Abstract
Antibiotic resistance is a major threat to human welfare. Inhibitors of multidrug efflux pumps (EPIs) are promising alternative therapeutics that could revive activities of antibiotics and reduce bacterial virulence. Identification of new druggable sites for inhibition is critical for the development of effective EPIs, especially in light of constantly emerging resistance. Here, we describe EPIs that interact with periplasmic membrane fusion proteins, critical components of efflux pumps that are responsible for the activation of the transporter and the recruitment of the outer-membrane channel. The discovered EPIs bind to AcrA, a component of the prototypical AcrAB-TolC pump, change its structure in vivo, inhibit efflux of fluorescent probes, and potentiate the activities of antibiotics in Escherichia coli and other Gram-negative bacteria. Our findings expand the chemical and mechanistic diversity of EPIs, suggest the mechanism for regulation of the efflux pump assembly and activity, and provide a promising path for reviving the activities of antibiotics in resistant bacteria.
Keywords: Gram-negative bacteria, antibiotic resistance, hyperporinated outer membrane, efflux pump inhibitors
Graphical Abstract
The threat of bacterial antibiotic resistance is real and broadly recognized. Multidrug efflux is a primordial mechanism of antibiotic resistance and is present in all microorganisms. By reducing antibiotic concentrations at targets, polyspecific drug efflux transporters not only provide intrinsic levels of antibiotic resistance but also enable development of class-specific resistance and often act cooperatively. Escherichia coli AcrAB-TolC is a prototype multidrug efflux pump and a major contributor to antibiotic resistance in clinical settings.1 Close homologues of this pump are conserved in the genomes of Gram-negative pathogens and share with it their structure and molecular mechanism. These transporters act synergistically with the low permeability barrier of the Gram-negative outer membrane (OM) by expelling drugs from the inner membrane and periplasm into the external medium. Such transport requires a concerted action of three proteins: the inner membrane transporter AcrB, the periplasmic membrane fusion protein (MFP) AcrA, and the outer membrane channel TolC. Together the three components form a protein conduit spanning the two membranes and the periplasm of E. coli.2 Inactivation of any of the three components completely abrogates drug efflux and leads to a dramatic increase in susceptibility to various antimicrobial agents.3
Efflux pump inhibitors (EPIs) are emerging alternative therapeutics that have the potential to revive the activities of existing antibiotics and to control the spread of antibiotic resistance.4 The inner membrane transporter AcrB has hitherto been a focus of efforts to identify EPIs.4a,5 The three previously described EPIs, DA-13-1809, MC207110 (PAβN), and MBX2319, were shown to bind to AcrB and have similar mechanisms of action.4a,5a,c,6 MC207110 stands somewhat apart because its mechanism also involves permeabilization of the outer membrane7 and interference with binding of other substrates.8 For different reasons, these inhibitors have not yet progressed to clinical trials, and continued efforts are needed to identify EPIs with different mechanisms of action. Chemical and mechanistic diversification of EPIs could facilitate the development of inhibitors suitable for clinical applications.
Here, we analyzed the periplasmic MFP, AcrA, as an allosteric target for multidrug efflux inhibition. AcrA and homologous MFPs enable the extrusion of antibiotics across the OM by assembling into a hexameric structure on the periplasmic surface of AcrB and creating a sealed tunnel for antimicrobials to reach TolC.2b,6,9 This assembly is highly dynamic and serves both structural and several functional roles. AcrA and other MFPs have been proposed to sense substrate-dependent conformational changes in the transporter and to interact with substrates directly, initiating engagement of the OM channel into the trans-envelope complex.9a Interactions with the OM channel trigger the assembly of the AcrA tunnel and the conformational changes in AcrA needed to activate the transporter and open the OM channel.
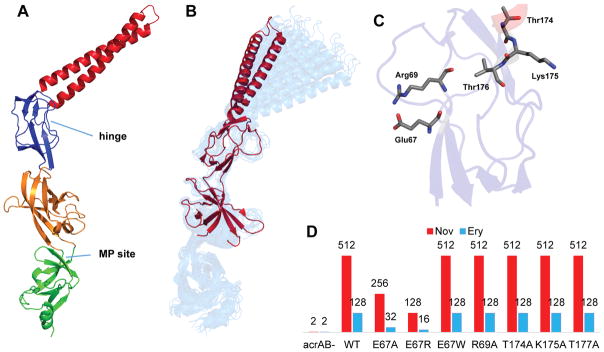
The structure of AcrA befits its diverse functions.10 AcrA and other MFPs form four linearly arranged domains connected by flexible linkers (Figure 1A). The docking site for TolC is the α-helical hairpin of AcrA. This domain assumes multiple conformations due to the flexibility of the hinge region linking it to the lipoyl domain.10a Although the exact functional role of the lipoyl domain remains unclear, analysis of mutations targeting this domain in AcrA suggests a role in stabilizing the complex.10a,11 The α–β-barrel domain is involved in ligand binding, as a Zn2+ ion, a substrate of the MFP ZneB, was found to bind to the α–β-barrel domain and cause a conformational change affecting the adjacent membrane proximal (MP) domain.12 It remains unclear whether or not substrates interact with AcrA or other MFPs. The MP domains are often unresolved in crystal structures of MFPs as they can be unstructured and proteolytically labile.10a,13 Ligand binding and modification with lipids and interprotomer interactions stabilize the MP domains in a β-roll structure, which varies substantially in different MFPs.10b,12 The relative positioning of the α–β-barrel and MP domains changes the protein from a crescent-like shape to a more extended conformation (Figure 1A,B).
Figure 1.
Structure of AcrA and binding sites for inhibition. (A) Structural model of AcrA with its four conserved domains highlighted: α-helical hairpin (red), lipoyl (blue), α–β-barrel (orange), and MP domain (green). Potential binding sites for efflux inhibition are labeled. (B) Hinge bending motion observed in molecular dynamics simulations (blue) superimposed onto the X-ray crystal structure of AcrA (red). (C) Positions of conserved residues in the hinge site (red). (D) Site-directed mutagenesis of conserved residues in the hinge site of AcrA. Numbers are MICs (μg/mL) of novobiocin and erythromycin in cells producing different AcrA variants.
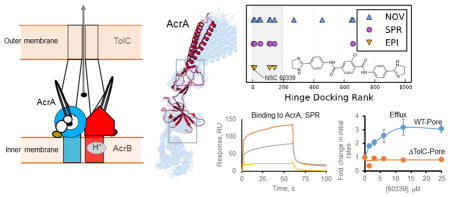
In the assembled AcrAB-TolC complex, AcrA functions as a trimer of dimers, in which each protomer of an AcrA dimer is functionally distinct and forms a unique interaction with AcrB and TolC.14 One protomer in each dimer is thought to be responsible for the stimulation of transporter turnover, whereas the second protomer is important for functional interactions with TolC (Figure 6). Hence, the oligomerization of AcrA is essential not only for the assembly of the trans-periplasmic drug tunnel but also for the coupling of transport processes in the two different membranes.
Figure 6.
