Abstract
Alternative polyadenylation (APA) is a process that changes the posttranscriptional regulation and translation potential of mRNAs via addition or deletion of 3′ untranslated region (3′ UTR) sequences. To identify posttranscriptional-regulatory events affected by APA in breast tumors, tumor datasets were analyzed for recurrent APA events. Motif mapping of the changed 3′ UTR regions found that APA-mediated removal of Pumilio regulatory elements (PRE) was unusually common. Breast tumor subtype–specific APA profiling identified triple-negative breast tumors as having the highest levels of APA. To determine the frequency of these events, an independent cohort of triple-negative breast tumors and normal breast tissue was analyzed for APA. APA-mediated shortening of NRAS and c-JUN was seen frequently, and this correlated with changes in the expression of downstream targets. mRNA stability and luciferase assays demonstrated APA-dependent alterations in RNA and protein levels of affected candidate genes. Examination of clinical parameters of these tumors found those with APA of NRAS and c-JUN to be smaller and less proliferative, but more invasive than non-APA tumors. RT-PCR profiling identified elevated levels of polyadenylation factor CSTF3 in tumors with APA. Overexpression of CSTF3 was common in triple-negative breast cancer cell lines, and elevated CSTF3 levels were sufficient to induce APA of NRAS and c-JUN. Our results support the hypothesis that PRE-containing mRNAs are disproportionately affected by APA, primarily due to high sequence similarity in the motifs utilized by polyadenylation machinery and the PUM complex.
Introduction
Cancer genome sequencing has identified driver mutations associated with disease. In addition, nongenetic changes such as epigenetic reprogramming and aberrant posttranscriptional regulation also impact tumor phenotypes. Posttranscriptional controls that affect mRNA processing and stability include miRNAs and RNA-binding proteins (RBP; ref. 1). These factors bind to seed sequences within the 3′ untranslated region (UTR) of transcripts (2) and are commonly misregulated in cancers (3). Tumor-specific fluctuations in the levels of RBPs and miRNAs may be particularly important, as transcripts from oncogenes and tumor suppressor genes contain a disproportionately high number of posttranscriptional regulatory motifs.
In line with these ideas, we recently discovered that the expression of the human oncogene E2F3 is posttranscriptionally controlled by the Pumilio complex (4). The Pumilio complex is comprised of two RBPs in humans, Pumilio (PUM) and NANOS (NOS), which bind to a conserved motif of UGUAXAUA (5–7) and a GU-rich element within the 3′ UTR of their substrates (2, 8). The Pumilio repressor has evolutionarily conserved roles in the regulation of proliferation and differentiation (6–9). PUM complexes function to inhibit translation (10) or induce the degradation of their substrates (4, 11–14). The PUM repressor regulates the translation potential of hundreds of transcripts including a large number of oncogenes and tumor suppressor genes (5).
While analyzing the role of PUM in modulating E2F3 levels in cancer cell lines, we found the 3′ UTR of E2F3 transcripts were often shortened. This change in mRNA length removed a number of regulatory elements, including the Pumilio regulatory elements (PRE) motifs (4). The shortened E2F3 mRNA in these cells was polyadenylated, indicating that they were generated by alternative polyadenylation (APA). APA is a poorly understood developmental process that leads to the use of noncanonical polyadenylation sites (15–17). APA affects the inclusion/exclusion of regulatory elements and can alter mRNA stability and translation potential (18). During normal development, APA is utilized to regulate differentiation (15, 16, 19–21). In cultured human cells, APA has been linked with genes responsible for cell proliferation (22, 23).
APA is also a likely contributor to oncogenesis (24), as APA has been correlated with poor patient prognosis in a variety of malignancies (25–27). Despite this, little is known about APA in tumor development. Our finding that the E2F3 mRNA is altered by APA in cancer cell lines, prompted us to investigate this phenomenon further. To do this, we looked for mRNAs that are altered by APA in human breast tumor samples compared with normal tissue. This analysis found that APA is widespread in breast tumors and that these APA events disproportionately remove putative PUM-regulatory elements.
Breast tumor subtype–specific APA profiles were then generated and unexpectedly, this revealed that triple-negative breast tumors have a distinct APA profile from other subtypes. Four candidate APA events were then selected and their frequency assayed in an independent cohort of triple-negative breast tumors. These results led us to focus on two recurrent APA-altered mRNAs, c-JUN and NRAS, for detailed analysis. These genes were selected as they displayed high levels of APA in tumors. Here, we show that these mRNAs are targets of PUM-mediated repression and demonstrate that APA circumvents this regulation. Strikingly, both c-JUN and NRAS were known to be deregulated in triple-negative breast tumors (28, 29), but the underlying mechanism was unclear. In addition, APA of NRAS and c-JUN altered the expression of downstream target genes. Intriguingly, unlike previous reports, APA in this tumor cohort correlated with smaller, slower growing but more invasive node-positive tumors. Collectively, these findings indicated that APA of PUM substrates is a common feature of triple-negative breast tumors. These results show that recurrent APA of PRE-containing mRNAs allows transcripts to circumvent PUM-mediated repression and provide a mechanistic explanation for the increased activity of NRAS and c-JUN in tumors.
Materials and Methods
Bioinformatics
We used the data from ref. 30 [Gene Expression Omnibus (31) (GSE3744)] to compare 38 tumor samples with 7 normal breast samples. Analysis of differential APA between tumors and normal tissues was performed as described previously (26). Briefly, the method exploits the 3′ bias and probe-set organization of 3′ IVT Affymetrix arrays to evaluate the relative expression of mRNA isoforms using alternative polyadenylation sites. APA sites were obtained from the Poly-A DB database.
RT-PCR
RNA was purified using the RNeasy Extraction Kit (Qiagen). Reverse transcription was performed using TaqMan Reverse Transcriptase and SYBR Green detection chemistry.
Cell culture, transfections, and luciferase expression constructs
Cell lines were provided and authenticated by The Center of Molecular Therapeutics, Massachusetts General Hospital (Charlestown, MA; July 2014). c-JUN plasmids were kind gifts from Dr. Lily Vardimon (Tel Aviv University, Israel; ref. 32). NRAS luciferase plasmids (33) were obtained from Addgene. Human cell lines were transfected for 48 hours with X-tremeGENE transfection reagent or Lipofectamine 2000. CSTF3 overexpression plasmid was a kind gift from B. Tian (34).
Antibodies
The following antibodies were used: Tubulin (DSH Bank/E7), PUM1 (Bethyl Laboratories/A300-201A), PUM2 (Bethyl Laboratories/A300-202A), PABPN1 (Abcam/AB75855), FOXO1 (Cell Signaling Technology/2880), JUN (Cell Signaling Technology/9165), PTEN (Cell Signaling Technology/9552), and NRAS (Abcam/ab77392).
