ABSTRACT
Prion-like domains are low complexity, intrinsically disordered domains that compositionally resemble yeast prion domains. Many prion-like domains are involved in the formation of either functional or pathogenic protein aggregates. These aggregates range from highly dynamic liquid droplets to highly ordered detergent-insoluble amyloid-like aggregates. To better understand the amino acid sequence features that promote conversion to stable, detergent-insoluble aggregates, we used the prediction algorithm PAPA to identify predicted aggregation-prone prion-like domains with a range of compositions. While almost all of the predicted aggregation-prone domains formed foci when expressed in cells, the ability to form the detergent-insoluble aggregates was highly correlated with glutamine/asparagine (Q/N) content, suggesting that high Q/N content may specifically promote conversion to the amyloid state in vivo. We then used this data set to examine cross-seeding between prion-like proteins. The prion protein Sup35 requires the presence of a second prion, [PIN+], to efficiently form prions, but this requirement can be circumvented by the expression of various Q/N-rich protein fragments. Interestingly, almost all of the Q/N-rich domains that formed SDS-insoluble aggregates were able to promote prion formation by Sup35, highlighting the highly promiscuous nature of these interactions.
KEYWORDS: yeast, prion, amyloid, protein aggregation, prion-like domains
INTRODUCTION
In the budding yeast Saccharomyces cerevisiae, at least 9 proteins have been identified that form self-propagating amyloid-based prions.1 Simple phenotypic assays have been developed to monitor prion activity, making the yeast prions useful model systems to study aggregation and prion activity. Most known yeast prion proteins contain a low-complexity, intrinsically-disordered prion-forming domain that is necessary for prion activity.2,3 These prion-forming domains tend to be glutamine/asparagine (Q/N) rich, and relatively lacking in charged and hydrophobic amino acids.4 Scrambling the sequence of Q/N-rich prion domains does not eliminate prion activity, suggesting that amino acid composition is the primary determinant of prion propensity.5-7
A variety of computational algorithms have been designed to identify proteins that are compositionally similar to known yeast prion proteins.8-10 Hundreds of proteins in the human genome contain such prion-like domains (PrLDs).11,12 Recently, mutations in several these PrLDs have been linked to degenerative disorders, including ALS.13,14 Emerging evidence suggests that these PrLDs may be designed to form dynamic, functional aggregates, and that disease-associated mutations can drive the proteins to form stable, detergent-insoluble amyloid-like aggregates.15-18 For example, stress granules are dynamic RNA-protein assemblies that form when translation is inhibited.19,20 Many RNA binding proteins found in stress granules contain PrLDs, and weak dynamic interactions between these PrLDs are thought to drive liquid-liquid phase separation, promoting granule formation.21,22 Disease-associated mutations in some of these PrLDs appear to drive conversion of the PrLDs into more stable amyloid-like aggregates, thereby disrupting stress granule dynamics.15-18
Therefore, there has been growing interest in understanding how amino acid sequence affects both PrLD aggregation propensity and the stability of these aggregates. As a first step toward addressing this question, we experimentally determined the prion propensity of each amino acid in the context of a yeast prion domain, and used these values to develop the prediction algorithm PAPA.23,24 PAPA scans proteins for regions of intrinsic disorder, and scores the prion propensities of these regions.25 PAPA has proven effective at predicting the prion-like activity of Q/N-rich PrLDs;14 designing mutations to modulate the aggregation activity of PrLDs;26-28 designing synthetic prion-forming domains;24 and predicting the effects of some disease-associated mutations in human PrLDs.29
However, PAPA still has substantial limitations. First, all of these previous validations of PAPA have been done on compositional homogenous data sets of Q/N-rich proteins. Therefore, it is less clear whether PAPA would be effective at identifying aggregation-prone PrLDs from a more compositionally diverse data set such as a whole proteome. In particular, it is surprising that Q/N residues dominate yeast prion domains, yet have relatively neutral prion propensities according to PAPA.4,11 Intrinsic disorder may provide a partial explanation for this discrepancy. The structural flexibility of yeast prion domains appears to be important for prion formation, likely because it increases accessibility of prion-nucleating regions.30 Q and N may therefore be over-represented in part because they balance intrinsic disorder and prion propensity.
However, this theory does not explain why the yeast prion domains tend to be specifically enriched in Q and N, and not amino acids like serine, threonine and glycine, which also promote intrinsic disorder and have similar aggregation propensities.23,31 This bias may in part be an artifact of how yeast prion proteins have been discovered. The first 2 prion proteins identified, Ure2 and Sup35, are both Q/N rich. Many of the subsequent prion proteins were identified either because they share similar sequence features to Ure2 and Sup35,3,10,32 or because they are able to promote prion formation by Sup35.33-35 Both methods may be biased toward Q/N residues.
Alternatively, the low predicted prion propensities of Q and N may be an artifact of the experiments used to develop PAPA. Prion propensity scores were derived by randomly mutagenizing a small portion of a Q/N-rich prion domain and examining the compositional biases of mutants that retained the ability to form prions. These experiments therefore report how small compositional changes affect prion propensity. In the context of a highly Q/N-rich domain, it appears that subtle changes in Q/N content have little effect on prion propensity. However, it remains possible that a threshold number of Q/N residues is required for some prion-promoting activity. For example, it has been proposed that, when present at high enough density, Q/N residues can promote the formation of polar zippers.36
A second major limitation of PAPA (and likely all available prion prediction algorithms) is that prion activity is a complex process, requiring a series of discrete steps, each of which may have distinct compositional requirement, yet PAPA does not separately assess the effects of amino acid composition on each of these steps. Specifically, for a protein to act as a prion in yeast, it needs to not only form prion aggregates, but also propagate these aggregates to daughter cells during cell division. We previously developed a method to separate the effects of composition on prion formation versus prion propagation, and found that PAPA predominantly measures prion formation propensity.37 Thus, PAPA could be more accurately characterized as an aggregation predictor for PrLDs, rather than a prion predictor. However, it is still not clear whether PAPA simply predicts aggregation propensity, or whether it specifically predicts the ability to form amyloid aggregates. This distinction is important in understanding how mutations affect the dynamics of PrLD-associated aggregates.
To begin to address both of these limitations of PAPA, we used PAPA to identify predicted aggregation-prone PrLDs with a range of compositions. Each domain was then tested for the ability to aggregate, and the ability to form stable, detergent insoluble aggregates. As a control, we identified Q/N-rich segments with low predicted aggregation propensity. Almost all of the predicted PrLDs formed foci when fused to GFP, while almost none of the control domains did; however, the ability to form the detergent-insoluble aggregates that characterize yeast prions was highly dependent on Q/N content. This suggests that high Q/N content has little effect on aggregation propensity, but promotes conversion of aggregates to an amyloid state. In most cases, the formation of detergent-insoluble aggregates was independent of [PIN+], a prion that is required for prion formation by the yeast prion protein Sup35.3,33 Strikingly, almost every protein from our data set that formed detergent-insoluble aggregates was also able to substitute for [PIN+] in stimulating prion formation by Sup35, highlighting the highly promiscuous nature of these interactions. Together these data aid in unraveling the complex biology and structural characteristics for a protein to form a prion in yeast.