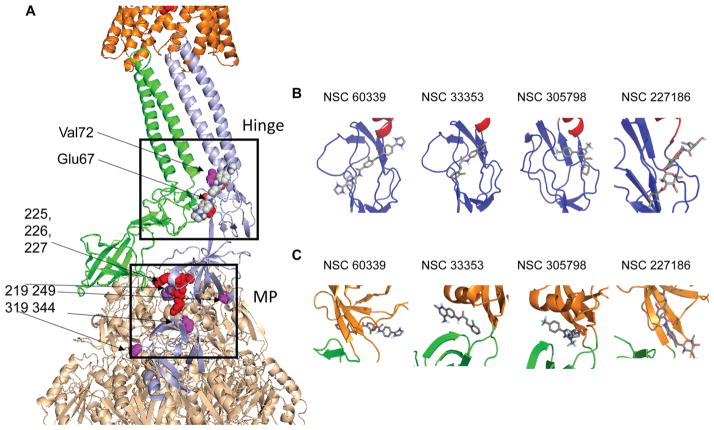
Predicted binding poses of NSC 60339, NSC 227186, NSC 33353, and NSC 305798 at the hinge and MP sites of AcrA. (A) AcrA dimer bound to TolC and AcrB. NSC 60339 bound to the hinge site of one AcrA protomer is shown as spheres. The amino acid residues in the docking sites of compounds are shown as red spheres, and the amino acid residues important for AcrA function are shown as pink spheres. (B) Docking poses of the EPIs in the hinge site of AcrA. The AcrA domains are colored as in Figure 1. (C) Docking poses of the EPIs in the MP site of AcrA.
The structural and functional studies discussed above suggest that a structure-based approach to ligand design specific to AcrA might be fruitful, in which allosteric and/or protein/protein interaction inhibitors are pursued that act at the interdomain interfaces or elsewhere to disrupt efflux pump assembly and/or function. In this study, we targeted AcrA to discover EPIs with desired biochemical activities. To achieve this, we developed a joint experimental–computational ligand screening approach of general applicability in the discovery of inhibitors for periplasmic targets. The experimental screen involved growth inhibition assays that were used previously to identify compounds acting synergistically with known antibiotics.5b,15 We modified this screen to increase its sensitivity and selectivity. In whole cell assays, it is difficult to separate the inhibition of efflux from the permeabilization of the OM because the two are synergistically linked by the efflux of antibiotics across the OM.5b Therefore, to increase the probability of selectively identifying EPIs, we used WT-Pore E. coli cells, which carry large, 2.4 nm, pores in their OM. These cells are susceptible to antibiotics that do not permeate the intact OM but they retain full efflux activity.16 Using this computational–experimental approach, we discovered new AcrA-binding EPIs that potentiate antibiotics in E. coli.
RESULTS AND DISCUSSION
Experimental Screening for EPIs
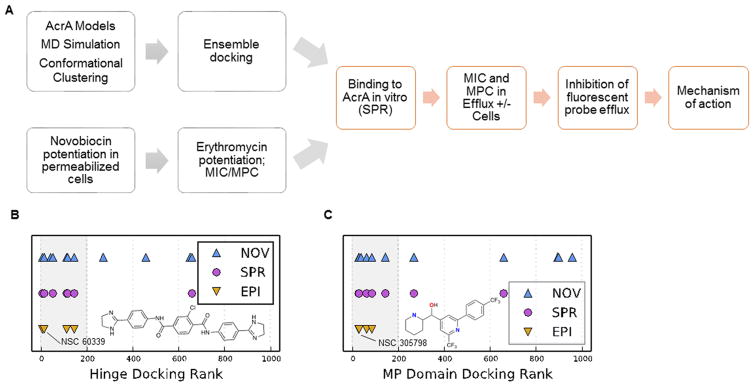
To identify AcrA-specific inhibitors of antibiotic efflux, we developed a protocol of parallel experimental and structure-based computational screens and counter-screens (Figure 2). The experimental screen involved the growth inhibition assay, in which antibacterial activities of various compounds were analyzed in the presence of subinhibitory concentrations of an antibiotic. For the computational screen, we used an ensemble docking approach to rank compounds on the basis of their binding to different AcrA conformers and sites. Top scorers from both types of screening were further analyzed to establish multidrug potentiation, to experimentally assess binding to AcrA, and to probe the mechanism of action.
Figure 2.
Flowchart and results of screening for AcrA-specific efflux pump inhibitors. (A) Flowchart of computational and experimental screenings. (B) NCI 5 library of compounds ranked by docking score for the hinge site. Symbols indicate compounds that scored high in the potentiation of the novobiocin activity (NOV), binding to AcrA (SPR), and counter-assays in ΔTolC-Pore cells (EPI). The structure of NSC 60339 and its rank position are also indicated. (C) The same as in (B) but the NCI 5 library of compounds was ranked by docking score for the MP site. The structure of NSC 305798 and its rank position are also indicated.
The National Cancer Institute (NCI) Diversity 5 set (1593 compounds) and the NCI Natural Product Set (117 compounds) (NCI 5) were screened for the potentiation of antimicrobial activity of the antibiotic novobiocin in WT-Pore cells.16 Screening in these cells counter-selects against compounds that permeabilize the OM and also increases the positive hit rate because more diverse compounds are able to reach the periplasmic AcrA. Novobiocin was selected for potentiation screening because the permeabilization of the OM in efflux-proficient WT-Pore cells decreased its minimal inhibitory concentrations (MICs) by only 4-fold, from 128 to 32 μg/mL. In contrast, inactivation of efflux decreased the MIC of novobiocin by 256-fold both in the ΔTolC-Pore cells with the intact OM and in permeabilized cells. This effect of active efflux on the activity of novobiocin creates a large potentiation window that is mostly insensitive to the permeability of the OM. Hence, to qualify as an EPI, a compound should demonstrate a concentration-dependent potentiation of the inhibitory activity of novobiocin in WT-Pore cells but not in ΔTolC-Pore cells lacking active efflux (Figure 2).
The screen identified 12 compounds that potentiated novobiocin inhibitory activity in WT-Pore cells in a concentration-dependent manner (0.7% hit rate) (Table 1). Checkerboard assays were carried out to determine the minimal potentiating concentration (MPC) of hits for novobiocin and erythromycin, another substrate of AcrAB-TolC. Four hits were found to potentiate both novobiocin and erythromycin equally well, with similar MPC4 values (MPC at which the inhibitory activity of an antibiotic is potentiated 4-fold) for both antibiotics (Table 1). Two hits potentiated erythromycin significantly better than novobiocin, and two potentiated the activity of novobiocin only. Of the 12 hits, 8 compounds did not potentiate the activities of antibiotics in the efflux-deficient ΔTolC-Pore cells. Therefore, these 8 compounds possess EPI-like properties.
Table 1.
