ABSTRACT
Many relevant applications have been demonstrated for chitinolytic enzymes. However, their successful exploitation depends upon the availability of strains and expression conditions that allow the production of active forms and large quantities of these enzymes. Escherichia coli has been commonly used to express and overproduce different proteins, among them chitinases. Improving the functional gene expression of chitinases is key to exploiting their potential. In a recent study, we described the effect of various parameters on the functional expression of 2 chitinases from different families, demonstrating that the effect of each of these parameters on the activity of both chitinases was specific to each enzyme. In this study, the expression of a Lactococcus lactis chitinase encoded by a new allele, ChiA1-2, was optimized. The results showed that not only the expression parameters seemed to influence protein production, solubility and activity but also the plasmid used for the expression. Herein, we describe the effect of 2 different promoters, tac and T7, on the expression of the active form of the chitinolytic enzyme.
KEYWORDS: heterologous expression, promoters, recombinant chitinase
Introduction
Chitinases are enzymes that hydrolyse β-1, 4-glycosidic bonds of chitin, a linear β-1, 4-linked homopolymer of N-acetylglucosamine that is very difficult to degrade.1,2 Chitinases are very diverse and belong to family 18 and 19 glycosyl hydrolases. They are classified based on the amino acid sequence of the catalytic modules, according to the CAZy database.3 A broad range of organisms, including bacteria, fungi, insects, plants and animals, produce these enzymes, and they are utilised in nutrition, morphogenesis or defense.1
Traditionally, chitinases have attracted attention because of their important potential applications, such as the production of specific sized bioactive chito-oligosaccharides of interest to the pharmaceutical and food industry, and their possible use as biocontrol agents against fungal, insect and nematode pests.2,4,5 Recently, new applications of chitinases, including uses in aquaculture for fish protection6 and bioethanol production, have been reported.7
The successful use of chitinases in the above-mentioned applications depends on the availability of strains and expression conditions that permit the production of active forms and large quantities of these enzymes.8 Improving the functional gene expression of chitinases is essential to achieve this objective. Escherichia coli is frequently used to express and overproduce many proteins, including chitinases,2 because of the characteristics of its expression system.9-11 Recently, we described the effect of the host strain, culture cell density, inducer concentration, post-induction time and induction temperature on the functional expression of 2 chitinases from different families and prokaryote domains and showed that the effect of each parameter on the activity of both chitinases was specific to each enzyme.12 This study describes the effect of 2 different promoters, tac and T7, on the expression of a Lactococcus lactis chitinolytic enzyme.
Results and discussion
In this work, optimization of the expression of a chitinase from L. lactis CECT 185 in E. coli was carried out. To this end, the ChiA1 gene in this strain was amplified using primers designed based on the gene sequence NP_268107.1 deposited in the database for L. lactis strain IL1403,13 whose chitinolytic activity had been previously demonstrated and characterized.14 A fragment of 1476 bp encoding a protein of 492 amino acids was amplified. The protein analysis using Pfam revealed the modular structure of the chitinolytic enzyme, as described by Vaaje-Kolstad et al.14 However, the sequence analysis of the protein showed that it differed in 4 amino acids from L. lactis strain IL1403. The first of these changes, an alanine (A) at position 12 instead of valine (V), is situated in the signal peptide of the protein. Two other changes, a valine (V) at position 149 instead of isoleucine (I) and threonine (T) at position 268 instead of serine (S), are located in the catalytic domain. Finally, in the case of the amino acid occupying position 479, glutamine (Q) is replaced by leucine (L) (i.e. a neutral and polar amino acid is replaced by other neutral and non-polar amino acids). This substitution is located in the chitin-binding domain. The alignment of the chitinase of the L. lactis CECT 185 strain with that of other strains of the same species showed that these changes also appeared in a strain involved in malolactic fermentation of wine isolated in our laboratory (VINI30) and in strains L. lactis KF147 (accession number ADA65702.1) and L. lactis NCDO 2118 (accession number AII13508.1). Due to these differences, the allele of the strain L. lactis CECT 185 was named ChiA1-2.