RESULTS
PAPA Predicts the Ability of PrLDs to Form Foci
Yeast prion domains are generally modular, meaning that they maintain aggregation and prion activity when transferred to other proteins.38 Alberti et al. previously scanned the yeast genome for domains that compositionally resembled known yeast prion domains, and taking advantage of this modularity, tested the top 100 PrLDs for aggregation and prion-like activity in a series of assays.10 PAPA was quite effective at predicting the ability of these PrLDs both to form foci when expressed as PrLD-GFP fusions, and to form SDS-insoluble aggregates (Fig. 1). For these PrLDs, there was only a modest correlation between aggregation activity and Q/N content;10 in both assays, Q content showed a slight negative correlation with aggregation activity, while N content showed a slight positive correlation, consistent with subsequent studies showing that N has a higher amyloid aggregation propensity.39
Figure 1.
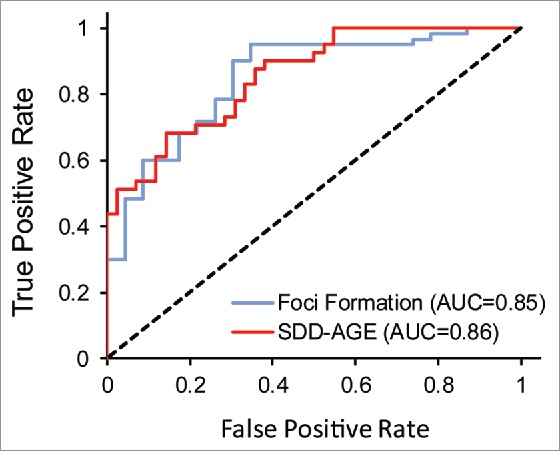
PAPA shows comparable accuracy in predicting foci formation and the formation of SDS-insoluble aggregates by Q/N-rich PrLDs. Alberti et al. tested 100 yeast PrLDs for the ability to form foci when fused to GFP, and the ability to form SDS-insoluble aggregates by SDD-AGE. Shown are ROC (receiver operator characteristic) plot assessing PAPA's effectiveness at distinguishing between positive and negative examples from these 2 data sets. The area under the curve (AUC) for each plot is indicated. The dotted line reflects the prediction accuracy which would be expected by random chance.
However, because the PrLDs were all identified based on compositional similarity to known yeast prion domains, this data set was reasonably compositionally homogenous. For example, all of the PrLDs that were tested in the full set of assays were at least 22% Q/N. By contrast, many of the PrLDs predicted to be aggregation-prone by PAPA have far lower Q/N-content. Therefore, while PAPA was accurate at predicting aggregation propensity for the Alberti data set, and while there was only a weak correlation between Q/N content and aggregation activity, it is unclear whether these trends would remain for a more compositionally diverse data set.
To address these questions, we searched the yeast proteome for PrLDs predicted by PAPA to be aggregation-prone. We identified 151 candidate PrLDs (PAPA score ≥ 0.05).24 We excluded any PrLDs that overlapped with the Alberti data set, and then selected 30 candidate PrLDs with a range of Q/N content (from 6–35% Q/N; Table 1). As a negative control, we additionally selected 10 Q/N-rich protein domains that scored well below PAPA's threshold.
TABLE 1.
Summary of results.
| Protein Name | PrLD Position | PAPA Score | %QN | %N | GFP Foci (%) | SDS-Insoluble Aggregates | [PIN+] proteins |
|---|---|---|---|---|---|---|---|
| Candidate prion-like domains | |||||||
| Swi4 | 177–380 | 0.09 | 35.3 | 25.5 | 100 | + | ++++ |
| Rpi1 | 192–306 | 0.05 | 33.0 | 27.8 | 96 | +c | + |
| Var1e | 191–349 | 0.20 | 31.0 | 30.4 | 100 | + | ++ |
| Mfg1 | 1–96 | 0.07 | 29.2 | 11.5 | 0 | - | - |
| Pam1 | 617–756 | 0.06 | 27.9 | 17.9 | 21 | + | + |
| Dat1 | 102–236 | 0.09 | 27.4 | 12.6 | 100 | + | +++ |
| YML053Cd | 34–148 | 0.05 | 26.5 | 25.7 | 97 | +c | ++ |
| Rna15 | 39–169 | 0.08 | 26.0 | 18.3 | 100 | - | - |
| Cdc39d | 966–1092 | 0.08 | 26.0 | 5.5 | 99 | - | - |
| Slf1 | 183–311 | 0.11 | 25.6 | 16.3 | 100 | - | - |
| Sky1 | 353–491 | 0.12 | 24.5 | 23.7 | 79 | + | +++ |
| Pin4 | 169–492 | 0.11 | 23.8 | 14.2 | 97 | + | +++ |
| Gis1 | 454–584 | 0.08 | 23.7 | 21.4 | 18 | + | - |
| Cln2 | 362–503 | 0.09 | 23.2 | 15.5 | 47.9 | +c | - |
| Fab1d | 427–552 | 0.06 | 23.0 | 22.2 | 97 | + | +++ |
| Mex67 | 1–95 | 0.14 | 21.1 | 14.7 | 0 | + | - |
| Q0255e | 341–472 | 0.07 | 20.6 | 19.8 | 100 | - | - |
| Tda7 | 513–636 | 0.10 | 20.2 | 12.9 | 42 | - | - |
| YGL036W | 270–478 | 0.15 | 20.1 | 15.8 | 100 | - | +++ |
| Bph1 | 1113–1243 | 0.10 | 19.1 | 14.5 | 100 | - | - |
| Ssn2 | 1025–1211 | 0.09 | 18.2 | 8.0 | 100 | - | - |
| AI3e | 228–387 | 0.17 | 17.6 | 15.7 | 100 | - | - |
| Lee1 | 151–301 | 0.12 | 17.2 | 11.3 | 19 | - | - |
| Vac14 | 690–818 | 0.09 | 17.1 | 8.5 | 97 | - | - |
| Cdc73 | 253–378 | 0.07 | 15.9 | 9.5 | 100 | - | - |
| Mdm1 | 745–864 | 0.10 | 14.2 | 8.3 | 89 | - | - |
| Pgs1 | 158–277 | 0.07 | 12.5 | 8.3 | 92 | - | - |
| Nte1 | 1–169 | 0.12 | 12.4 | 8.3 | 0 | - | - |
| Cos111 | 336–465 | 0.10 | 8.5 | 6.9 | 0 | - | - |
| Izh3 | 176–492 | 0.13 | 6.0 | 3.8 | 100 | - | - |
| Negative control Q/N-rich domains | |||||||
| Dal81 | 4–168 | −0.06 | 35.2 | 20.0 | 3 | − | + |
| Yck2 | 369–533 | −0.15 | 35.2 | 9.7 | 0 | − | − |
| Hrk1 | 483–647 | −0.12 | 30.3 | 15.8 | 0 | − | − |
| Grr1 | 3–167 | −0.02 | 29.7 | 21.2 | 3 | + | +++ |
| Apg13 | 250–414 | −0.02 | 29.1 | 6.7 | 100 | − | − |
| Siz1 | 390–554 | −0.10 | 28.5 | 24.2 | 0 | − | − |
| Crz1 | 15–179 | −0.11 | 28.5 | 10.3 | 0 | − | − |
| Vac7 | 377–541 | −0.05 | 27.9 | 18.8 | 0 | + | − |
| Tbs1 | 898–1062 | −0.14 | 25.5 | 23.6 | 3 | − | − |
| Vid22 | 641–806 | −0.04 | 23.0 | 18.2 | 0 | − | − |
Percentage of cells with GFP foci. At least 50 cells were counted for each strain.