Minimal Inhibitory Concentrations and Potentiating Activities of Top Hits
| MIC WT, μM | MIC WT-Pore, μM | MIC ΔTolC, μM | MIC ΔTolC-Pore, μM | MPC4 NOV,a μM | MPC4 ERY, a μM | AcrAb binder | ΔTolC-Porec | |
|---|---|---|---|---|---|---|---|---|
| NSC 60339 | >200 | 200 | 12.5 | 12.5 | 25 | 12.5 | yes | no |
| NSC 227186 | >50 | 25 | 6.25 | 3.13 | 6.25 | 3.13 | yes | no |
| NSC 33353 | 200 | 100 | 12.5 | 12.5 | 1.56 | 3.13 | yes | no |
| NSC 305798 | 50 | 25–50 | 25 | 12.5 | 12.5 | 3.13 | yes | no |
| NSC 50651 | >50 | 3.13 | 6.25 | 0.8 | 1.56 | 6.25 | yes | no |
| NSC 260594 | >50 | 3.13 | 0.8 | 0.1 | 0.39 | 3.13 | no | yes |
| NSC 354844 | >100 | 6.25 | 50 | 6.25 | 1.56 | NAd | yes | no |
| NSC 325014 | 25 | 3.13 | 12.5 | 3.13 | 3.1 | 0.39 | no | no |
| NSC 207895 | 25 | 6.25 | 25 | 12.5 | 0.78 | 1.56 | yes | no |
| NSC 56410 | 12.5 | 3.13 | 1.56 | 1.56 | 0.39 | NA | no | yes |
| NSC 26980 | 3.13 | 3.13 | 0.19 | 0.19 | 0.39 | 0.39 | no | yes |
Potentiation in WT-Pore; NOV, novobiocin; ERY, erythromycin.
See Figure S5 for experimental data and KD values of selected hits.
Potentiation in efflux-deficient ΔTolC-Pore cells.
NA, no activity.
Validation of Functional Binding Sites and Computational Screening
For the computational screens, we generated structural models of full-length AcrA from E. coli K-12 substrate MG1655 on the basis of homology with crystallographically determined structures (see the Supporting Information). Consistent with previous studies,10a,17 molecular dynamics (MD) simulations indicated the presence of a hinge-bending motion between the lipoyl and α–β-barrel domains of AcrA (Figure 1B). The “hinge bending” defines the orientation of the hairpin domain, and the hinge region contains a well-defined groove (Figure 1C).
A multiple sequence alignment of 1670 AcrA homologues showed that the hinge site is lined by highly conserved residues of the lipoyl domain of AcrA (Figure 1C; Figure S1). To analyze the functional significance of this site, Glu67, Arg69, Thr174, Lys175, and Thr177 were substituted with alanine in plasmid-borne AcrA coexpressed with AcrB. Expression of all the constructed AcrA variants was similar to that of the wild-type protein. On the basis of MIC measurements with novobiocin and erythromycin, R69A, T174A, K175A, and T177A mutants fully complemented the susceptible phenotype of E. coli ΔacrAB cells. In contrast, the E67A variant was only partially active, and the reversal of charge in the E67R variant further reduced the ability of AcrA to complement the drug susceptibility phenotype (Figure 1D). Thus, both the size and the charge of the residue at position 67 are important for AcrA function. Therefore, the hinge site (hinge) of AcrA is functionally important and was selected as a potential binding site for small molecules. Another potential binding site was identified on the conformationally flexible MP domain. This site, which again contains functionally important and conserved residues,18 is located at the interface of the MP domain and the adjacent α–β-barrel domain (Figure 1A).
To identify compounds that could potentially bind to the hinge and/or MP sites, we performed short MD simulations of monomeric AcrA and clustered the structures to extract an ensemble of ~30 representative conformations. Using an ensemble docking approach, we carried out in silico high-throughput docking screens on each conformation19 (Figure S2). This screen was performed with the NCI 5 library. The compounds were ranked on the basis of their docking scores. Among the 50 top-scoring compounds for each site, 21 were predicted to bind specifically at the hinge site, 21 were predicted to bind specifically at the MP site, and 29 were predicted to bind at both sites (Table S1).
Identification of AcrA Inhibitors
The 12 primary hits from the experimental screen and the 18 top scorers from the virtual screening were then experimentally counter-screened for physical interactions with purified AcrA using surface plasmon resonance (SPR). The purified AcrA protein has a very low propensity to oligomerize, and it is the association with AcrB and cooperative interactions with TolC that are believed to drive AcrA oligomerization and formation of the trans-envelope tunnel.2a,6,9b For SPR, purified AcrA was immobilized onto a CM5 chip at three protein densities, and ligands were injected at 50 μM concentration. A total of 11 AcrA binders (36%) were identified in this counter-screen, of which 7 compounds (23%) were potentiators of antibiotic activities in E. coli cells (Table 1 and Figure 2), suggesting that they may be acting on AcrA.
Seven of the experimentally validated AcrA binder–potentiators were in the top 10% of the rankings in the computational screen at the hinge site, MP site, or both (Figure 2B,C; Figures S3 and S4). Of these, NSC 60339, NSC 227186, NSC 33353, and NSC 305798 were all ranked in the top 5% of the computational hits, were shown to bind AcrA in SPR experiments, and potentiated the inhibitory activity of novobiocin. The confluence of computation and experiment led us to select these four compounds for further investigation.
NSC 227186, also called chlorobiocin, is an aminocoumarin antibiotic that differs from novobiocin by a chloro substitution of the C-8′ methyl group and a pyrrole substitution of the 3-O″-carbamoyl group in novobiocin20 (Figure S5). Similar to novobiocin, NSC 227186 is an excellent substrate for the AcrAB-TolC efflux pump, as seen from the ~10-fold decrease in MIC caused by deletion of TolC (Table 1). Thus, NSC 227186 may act as a competitive inhibitor. To evaluate this notion, we next compared the properties of novobiocin and NSC 227186 in potentiating the activity of erythromycin.
The checkerboard assay showed that NSC 227186 interacts synergistically with both novobiocin and erythromycin. Because MICs of NSC 60339 and NSC 227186 were too high to be measured in E. coli WT cells, all drug combinations were analyzed in WT-Pore cells. In these cells, the MPC4 of NSC 227186 was found to be 3 μM for novobiocin and 6 μM for erythromycin, and the fractional inhibitory concentration (FIC) indices were 0.325 and 0.375 for novobiocin and erythromycin, respectively. An FIC index ≤0.5 indicates synergistic interactions between antibiotics, and an FIC of 1 indicates an additive effect between the drugs.21 No synergistic interactions between NSC 227186 and antibiotics were found in the efflux-deficient ΔTolC-Pore cells (Table 1). Importantly, novobiocin did not potentiate the inhibitory activity of erythromycin in the WT-Pore cells (FIC index = 2, indicating an indifferent effect) and the combination of the two antibiotics was additive in ΔTolC-Pore cells, with an FIC index of 0.75. Thus, NSC 227186, but not novobiocin, has an EPI-like activity in E. coli cells.
NSC 60339 is a substituted phthalanilide (2-chloro-4′,4″-bis (2-imidazolin-2-yl)terephthalanilide) with chemotherapeutic activity against a wide range of cancers.22 This compound has a weak antibacterial activity against WT-Pore E. coli with an MIC of 200 μM. The MIC decreases to 12.5 μM in ΔTolC-Pore cells, suggesting that NSC 60339 is a substrate of AcrAB-TolC (Table 1). NSC 60339 potentiates both NOV and erythromycin in WT-Pore E. coli cells with an MPC4 of 25 μM for novobiocin and 12.5 μM for erythromycin, but has no effect on these antibiotics in ΔTolC-Pore cells (FIC index = 2).