To optimize the production of this chitinase, the ChiA1-2 gene was cloned into E. coli expression vectors pGEX4T-2 (carrier of the tac promoter) and pET41 EK/LIC (carrier of the T7 promoter) generating, by transformation, the recombinant E. coli strains BL21ChiA1-2Ll1 (in which the gene is expressed under the control of the tac promoter) and BL21ChiA1-2Ll2 (in which the gene is expressed under the control of the T7 promoter).
The level of recombinant protein expression in E. coli is affected by the strength of the expression system, which depends on the strength of the promoter used and the number of copies of the plasmid15 as well as other parameters that affect the expression, mainly the cell density, temperature, inducer concentration and induction time.16-18 However, very high levels of expression can lead to large amounts of proteins that cannot be processed correctly and that accumulate in inclusion bodies in an inactive form.19,20 As a result, a reduction in expression levels frequently results in a higher amount of active protein.21 Therefore, for optimum production, the expression levels must be modulated and adjusted.22,23
The enzymatic activity of the protein is indicative of proper folding and solubility. In this study, the conditions required to obtain the greatest amount of active protein in the most efficient manner were established. To do this, the expression levels of the enzyme were evaluated by testing 3 different cell densities at the time of induction, 3 concentrations of inducer and 3 post-induction times. The assays were carried out by varying one of the parameters and maintaining the others under the following standard conditions: OD600 of 0.8, isopropyl thio-β-D-galactoside (IPTG) concentration of 0.5 mM, induction time of 4 h, temperature of 37°C and agitation of 250 rpm. The results obtained with each of the 2 recombinant strains constructed are shown in Table 1. The results confirmed that the ChiA1-2 allele encoded a functional protein. They also demonstrated that the production levels of the active protein were dependent on the initial concentration of the cells, inducer concentration and induction time, all of which were optimized for each chitinase, as described and discussed for other chitinases in our previous work.12 Importantly, the findings also highlighted that under all the conditions tested, the levels of active protein were considerably higher when using the BL21ChiA1-2Ll1 strain (in which the gene was expressed from the tac promoter) than when using the BL21ChiA1-2Ll2 strain (in which the gene was expressed from the strong T7 promoter). These results contrast with the findings of the sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis showed in Fig. 1, which revealed higher production of the recombinant protein when using the BL21ChiA1-2Ll2 strain than the BL21ChiA1-2Ll1 strain. The findings indicated that less protein was synthesized with a weaker promoter due to lower levels of mRNA but that correct folding of the protein was more efficient under these conditions, resulting in higher levels of active enzyme. They also confirmed that the effects of the plasmid used on the synthesized active protein were more important than variations in other parameters that affected protein expression.
Table 1.
Effect of the promoters and different induction conditions on the expression of the active form of the chitinase encoded by the ChiA1-2 gene.
| Activity (U/mL) | ||
|---|---|---|
| Parameter analyzed | BL21ChiA1-2Ll1a | BL21ChiA1-2Ll2b |
| OD600 nm | ||
| 0.5 | 240.7 ± 0.4 | 38.4 ± 2.7 |
| 0.8 | 98.7 ±2 .4 | 55.8 ± 1.1 |
| 1.2 | 95.1 ± 0.8 | 66.3 ± 4 |
| IPTG concentration (mM) | ||
| 0.12 | 89.4 ± 5 | 62 ± 4.3 |
| 0.25 | 108 ± 0.4 | 56.8 ± 1.2 |
| 0.5 | 98.7 ± 1.5 | 55.8 ± 1.1 |
| Post-induction time (h) | ||
| 1 | 152.7 ± 6 | 45.2 ± 0.9 |
| 5 | 98.6 ± 0.5 | 55.8 ± 2.8 |
| 24 | 66.2 ± 5.2 | 33 ± 0.4 |
Expressing the gene from the tac promoter.
Expressing the gene from the T7 promoter.
Figure 1.