Luciferase assays
Cells were transfected and luciferase levels were measured 48 hours after transfection (data are expressed as mean ± SD, n = 3). Luciferase readings were taken using the Dual-Luciferase Reporter Assay System (Promega).
Lentiviral shRNA
The DNA preparation, transfections, and virus preparation methods have been published elsewhere (4, 35).
siRNA transfection
Cells were transfected with siRNAs targeting PABPN1 and a scrambled control (Dharmacon) using Lipofectamine RNAiMAX. Cells were analyzed using RT-PCR and Western blotting 2 days after transfection. Individual siRNAs targeting PUM1, PUM2, and CSTF3 were from Life Technologies (Sequences in Supplementary Table S2).
RNA stability assays
Cells were treated with actinomycinD (5 µmol/L) and collected every hour. Transcript levels were assayed using RT-PCR. Cells were depleted using shRNA specific to PUM1, PUM2, and scrambled controls and placed under puromycin selection for 4 days. Knockdown was validated by Western blot analysis.
3′RACE RT-PCR
Cells were lysed after selection for lentiviral infection and RNA was extracted using the Qiagen RNeasy kit. cDNA was generated using an extended oligo-dT primer (P7 oligo-dT). This cDNA was used in a standard RT-PCR reaction using a gene and poly-A site–specific primer as a reverse P7 primer (36).
Patients and tumor samples
All triple-negative breast cancer specimens were formalin-fixed paraffin-embedded (FFPE) biopsy samples derived from patients diagnosed at the Massachusetts General Hospital (Charlestown, MA). The diagnostic criteria/tumor grading were described previously (37). The study was approved by the Massachusetts General Hospital human research committee in accordance with the NIH human research study guidelines.
Quantitative RT-PCR from tumor samples
RNA was isolated from tumor samples isolated using the Recoverall Total Nucleic Acid Isolation Kit. KI-67 expression level was assessed using TaqMan Gene Expression Assay (4331182), and the expression of the two reference genes was assessed using custom primers/probes.
Primers
For primer sequences, see Supplementary Table S2.
Results
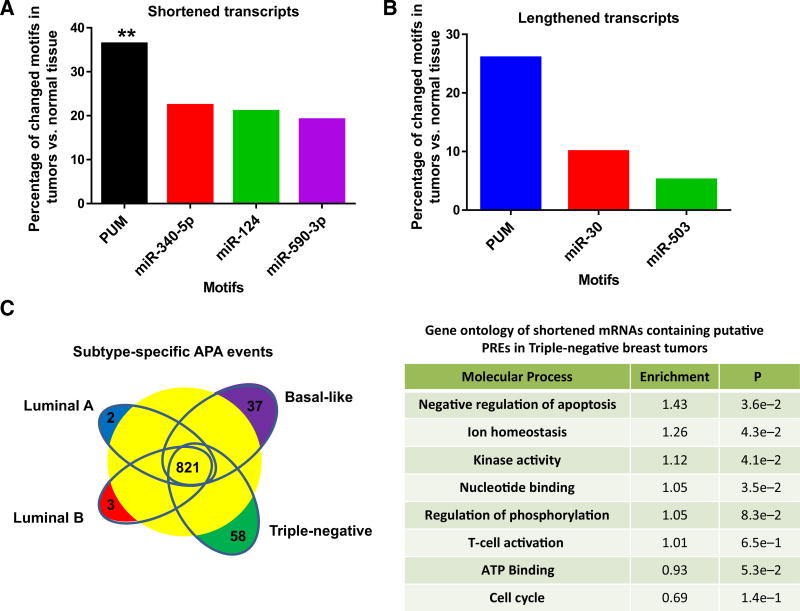
To identify posttranscriptional regulatory motifs that were disproportionately affected by APA in tumor samples, we analyzed breast tumor datasets for APA events. Breast tumors were chosen for this study as APA of E2F3 was common in breast cancer cells (4) and APA has previously been detected in this tumor type (26). Microarray data from 38 breast tumors and 7 normal breast tissue samples were examined for APA events. As described in ref. 26, microarray data obtained from the Affymetrix 3′ IVT series were used to assess APA using the 3′ bias of their probes. This revealed mRNAs that showed a significantly different prevalence of 3′ UTR isoforms in tumors compared with normal tissue. This analysis identified 921 shortened mRNAs and 189 lengthened mRNAs in human breast tumors at a false discovery rate of 5% (26). To understand how APA of these transcripts changed the capacity of the posttranscriptional regulatory machinery to target these mRNAs, an unbiased motif mapping examination of the affected 3′ UTRs was conducted. This computational search looked for posttranscriptional regulatory motif(s) that were enriched among the 3′ UTR regions that underwent shortening or lengthening in cancer. This work found that the PRE (UGUAXAUA) was the most frequently lost motif (P = 0.01) in 3′ UTR sequences shortened in cancer (Fig. 1A; Supplementary Table S1A). In addition, the PRE was also the motif most often gained through APA (Fig. 1B; Supplementary Table S1B). These results suggested that PRE-containing transcripts are frequently altered by APA in tumors including numerous genes that are known to be important for oncogenesis.
Figure 1.
PRE-containing mRNAs are disproportionately targeted by alternative polyadenylation. A, Percentage of the 921 mRNAs shortened by alternative polyadenylation in breast tumors that lose PRE (PUM) or miRNA seed sequences (**, P < 0.01). B, Percentage of the 189 mRNAs lengthened by alternative polyadenylation in breast tumors that gain PRE (PUM) or miRNA seed sequences. C, Breast tumor subtype shared and unique APA events. Table, Gene ontology of PRE-containing mRNAs shortened in triple-negative breast tumors.
Breast tumors can be subdivided into subtypes based on distinct molecular alterations (38, 39). To understand how APA impacts these subtypes, we constructed APA profiles for each breast tumor group. This analysis revealed that although the majority of APA events occur in all breast tumor subtypes (821), basal-like and triple-negative breast tumors display more extensive and exclusive patterns of APA than luminal derived tumors (Fig. 1C). Gene ontology analysis of APA-specific events in triple-negative breast tumors showed that APA affected mRNAs involved in the negative regulation of apoptosis, kinase activity, and nucleotide binding (Fig. 1C).