NSC 33353 is a substituted aminoquinoline with a chemokine receptor-modulating activity.23 This compound has weak antimicrobial activity with an MIC in E. coli WT-Pore cells of 100 μM. It is a substrate of AcrAB-TolC as seen from the 8-fold decrease in MIC to 12.5 μM in ΔTolC-Pore cells (Table 1). NSC 33353 potentiates equally well the activities of novobiocin and erythromycin with MPC4 for both antibiotics in the 1.56–3.125 μM range. This compound has the largest fold potentiation window (MIC/MPC4 = 32–64) among the top four compounds.
Lastly, NSC 305798 is an antimalarial agent, enpiroline ((R)-piperidin-2-yl(2-trifluoromethyl)-6-[4-(trifluoromethyl)-pyridine-4-yl]methanol), with an unknown antimalarial mechanism of action.24 The MIC of enpiroline is 25–50 μM in E. coli WT, WT-Pore, and efflux-deficient ΔTolC cells and 12.5 μM in ΔTolC-Pore cells. Thus, among the top four hits, NSC 305798 is the only compound for which activity is not affected significantly by the permeability of the OM or AcrAB-TolC-mediated efflux. NSC 305798 potentiates erythromycin with an MPC4 of 1.56–3.125 μM (MIC/MPC4 = 16–32) but has a weaker potentiating activity in combination with novobiocin with MPC4 at 12.5 μM (MIC/MPC4 = 2–4).
The Four Top Hits Are Inhibitors of AcrAB-TolC with Different Mechanisms of Action
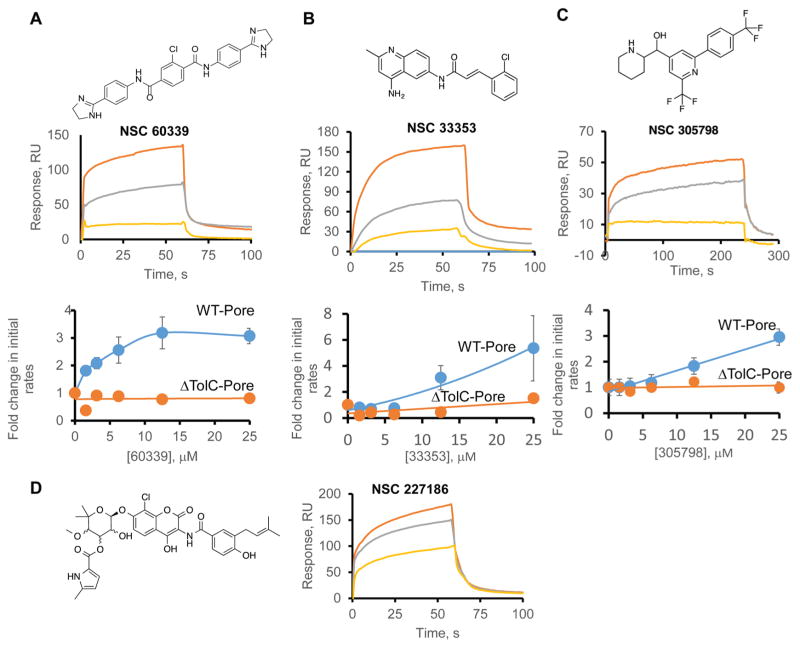
To further characterize the mechanism(s) of inhibition, we compared the biochemical activities of the top four hits to two previously characterized EPIs, phenylalanyl-β-naphthylamide (MC207110) and the pyranopyridine MBX2319, both of which are thought to act on the transporter AcrB.8,25 MC207110, but not MBX2319, is also a substrate of the AcrAB-TolC pump.5b,8 We first compared the affinities of the hits and EPIs, as well as those of novobiocin and erythromycin, to purified AcrA protein. For this purpose, SPR sensorgrams were collected with immobilized AcrA and increasing concentrations of ligands from 6 to 100 μM. We found that NSC 227186, NSC 60339, NSC 33353, NSC 305798, MC207110, and novobiocin bind AcrA with low-to-mid micromolar affinities (Table 1 and Figure 3; Figure S5), suggesting that all of these compounds may interact with AcrA in vivo. In contrast, MBX2319 and erythromycin do not bind AcrA with detectable affinities.
Figure 3.
Top experimental and computational hits bind to AcrA and inhibit efflux of HT. (A) (Top) Structure of NSC 60339. (Middle) Representative sensorgram of NSC 60339 bound to AcrA immobilized at three different surface densities: 1261 RU (yellow), 3202 RU (gray), and 5079 RU (orange). The concentration of NSC 60339 was 100 μM. Here and below, compounds were injected in the binding buffer (pH 6.0) supplemented with 5% DMSO. (Bottom) Fold change in initial rates of HT uptake into permeabilized WT-Pore and ΔTolC-Pore cells in the presence of increasing concentrations of NSC 60339. Initial rates were determined by fitting time-dependent changes in the HT fluorescence in the presence and absence of the inhibitor (Figure S7). (B) (Top) Structure of NSC 33353. (Middle) Sensorgram of 50 μM NSC 33353 injected onto the same AcrA chip as in (A). (Bottom) Fold change in initial rates of HT into WT-Pore and ΔTolC-Pore cells pretreated with increasing concentrations of NSC 33353. (C) (Top) Structure of NSC 305798. (Middle) Sensorgram of 50 μM NSC 305798 bound to AcrA immobilized at three different surface densities: 807 RU (yellow), 5732 RU (gray), and 8908 RU (orange). (Bottom) Fold change in initial rates of HT into WT-Pore and ΔTolC-Pore cells pretreated with increasing concentrations of NSC 305798. (D) (Left) Structure of NSC 227186. (Right) Sensorgram of 100 μM NSC 227186 injected onto the same AcrA chip as in (A).
We next investigated whether the top hits inhibit efflux of the fluorescent drug bisbenzimide H33342 (HT). The fluorescence of HT increases dramatically when it intercalates into membranes and intracellular DNA. When added to efflux-proficient cells, HT fluorescence increases very slowly because it is an excellent substrate of AcrAB-TolC, but inactivation of TolC leads to a rapid increase in HT fluorescence.26 Similar fluorescence-based assays have been used previously to test the abilities of candidate EPIs to increase intracellular concentrations of compounds.15,27 In agreement with previous studies, treatment of cells with increasing concentrations of either MBX2319 or MC207110 led to increased fluorescence and hence inhibition of HT efflux (Figure S6). Novobiocin and NSC 227186 are intrinsically fluorescent, and their ability to inhibit the efflux of HT could not be analyzed using this approach. However, NSC 60339, NSC 33353, and NSC 305798 all demonstrated efflux-inhibitory activities, albeit with different concentration dependencies (Figure 3A–C). In the presence of NSC 60339, the initial rates of HT uptake into cells increased by ~3 fold and remained constant at or above 12 μM NSC 60339. In contrast, the rates of HT uptake increase linearly with increasing concentrations of NSC 305798. The concentration dependence of NSC 33353 inhibition is more complex and at high (≥25 μM) concentrations, some enhancement of HT uptake was also observed in ΔTolC-Pore cells (Figure S7). Thus, all three top hits are EPIs, but with different mechanisms of inhibition. NSC 33353 is likely to have additional interactions in cells.
