage assays were performed with etoposide, the catechol and quinone metabolites of etoposide, and dactinomycin. The results provide functional evidence that the repair of stable, etoposide- or etoposide metabolite-induced DNA topoisomerase II-mediated breakage in MLL and AF-4 could have formed both genomic breakpoint junctions. The microhomologies at and near the der(11) and der(4) junctions in this treatment-related leukemia, like those at MLL genomic breakpoint junctions in de novo cases (20, 21, 23, 24), are characteristic of nonhomologous end-joining (23), which possibly may refute DNA topoisomerase II subunit exchange as the mechanism of interchromosomal DNA recombination.
Based on in vitro observations suggesting that calf thymus DNA topoisomerase II could mediate illegitimate recombination in λ-DNA, Bae et al. (26) proposed in 1988 that DNA topoisomerase II could exchange DNA strands through the exchange of enzyme subunits. Shortly afterward, experiments in Chinese hamster ovary cells were interpreted as evidence that DNA topoisomerase II could mediate in vivo recombination (27). However, the data on DNA topoisomerase II subunit exchange are controversial. Tennyson and Lindsley (28) suggested that the high stability of the DNA topoisomerase II homodimer to dissociation when covalently attached to DNA might prevent catalysis of DNA recombination by subunit exchange, whereas Zhou et al. (29) suggested that recombination between the aprt locus and an unrelated sequence in Chinese hamster ovary cells was consistent with this mechanism.
In the present study, the combined molecular and biochemical data are inconsistent with a simple reciprocal exchange of enzyme subunits and DNA-strand transfer, because further processing of the staggered nicks induced by DNA topoisomerase II including exonucleolytic deletion and template-directed polymerization would have been required to generate the observed genomic breakpoint junctions before ligation of the ends. However, the data are consistent with reciprocal DNA topoisomerase II cleavage events in MLL and AF-4 in which both breaks became stable and the DNA ends were processed and underwent ligation. Alternatively, the data may not be inconsistent with a translocation mechanism involving DNA topoisomerase II subunit exchange that allows nick processing. Zhou et al. (29) suggested that correction of mismatches in cohesive DNA ends was a component of subunit exchange. The active-site cleft of DNA topoisomerase II (30) would have had to accommodate nick processing for the t(4;11) translocation to have occurred by this means. It is unknown what governs the exonucleolytic nibbling and template-directed polymerization of the staggered nicks formed by DNA topoisomerase II, but the degree of processing may be sequence-dependent.
To determine the relevant drug exposure, the relationship of functional DNA topoisomerase II cleavage sites in the MLL and AF-4 substrates to the genomic translocation breakpoints was evaluated in the presence of etoposide, its metabolites, and dactinomycin. The association between cytochrome P450 (CYP) 3A4 genotype and epipodophyllotoxin-related leukemias with MLL translocations (31) provided the rationale to examine DNA topoisomerase II cleavage of MLL and AF-4 induced by etoposide catechol and etoposide quinone. CYP3A converts epipodophyllotoxin to a catechol metabolite, which is further oxidized to a quinone (32–34). We recently reported that, like etoposide, etoposide catechol and etoposide quinone induce DNA topoisomerase II cleavage in the MLL breakpoint cluster region, and that the N7 position of guanine is important for metabolite-induced cleavage (17). A relationship between several other MLL translocation breakpoints and DNA topoisomerase II in vitro cleavage sites induced by etoposide and these etoposide metabolites also was observed (17). The metabolites are of further interest because both adult and pediatric pharmacokinetic data suggest increasing plasma levels of the potentially genotoxic catechol from the first day to the last day of multiple-day bolus etoposide infusions (35, 36). In the MLL and AF-4 substrates shown in Fig. 2, the etoposide metabolites, especially the quinone, generally induced greater DNA topoisomerase II cleavage than etoposide and enhanced many similar but some unique cleavage sites. Cleavage at most sites was more stable in the presence of the metabolites compared with etoposide. Nucleotide 2,160 in the MLL substrate, which coincided with the t(4;11) breakpoint, was the strongest cleavage site in assays performed without drug and with etoposide quinone; it was a preferred cleavage site with etoposide and etoposide catechol. The covalent complexes formed at this site were particularly heat-stable. There was detectable cleavage at the translocation breakpoint at position 7,403 in the AF-4 substrate and the cleavage was heat-stable. In contrast, dactinomycin did not induce DNA topoisomerase II cleavage at either of these sites, possibly suggesting that etoposide and/or its metabolites, but not dactinomycin, were the relevant drug exposures for formation of the translocation in the leukemia of this patient. Leukemia occurs less often after anthracyclines and dactinomycin than after etoposide (37).