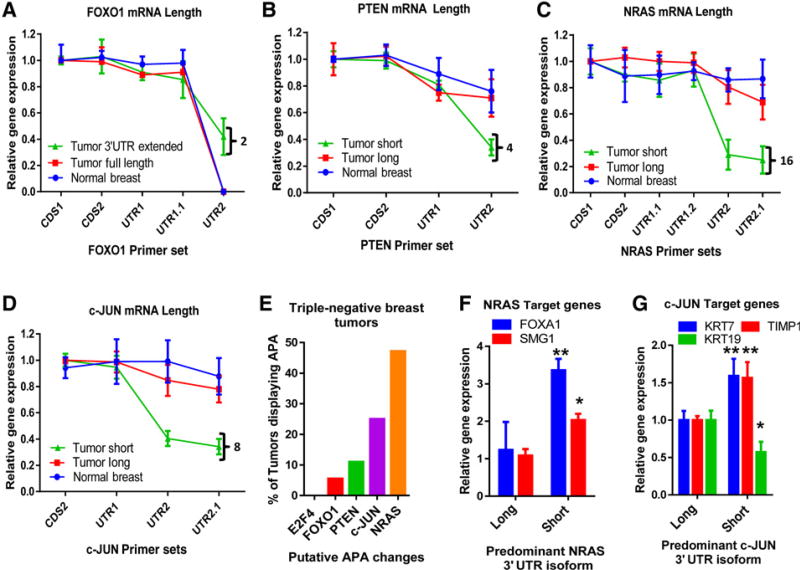
To determine the frequency of candidate 3′ UTR alterations in tumors, an independent cohort of 34 triple-negative breast tumors and 8 samples of control breast tissue were examined. Triple-negative breast tumors were selected for this analysis, as this group had the most extensive APA profiles (Fig. 1C) and exhibits the worst patient prognosis (40). RT-PCR was used to determine the relative expression levels of each 3′ UTR isoform. Five candidate genes were selected for this analysis. Forkhead Box O1 (FOXO1) is the sole candidate with an extended 3′ UTR sequence in tumor samples. PTEN, Neuroblastoma RAS viral (v-Ras) oncogene homolog (NRAS), and the Jun proto-oncogene (c-JUN), all showed recurrent 3′ UTR shortening in our database analysis. E2F4 was used as a negative control and showed no detectable level of APA. In this cohort of tumors, E2F4 was not altered by APA (Supplementary Fig. S1A – C) and 3′ UTR extensions of FOXO1 occurred only rarely (twice) (Fig. 2A and Supplementary Fig. S1D and S1E). In contrast, 3′ UTR shortening was much more prevalent. Using RT-PCR, we identified tumors containing short 3′ UTR isoforms of PTEN, NRAS, and c-JUN (Fig. 2B – D and Supplementary Fig. S1F – H and S2 and S3). In particular, shortening of the NRAS (16/34) and c-JUN 3′ UTRs (8/34) were frequent events (Fig. 2C – E).
Figure 2.
Candidate gene alternative polyadenylation status in an independent cohort of triple-negative breast tumors. A, RT-PCR of FOXO1 from tumor and control samples. Tumors were subdivided on the basis of FOXO1 length at UTR (>50% or <50%), full length and 3'UTR extended. Black bracket indicates lengthened 3′ UTRs in tumors and the number represents the number of tumors displaying this APA event. B, RT-PCR of PTEN from tumor and control samples. Black bracket, number of shortened 3′ UTRs. C, RT-PCR of NRAS from tumor and control samples. Black bracket, number of shortened 3′ UTRs. Tumors were subdivided on the basis of 3' UTR length (>50% or <50%), short and long. D, RT-PCR of the c-JUN from tumors and control samples. Black bracket, number of shortened 3′ UTRs. E, Percentage of tumors displaying APA of each transcript. E2F4 was used as a negative control. F, NRAS target gene expression (FOXA1 and SMG1) in tumors expressing either the long or short 3′ UTR isoform of NRAS (*, P < 0.05; **, P < 0.01). G, c-JUN target gene expression (KRT7 and TIMP1, activated; KRT19, repressed) in tumors expressing either the long or short 3′ UTR isoform of c-JUN (*, P < 0.05; **, P < 0.01).
Because of the frequency of these APA events, we focused on understanding how APA changed NRAS and c-JUN mRNA regulation. We asked whether changes in 3′ UTR length of NRAS and c-JUN correlated with changes in downstream target gene expression. Tumors expressing the shortened isoform of the NRAS 3′ UTR (<50%) displayed elevated levels of Forkhead Box A1 (FOXA1) and SMG1 phosphatidylinositol 3-kinase-related Kinase (SMG1) genes, previously shown to respond to NRAS signaling (Fig. 2F; ref. 41). Tumors expressing the shortened form of c-JUN had higher expression of the c-JUN–activated genes, cytokeratin 7 (KRT7) and TIMP Metallopeptidase Inhibitor 1 (TIMP1; ref. 42), and lower levels of the c-JUN–repressed gene, Cytokeratin 19 (KRT19; Fig. 2G; ref. 42). These results suggest that APA affects the levels and activity of its targets and leads to changes in the patterns of downstream transcriptional programs.
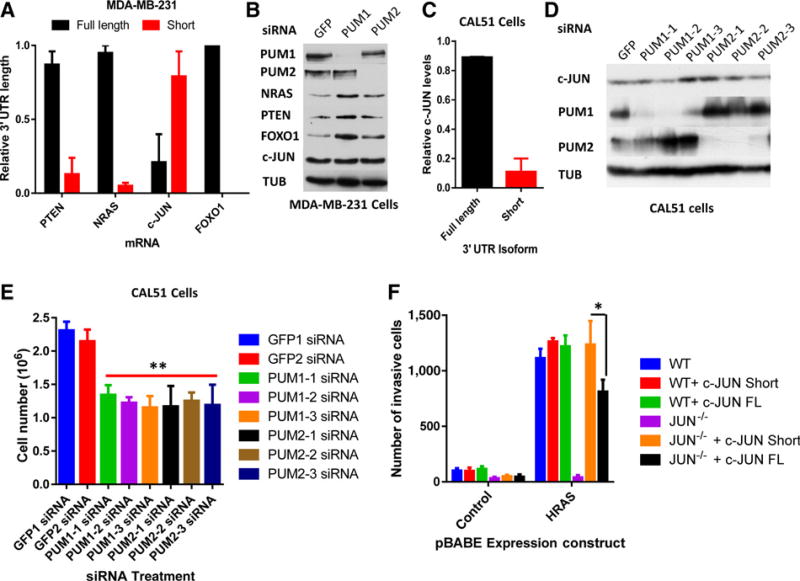
To examine the possible role for PUM in regulating our candidate genes (FOXO1, PTEN, NRAS, and c-JUN), we assayed the consequence of depleting PUM1 or PUM2 from MDA-MB-231 triple-negative breast cancer (TNBC) cells. As a control, the length of each candidate mRNA was measured using RT-qPCR. This analysis found that the shortened 3′ UTR isoform of c-JUN was expressed at high levels in MDA-MB-231 cells (Fig. 3A). siRNA-mediated knockdown of PUM1 or PUM2 in MDA-MB-231 cells increased the protein levels of PTEN, NRAS, and FOXO1 but not c-JUN (Fig. 3B). The triple-negative breast cancer cell line with the lowest levels of c-JUN APA, CAL51 (Fig. 3C), was then used to test how depleting PUM1 or PUM2 changed c-JUN protein levels. As shown in Fig. 3D, reduced PUM levels modestly increased c-JUN protein levels.