This conclusion was further confirmed by studies of the potentiating activities of NSC 33353 and NSC 60339 in different Gram-negative bacteria (Table 2). In addition to E. coli, NSC 33353 potentiated activities of novobiocin and erythromycin in Acinetobacter baumannii, Enterobacter cloacae, and Klebsiella pneumoniae and of a fluoroquinolone levofloxacin only in E. coli and K. pneumoniae (Table 2). In contrast, NSC 60339 was active only in E. coli but potentiated activities of not only novobiocin and erythromycin but also levofloxacin. Thus, NSC 60339 is a species-specific EPI. None of the EPIs was active in Pseudomonas aeruginosa PAO1, presumably because they cannot penetrate the outer membrane of this bacterium.
Table 2.
Minimal Inhibitory Concentrations and Potentiating Activities of NSC 60339 and NSC 33353 in Gram-Negative Bacteriaa
| strain | MIC, μM | MPC4 NOV, μM | MPC4 ERY, μM | MPC4 LEVO, μM | ||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||
| 60339 | 33353 | 60339 | 33353 | 60339 | 33353 | 60339 | 33353 | |
| E. coli BW25113 | >200 | 200 | 100 | 25 | 50 | 25 | 50 | 50 |
| P. aeruginosa PAO1 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| A. baumannii ATCC 17798 | >100 | >100 | >100 | 6.25 | >100 | 50 | >100 | >100 |
| E. cloacae ATCC 13047 | >100 | >100 | >100 | 25 | >100 | 25 | >100 | >100 |
| K. pneumoniae ATCC 13883 | >100 | >100 | >100 | 12.5 | >100 | 12.5 | >100 | 25 |
NOV, novobiocin; ERY, erythromycin, LEVO, levofloxacin.
NSC 60339 Changes the Structure of AcrA in Vivo
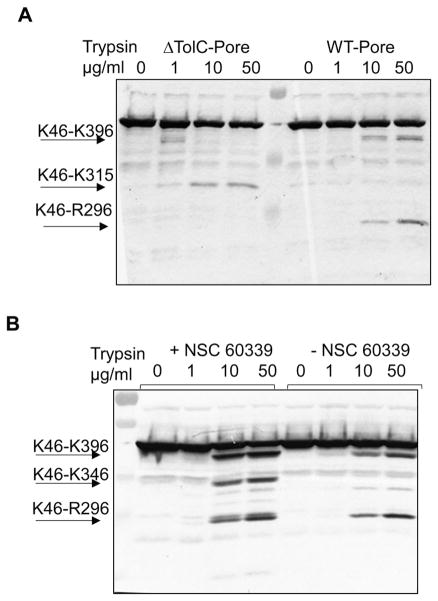
To determine whether or not interactions of the EPIs with AcrA affect its structure or ability to form a functional AcrAB-TolC complex in vivo, we used an in vivo proteolysis approach. In agreement with previous results,18 the proteolytic cleavage profile of AcrA in the absence of TolC (ΔTolC-Pore cells) accumulated a K46–R315 fragment that was absent in the profile of AcrA engaged in the tripartite complex (WT-Pore cells) (Figure 4A). The assembly of the AcrAB-TolC complex leads to cleavage in the MP domain of AcrA, as seen in the accumulation of the K46–R294/296 fragment that dominates the AcrA proteolytic profile in the WT-Pore cells. Pretreatment of WT-Pore cells with NSC 227186, NSC 305798, NSC 33353, MC207110, or MBX2319 did not result in notable changes in the proteolytic cleavage of AcrA, consistent with these inhibitors not significantly affecting the structure of AcrA or the assembly of the AcrAB-TolC complex (Figure S6). In contrast, two stable AcrA fragments were accumulated in WT-Pore cells treated with NSC 60339: the K46–R294/296 fragment characteristic for the assembled AcrAB-TolC complex and a new fragment corresponding to residues K46–K346 of AcrA (Figure 4B). Thus, the structure of AcrA is different in the presence of NSC 60339 from that in its absence. Taken together, these findings suggest that the mechanism of inhibition by NSC 60339 may involve structural changes in AcrA.
Figure 4.
NSC 60339 is an EPI with a novel mechanism of action. (A) Proteolytic profiles of AcrA in WT-Pore and ΔTolC-Pore cells. Cells were osmotically shocked with sucrose and treated with the indicated concentrations of trypsin for 30 min at 37 °C. Reactions were terminated by SDS sample buffer and boiling the samples for 5 min. Proteins and proteolytic fragments were separated by 12% SDS-PAGE, and AcrA fragments were visualized using polyclonal anti-AcrA antibody. The characteristic proteolytic fragments of AcrA as identified in ref 18a are indicated. (B) WT-Pore cells were pretreated with NSC 60339 and proteolysis was carried out as in (A).
Binding to AcrA Is Required for the Inhibitory Activities of EPIs
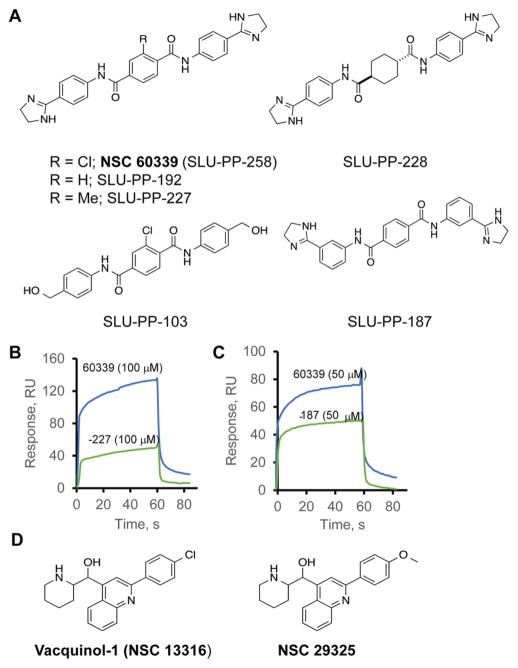
The results described above show that NSC 60339 is an EPI that binds to and changes the structure of AcrA in vivo. In a preliminary investigation of structure–activity relationships (SARs), we prepared several analogues of NSC 60339 (Figure 5A). Compounds in which the chlorine of the central 2-chlorophthalamide ring was replaced, that is, R = Me (SLU-PP-227) or R = H (SLU-PP-192), still demonstrated binding to AcrA and potentiation of novobiocin and erythromycin, albeit with reduced activities (Figure 5B and Table 3). However, the analogue SLU-PP-228, in which the central ring has been converted to a saturated cyclohexyl ring, was found to have no binding with AcrA or potentiation (Table 3). Interestingly though, this compound did possess the greatest antimicrobial activity of the new analogues, with an MIC of 50 μM in WT-Pore cells.
Figure 5.