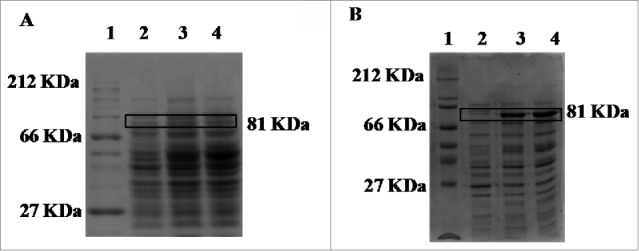
SDS-PAGE showing the expression of the ChiA1-2 gene using BL21ChiA1-2Ll1 (A) and BL21ChiA1-2Ll2 (B) strains induced with IPTG. Lane 1: molecular weight protein marker (A: Hide Range Sigma Marker™, Sigma-Aldrich; B: Protein Marker, Broad Range, Biolabs); lane 2: cell extract before induction; lanes 3–4: cell extract 2 and 4 h, respectively, after induction. The recombinant protein fused with the GST tag is boxed.
As shown by the comparison of the results obtained with the 2 recombinant strains (Table 1), the highest levels of active enzyme using plasmid pGEX4-T2 (with the tac promoter) were achieved under conditions of lower cell densities and shorter induction times than under optimized expression conditions using the pET41 EK/LIC vector (with the T7 promoter). In addition, none of the the optimized expression conditions using the BL21ChiA1-2Ll2 strain (expressing the gene from the T7 promoter) led to expression levels of active protein comparable to those obtained with the BL21ChiA1-2Ll1 strain (expressing the gene from the tac promoter). Under the optimum conditions employed in this study, this allowed 3.6 times more active protein to be obtained using the BL21ChiA1-2Ll1 strain than using the BL21ChiA1-2Ll2 strain. These results suggest that processes conducted with the recombinant BL21ChiA1-2Ll1 strain would result in a higher yield of active enzyme in shorter times, making this process economically more profitable.
Materials and methods
Plasmids, bacterial strains and culture conditions
The plasmids and strains used in this study are shown in Table 2.
Table 2.
Strains and plasmids used in this study.
| Strain | Source | Genotype/characteristics |
|---|---|---|
| L. lactis sp. lactis CECT 185 | CECTa | Wild type |
| E. coli One Shot® TOP10 | Invitrogen | F− mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15ΔlacX74 recA1 araD139Δ(ara-leu)7697 gal/U galK rpsL (StrR) endA1 nupG |
| E. coli BL21 (DE3) | Invitrogen | F− ompT gal dcm lon hsdSB(rB− mB−) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) |
| E. coli BL21 Star (DE3) | Invitrogen | F− ompT hsdSB (rB− mB−) gal dcm rne131 (DE3) |
| BL21ChiA1-2Ll1 | This study | E. coli BL21 (DE3) transformed with pGEX-4T-2-ChiA1-2Llp recombinant vector |
| BL21ChiA1-2Ll2 | This study | E. coli BL21 Star (DE3) transformed with pET41-ChiA1-2Llp recombinant vector |
| Plasmid | ||
| pCR®- Blunt | Invitrogen | Cloning vector. Confers zeocin and kanamycin resistance |
| pGEX4T-2 | Amersham | IPTG inducible expression vector under control of the tac promoter. Introduces a GST tag at the N-terminus of the expression protein. Confers ampicillin resistance |
| pET41 EK/LIC | Novagen | IPTG inducible expression vector under control of the T7 promoter. Introduces a GST tag at the N-terminus of the expression protein. Confers kanamycin resistance |
| pTOPChiA1-2Llp | This study | pCR®- Blunt vector containing the ChiA1-2 gene |
| pGEX-4T-2-ChiA1-2Llp | This study | pGEX4T-2 vector containing the ChiA1-2 gene. |
| pET41-ChiA1-2Llp | This study | pET41 EK/LIC vector containing the ChiA1-2 gene. |
CECT: Spanish Type Culture Collection.
E. coli strains were cultured at 37° C, with shaking in Luria–Bertani (LB) medium. The media were supplemented with ampicillin (100 µg/mL) or kanamycin (50 µg/mL) when required. The media were solidified by adding 20 g/L of agar when necessary.