The genomic breakpoint-junction sequences on both derivative chromosomes have been examined in only a few primary leukemias with MLL translocations after chemotherapy with DNA topoisomerase II inhibitors (25, 38). Whether additional MLL genomic breakpoint-junction sequences in treatment-related leukemia will reveal precise interchromosomal DNA recombination at the sequence level remains to be determined. In conjunction with the genomic-sequence information, the DNA topoisomerase II cleavage assay used in this analysis becomes a tool not only to investigate the MLL translocation mechanism in specific cases but also to link DNA topoisomerase II-induced cleavage with clinical drug exposure. The combined molecular and biochemical data on the ALL described herein are consistent with a translocation mechanism involving either interchromosomal recombination by DNA topoisomerase II subunit exchange after processing of staggered nicks or, more likely, DNA topoisomerase II-mediated chromosomal breakage, processing of DNA ends, and recombination by another means. Although etoposide, etoposide metabolites, and dactinomycin all interfere with the cleavage-religation equilibrium of DNA topoisomerase II, the in vitro cleavage assays implicate etoposide and/or its metabolites in the leukemia of this patient. Associations of DNA topoisomerase II-targeted drugs with chemotherapy-induced leukemia have suggested a role of DNA topoisomerase II in the translocation process. These results support involvement of DNA topoisomerase II in MLL translocations. Genomic translocation breakpoint cloning and in vitro DNA topoisomerase II cleavage assays together support a model for treatment-related leukemia in which DNA topoisomerase II causes chromosomal breakage and translocations form when the breakage is repaired.
Acknowledgments
We thank Christine Richardson for helpful review and suggestions. C.A.F. is supported by National Institutes of Health Grants CA80175, CA77683, and CA85469 and a Leukemia and Lymphoma Society Scholar Award. N.O. is supported by National Institutes of Health Grant GM33944.
Abbreviation
- ALL
acute lymphoblastic leukemia
Footnotes
References
- 1.Smith M A, McCaffrey R P, Karp J E. J Natl Cancer Inst. 1996;88:407–418. doi: 10.1093/jnci/88.7.407. [DOI] [PubMed] [Google Scholar]
- 2.Felix C A. Biochim Biophys Acta. 1998;1400:233–255. doi: 10.1016/s0167-4781(98)00139-0. [DOI] [PubMed] [Google Scholar]
- 3.Felix C. In: Multiple Primary Cancers: Incidence, Etiology, Diagnosis and Prevention. Neugut A I, Meadows A T, Robinson E, editors. Baltimore: Williams & Wilkins; 1999. pp. 137–164. [Google Scholar]
- 4.Felix C A. Med Pediatr Oncol. 2001;36:525–535. doi: 10.1002/mpo.1125. [DOI] [PubMed] [Google Scholar]
- 5.Felix C A, Megonigal M D. In: American Society of Clinical Oncology 2001 Educational Book. Perry M C, editor. Alexandria, VA: Am. Soc. Clin. Oncol.; 2001. pp. 578–590. [Google Scholar]
- 6.Burden D A, Osheroff N. Biochim Biophys Acta. 1998;1400:139–154. doi: 10.1016/s0167-4781(98)00132-8. [DOI] [PubMed] [Google Scholar]
- 7.Corbett A H, Osheroff N. Chem Res Toxicol. 1993;6:585–597. doi: 10.1021/tx00035a001. [DOI] [PubMed] [Google Scholar]
- 8.Pommier Y, Bertrand R. In: The Causes and Consequences of Chromosomal Aberrations. Kirsch I R, editor. Boca Raton, FL: CRC Press; 1993. pp. 277–309. [Google Scholar]
- 9.Capranico G, Kohn K W, Pommier Y. Nucleic Acids Res. 1990;18:6611–6619. doi: 10.1093/nar/18.22.6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pommier Y, Capranico G, Orr A, Kohn K W. J Mol Biol. 1991;222:909–924. doi: 10.1016/0022-2836(91)90585-t. [DOI] [PubMed] [Google Scholar]
- 11.Pommier Y, Capranico G, Orr A, Kohn K W. Nucleic Acids Res. 1991;19:5973–5980. doi: 10.1093/nar/19.21.5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pommier Y, Kohn K W, Capranico G, Jaxel C. In: Molecular Biology of DNA Topoisomerases and its Application to Chemotherapy. Andoh T, Ikeda H, Oguro M, editors. Boca Raton, FL: CRC Press; 1993. pp. 215–227. [Google Scholar]
- 13.Megonigal M D, Rappaport E F, Jones D H, Kim C S, Nowell P C, Lange B J, Felix C A. Proc Natl Acad Sci USA. 1997;94:11583–11588. doi: 10.1073/pnas.94.21.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leahey A M, Friedman D L, Bunin N J. Bone Marrow Transplant. 1999;23:21–25. doi: 10.1038/sj.bmt.1701517. [DOI] [PubMed] [Google Scholar]