NSC 60339 analogues and their activities. (A) Structures of NSC 60339 analogues. (B) Sensorgrams of 100 μM NSC 60339 and 100 μM SLU-PP-227 collected over AcrA immobilized at a density of 5077 RU. Compounds were injected in the binding buffer (pH 6.0) supplemented with 5% DMSO. (C) The same as in (B), but 50 μM NSC 60339 and 50 μM SLU-PP-187 were injected over AcrA. (D) Structures of NSC 305798 analogues.
Table 3.
Potentiation of Antibiotic Activities and Binding to AcrA by NSC 60339 Analogues
| compound | MIC WT-Pore, μM | MPC4 NOV, μM | MPC4 ERY, μM | binding to AcrA |
|---|---|---|---|---|
| SLU-PP-258 (NSC 60339) | 100–200 | 25 | nda | yes |
| SLU-PP-227 | 100–200 | 50 | 25 | weak |
| SLU-PP 187 | 200 | 25 | 25 | yes |
| SLU-PP-192 | 100–200 | 50 | 12.5 | nd |
| SLU-PP-228 | 50 | NAa | NA | no |
| SLU-PP-103 | >400 | >200 | NA | no |
| SLU-PP-194 (NSC 33353) | 100 | 1.56–3.125 | nd | yes |
| NSC 23925 | 400 | 100 | 50 | weak |
| vacquinol-1 | 100 | 25 | NA | weak |
NA, no activity; nd, no data.
We also prepared analogues by modifying the flanking imidazoline rings. The replacement of these groups with hydroxymethyl substituents (SLU-PP-103) resulted in a complete loss of both binding to AcrA and potentiation. We prepared another analogue in which the imidazoline group is migrated to the 3-position, SLU-PP-187. This compound retains most of the binding and potentiation exhibited by NSC 60339 and similarly has low antimicrobial activity (MIC = 200 μM) on its own (Figure 5C and Table 3). This finding suggests that the imidazoline group is critical for binding and that the AcrA binding site is able to accommodate a less linear molecule, such as SLU-PP-187. These findings also show that the interaction between NSC 60339 and AcrA is specific and that there is a clear relationship between the changes in binding to AcrA and the EPI activities of the NSC 60339 analogues.
In addition, two commercially available structural analogues of NSC 305798 were identified and tested in AcrA binding and potentiation assays (Figure 5D). Both compounds are weak antimicrobial agents with MICs of 400 and 100 μM in the WT-Pore cells for NSC 29375 and vacquinol-1, respectively (Table 3). Efflux inactivation reduced the MICs by 2–4-fold, which suggests that both compounds are weak substrates of AcrAB-TolC. NSC 29375 showed weak AcrA binding activity and weak potentiation activity for both novobiocin and eryth-romycin. Similarly, vacquinol-1 binds AcrA and potentiates the activity of novobiocin (MPC4 = 25 μM), albeit with weaker activities than those of NSC 305798. Thus, as is the case with the NSC 60339 series, there is a relationship between the activity of the EPI and binding to AcrA. In agreement, among the four primary hits that potentiated the activity of novobiocin but did not bind AcrA, three compounds, NSC56410, NSC 260594, and NSC 26980, potentiated the activity of novobiocin in an efflux-independent manner in ΔTolC-Pore cells and, therefore, were not EPIs (Table 1).
Inhibitors with Different Mechanisms May Target Different Sites on AcrA
We cannot exclude the possibility that in addition to binding to AcrA, interactions with AcrB or other intracellular targets could also contribute to the observed EPI activities. However, our results suggest that the targeting of two different sites on AcrA may yield inhibitors with different mechanisms of actions. Two binding sites on AcrA were used for computational molecular docking: the hinge and the MP sites. The hinge site is important for the assembly of the hexameric AcrA tunnel.2a,28 In addition, the conformations of the hinge site in the crystal structure of a soluble AcrA fragment10a and in the assembled hexameric AcrA tunnel2 are slightly different. Among the identified EPIs, NSC 60339 and NSC 227186 were ranked highest in docking to the hinge site and their computed molecular contacts with AcrA are similar (Figure 6B). In these models of the docked complex Glu67, which is important for AcrA function, interacts electrostatically with one of the dihydroimidazole rings of NSC 60339 and with a hydroxy group of NSC 227186 (Figure 1).
To probe possible interactions between NSC 60339 and AcrA in the efflux pump, we superimposed our model of monomeric AcrA with NSC 60339 bound in its highest ranking docked pose at the hinge site onto one of the protomers of hexameric AcrA in a pseudoatomic model of the efflux complex.2b This procedure placed the inhibitor at an interprotomer interface such that it would interfere with the AcrB- and TolC-driven hexamerization of AcrA required to assemble the functional complex (Figure 6A). Thus, it is plausible that binding of NSC 60339 to AcrA may prevent or disrupt the structure of the pump. The in vivo proteolysis supports such a mechanism for NSC 60339 (Figure 4). Pretreatment of WT-Pore cells with this compound changed the proteolytic pattern of AcrA, suggesting that NSC 60339 binding indeed changes the structure of AcrA.
In the assembled AcrAB-TolC complex, the MP site of AcrA has two different interfaces with AcrB.2a One site occupies an interprotomer cavity of the AcrB trimer, whereas the MP site of another AcrA subunit interfaces with the porter domain of AcrB, the site of substrate capture and conformational changes in AcrB needed for substrate extrusion (Figure 6A). The top four hits scored relatively high in docking to the MP site, but the docked molecules have different poses and molecular contacts in this site. Several residues predicted to contact NSC 305798 and NSC 33353 or to be in immediate proximity to their binding sites (Figure 6C) were previously identified by mutagenesis and chemical cross-linking as essential for interactions between AcrA and AcrB, as well as in their respective homologues MexA and MexB from P. aeruginosa.28,29 In particular, cysteine residues introduced at positions 219, 249, 319, and 344 in AcrA can be cross-linked directly to AcrB,10b whereas substitutions at G240, V244, and S249 in AcrA enabled its function with a noncognate MexB transporter.29c Thus, binding of compounds at the MP site could interfere with functional interactions between AcrA and AcrB or by preventing conformational changes in AcrA needed for stimulation of the AcrB transporter.
Differences in Functionalities and Molecular Contacts Distinguish Substrates from Inhibitors
With the exception of NSC 305798, the described inhibitors are excellent substrates of the AcrAB-TolC pump, and hence their mechanisms of action could involve interactions with the transporter as well. Surprisingly, the substrate, novobiocin, which does not have an EPI-like activity, also interacts with AcrA (Figure S5), leading to the possibility that AcrA binding sites might be shared by both substrates and inhibitors. Comparison of novobiocin and clorobiocin binding to AcrA by ensemble docking analyses suggests that in both the hinge and MP sites clorobiocin binds more strongly than novobiocin and that the predicted poses and contacts for these two molecules are different (Figure S8). The pyrrole moiety of clorobiocin could play the same functional role in AcrA binding as the imidazoline ring of NSC 60339 (Figure 5). Hence, the distinction between substrate and inhibitor could arise from small differences in molecular contacts of compounds with AcrA and/or AcrB binding sites.