Cloning of L. lactis ChiA1-2 gene
Recombinant DNA was performed as described by Sambrook and Russell.24 DNA from L. lactis was isolated using a GFX Genomic Blood DNA Purification kit (Amersham Biosciences UK, Ltd.). The ChiA1-2 gene of L. lactis was amplified by PCR using the genomic DNA as a template and primers based on the sequence of the open reading frame NP_268107.1. To amplify the gene cloned into pET-41 Ek/LIC, i) the forward primer LLacchiA1FW (5′-GACGACGACAAGATGATTTCAGTGAAAAAACGTAGAGA-3′) and reverse primer LLacchiA1REV (5′-GAGGAGAAGC CCGGTTATAGCTTTTTCCATGGACCAAAAT-3′) were employed, using a kit from Novagen (Darmstadt, Germany), following the manufacturer's instructions; ii) the forward primer LLchitFW (5′-GGATCC ATGATTTCAGTGAAAAAACGTAGAGA-3′) and reverse primer LLchitREV (5′-CTCGAGTTATAGCTTTTTCCATGGACCAAAAT-3′) were used to amplify the gene cloned into pGEX-4T-2. The underlined bases are the BamHI and XhoI restriction recognition sites, respectively. The amplified product was purified using a Geneclean kit (Qbiogene/MP Biomedicals, LLC) and cloned first into a pCR®- Blunt vector (Invitrogen, Carlsbad, CA) following the instructions of the manufacturer, generating the recombinant plasmid pTOPChiA1-2Llp. The BamHI and XhoI fragment from the vector pTOPChiA1-2Llp was ligated into a pGEX-4T-2 vector digested with the same enzymes. The thermal conditions for PCR amplification were an initial denaturation step for 1 min at 95°C followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 60°C for 90 s, extension at 72°C for 1 min and finished with a final extension at 72°C for 5 min. The two recombinant constructs obtained (pGEX-4T-2-ChiA1-2Llp and pET41-ChiA1-2Llp) were used to transform the BL21 (DE3) and BL21 Star (DE3) E. coli strains, respectively, and the recombinant strains selected in LB medium supplemented with the appropriate antibiotic (Table 2).
DNA and protein sequence analysis
DNA and protein sequences comparison were done using the BLAST 2.0 program and the NCBI database. Protein alignments were carried out using the ClustalW 2.0.12 program in EMBL-EBI. Protein sequence analysis was done with Pfam and ExPASy proteomic tools.
Protein expression and chitinolytic activity
Protein expression and analysis, as well as the determination of chitinase activity, were carried out as described previously.12 The samples were dialysed against 50 mM phosphate buffer (pH 7.6). One unit (U) of enzyme activity was defined as the amount of enzyme releasing 1 nmol of 4-nitrophenol in 1 h. Expression experiments and enzymatic assays were carried out in triplicate, and the results were expressed as the mean ± SD.
Disclosure of potential conflict of interest
No potential conflicts of interest were disclosed.
Funding
This work was funded by Grant 10PXIB310278PR (Xunta de Galicia, Spain).