- 15.Wasserman R A, Austin C A, Fisher L M, Wang J C. Cancer Res. 1993;53:3591–3596. [PubMed] [Google Scholar]
- 16.Kingma P S, Greider C A, Osheroff N. Biochemistry. 1997;36:5934–5939. doi: 10.1021/bi970507v. [DOI] [PubMed] [Google Scholar]
- 17.Lovett B D, Strumberg D, Blair I A, Pang S, Burden D A, Megonigal M D, Rappaport E F, Osheroff N, Pommier Y, Felix C A. Biochemistry. 2001;40:1159–1170. doi: 10.1021/bi002361x. [DOI] [PubMed] [Google Scholar]
- 18.Nilson I, Reichel M, Ennas M G, Greim R, Knorr C, Siegler G, Greil J, Fey G H, Marschalek R. Br J Haematol. 1997;98:157–169. doi: 10.1046/j.1365-2141.1997.1522966.x. [DOI] [PubMed] [Google Scholar]
- 19.Felix C A, Lange B J, Hosler M R, Fertala J, Bjornsti M-A. Cancer Res. 1995;55:4287–4292. [PubMed] [Google Scholar]
- 20.Megonigal M D, Rappaport E F, Jones D H, Williams T M, Lovett B D, Kelly K M, Lerou P H, Moulton T, Budarf M L, Felix C A. Proc Natl Acad Sci USA. 1998;95:6413–6418. doi: 10.1073/pnas.95.11.6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felix C, Kim C, Megonigal M, Slater D, Jones D, Spinner N, Stump T, Hosler M, Nowell P, Lange B, Rappaport E. Blood. 1997;90:4679–4686. [PubMed] [Google Scholar]
- 22.Atlas M, Head D, Behm F, Schmidt E, Zeleznik-Le N J, Roe B A, Burian D, Domer P H. Leukemia. 1998;12:1895–1902. doi: 10.1038/sj.leu.2401223. [DOI] [PubMed] [Google Scholar]
- 23.Gillert E, Leis T, Repp R, Reichel M, Hosch A, Breitenlohner I, Angermuller S, Borkhardt A, Harbott J, Lampert F, et al. Oncogene. 1999;18:4663–4671. doi: 10.1038/sj.onc.1202842. [DOI] [PubMed] [Google Scholar]
- 24.Felix C A, Hosler M R, Slater D J, Megonigal M D, Lovett B D, Williams T M, Nowell P C, Spinner N B, Owens N L, Hoxie J, et al. Mol Diagn. 1999;4:269–283. doi: 10.1016/s1084-8592(99)80002-2. [DOI] [PubMed] [Google Scholar]
- 25.Megonigal M D, Cheung N-K V, Rappaport E F, Nowell P C, Wilson R B, Jones D H, Addya K, Leonard D G B, Kushner B H, Williams T M, et al. Proc Natl Acad Sci USA. 2000;97:2814–2819. doi: 10.1073/pnas.050397097. . (First Published March 7, 2000; 10.1073/pnas.050397097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bae Y-S, Kawasaki I, Ikeda H, Liu L F. Proc Natl Acad Sci USA. 1988;85:2076–2080. doi: 10.1073/pnas.85.7.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charron M, Hancock R. Chromosoma. 1991;100:97–102. doi: 10.1007/BF00418242. [DOI] [PubMed] [Google Scholar]
- 28.Tennyson R B, Lindsley J E. Biochemistry. 1997;36:6107–6114. doi: 10.1021/bi970152f. [DOI] [PubMed] [Google Scholar]
- 29.Zhou R-H, Wang P, Zou Y, Jackson-Cook C, Povirk L. Cancer Res. 1997;57:4699–4702. [PubMed] [Google Scholar]
- 30.Berger J M, Gamblin S J, Harrison S C, Wang J C. Nature (London) 1996;379:225–233. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 31.Felix C A, Walker A H, Lange B J, Williams T M, Winick N J, Cheung N-K V, Lovett B D, Nowell P C, Blair I A, Rebbeck T R. Proc Natl Acad Sci USA. 1998;95:13176–13181. doi: 10.1073/pnas.95.22.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Maanen J M S, Retel J, de Vries J, Pinedo H M. J Natl Cancer Inst. 1988;80:1526–1533. doi: 10.1093/jnci/80.19.1526. [DOI] [PubMed] [Google Scholar]
- 33.Mans D R A, Retel J, van Maanen J M S, Lafleur M V M, van Schaik M A, Pinedo H M, Lankelma J. Br J Cancer. 1990;62:54–60. doi: 10.1038/bjc.1990.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Relling M V, Nemec J, Schuetz E G, Schuetz J D, Gonzalez F J, Korzekwa K R. Mol Pharmacol. 1994;45:352–358. [PubMed] [Google Scholar]
- 35.Stremetzne S, Jaehde U, Kasper R, Beyer J, Siegert W, Schunack W. Eur J Cancer. 1997;33:978–979. doi: 10.1016/s0959-8049(97)00087-7. [DOI] [PubMed] [Google Scholar]
- 36.Pang, S., Zheng, N., Felix, C. A., Boston, R. & Blair, I. A., (2001) J. Mass Spectrometry, in press. [DOI] [PubMed]
- 37.Sandoval C, Pui C-H, Bowman L C, Heaton D, Hurwitz C A, Raimondi S C, Behm F G, Head D R. J Clin Oncol. 1993;11:1039–1045. doi: 10.1200/JCO.1993.11.6.1039. [DOI] [PubMed] [Google Scholar]
- 38.Domer P H, Head D R, Renganathan N, Raimondi S C, Yang E, Atlas M. Leukemia. 1995;9:1305–1312. [PubMed] [Google Scholar]