METHODS
Microbiological and Biochemical Assays
E. coli WT-Pore and ΔTolC-Pore strains are derivatives of BW 25113 and GD102.16 Plasmid pAcrABHis carrying the acrAB operon under the native promoter30 was used for construction of AcrA mutants and in the complementation assays. Plasmid pEZ11 expressing a soluble AcrAHis variant under the IPTG-inducible T7-promoter was used for purification of AcrA.31
MICs were analyzed using a 2-fold broth dilution method.32 For the checkerboard assay, an antibiotic and a test compound were serially diluted into 96-well plates as described previously.5b SPR experiments were carried out with the purified AcrA immobilized onto a CM5 chip (Biacore).9b For the in vivo proteolysis, exponentially grown WT-Pore and ΔTolC-Pore cells were permeabilized by inducing the expression of the pore with 0.1% arabinose for 3 h. Cells were treated with increasing concentrations of trypsin, and AcrA fragments were visualized by immunoblotting of total cellular protein.18a The HT uptake assay was performed in a temperature-controlled microplate reader (Tecan Spark 10M) equipped with a sample injector, in fluorescence mode. Data were fitted to extract initial rates as described previously (Krishnamoorthy et al., submitted for publication).
Supplementary Material
Acknowledgments
Funding
This work was supported by National Institutes of Health Grant AI052293 to H.I.Z. and the award for Project HR14-042 from the Oklahoma Center for Advancement of Science and Technology to V.V.R. This research used the TITAN supercomputer at the Oak Ridge Leadership Computing Facility (OLCF) at Oak Ridge National Laboratory, which is supported by the Office of Science of the U.S. Department of Energy under Contract DE-AC05-00OR22725. This paper has been coauthored by UT-Battelle, LLC, under Contract DE-AC05-00OR22725 with the U.S. Department of Energy.
We thank Nam-Chul Ha for sharing the pseudoatomic model of the AcrAB-MacA-TolC hybrid complex and Timothy Opperman (Microbiotix) for MBX2319. We acknowledge Khushboo Bafna for her help with initial AcrA models.
Footnotes
Notes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfec-dis.6b00167.
Computational methods and chemical synthesis; details of docking analyses; expanded results of compound binding to AcrA, in vivo proteolysis, and fluorescence-based efflux assays (PDF)
References
- 1.Nikaido H, Zgurskaya HI. AcrAB and related multidrug efflux pumps of Escherichia coli. J Mol Microbiol Biotechnol. 2001;3(2):215–218. [PubMed] [Google Scholar]
- 2.(a) Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, Venter H, Chiu W, Luisi BF. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509(7501):512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Jeong H, Kim JS, Song S, Shigematsu H, Yokoyama T, Hyun J, Ha NC. Pseudoatomic Structure of the Tripartite Multidrug Efflux Pump AcrAB-TolC Reveals the Intermeshing Cogwheel-like Interaction between AcrA and TolC. Structure. 2016;24(2):272–276. doi: 10.1016/j.str.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 3.(a) Ma D, Cook DN, Alberti M, Pon NG, Nikaido H, Hearst JE. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16(1):45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]; (b) Fralick JA. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178(19):5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Opperman TJ, Nguyen ST. Recent advances toward a molecular mechanism of efflux pump inhibition. Front Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lomovskaya O, Zgurskaya HI, Bostian KA, Lewis K. Multidrug Efflux Pumps: Structure, Mechanism, and Inhibition. In: Wax RG, Lewis K, Salyers AA, Taber H, editors. Bacterial Resistance to Antimicrobials. CRC Press; Boca Raton, FL, USA: 2008. pp. 45–69. [Google Scholar]
- 5.(a) Lomovskaya O, Bostian KA. Practical applications and feasibility of efflux pump inhibitors in the clinic—a vision for applied use. Biochem Pharmacol. 2006;71(7):910–918. doi: 10.1016/j.bcp.2005.12.008. [DOI] [PubMed] [Google Scholar]; (b) Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, Blais J, Cho D, Chamberland S, Renau T, Leger R, Hecker S, Watkins W, Hoshino K, Ishida H, Lee VJ. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob Agents Chemother. 2001;45(1):105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Naka-shima R, Sakurai K, Yamasaki S, Hayashi K, Nagata C, Hoshino K, Onodera Y, Nishino K, Yamaguchi A. Structural basis for the inhibition of bacterial multidrug exporters. Nature. 2013;500(7460):102–106. doi: 10.1038/nature12300. [DOI] [PubMed] [Google Scholar]
- 6.Tikhonova EB, Yamada Y, Zgurskaya HI. Sequential mechanism of assembly of multidrug efflux pump AcrAB-TolC. Chem Biol. 2011;18(4):454–463. doi: 10.1016/j.chembiol.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsumoto Y, Hayama K, Sakakihara S, Nishino K, Noji H, Iino R, Yamaguchi A. Evaluation of Multidrug Efflux Pump Inhibitors by a New Method Using Microfluidic Channels. PLoS One. 2011;6(4):e18547. doi: 10.1371/journal.pone.0018547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinana AD, Vargiu AV, May T, Nikaido H. Aminoacyl beta-naphthylamides as substrates and modulators of AcrB multidrug efflux pump. Proc Natl Acad Sci U S A. 2016;113(5):1405–1410. doi: 10.1073/pnas.1525143113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Zgurskaya HI, Weeks JW, Ntreh AT, Nickels LM, Wolloscheck D. Mechanism of coupling drug transport reactions located in two different membranes. Front Microbiol. 2015;6:100. doi: 10.3389/fmicb.2015.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tikhonova EB, Dastidar V, Rybenkov VV, Zgurskaya HI. Kinetic control of TolC recruitment by multidrug efflux complexes. Proc Natl Acad Sci U S A. 2009;106(38):16416–16421. doi: 10.1073/pnas.0906601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Mikolosko J, Bobyk K, Zgurskaya HI, Ghosh P. Conformational Flexibility in the Multidrug Efflux System Protein AcrA. Structure. 2006;14(3):577–587. doi: 10.1016/j.str.2005.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc Natl Acad Sci U S A. 2009;106(17):7173–7178. doi: 10.1073/pnas.0900693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Xu Y, Lee M, Moeller A, Song S, Yoon BY, Kim HM, Jun SY, Lee K, Ha NC. Funnel-like Hexameric Assembly of the Periplasmic Adapter Protein in the Tripartite Multidrug Efflux Pump in Gram-negative Bacteria. J Biol Chem. 2011;286(20):17910–17920. doi: 10.1074/jbc.M111.238535. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Staron P, Forchhammer K, Maldener I. Structure-function analysis of the ATP-driven glycolipid efflux pump DevBCA reveals complex organization with TolC/HgdD. FEBS Lett. 2014;588(3):395–400. doi: 10.1016/j.febslet.