Figure 3.
PUM regulation of candidate gene protein levels. A, RT-PCR from triple-negative breast cancer cells MDA-MB-231 for PTEN, NRAS, c-JUN, and FOXO1. B, Western blots from siRNA-treated MDA-MB-231 cells. C, 3′ RACE RT-PCR from CAL51 cells for c-JUN. D, Western blots of CAL51 cells transfected with individual siRNAs targeting GFP, PUM1, and PUM2. E, Cell number assay from CAL51 cells transfected with individual siRNAs. F, Number of HRAS-mediated invasive cells from wild-type (WT) or c-JUN-null MEFs (JUN−/−), infected with the c-JUN coding sequences and either 3′ UTR isoform (*, P < 0.05).
To determine how PUM affected cell proliferation, the effect of PUM depletion on CAL51 cells was assayed. As shown in Fig. 3E, reduced PUM1 or PUM2 levels modestly slowed CAL51 growth rates, suggesting that PUM regulation may help to maintain proliferation in TNBC cells. To assay how APA-mediated shortening of c-JUN affected the invasive potential of cells, the capacity of the c-JUN coding sequence and either the short- or full-length 3′ UTR to rescue the invasive capacity of c-JUN−/− mouse embryonic fibroblasts (MEF) was tested. The ability of HRAS to promote cellular invasion was dependent on c-JUN (compare wild-type vs. c-JUN−/− MEFs + HRAS; Fig. 3F) and provided an ideal model to evaluate APA. In this system, the overexpression of the c-JUN CDS + short 3′ UTR greatly improved the invasive capacity of cells, compared with the expression of the c-JUN CDS + full-length 3′ UTR (Fig. 3F). These results suggest that PUM and APA regulation of c-JUN may contribute to the proliferation and invasion potential of cells.
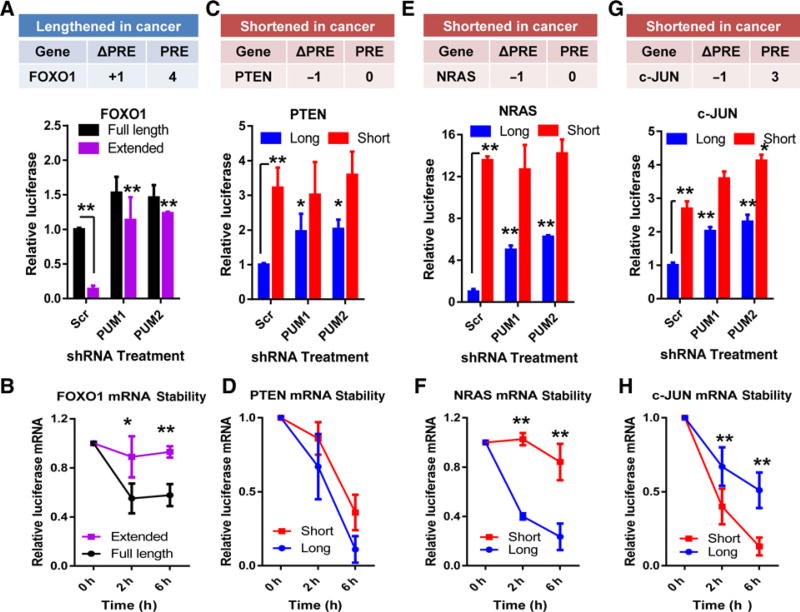
To examine how APA modified PUM-mediated control, the short and long 3′ UTR isoform of each candidate was cloned downstream of luciferase reporter genes. These plasmids were then transfected into MDA-MB-231 cells depleted of PUM1, PUM2, or scrambled control sequences (Supplementary Fig. S4A). Lengthening of the FOXO1 3′ UTR increased the number of PRE elements to 4 (Fig. 4A). Extension of the FOXO1 3′ UTR decreased luciferase levels (full-length vs. extended in Scr; Fig. 4A). This effect is partially dependent on PUM activity (Fig. 4A). To examine how APA changes mRNA degradation, mRNA stability assays were conducted. Cells were treated with the polymerase inhibitor, actinomycin D, and luciferase mRNA levels measured over time. Extension of the FOXO1 3′ UTR increased the stability of the luciferase mRNA (Fig. 4B). To determine PUM's contribution to these effects, these experiments were repeated in MDA-MB-231 cells depleted of PUM. Reduced PUM activity increased the mRNA stability of the full-length 3′ UTR but not the extended 3′ UTR isoform (Supplementary Fig. S4B). These results show that the PUM protein complex modulates FOXO1 levels and that APA-mediated lengthening increased PUM regulation.
Figure 4.
The effect of APA on mRNA stability and protein production on candidate genes. A, Table, change in PRE number (Δ in PRE) and total PRE after APA of FOXO1. Graph, luciferase assays comparing FOXO1 full-length and extended 3′ UTR isoforms in MDA-MB-231 cells depleted of PUM1, PUM2, or control Scrambled sequence (Scr; **, P < 0.01). Full-length 3′ UTR isoform set to 1. B, mRNA stability assays of the FOXO1 full-length or extended 3′ UTR isoforms from MDA-MB-231 cells treated with the polymerase inhibitor, actinomycin D (**, P < 0.01; *, P < 0.05). C, Table, transcript information for the shortened 3′ UTR of PTEN. Graph, luciferase assays comparing PTEN full-length and short 3′ UTR isoforms in MDA-MB-231 cells depleted of PUM1, PUM2, or Scrambled (**, P < 0.01). D, mRNA stability assays of the PTEN short or long 3′ UTR isoforms from MDA-MB-231 cells treated with actinomycin D (**, P < 0.01). E, Table, transcript information for the shortened 3′ UTR of NRAS. Graph, luciferase assays comparing NRAS full-length and short 3′ UTR isoforms in MDA-MB-231 cells depleted of PUM1, PUM2, or Scrambled (*, P < 0.05). F, mRNA stability assays of the NRAS short or long 3′ UTR isoforms from MDA-MB-231 cells treated with actinomycin D. G, Table, transcript information for the shortened 3′ UTR of c-JUN. Graph, luciferase assays comparing c-JUN full-length and short 3′ UTR isoforms in MDA-MB-231 cells depleted of PUM1, PUM2, or Scrambled (*, P < 0.05; **, P < 0.01). H, mRNA stability assays of the c-JUN short or long 3′ UTR isoforms from MDA-MB-231 cells treated with actinomycin D (**, P < 0.01).