Cells were assessed for the ability to substitute for [PIN+] in supporting [PSI+] formation. +, ++, +++, and ++++ correspond to the number of spots in the dilution series with at least 10 colonies.
SDS-insoluble aggregates forming after 48hrs
Sequence polymorphism; see Methods
Synthetically built; see Methods
To test for the ability to form foci, we generated PrLD-GFP fusions under control of the GAL1 promoter. Although there was some variability in efficiency of expression among the fusions, most of the fusions showed efficient expression upon growth in galactose-containing medium (Figure S1). Likewise, while a few of the fusions showed some degradation, in most cases the predominant band corresponded to the approximate expected size of the fusion (Figure S1). Almost all of the candidate PrLDs formed distinct cytoplasmic foci (Fig. 2, Table 1), regardless of Q/N content. This result suggests that high Q/N content is not critical for PrLD aggregation, and that PAPA is effective at identifying aggregation-prone domains, regardless of Q/N content. Additionally, all but one of the negative control PrLDs showed diffuse cytoplasmic signal (Fig. 2, Table 1), confirming PAPA's ability to distinguish between aggregation-prone and non-aggregation-prone Q/N-rich domains.
Figure 2.
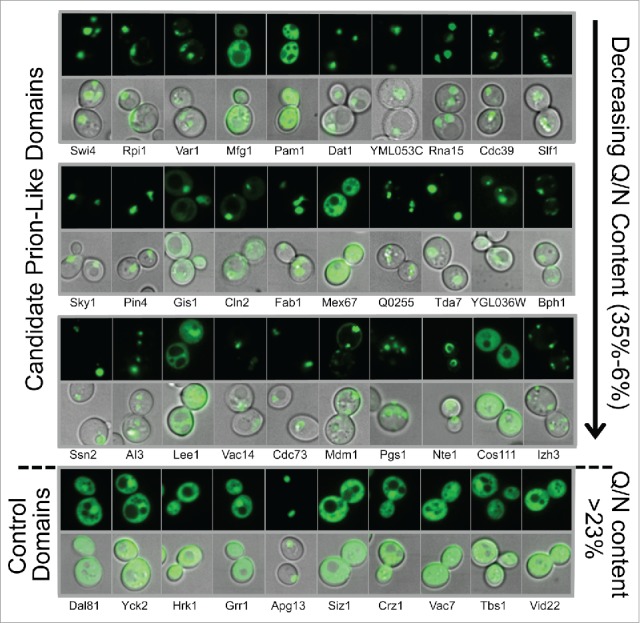
Prion-like domains form distinct foci in the cytoplasm. The [PIN+] strain yER632 was transformed with plasmids expressing PrLD-GFP fusions under control of the GAL1 promoter. Cells were grown in galactose/raffinose dropout medium for 24 h, and then visualized by fluorescence microscopy and differential interference contrast (DIC). The first 3 rows contain PrLDs that are predicted by PAPA to be aggregation prone (PAPA score >0.05), ordered by Q/N content. The bottom row contains Q/N-rich domains predicted by PAPA not to be aggregation-prone. Representative images are shown. See Table 1 for quantification.
Prion-Like Domains Form SDS-Insoluble Aggregates
Foci formation, while common to the yeast prion proteins, is also seen with many non-prion proteins. Protein aggregates can range from amorphous aggregates to the highly ordered, detergent-insoluble amyloid aggregates that characterize yeast prions. Therefore, we used semi-denaturing detergent-agarose gel electrophoresis (SDD-AGE)40 as a more stringent approach to determine if the PrLDs had the propensity to form SDS-insoluble aggregates in vivo.
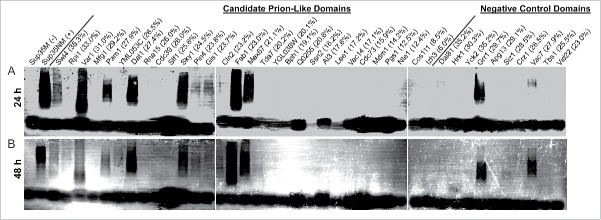
HA-tagged PrLDs were transiently expressed from the GAL1 promoter for 24 or 48 hours. Cells were then harvested, and cell lysates were examined by SDD-AGE. Many of the PrLDs formed high molecular weight SDS-insoluble aggregates after 24 hours of expression (Fig. 3A). It should be noted that Fig. 3A is overexposed to allow for detection of inefficient oligomer formation, and that for some of the PrLDs that formed SDS-insoluble aggregates (Swi4, Pin4 and Gis1), the majority of the protein was monomeric on SDD-AGE.
Figure 3.
Q/N-rich PrLDs more likely to form SDS-insoluble aggregates. The [PIN+] strain yER632 was transformed with plasmids expressing PrLD-GFP fusions under control of the GAL1 promoter. Cells were grown in galactose/raffinose dropout medium for 24 h (A) or 48 h (B) and analyzed by SDD-AGE. Q/N-content for each PrLD is indicated.
For all PrLDs that formed SDS-insoluble aggregates at 24 hours, aggregates were still observed at 48 hours; additionally, new SDS-insoluble aggregates for Cln2, YML053C and Rpi1 appeared, suggesting a longer lag phase (Fig. 3B).
Strikingly, among the PrLDs with greater than 21% Q/N content, over 75% formed SDS-insoluble aggregates, while all of the PrLDs with less than 21% Q/N content failed to form SDS-insoluble aggregates. Additionally, only 2 of the negative control Q/N-rich domains formed SDS-insoluble aggregates. Thus, as was seen for the Alberti data set (Fig. 1), if our data set is limited to Q/N-rich proteins, PAPA is reasonably effective at predicting which PrLDs will form detergent-insoluble aggregates (Fig. 4A); however, PAPA is not effective for domains with lower Q/N content (Fig. 4A). By contrast, PAPA was equally effective at predicting foci formation for the full data set and for the Q/N-rich subset (Fig. 4B).
Figure 4.
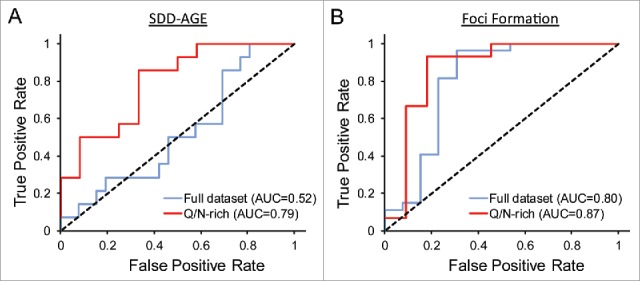
PAPA accuracy for the tested proteins. A) ROC plot examining the ability of PAPA to predict formation of SDS-insoluble aggregates. Among the full data set, PAPA shows almost no ability to distinguish between positive and negative examples (AUC = 0.52), but among the subset of domains with greater than 21% Q/N content, PAPA shows reasonably accurate predictions (AUC = 0.79). B) ROC plot examining the ability of PAPA to predict foci formation. For foci formation, PAPA shows roughly equivalent ability to distinguish between positive and negative examples among the full data set and for the Q/N-rich subset.