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 12.De Angelis F, Lee JK, O’Connell JD, Miercke LJW, Verschueren KH, Srinivasan V, Bauvois C, Govaerts C, Robbins RA, Ruysschaert JM, Stroud RM, Vandenbussche G. Metal-induced conformational changes in ZneB suggest an active role of membrane fusion proteins in efflux resistance systems. Proc Natl Acad Sci U S A. 2010;107(24):11038–11043. doi: 10.1073/pnas.1003908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgins MK, Bokma E, Koronakis E, Hughes C, Koronakis V. Structure of the periplasmic component of a bacterial drug efflux pump. Proc Natl Acad Sci U S A. 2004;101(27):9994–9999. doi: 10.1073/pnas.0400375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weeks JW, Nickels LM, Ntreh AT, Zgurskaya HI. Non-equivalent roles of two periplasmic subunits in the function and assembly of triclosan pump TriABC from Pseudomonas aeruginosa. Mol Microbiol. 2015;98(2):343–356. doi: 10.1111/mmi.13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Opperman TJ, Kwasny SM, Kim HS, Nguyen ST, Houseweart C, D’Souza S, Walker GC, Peet NP, Nikaido H, Bowlin TL. Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob Agents Chemother. 2014;58(2):722–733. doi: 10.1128/AAC.01866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnamoorthy G, Wolloscheck D, Weeks JW, Croft C, Rybenkov VV, Zgurskaya HI. Breaking the permeability barrier of Escherichia coli by controlled hyperporination of the outer membrane. Antimicrob Agents Chemother. 2016 doi: 10.1128/AAC.01882-16. AAC.01882-16. [DOI] [PMC free article] [PubMed]
- 17.Vaccaro L, Koronakis V, Sansom MS. Flexibility in a drug transport accessory protein: molecular dynamics simulations of MexA. Biophys J. 2006;91(2):558–564. doi: 10.1529/biophysj.105.080010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.(a) Ge Q, Yamada Y, Zgurskaya H. The C-terminal domain of AcrA is essential for the assembly and function of the multidrug efflux pump AcrAB-TolC. J Bacteriol. 2009;191(13):4365–4371. doi: 10.1128/JB.00204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Modali SD, Zgurskaya HI. The periplasmic membrane proximal domain of MacA acts as a switch in stimulation of ATP hydrolysis by MacB transporter. Mol Microbiol. 2011;81(4):937–951. doi: 10.1111/j.1365-2958.2011.07744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(a) Ellingson SR, Miao Y, Baudry J, Smith JC. Multi-conformer ensemble docking to difficult protein targets. J Phys Chem B. 2015;119(3):1026–1034. doi: 10.1021/jp506511p. [DOI] [PubMed] [Google Scholar]; (b) Ellingson SR, Smith JC, Baudry J. VinaMPI: Facilitating multiple receptor high-throughput virtual docking on high-performance computers. J Comput Chem. 2013;34(25):2212–2221. doi: 10.1002/jcc.23367. [DOI] [PubMed] [Google Scholar]
- 20.(a) Eustaquio AS, Gust B, Luft T, Li S-M, Chater KF, Heide L. Clorobiocin Biosynthesis in Streptomyces: Identification of the Halogenase and Generation of Structural Analogs. Chem Biol. 2003;10(3):279–288. doi: 10.1016/s1074-5521(03)00051-6. [DOI] [PubMed] [Google Scholar]; (b) Anderle C, Stieger M, Burrell M, Reinelt S, Maxwell A, Page M, Heide L. Biological Activities of Novel Gyrase Inhibitors of the Aminocoumarin Class. Antimicrob Agents Chemother. 2008;52(6):1982–1990. doi: 10.1128/AAC.01235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorian V. Antibiotic in Laboratory Medicine. 4. Williams & Wilkins; Baltimore, MD, USA: 1996. [Google Scholar]
- 22.Yesair DW, Kohner FA, Rogers WI, Baronowsky PE, Kensler CJ. Relationship of Phthalanilide-Lipid Complexes to Uptake and Retention of 2-Chloro-4′,4″-di(2-imidazolin-2-yl)terephthalanilide (NSC 60339) by Sensitive and Resistant P388 Leukemia Cells. Cancer Res. 1966;26(2 Part 1):202–207. [PubMed] [Google Scholar]
- 23.Hagmann WK, Springer MS. Substituted aminoquinolines as modulators of chemokine receptor activity. Google Patents 1999 [Google Scholar]
- 24.Karle JM, Karle IL. Crystal and molecular structure of the antimalarial agent enpiroline. Antimicrob Agents Chemother. 1989;33(7):1081–1089. doi: 10.1128/aac.33.7.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sjuts H, Vargiu AV, Kwasny SM, Nguyen ST, Kim HS, Ding X, Ornik AR, Ruggerone P, Bowlin TL, Nikaido H, Pos KM, Opperman TJ. Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc Natl Acad Sci U S A. 2016;113:3509–3514. doi: 10.1073/pnas.1602472113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coldham NG, Webber M, Woodward MJ, Piddock LJV. A 96-well plate fluorescence assay for assessment of cellular permeability and active efflux in Salmonella enterica serovar Typhimurium and Escherichia coli. J Antimicrob Chemother. 2010;65(8):1655–1663. doi: 10.1093/jac/dkq169. [DOI] [PubMed] [Google Scholar]
- 27.Stavri M, Piddock LJ, Gibbons S. Bacterial efflux pump inhibitors from natural sources. J Antimicrob Chemother. 2007;59(6):1247–1260. doi: 10.1093/jac/dkl460. [DOI] [PubMed] [Google Scholar]
- 28.Weeks JW, Bavro VN, Misra R. Genetic assessment of the role of AcrB beta-hairpins in the assembly of the TolC-AcrAB multidrug efflux pump of Escherichia coli. Mol Microbiol. 2014;91(5):965–975. doi: 10.1111/mmi.12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.(a) Weeks JW, Celaya-Kolb T, Pecora S, Misra R. AcrA suppressor alterations reverse the drug hypersensitivity phenotype of a TolC mutant by inducing TolC aperture opening. Mol Microbiol. 2010;75(6):1468–1483. doi: 10.1111/j.1365-2958.2010.07068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Nehme D, Poole K. Interaction of the MexA and MexB components of the MexAB-OprM multidrug efflux system of Pseudomonas aeruginosa: identification of MexA extragenic suppressors of a T578I mutation in MexB. Antimicrob Agents Chemother. 2005;49(10):4375–4378. doi: 10.1128/AAC.49.10.4375-4378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Krishnamoorthy G, Tikhonova EB, Zgurskaya HI. Fitting periplasmic membrane fusion proteins to inner membrane transporters: mutations that enable Escherichia coli AcrA to function with Pseudomonas aeruginosa MexB. J Bacteriol. 2008;190(2):691–698. doi: 10.1128/JB.01276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tikhonova EB, Zgurskaya HI. AcrA, AcrB, and TolC of Escherichia coli Form a Stable Intermembrane Multidrug Efflux Complex. J Biol Chem. 2004;279(31):32116–32124. doi: 10.1074/jbc.M402230200. [DOI] [PubMed] [Google Scholar]
- 31.Zgurskaya HI, Nikaido H. AcrA is a highly asymmetric protein capable of spanning the periplasm. J Mol Biol. 1999;285:409–420. doi: 10.1006/jmbi.1998.2313. [DOI] [PubMed] [Google Scholar]
- 32.Tikhonova EB, Wang Q, Zgurskaya HI. Chimeric Analysis of the Multicomponent Multidrug Efflux Transporters from Gram-Negative Bacteria. J Bacteriol. 2002;184(23):6499–6507. doi: 10.1128/JB.184.23.6499-6507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.