This dual approach, luciferase reporter and mRNA stability assays, was used to test how shortening of the candidate genes might affect luciferase production. APA of PTEN removed the sole PRE sequence from the 3′ UTR (Fig. 4C) and increased luciferase levels (Fig. 4C).Knockdown of PUM1 or PUM2 modestly elevated luciferase production from the short 3′ UTR isoform (Fig. 4C). Surprisingly, APA of PTEN or PUM depletion did not change the overall stability of the mRNA (Fig. 4D and Supplementary Fig. S4C). These results suggested that PUM-mediated regulation of PTEN is only partially responsible for constraining PTEN levels.
Shortening of the NRAS 3′ UTR circumvents PUM repression by removing the sole PRE (Fig. 4E) and significantly increased luciferase production (Fig. 4E). Depletion of PUM1 or PUM2 only moderately elevated protein levels from the full-length 3′ UTR (Fig. 4E, Scr vs. PUM). This suggested that other regulatory mechanisms are also circumvented upon NRAS 3′ UTR shortening. Site-directed mutagenesis was used to inactivate the sole PRE within the NRAS 3′ UTR (UGUAXAUA-UCCAXAUA). This lucif-erase construct was then transfected into MDA-MB-231 andMCF7 cells depleted of PUM1 or PUM2 (Supplementary Fig. S4D). As shown in Supplementary Fig. S4E and S4F, inactivation of the PRE within the NRAS 3′ UTR only partially rescued protein levels. To examine how APA and PUM modulate the stability of both 3′ UTR isoforms of NRAS, we conducted RNA stability assays. These results showed that the half-life of the short 3′ UTR of NRAS was greatly increased, compared with full-length (Fig. 4F) and that this increase was due to reduced PUM regulation (Supplementary Fig. S5A). In support of this, depletion of PUM1 or PUM2 increased the stability of the NRAS full-length 3′ UTR (Supplementary Fig. S5A). These results indicate that PUM regulates the stability of NRAS mRNA.
Unlike the other shortened candidates, APA of c-JUN only removes 1 of the 4 PRE motifs (Fig. 4G). In the luciferase assay system, shortening the c-JUN 3′ UTR increased luciferase levels. In addition, both 3′ UTR isoforms of c-JUN were sensitive to PUM activity (Fig. 4G). RNA stability assays surprisingly showed that the shortened 3′ UTR was more unstable than the full-length 3′ UTR (Fig. 4H) and that this was in part PUM dependent (Supplementary Fig. S5B). These findings suggest that PUM tightly modulates both the stability and the translation of c-JUN mRNA.
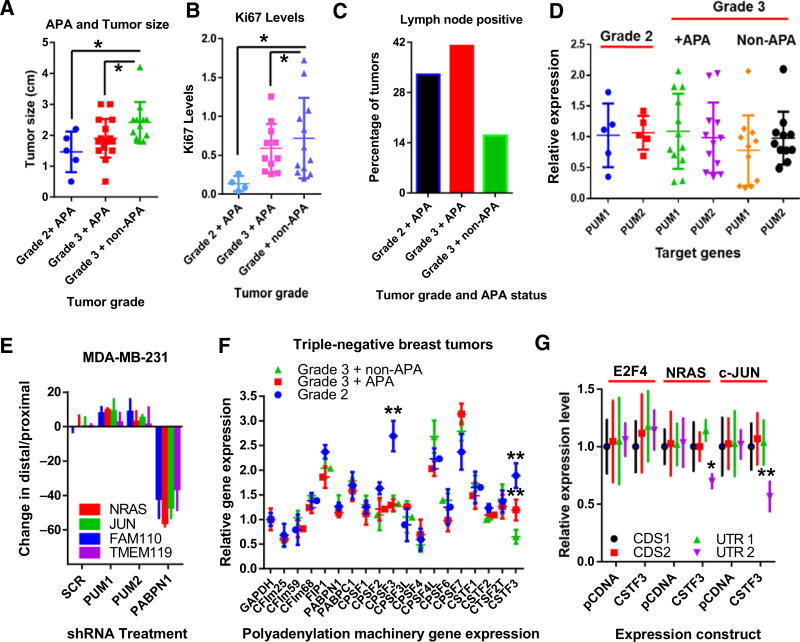
To investigate the clinical characteristics of tumors with APA in this cohort, tumors were subdivided on the basis of the APA status and clinical parameters. Tumors containing APA of c-JUN and/or NRAS were split from those expressing the full-length versions. Tumors were then divided into groups based on pathologic tumor grade. This produced 3 groups: grade 2 tumors, all of which had APA (n = 6), and grade 3 tumors, with (n = 12) and without APA (n = 12). In these tumors, every grade 2 tumor displayed APA, and so no grade 2 without APA group was formed. This analysis revealed a number of unexpected results. First, APA was more prevalent in smaller, lower grade tumors (Fig. 5A). Second, APA of NRAS and c-JUN was evident in less-proliferative tumors, as measured by Ki67 RT-PCR (Fig. 5B). Third, triple-negative breast tumors utilizing APA appear more invasive, as they were more frequently associated with positive lymph node status, compared with non-APA tumors (Fig. 5C). These results support the idea that APA of PUM substrates may contribute to tumor phenotypes and demonstrate that APA in triple-negative breast tumors does not directly correlate with proliferation.
Figure 5.
APA in triple-negative breast tumors correlates with slower growth, smaller yet more invasive tumors. A, Tumor size of triple-negative breast tumors, divided by grade and APA status (*, P < 0.05). Grade 2 tumors (n = 6), grade 3 APA tumors (n = 12), and grade 3 non-APA tumors (n = 12). B, Ki67 levels of triple-negative breast tumors, divided by grade and APA status (*, P < 0.05). C, Lymph node status (positive or negative) from patients with triple-negative breast tumors, divided by grade and APA status. D, RT-PCR analysis of PUM1 and PUM2 from triple-negative breast tumors, divided by grade and APA status. E, 3′ RACE experiments from MDA-MB-231 cells depleted of PUM1, PUM2, PABPN1, or Scrambled control sequence by shRNA. Change in 3′ UTR length calculated by dividing distal values with proximal readings. F, RT-PCR analysis of the polyadenylation machinery from triple-negative breast tumors, divided by grade and APA status (**, P < 0.01). G, RT-PCR measuring mRNA length from cells transfected with either the pCDNA-empty vector or pCDNA-CSTF3 (**, P < 0.01; *, P < 0.05).