These data suggest that high Q/N content promotes the formation of SDS insoluble aggregates. In particular, formation of SDS-insoluble aggregates was correlated with N content, consistent with previous results suggesting that N more efficiently promotes conversion to an amyloid state.39 Among the predicted aggregation-prone PrLDs with >21% Q/N content, 4 failed to form SDS-insoluble aggregates: Mfg1, Rna15, Cdc39, and Slf1. Two of these (Mfg1 and Cdc39) had the lowest ratio of N:Q, and lowest N content of the PrLDs with >21% Q/N (Table 1). It is less clear why Rna15 and Slf1 failed to form SDS-insoluble aggregates.
Most PrLDs Are Rnq1 Independent, but Hsp104 Dependent
[PIN+] and [PSI+] are the prion forms of the yeast prion proteins Rnq1 and Sup35, respectively. [PIN+] is required for de novo [PSI+] formation, and for formation of SDS-insoluble aggregates by Sup35.3,33,41 [PIN+] is thought to act as a template to cross-seed amyloid formation by Sup35,42 although it remains possible that [PIN+] may promote [PSI+] formation by an indirect mechanism, such as titrating away an inhibitor of [PSI+] formation. [PIN+] also promotes, but is not required for, prion formation by the prion protein Ure2.43 If [PIN+] specifically promotes amyloid formation by Q/N-rich proteins, it could explain why the Q/N-rich proteins in our data set were more likely to form SDS-insoluble aggregates. However, most of the PrLDs that efficiently formed SDS-insoluble aggregates still formed SDS-insoluble aggregates in the absence of Rnq1 (Fig. 5A). Thus, [PIN+] is not responsible for the observed bias toward Q/N-rich proteins among the PrLDs that formed detergent-insoluble aggregates.
Figure 5.
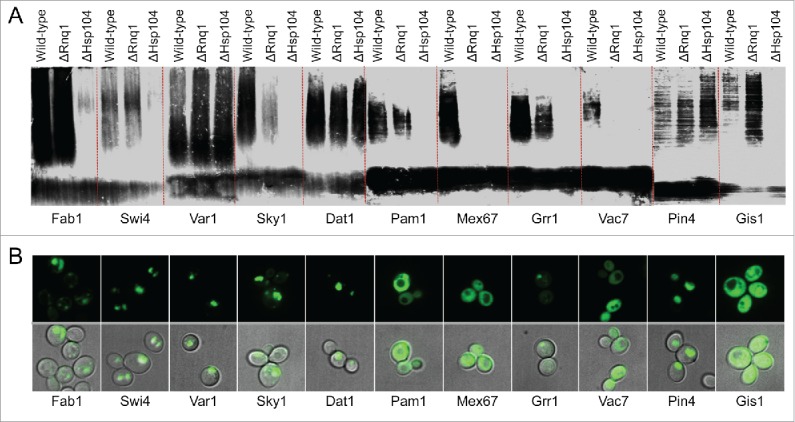
Effect of [PIN+] and Hsp104 on PrLD aggregation. A) Plasmids expressing PrLD-HA fusions that formed SDS-insoluble aggregates were transformed into yER1017 (rnq1Δ) and yER1018 (hsp104Δ). Cells were grown for 24 h in galactose/raffinose dropout medium, and analyzed by SDD-AGE. B) Plasmids expressing PrLD-GFP fusions were transformed into the hsp104Δ strain yER1615. Cells were grown for 24 h in galactose/raffinose dropout medium, and visualized by fluorescence microscopy and differential interference contrast.
By contrast, most of the PrLDs were dependent on Hsp104 for efficient formation of SDS-insoluble aggregates (Fig. 5A). Hsp104 is a chaperone required for the maintenance of almost all yeast prions.1,44 Hsp104 is a homohexameric AAA+ ATPase that fragments prion fibers, creating new prion seeds to offset dilution by cell division.45 Additionally, Hsp104 promotes de novo aggregation by Sup35; by contrast, Hsp104 is not required for de novo aggregation of the prion-like protein Pin4, one of the proteins in our data set.46 Hsp104 deletion eliminated or substantially diminished formation of SDS insoluble aggregates for all of the PrLDs except Pin4s and Var1s. While Hsp104 deletion results in loss of [PIN+], the fact that the PrLDs all formed SDS-insoluble aggregates in the absence of [PIN+] suggests that Hsp104 promotes formation of SDS-insoluble aggregates by a mechanism independent of [PIN+].
Interestingly, for the Fab1, Swi4, Sky1, and Grr1 PrLDs, Hsp104 deletion substantially reduced or eliminated formation of SDS-insoluble aggregates, but did not prevent the formation of foci, suggesting that Hsp104 may specifically promote conversion to a stable amyloid-like state (Fig. 5B). The nature of these non-amyloid foci is unclear. None of the respective full-length proteins has been reported to form foci; in a large-scale screen of GFP fusions expressed at endogenous levels, Fab1 localized to the vacuolar membrane, Sky1 showed diffuse cytoplasmic localization, and Swi4 was diffusely localized to the cytoplasm and nucleus (Grr1 was not visualized in this screen).47 Nevertheless, foci formation could reflect localization of the PrLD to a subcellular compartment rather than aggregation per se.
Pin+ Activity of Q/N-Rich Prion-Like Domains
Although [PIN+] is generally required for prion formation by Sup35, overexpression of either poly-Q or various Q/N-rich PrLDs can substitute for [PIN+] in promoting [PSI+] formation.33,42 However, it is not known whether every aggregation-prone Q/N-rich domain has Pin+ activity (i.e., can substitute for [PIN+] in promoting de novo [PSI+] formation), or whether this property is unique to only a subset of Q/N-rich aggregation-prone domains. Because many of the tested PrLDs were able to form SDS-insoluble aggregates independent of Rnq1, we examined whether these PrLDs could substitute for [PIN+] in promoting [PSI+] formation.
[PSI+] formation was detected by monitoring nonsense suppression of the ade2–1 allele.48 Sup35 is a GTP binding protein that interacts with Sup45 to form the release factor that recognizes in-frame stop codons in mRNAs.49 [PSI+] formation reduces the pool of active Sup35, increasing stop codon read-through, and allowing ade2–1 cells to grow in the absence of adenine. In a strain lacking RNQ1, we monitored the formation of Ade+ colonies when each PrLD was co-overexpressed with Sup35NM (Fig. 6A and data not shown). Interestingly, almost all of the PrLDs that formed SDS-insoluble aggregates in the absence of Rnq1 were able to substitute [PIN+] in promoting Ade+ colony formation by Sup35, highlighting the lack of sequence specificity for this activity. By contrast, only 2 PrLDs that failed to form SDS-insoluble aggregates were able to promote Ade+ colony formation by Sup35 (YGL036W and Dal81). In all cases, formation of Ade+ colonies was associated with [PIN+]-independent foci formation by the PrLDs (Fig. 6B).
Figure 6.
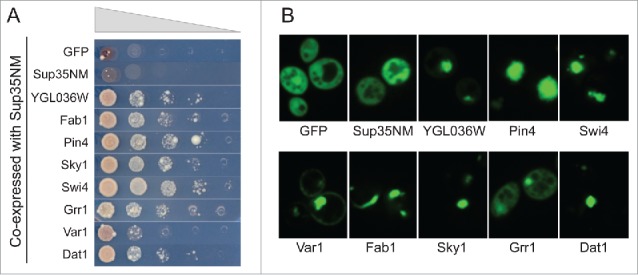
Q/N-rich prion-like domains have the ability to act like [PIN+]. Yeast strain yER1019 (rnq1Δ) expressing Sup35NM from the GAL1 promoter was transformed with plasmids expressing PrLD-GFP fusions from the GAL1 promoter. A) Cells were grown for 72 h in galactose/raffinose dropout medium, and then 10-fold serial dilutions were plated onto medium lacking adenine to select for [PSI+] formation. B) Cells were grown for 24 h in galactose/raffinose dropout medium, and visualized by microscopy to test for [PIN+]-independent foci formation.
The ability to promote Ade+ colony formation was not limited to naturally-occurring yeast PrLDs. We previously used PAPA to design 2 synthetic Q/N-rich prion domains.24 Both formed foci when expressed as PrLD-GFP fusions (Fig. 7A), and formed SDS-insoluble aggregates (Fig. 7B). By contrast, synthetic Q/N-rich PrLDs designed to have low aggregation activity remained soluble (Fig. 7A,B). Consistent with what was observed for naturally-occurring yeast PrLDs, both of the synthetic PrLDs were able to promote [PSI+] formation, while the negative control Q/N-rich domains were not (Fig. 7C).
Figure 7.
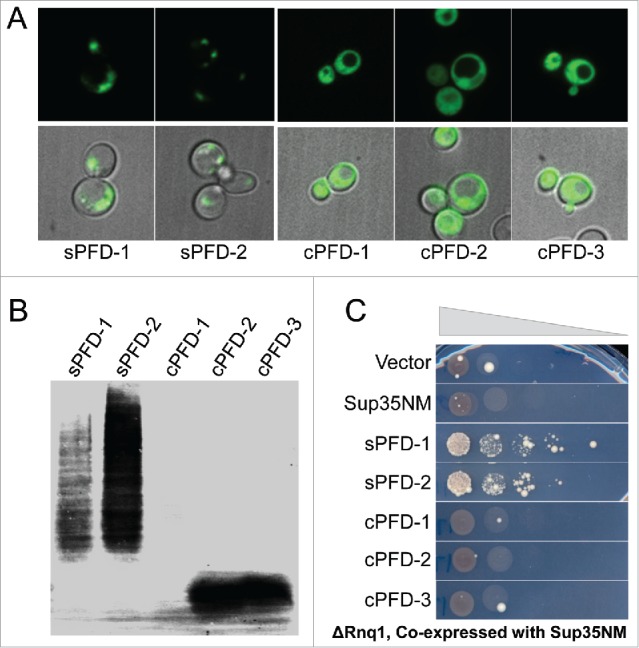
Q/N-rich synthetic PrLDs have the ability to act as [PIN+]. A) Plasmids expressing synthetic PrLDs (sPFD) and negative control Q/N-rich domains (cPFD) fused to GFP were transformed into the [PIN+] strain yER632. Cells were grown 24 h in galactose/raffinose dropout medium and visualized by microscopy. B) SDD-AGE of sPFD-HA and cPFD-HA fusions in [PIN+] strain yER632. C) sPFDs can substitute for [PIN+]. Plasmids expressing PrLD-GFP and Sup35NM were transformed into the rnq1Δ strain yER1019. Cells were grown for 72 h in galactose/raffinose dropout medium and plated onto medium lacking adenine to select for [PSI+] formation.
DISCUSSION
There appears to be distinct classes of amyloid-forming proteins. For many amyloid-forming proteins, amyloid formation is thought to be driven by short, generally hydrophobic segments.31,50 This class of amyloid proteins includes amyloid β and the human prion protein PrP. By contrast, other amyloid-forming proteins, including many yeast prion proteins, lack these short, highly amyloidogenic segments.51 Instead, amyloid propensity is more diffusely spread across a long, intrinsically disordered low-complexity domain. For example, the entire prion domain of both Ure2 and Sup35 can be scrambled without disrupting prion formation,5,6 and no single segment of these scrambled prion domains is required for prion formation.6,23 This second class of amyloid-forming proteins includes proteins that are mutated in ALS and FTLD.11,52 Here, we are specifically examining the sequence features that promote amyloid formation by this second class of proteins.
While it is clear that PrLDs are important both in normal biology and in pathology, the exact definition of what constitutes a PrLD has never been rigorously defined. Almost all yeast prion proteins contain a Q/N-rich prion domain. Various studies suggest that glutamine and asparagine residues have relatively low aggregation propensities, so the importance of Q/N content is unclear.31,53 Yeast prion domains are also intrinsically disordered, so we previously hypothesized that Q/N content is common simply because Q and N balance aggregation propensity and intrinsic disorder. However, other polar residues (serine, glycine, and threonine) have similar characteristics,23 making it unclear why aggregation-prone PrLDs tend to be specifically enriched in Q and N.
Our current results suggest an answer. Q/N content does not appear to be important for PrLD aggregation, as there was little correlation between Q/N content and foci formation among the tested PrLDs (Fig. 2). However high Q/N content, particularly N content, was highly correlated with formation of SDS-insoluble aggregates (Table 1). This result suggests a challenge in predicting PrLD aggregation propensity: the effect of a given mutation will be dependent on context. For example, we previously showed that small changes in Q/N content have little effect on the aggregation propensity of highly Q/N-rich yeast prion proteins.23 However, our current results showed a threshold effect at about 20% Q/N content, where above this level PrLDs were much more likely to form SDS-resistant aggregates. This suggests that near this threshold, small changes in Q/N content could significantly affect formation of the SDS-insoluble aggregates that characterize both the yeast prions and the disease-associated PrLD aggregates. This finding is important, because many of the disease-associated human PrLDs have Q/N content near this threshold.11
Our results also suggest an additional challenge for PrLD prediction: that the prediction method needs to be optimized for the desired task. We have shown PAPA is sufficient if the goal is to identify aggregation-prone low complexity domains. However, for identifying proteins that form SDS-insoluble aggregates, PAPA needs to be coupled with a pre-selection for Q/N content. To identify bona fide prions, sequence features that promote chaperone-dependent cleavage need to be accounted for. These factors have not been rigorously defined, but aromatic residues seem to promote this process.37,54
Our results also provide insight into the requirements for Pin+ activity. Previous studies have shown that various Q/N-rich domains can substitute for [PIN+] in promoting [PSI+] formation, but it was unclear whether this was a universal feature of aggregation-prone Q/N-rich domains. We found that almost every PrLD that formed [PIN+]-independent SDS-insoluble aggregates was able to substitute for [PIN+] in supporting [PSI+] formation. These results highlight the lack of sequence specificity of this activity, and suggest that a diverse array of proteins may influence [PSI+] formation. However, it should be noted that the mechanistic basis for this effect remains unclear. It is not known why Sup35, which efficiently forms amyloid in vitro, requires [PIN+] for prion formation in vivo. Additionally, although a variety of evidence suggests that [PIN+] promotes [PSI+] formation at least in part through direct cross-seeding,1 it remains possible that proteins with Pin+ activity may support [PSI+] formation by an indirect mechanism, such as titration of an inhibitor of [PSI+] formation.1,46
Although our current study addresses the sequence features that support PrLD aggregation and formation of SDS-insoluble aggregates, and provides insights into the promiscuity of Q/N-rich proteins, whether the identified PrLDs can support formation of bona fide prions is still unclear. We performed preliminary tests using the well characterized Sup35 fusion assay,38 but it proved inconclusive (data not shown). In this assay, potential prion domains are inserted in the place of the Sup35 prion domain; cells expressing the fusion protein are then tested for prion formation using the ade2–1 reporter described above. Unfortunately, most of our PrLD-Sup35 fusions had a constitutive Ade+ phenotype (data not shown). This suggests that these fusion proteins were non-functional, either because the PrLDs interfered with Sup35 activity, or because the fusion proteins aggregated so rapidly that they lacked a stable soluble state. Because of the substantial limitations of this assay, we opted not to pursue it further. Specifically, the Sup35 fusion assay is prone to both false positives and false negatives. The majority of the PrLDs that show prion activity in this assay have not been demonstrated to support prion activity in their native context, and the prion domains from 2 known yeast prion proteins, Cyc8 and Mot3, are unable to form prions in this assay.10,35,55 Because of these limitations, we are instead focusing on developing assays to test interesting PrLDs in their native context.