To understand why APA disproportionately affected transcripts containing PRE motifs, we asked whether PUM proteins have a role in regulating APA. RT-PCR was used to compare PUM levels between tumors but showed no variation in expression (Fig. 5D). To directly evaluate PUM's role in regulating APA, PUM1, PUM2, or the APA regulator Poly-A Binding Protein Nuclear 1 (PABPN1) (36) were depleted from MDA-MB-231 cells and APA levels compared. Reduced PUM levels did not modify previously characterized APA substrates, compared with PABPN1 (Fig. 5E and Supplementary Fig. S6A–S6D). These findings are in agreement with previous work discounting PUM as a direct regulator of APA in cancer cell lines (36).
To investigate the molecular drivers of APA in these tumors, the expression levels of the polyadenylation machinery was analyzed using RT-PCR. These factors represent prime APA-regulating candidates, as they are directly responsible for poly-A site selection/catalysis. This analysis identified elevated levels of two components of the polyadenylation machinery in tumors with APA; cleavage stimulation factor 3 (CSTF3) and cleavage polyadenylation specificity factor 3 (CPSF3; Fig. 5F). These observations suggested that APA in triple-negative breast tumors may be regulated by fluctuations in the levels of the polyadenylation machinery.
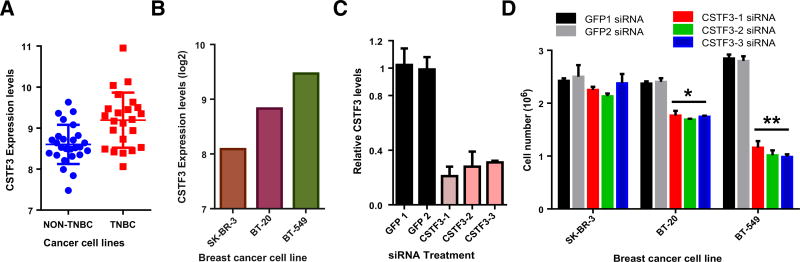
As CSTF3 levels are elevated in both tumor grades with APA, we examined its potential role in modulating APA. To do this RPE cells (which have low levels of APA), were transfected with plasmids expressing either CSTF3 (34) or an empty plasmid. In RPE cells, the overexpression of CSTF3 (Supplementary Fig. S6E) was sufficient to induce APA-mediated 3′ UTR shortening of c-JUN and NRAS but not E2F4 (Fig. 5G). To ask whether the overexpression of CSTF3 was common in triple-negative breast cancer cell lines, we analyzed mRNA levels of CSTF3. As shown in Fig. 6A, triple-negative breast cancer cell lines (TNBC) express higher levels of CSTF3 than other breast cancer cell lines (non-TNBC). To determine the contribution of CSTF3 to proliferation in breast cancer cells, CSTF3 was depleted from SK-BR-3 (non-TNBC), BT-20 (TNBC), and BT-549 (TNBC) cells using single siRNAs. Each of these cell lines expressed different levels of CSTF3 (Fig. 6B) however siRNA-treatment resulted in strong CSTF3 depletion (Fig. 6C). Proliferation assays from each cell line indicated that CSTF3 silencing reduced the proliferation TNBC but not non-TNBC cells (Fig. 6D). These results suggest that fluctuations in the expression of CSTF3 can modify the 3′ UTR length of NRAS and c-JUN and contribute to proliferation.
Figure 6.
Triple-negative breast cancer cell lines are more dependent on CSTF3. A, Expression of CSTF3 from triple-negative breast cancer cell lines (TNBC) versus breast cancer cells from other breast cancer subtypes. B, CSTF3 expression levels from SK-BR-3 (non-TNBC), BT-20 (TNBC), and BT-549 (TNBC) cells. C, Relative CSTF3 depletion from the three cell lines using independent siRNAs. D, Cell numbers from SK-BR-3, BT-20, and BT-549 depleted of GFP or CSTF3 by siRNA (**, P < 0.01; *, P < 0.05).
Discussion
These results show that divergent patterns of APA occur in different subtypes of breast cancers and highlight the significance of APA as a modulator of NRAS and c-JUN gene expression in triple-negative breast tumors/cells. We propose that APA is an adaptive mechanism that alters gene expression without requiring genetic rearrangements. In agreement with this hypothesis, previous studies have identified aberrant c-JUN and NRAS activity in triple-negative breast tumors without detecting a mechanism of activation (28). In addition, the overexpression of the c-JUN and NRAS target genes, KRT7 and FOXA1, and the silencing of KRT19, which is directly repressed by c-JUN, have been previously correlated to increased local recurrence in triple-negative breast cancer patients (43, 44). These findings suggest that nongenetic changes in c-JUN and NRAS may alter their respective transcriptional profiles and contribute to localized tumor relapse.
The prevalence of APA in cancer cells has been linked to multiple parameters including chromatin structure (45, 46), silencing of APA-inhibiting proteins (36, 47, 48) and the aberrant expression of the polyadenylation machinery (25, 27). One emerging theme from studies investigating APA, is that imbalances in the levels of the polyadenylation machinery, such as the downregulation of CFIm25 (25) or the increased expression of CSTF2 (27), changes the APA spectrum. In our study, APA levels in tumors correlated with the elevated expression of polyadenylation factor, CSTF3. These results show that increased CSTF3 expression is sufficient to trigger APA shortening of both NRAS and c-JUN. Our results are consistent with previous work and suggest that changes in the expression of the polyadenylation machinery promote APA.