It is also worth noting that many PrLDs form biologically-relevant aggregates that are not prions. In mammalian cells, a segment of a melanosome protein, Pmel17 forms amyloid fibers in order for proper melanin production.56 In addition, yeast also have several examples of amyloid formation serving a beneficial role to the cell. Among these include a RNA-binding protein, Rim4, which forms amyloid-like aggregates that regulate gametogenesis.57,58 Likewise, the amyloid-like aggregation of Whi3, an RNA-binding protein, allows the cell to permanently escape pheromone-induced cell cycle arrest.59 Furthermore, as previously discussed, many PrLDs are thought to support formation of non-amyloid aggregates, such as the dynamic liquid-liquid phase separation that is seen with stress granules. Overall, cells exploit PrLDs because they have a propensity to form various types of protein aggregates, providing an increase in concentration of the prion-like protein, as well as its binding partners, at sites of interest. These interactions can be regulated based on environmental cues like stress, changing salt or ion concentration, etc. Therefore, our results revealed several promising candidates for PrLDs that may form functional amyloid or non-amyloid aggregates.
MATERIALS AND METHODS
Predicting Prion-Like Domains in S. Cerevisiae
The S. cerevisiae proteome was scanned using PAPA.23 PAPA effectively uses an 81-aa window, but weights each amino acid inverse proportion to its distance from the center of the window. The predicted prion propensities of all 81-aa windows across the proteome were calculated. PrLDs were excluded if they had been previously tested for prion-like activity.10 Each PrLD contains the core 81 amino acids predicted by PAPA; the PrLDs were extended to include all flanking amino acids that were scored positive (> 0) by PAPA. If 2 high scoring PrLD were separated by a small region (less than the length of an average prion domain) that scored below 0, the entire region was included. Specifically, the PrLD of Pin4 contains 3 high scoring segments, separated by 66 and 51 amino acid low scoring segments, so the entire region was included. PrLDs of Var1 and Swi4 contains 2 high scoring segments separated by 10 and 31 amino acid low scoring segments, respectively, so the entire segment was included. The average length of the PAPA positive PrLD was 165 amino acids. Therefore, PAPA negative domains were chosen by identifying 165 amino acid Q/N-rich segments with low PAPA scores.
Plasmid Construction
To generate the PrLD-GFP fusions, the PrLDs were amplified from strain yER632,26 adding BamHI and XhoI restriction sites, as well as a start codon at the beginning of the PrLD and a flexible linker at the end of the PrLD (see Tables S1 and S2 for primer sequences and plasmid names, respectively). PCR products were digested with BamHI and XhoI and ligated into pER760, a TRP1 2 µm plasmid containing GFP under control of the GAL1 promoter.26 To generate PrLD-HA fusions for SDD-AGE, PrLDs were amplified from the respective GFP plasmids, using the same sense primer as for construction of the GFP plasmids, paired with a common antisense primer that added a C-terminal HA tag, a stop codon, and a SalI restriction site for cloning into pER687 (a TRP1 2 µm plasmid containing the GAL1 promoter and ADH1 terminator).26 All sequences were confirmed by DNA sequencing. Three PrLD sequences (from YML053C, Cdc39, and Fab1) contained minor polymorphisms that altered the amino acid sequence from the reference strains in the Saccharomyces Genome Database; see Table S3 for protein sequences.
Mitochondria use different codons; therefore, the mitochondrial PrLDs (from Var1, Q0255, AI3) were built synthetically using overlapping primers, followed by primers to add the restriction sites for cloning. We omitted single cysteine residues that were present in each mitochondria PrLD to prevent disulfide bond formation; see Table S3 for protein sequences.
Yeast Strains and Media
Standard yeast media and methods were used as described previously.60 In all experiments, yeast were grown at 30°C. See Table 2 for a list of strains used in this study.
TABLE 2.
Strain list.
| Strain | Genotype |
|---|---|
| yER632 | MATα kar1–1 SUQ5 ade2–1 his3 leu2 trp1 ura3 sup35::KanMx [psi−] [PIN+] pJ533(URA3)a |
| yER1017 | MATα kar1–1 SUQ5 ade2–1 his3 leu2 trp1 ura3 sup35::KanMx [psi−] rnq1::HIS3 pJ533(URA3) |
| yER1018 | MATα kar1–1 SUQ5 ade2–1 his3 leu2 trp1 ura3 sup35::KanMx hsp104::HIS3 pJ533(URA3) |
| yER1019 | MATα kar1 SUQ5 ade2–1 his3 leu2 trp1 ura3 rnq1::KanMx [psi−] |
| yER1615 | MATα kar1 SUQ5 ade2–1 his3 leu2 trp1 ura3 hsp104::KanMX |
pJ533 is a cen plasmid expressing Sup35 from the SUP35 promoter.26
Foci Formation
Foci formation assays were performed as described previously.27 Briefly, yeast strains yER632, yER1019, and yER1615 were transformed with TRP1 plasmids expressing each PrLD-GFP from the GAL1 promoter. Strains were grown in galactose/raffinose dropout medium lacking tryptophan for 24 hours, and then imaged by confocal microscopy.
To examine the ability of PrLDs to promote Sup35N foci formation in an rnq1Δ strain, yER1019 was transformed with both a TRP1 plasmid expressing PrLD-GFP fusion from the GAL1 promoter and a LEU2 plasmid expressing Sup35N from the GAL1 promoter. Cells were grown in galactose/raffinose dropout medium lacking leucine and tryptophan for 24 hours.
Semi-Denaturing Detergent-Agarose Gel Electrophoresis (SDD-AGE)
For SDD-AGE, yER632, yER1017 and yER1018 were transformed with TRP1 plasmids expressing each PrLD-HA from the GAL1 promoter. Strains were grown for 24 or 48 hours in galactose/raffinose dropout medium lacking tryptophan. Cells were harvested and lysed as previously reported.61 80 ug of total protein lysate was incubated in 2% SDS loading buffer for 7 minutes at room temperature before loading onto a 1.5% agarose gel containing 0.1% SDS and 1 × TAE. The gel was run in running buffer (1 × TAE, 0.1% SDS) at 60 V for 3 hours. Protein was transferred to a PVDF membrane by capillary transfer for 24 hours using 1 × PBS at room temperature. The membrane was probed with an anti-HA primary antibody (HA.11 16B12, Covance), and Alexa Fluor IR800 goat anti-mouse secondary antibody (Rockland).
[PIN+] Assays
To examine the ability of PrLDs to substitute for [PIN+] in promoting [PSI+] formation, yER1019 was transformed with both a TRP1 plasmid expressing PrLD-GFP fusion from the GAL1 promoter and a LEU2 plasmid expressing Sup35N from the GAL1 promoter. Cells were grown in galactose/raffinose dropout medium lacking leucine and tryptophan for 72 hours, and 10-fold serial dilutions were plated on medium lacking adenine.
Supplementary Material
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
FUNDING
This work was supported by the National Science Foundation under Grant MCB-1517231.
REFERENCES
- [1].Liebman SW, Chernoff YO. Prions in yeast. Genetics 2012; 191:1041-72; PMID:22879407; https://doi.org/ 10.1534/genetics.111.137760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Edskes HK, Gray VT, Wickner RB. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc Natl Acad Sci U S A 1999; 96:1498-503; PMID:9990052; https://doi.org/ 10.1073/pnas.96.4.1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sondheimer N, Lindquist S. Rnq1: An epigenetic modifier of protein function in yeast. Mol Cell 2000; 5:163-72; PMID:10678178; https://doi.org/ 10.1016/S1097-2765(00)80412-8 [DOI] [PubMed] [Google Scholar]
- [4].Du Z. The complexity and implications of yeast prion domains. Prion 2011; 5:311-6; PMID:22156731; https://doi.org/ 10.4161/pri.18304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ross ED, Baxa U, Wickner RB. Scrambled prion domains form prions and amyloid. Mol Cell Biol 2004; 24:7206-13; PMID:15282319; https://doi.org/ 10.1128/MCB.24.16.7206-7213.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci U S A 2005; 102:12825-30; PMID:16123127; https://doi.org/ 10.1073/pnas.0506136102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Afsar Minhas FUA, Ross ED, Ben-Hur A. Amino acid composition predicts prion activity. PLOS Comput Biol 2017; 13:e1005465; PMID:28394888; https://doi.org/ 10.1371/journal.pcbi.1005465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: Implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci U S A 2000; 97:11910-5; PMID:11050225; https://doi.org/ 10.1073/pnas.97.22.11910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Harrison PM, Gerstein M. A method to assess compositional bias in biological sequences and its application to prion-like glutamine/asparagine-rich domains in eukaryotic proteomes. Genome Biol 2003; 4:R40; PMID:12801414; https://doi.org/ 10.1186/gb-2003-4-6-r40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 2009; 137:146-58; PMID:19345193; https://doi.org/ 10.1016/j.cell.2009.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cascarina SM, Ross ED. Yeast prions and human prion-like proteins: Sequence features and prediction methods. Cell Mol Life Sci 2014; 71:2047-63; PMID:24390581; https://doi.org/ 10.1007/s00018-013-1543-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].March ZM, King OD, Shorter J. Prion-like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease. Brain Res 2016; 1647:9-18; PMID:26996412; https://doi.org/ 10.1016/j.brainres.2016.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].King OD, Gitler AD, Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res 2012; 1462:61-80; PMID:22445064; https://doi.org/ 10.1016/j.brainres.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol 2013; 201:361-72; PMID:23629963; https://doi.org/ 10.1083/jcb.201302044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 2015; 163:123-33; PMID:26406374; https://doi.org/ 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Murakami T, Qamar S, Lin JQ, Schierle GSK, Rees E, Miyashita A, Costa AR, Dodd RB, Chan FTS, Michel CH, et al.. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron 2015; 88:678-90; PMID:26526393; https://doi.org/ 10.1016/j.neuron.2015.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lin Y, Protter DSW, Rosen MK, Parker R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell 2015; 60:208-19; PMID:26412307; https://doi.org/ 10.1016/j.molcel.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al.. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 2015; 162:1066-77; PMID:26317470; https://doi.org/ 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- [19].Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell 2007; 25:635-46; PMID:17349952; https://doi.org/ 10.1016/j.molcel.2007.02.011 [DOI] [PubMed] [Google Scholar]
- [20].Anderson P, Kedersha N. RNA granules: Post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 2009; 10:430-6; PMID:19461665; https://doi.org/ 10.1038/nrm2694 [DOI] [PubMed] [Google Scholar]
- [21].Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al.. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012; 149:753-67; PMID:22579281; https://doi.org/ 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kroschwald S, Maharana S, Mateju D, Malinovska L, Nüske E, Poser I, Richter D, Alberti S. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife 2015; 4:e06807; PMID:26238190; https://doi.org/ 10.7554/eLife.06807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Toombs JA, McCarty BR, Ross ED. Compositional determinants of prion formation in yeast. Mol Cell Biol 2010; 30:319-32; PMID:19884345; https://doi.org/ 10.1128/MCB.01140-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Toombs JA, Petri M, Paul KR, Kan GY, Ben-Hur A, Ross ED. De novo design of synthetic prion domains. Proc Natl Acad Sci U S A 2012; 109:6519-24; PMID:22474356; https://doi.org/ 10.1073/pnas.1119366109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ross ED, MacLea KS, Anderson C, Ben-Hur A. A bioinformatics method for identifying Q/N-rich prion-like domains in proteins. Methods Mol Biol 2013; 1017:219-28; PMID:23719919; https://doi.org/ 10.1007/978-1-62703-438-8_16 [DOI] [PubMed] [Google Scholar]