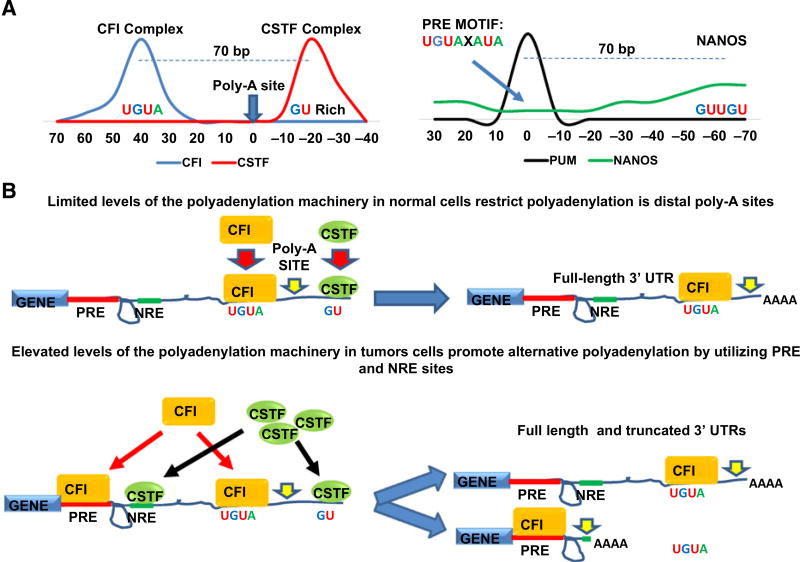
These findings pose the question: why are mRNAs that contain binding sites for PUM, a cytoplasmic RBP, disproportionately affected by the nuclear process of poly-A site selection? Insight into this relationship was provided when we compared the motifs utilized by the PUM complex to those used by the polyadenylation machinery. This comparison revealed that the two central polyadenylation complexes, the cleavage factor I (CFI) complex and the cleavage stimulation factor (CSTF) complex, interact with GU-rich sequences highly similar to the PRE and the sequence bound by the PUM-interacting protein, NANOS. The CFI complex, which functions upstream of the Poly-A site, to mediate mRNA cleavage, interacts with the same RNA sequence as the PRE, CFI: UGUA (25, 49) and PRE: UGUAXAUA (Fig. 7A; ref. 5). In addition, the CSTF complex, which functions downstream of the Poly-A site, binds to a GU-rich region (49), homologous to the GUUGU repeats favored by the PUM-interacting protein, NANOS (Fig. 7A; refs. 2, 8). These findings support a simple model: that changes in the expression of the polyadenylation machinery promote APA through the preferential use of sequences that are also recognized by PUM and the PUM-interacting protein, NANOS (Fig. 7B).
Figure 7.
The polyadenylation machinery utilizes GU-rich sequences of the PUM complex to catalyze APA. A, Schematic of the positioning and sequence composition of motifs recognized by the CFI, CSTF, and PUM complexes. B, Model describing how the CFI and CSTF complex can use motifs of the PUM complex for APA.
Our research reveals a surprisingly disproportionate level of APA on PRE-containing mRNAs in triple-negative breast tumors. It is possible that this is a unique feature of these tumors, but given the general similarity between the motifs used by the polyadenylation machinery and by PUM complexes, we anticipate that this association is likely to hold true in other contexts. Indeed, we note that PUM2 motifs were present in putative APA substrates identified in glioblastoma cells (50) and that several of the APA targets identified in developmental studies are, in fact, PRE-containing substrates (16, 20). Thus, due to the similarity of the sequences used by the PUM complex and by the polyadenylation machinery, it seems likely that PRE-containing substrates are often disproportionately affected by APA.
In summary, our results suggest that in triple-negative breast tumors, APA is reactivated by the aberrant overexpression of the polyadenylation machinery. APA of PUM targets, NRAS and c-JUN, leads to the removal of functional PRE motifs and the aberrant activation of these substrates, which contributes to changes in the expression of their downstream targets. On the basis of our findings, we propose that APA is a form of gene regulation that is exploited by triple-negative breast tumors to promote changes in the transcriptome and proteome that are conducive to tumor growth.
Supplementary Material
Acknowledgments
We would like to thank Shyamala Maheswaran, Leif Ellisen, and Lee Zou for their valuable insights.
Grant Support
D. Sgroi is supported by NIH grant R01 CA112021, the Avon Foundation, DOD Breast Cancer Research Program, and the Susan G. Komen for the Cure. N.J. Dyson is supported by NIH grants R01 CA163698 and R01 CA64402. N. Dyson is the James and Shirley Curvey MGH Research Scholar.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors' Contributions
Conception and design: W.O. Miles, A. Lembo, N. Dyson Development of methodology: W.O. Miles, A. Lembo, N. Dyson
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): W.O. Miles, A. Volorio, E. Brachtel, D. Sgroi, N. Dyson
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): W.O. Miles, A. Lembo, P. Provero, N. Dyson
Writing, review, and/or revision of the manuscript: W.O. Miles, N. Dyson
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): E. Brachtel, B. Tian, D. Sgroi
References
- 1.Kechavarzi B, Janga SC. Dissecting the expression landscape of RNA-binding proteins in human cancers. Genome Biol. 2014;15:R14. doi: 10.1186/gb-2014-15-1-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miles WO, Korenjak M, Griffiths LM, Dyer MA, Provero P, Dyson NJ. Post-transcriptional gene expression control by NANOS is up-regulated and functionally important in pRb-deficient cells. EMBO J. 2014;33:2201–15. doi: 10.15252/embj.201488057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciafre SA, Galardi S. microRNAs and RNA-binding proteins: a complex network of interactions and reciprocal regulations in cancer. RNA Biol. 2013;10:935–42. doi: 10.4161/rna.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miles WO, Tschop K, Herr A, Ji JY, Dyson NJ. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev. 2012;26:356–68. doi: 10.1101/gad.182568.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One. 2008;3:e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genomewide identification of mRNAs associated with the translational regulator PUMILIO in Drosophilamelanogaster. Proc Natl Acad Sci U S A. 2006;103:4487–92. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonoda J. Wharton RPRecruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13:2704–12. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaruzelska J, Kotecki M, Kusz K, Spik A, Firpo M, Reijo Pera RA. Conservation of a Pumilio-Nanos complex from Drosophila germ plasm to human germ cells. Dev Genes Evol. 2003;213:120–6. doi: 10.1007/s00427-003-0303-2. [DOI] [PubMed] [Google Scholar]
- 10.Friend K, Campbell ZT, Cooke A, Kroll-Conner P, Wickens MP, Kimble J. A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat Struct Mol Biol. 2012;19:176–83. doi: 10.1038/nsmb.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Q, Padmanabhan K, Richter JD. Pumilio 2 controls translation by competing with eIF4E for 7-methyl guanosine cap recognition. RNA. 2010;16:221–7. doi: 10.1261/rna.1884610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Etten J, Schagat TL, Hrit J, Weidmann CA, Brumbaugh J, Coon JJ, et al. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J Biol Chem. 2012;287:36370–83. doi: 10.1074/jbc.M112.373522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27–3' UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12:1014–20. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 14.Nolde MJ, Saka N, Reinert KL, Slack FJ. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3'UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Dev Biol. 2007;305:551–63. doi: 10.1016/j.ydbio.2007.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilgers V, Perry MW, Hendrix D, Stark A, Levine M, Haley B. Neural-specific elongation of 3' UTRs during Drosophila development. Proc Natl Acad Sci U S A. 2011;108:15864–9. doi: 10.1073/pnas.1112672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3' untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 2009;106:7028–33. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Sun Y, Fu Y, Li M, Huang G, Zhang C, et al. Dynamic landscape of tandem 3' UTRs during zebrafish development. Genome Res. 2012;22:1899–906. doi: 10.1101/gr.128488.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta I, Clauder-Munster S, Klaus B, Jarvelin AI, Aiyar RS, Benes V, et al. Alternative polyadenylation diversifies post-transcriptional regulation by selective RNA-protein interactions. Mol Syst Biol. 2014;10:719. doi: 10.1002/msb.135068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–12. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smibert P, Miura P, Westholm JO, Shenker S, May G, Duff MO, et al. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep. 2012;1:277–89. doi: 10.1016/j.celrep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon OK, Hsu TY, Im JH, Brem RB. Genetics and regulatory impact of alternative polyadenylation in human B-lymphoblastoid cells. PLoS Genet. 2012;8:e1002882. doi: 10.1371/journal.pgen.1002882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer micro-RNA target sites. Science. 2008;320:1643–7. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, et al. Differential genome-wide profiling of tandem 3' UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011;21:741–7. doi: 10.1101/gr.115295.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayr C, Bartel DP. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–84. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masamha CP, Xia Z, Yang J, Albrecht TR, Li M, Shyu AB, et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510:412–16. doi: 10.1038/nature13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lembo A, Di Cunto F, Provero P. Shortening of 3'UTRs correlates with poor prognosis in breast and lung cancer. PLoS One. 2012;7:e31129. doi: 10.1371/journal.pone.0031129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aragaki M, Takahashi K, Akiyama H, Tsuchiya E, Kondo S, Nakamura Y, et al. Characterization of a cleavage stimulation factor, 3' pre-RNA, subunit 2, 64 kDa (CSTF2) as a therapeutic target for lung cancer. Clin Cancer Res. 2011;17:5889–900. doi: 10.1158/1078-0432.CCR-11-0240. [DOI] [PubMed] [Google Scholar]
- 28.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parikh RR, Housman D, Yang Q, Toppmeyer D, Wilson LD, Haffty BG. Prognostic value of triple-negative phenotype at the time of locally recurrent, conservatively treated breast cancer. Int J Radiat Oncol Biol Phys. 2008;72:1056–63. doi: 10.1016/j.ijrobp.2008.02.066. [DOI] [PubMed] [Google Scholar]
- 30.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–32. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41:D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polak P, Oren A, Ben-Dror I, Steinberg D, Sapoznik S, Arditi-Duvdevany A, et al. The cytoskeletal network controls c-Jun translation in a UTR-dependent manner. Oncogene. 2006;25:665–76. doi: 10.1038/sj.onc.1209114. [DOI] [PubMed] [Google Scholar]
- 33.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 34.Luo W, Ji Z, Pan Z, You B, Hoque M, Li W, et al. The conserved intronic cleavage and polyadenylation site of CstF-77 gene imparts control of 3' end processing activity through feedback autoregulation and by U1 snRNP. PLoS Genet. 2013;9:e1003613. doi: 10.1371/journal.pgen.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miles WO, Korenjak M, Griffiths LM, Dyer MA, Provero P, Dyson NJ. Post-transcriptional gene expression control by NANOS is up-regulated and functionally important in pRb-deficient cells. EMBO J. 2014;33:2201–15. doi: 10.15252/embj.201488057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kuhn U, Menzies FM, et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149:538–53. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 37.Sgroi DC, Carney E, Zarrella E, Steffel L, Binns SN, Finkelstein DM, et al. Prediction of late disease recurrence and extended adjuvant letrozole benefit by the HOXB13/IL17BR biomarker. J Natl Cancer Inst. 2013;105:1036–42. doi: 10.1093/jnci/djt146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertucci F, Finetti P, Rougemont J, Charafe-Jauffret E, Cervera N, Tarpin C, et al. Gene expression profiling identifies molecular subtypes of inflammatory breast cancer. Cancer Res. 2005;65:2170–8. doi: 10.1158/0008-5472.CAN-04-4115. [DOI] [PubMed] [Google Scholar]
- 39.Jacquemier J, Ginestier C, Rougemont J, Bardou VJ, Charafe-Jauffret E, Geneix J, et al. Protein expression profiling identifies subclasses of breast cancer and predicts prognosis. Cancer Res. 2005;65:767–79. [PubMed] [Google Scholar]
- 40.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–9. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neben K, Schnittger S, Brors B, Tews B, Kokocinski F, Haferlach T, et al. Distinct gene expression patterns associated with FLT3- and NRAS-activating mutations in acute myeloid leukemia with normal karyotype. Oncogene. 2005;24:1580–8. doi: 10.1038/sj.onc.1208344. [DOI] [PubMed] [Google Scholar]
- 42.Rinehart-Kim J, Johnston M, Birrer M, Bos T. Alterations in the gene expression profile of MCF-7 breast tumor cells in response to c-Jun. Int J Cancer. 2000;88:180–90. doi: 10.1002/1097-0215(20001015)88:2<180::aid-ijc6>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 43.Parikh RR, Yang Q, Higgins SA, Haffty BG. Outcomes in young women with breast cancer of triple-negative phenotype: the prognostic significance of CK19 expression. Int J Radiat Oncol Biol Phys. 2008;70:35–42. doi: 10.1016/j.ijrobp.2007.05.066. [DOI] [PubMed] [Google Scholar]
- 44.Fujisue M, Nishimura R, Okumura Y, Tashima R, Nishiyama Y, Osako T, et al. Clinical significance of CK19 negative breast cancer. Cancers. 2012;5:1–11. doi: 10.3390/cancers5010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee CY, Chen L. Alternative polyadenylation sites reveal distinct chromatin accessibility and histone modification in human cell lines. Bioinformatics. 2013;29:1713–7. doi: 10.1093/bioinformatics/btt288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang H, Chen J, Liu H, Sun X. The nucleosome regulates the usage of polyadenylation sites in the human genome. BMC Genomics. 2013;14:912. doi: 10.1186/1471-2164-14-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–8. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin G, Gruber AR, Keller W, Zavolan M. Genome-wide analysis of pre-mRNA3' end processing reveals a decisive role of human cleavage factor I in the regulation of 3' UTR length. Cell Rep. 2012;1:753–63. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 50.Shao J, Zhang J, Zhang Z, Jiang H, Lou X, Huang B, et al. Alternative polyadenylation in glioblastoma multiforme and changes in predicted RNA binding protein profiles. OMICS. 2013;17:136–49. doi: 10.1089/omi.2012.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.