- [26].Gonzalez Nelson AC, Paul KR, Petri M, Flores N, Rogge RA, Cascarina SM, Ross ED. Increasing prion propensity by hydrophobic insertion. PLoS One 2014; 9:e89286; PMID:24586661; https://doi.org/ 10.1371/journal.pone.0089286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Paul KR, Hendrich CG, Waechter A, Harman MR, Ross ED. Generating new prions by targeted mutation or segment duplication. Proc Natl Acad Sci U S A 2015; 112:8584-9; PMID:26100899; https://doi.org/ 10.1073/pnas.1501072112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Paul KR, Molliex A, Cascarina S, Boncella AE, Taylor JP, Ross ED. The effects of mutations on the aggregation propensity of the human prion-like protein hnRNPA2B1. Mol Cell Biol 2017; 37:e00652-16; PMID:28137911; https://doi.org/ 10.1128/MCB.00652-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kim HJ, Kim NC, Wang Y-D, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al.. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 2013; 495:467-73; PMID:23455423; https://doi.org/ 10.1038/nature11922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Scheibel T, Lindquist SL. The role of conformational flexibility in prion propagation and maintenance for Sup35p. Nat Struct Biol 2001; 8:958-62; PMID:11685242; https://doi.org/ 10.1038/nsb1101-958 [DOI] [PubMed] [Google Scholar]
- [31].Pawar AP, DuBay KF, Zurdo J, Chiti F, Vendruscolo M, Dobson CM. Prediction of “Aggregation-prone” and “Aggregation-susceptible” regions in proteins associated with neurodegenerative diseases. J Mol Biol 2005; 350:379-92; PMID:15925383; https://doi.org/ 10.1016/j.jmb.2005.04.016 [DOI] [PubMed] [Google Scholar]
- [32].Halfmann R, Wright JR, Alberti S, Lindquist S, Rexach M. Prion formation by a yeast GLFG nucleoporin. Prion 2012; 6:391-9; PMID:22561191; https://doi.org/ 10.4161/pri.20199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: The story of [PIN(+)]. Cell 2001; 106:171-82; PMID:11511345; https://doi.org/ 10.1016/S0092-8674(01)00427-5 [DOI] [PubMed] [Google Scholar]
- [34].Du Z, Park K-W, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet 2008; 40:460-5; PMID:18362884; https://doi.org/ 10.1038/ng.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol 2009; 11:344-9; PMID:19219034; https://doi.org/ 10.1038/ncb1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Perutz MF, Pope BJ, Owen D, Wanker EE, Scherzinger E. Aggregation of proteins with expanded glutamine and alanine repeats of the glutamine-rich and asparagine-rich domains of Sup35 and of the amyloid beta-peptide of amyloid plaques. Proc Natl Acad Sci U S A 2002; 99:5596-600; PMID:11960015; https://doi.org/ 10.1073/pnas.042681599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].MacLea KS, Paul KR, Ben-Musa Z, Waechter A, Shattuck JE, Gruca M, Ross ED. Distinct amino acid compositional requirements for formation and maintenance of the [PSI+] prion in yeast. Mol Cell Biol 2015; 35:899-911; PMID:25547291; https://doi.org/ 10.1128/MCB.01020-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Li L, Lindquist S. Creating a protein-based element of inheritance. Science 2000; 287:661-4; PMID:10650001; https://doi.org/ 10.1126/science.287.5453.661 [DOI] [PubMed] [Google Scholar]
- [39].Halfmann R, Alberti S, Krishnan R, Lyle N, O'Donnell CW, King OD, Berger B, Pappu RV, Lindquist S. Opposing effects of glutamine and asparagine govern prion formation by intrinsically disordered proteins. Mol Cell 2011; 43:72-84; PMID:21726811; https://doi.org/ 10.1016/j.molcel.2011.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bagriantsev SN, Kushnirov VV, Liebman SW. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol 2006; 412:33-48; PMID:17046650; https://doi.org/ 10.1016/S0076-6879(06)12003-0 [DOI] [PubMed] [Google Scholar]
- [41].Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 1997; 147:507-19; PMID:9335589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci U S A 2004; 101:12934-9; PMID:15326312; https://doi.org/ 10.1073/pnas.0404968101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion "strains" in yeast. Proc Natl Acad Sci U S A 2002; 99(Suppl 4):16392-9; PMID:12149514; https://doi.org/ 10.1073/pnas.152330699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 1995; 268:880-4; PMID:7754373; https://doi.org/ 10.1126/science.7754373 [DOI] [PubMed] [Google Scholar]
- [45].Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol 2001; 21:4656-69; PMID:11416143; https://doi.org/ 10.1128/MCB.21.14.4656-4669.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Arslan F, Hong JY, Kanneganti V, Park S-K, Liebman SW. Heterologous aggregates promote de novo prion appearance via more than one mechanism. PLoS Genet 2015; 11:e1004814; PMID:25568955; https://doi.org/ 10.1371/journal.pgen.1004814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature 2003; 425:686-91; PMID:14562095; https://doi.org/ 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- [48].Cox BS. Ψ, A cytoplasmic suppressor of super-suppressor in yeast. Heredity (Edinb) 1965; 20:505-21; https://doi.org/ 10.1038/hdy.1965.65 [DOI] [Google Scholar]
- [49].Tuite MF, Marchante R, Kushnirov V. Fungal prions: structure, function and propagation. Top Curr Chem 2011; 305:257-98; PMID:21717344; https://doi.org/ 10.1007/128_2011_172 [DOI] [PubMed] [Google Scholar]
- [50].Esteras-Chopo A, Serrano L, de la Paz ML. The amyloid stretch hypothesis: Recruiting proteins toward the dark side. Proc Natl Acad Sci 2005; 102:16672-7; PMID:16263932; https://doi.org/ 10.1073/pnas.0505905102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ross ED, Toombs JA. The effects of amino acid composition on yeast prion formation and prion domain interactions. Prion 4:60-5; PMID:20495349; https://doi.org/ 10.4161/pri.4.2.12190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell 2013; 154:727-36; PMID:23953108; https://doi.org/ 10.1016/j.cell.2013.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature 2003; 424:805-8; PMID:12917692; https://doi.org/ 10.1038/nature01891 [DOI] [PubMed] [Google Scholar]
- [54].Alexandrov AI, Polyanskaya AB, Serpionov GV, Ter-Avanesyan MD, Kushnirov VV. The effects of amino acid composition of glutamine-rich domains on amyloid formation and fragmentation. PLoS One 2012; 7:e46458; PMID:23071575; https://doi.org/ 10.1371/journal.pone.0046458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Holmes DL, Lancaster AK, Lindquist S, Halfmann R. Heritable remodeling of yeast multicellularity by an environmentally responsive prion. Cell 2013; 153:153-65; PMID:23540696; https://doi.org/ 10.1016/j.cell.2013.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Berson JF, Theos AC, Harper DC, Tenza D, Raposo G, Marks MS. Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis. J Cell Biol 2003; 161:521-33; PMID:12732614; https://doi.org/ 10.1083/jcb.200302072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Berchowitz LE, Kabachinski G, Walker MR, Carlile TM, Gilbert W V, Schwartz TU, Amon A. Regulated formation of an amyloid-like translational repressor governs gametogenesis. Cell 2015; 163:406-18; PMID:26411291; https://doi.org/ 10.1016/j.cell.2015.08.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ford AF, Shorter J. Fleeting amyloid-like forms of Rim4 ensure meiotic fidelity. Cell 2015; 163:275-6; PMID:26451477; https://doi.org/ 10.1016/j.cell.2015.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Caudron F, Barral Y. A super-assembly of Whi3 encodes memory of deceptive encounters by single cells during yeast courtship. Cell 2013; 155:1244-57; PMID:24315096; https://doi.org/ 10.1016/j.cell.2013.10.046 [DOI] [PubMed] [Google Scholar]
- [60].Sherman F. Getting started with yeast. Methods Enzymol 1991; 194:3-21; PMID:2005794 [DOI] [PubMed] [Google Scholar]
- [61].Halfmann R, Lindquist S. Screening for amyloid aggregation by semi-denaturing detergent-agarose gel electrophoresis. J Vis Exp 2008; (17):838; PMID:19066511; https://doi.org/ 10.3791/838 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.