References
- [1].Adrangi S, Faramarzi MA. From bacteria to human: a journey into the world of chitinases. Biotechnol Adv 2013; 31:1786-95; PMID:24095741; https://doi.org/ 10.1016/j.biotechadv.2013.09.012 [DOI] [PubMed] [Google Scholar]
- [2].Hamid R, Khan MA, Ahmad M, Ahmad MM, Abdin MZ, Musarrat J, Javed S. Chitinases: an update. J Pharm Bioallied Sci 2013; 5:21-9; PMID:23559820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 2009; 37:D233-8; PMID:18838391; https://doi.org/ 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yang J, Liang L, Li J, Zhang KQ. Nematicidal enzymes from microorganisms and their applications. Appl Microbiol Biotechnol 2013; 97:7081-95; PMID:23832084; https://doi.org/ 10.1007/s00253-013-5045-0 [DOI] [PubMed] [Google Scholar]
- [5].Nagpure A, Choudhary B, Gupta RK. Chitinases: In agriculture and human healthcare. Crit Rev Biotechnol 2014; 34:215-32; PMID:23859124; https://doi.org/ 10.3109/07388551.2013.790874 [DOI] [PubMed] [Google Scholar]
- [6].Zhang Y, Zhou Z, Liu Y, Cao Y, He S, Huo F, Qin C, Yao B, Ringo E. High-yield production of a chitinase from Aeromonas veronii B565 as a potential feed supplement for warm-water aquaculture. Appl Microbiol Biotechnol 2014; 98:1651-62; PMID:23775269; https://doi.org/ 10.1007/s00253-013-5023-6 [DOI] [PubMed] [Google Scholar]
- [7].Inokuma K, Takano M, Hoshino K. Direct ethanol production from N-acetylglucosamine and chitin substrates by Mucor species. Biochem Eng J 2013; 72:24-32; https://doi.org/ 10.1016/j.bej.2012.12.009 [DOI] [Google Scholar]
- [8].Liu L, Yang H, Shin H, Chen RR, Li J, Du G, Chen J. How to achieve high-level expression of microbial enzymes. Strategies and perspectives. Bioengineered 2013; 4:212-23; PMID:23686280; https://doi.org/ 10.4161/bioe.24761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hannig G, Makrides SC. Strategies for optimizing heterologous protein expression in Escherichia coli. Trends Biotechnol 1998; 16:54-60; PMID:9487731; https://doi.org/ 10.1016/S0167-7799(97)01155-4 [DOI] [PubMed] [Google Scholar]
- [10].Waegeman H, Soetaert W. Increasing recombinant protein production in Escherichia coli through metabolic and genetic engineering. J Ind Microbiol Biotechnol 2011; 38:1891-910; PMID:21901404; https://doi.org/ 10.1007/s10295-011-1034-4 [DOI] [PubMed] [Google Scholar]
- [11].Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol 2014; 5:1-17; PMID:24478763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].García-Fraga B, da Silva AF, López-Seijas J, Sieiro C. Optimized expression conditions for enhancing production of two recombinant chitinolytic enzymes from different prokaryote domains. Bioprocess Biosyst Eng 2015; 38:2477-86; https://doi.org/ 10.1007/s00449-015-1485-5 [DOI] [PubMed] [Google Scholar]
- [13].Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, Ehrlich SD, Sorokin A. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res 2001; 11:731-53; PMID:11337471; https://doi.org/ 10.1101/gr.GR-1697R [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Vaaje-Kolstad G, Bunaes AC, Mathiesen G, Eijsink VG. The chitinolytic system of Lactococcus lactis ssp. lactis comprises a nonprocessive chitinase and a chitin-binding protein that promotes the degradation of α- and β-chitin. FEBS J 2009; 276:2402-15; PMID:19348025; https://doi.org/ 10.1111/j.1742-4658.2009.06972.x [DOI] [PubMed] [Google Scholar]
- [15].Keasling JD. Gene-expression tools for the metabolic engineering of bacteria. Trends Biotechnol 1999; 17:452-60; PMID:10511704; https://doi.org/ 10.1016/S0167-7799(99)01376-1 [DOI] [PubMed] [Google Scholar]
- [16].Balbás P. Understanding the art of producing protein and nonprotein molecules in Escherichia coli. Mol Biotechnol 2001; 19:251-67; https://doi.org/ 10.1385/MB:19:3:251 [DOI] [PubMed] [Google Scholar]
- [17].Tolia NH, Joshua-Tor L. Strategies for protein coexpression in Escherichia coli. Nat Methods 2006; 3:55-64; PMID:16369554; https://doi.org/ 10.1038/nmeth0106-55 [DOI] [PubMed] [Google Scholar]
- [18].Rodríguez-Carmona E, Cano-Garrido O, Dragosits M, Maurer M, Mader A, Kunert R, Mattanovich D, Villaverde A, Vázquez F. Recombinant Fab expression and secretion in Escherichia coli continuous culture at medium cell densities: Influence of temperature. Process Biochem 2012; 47:446-52; https://doi.org/ 10.1016/j.procbio.2011.11.024 [DOI] [Google Scholar]
- [19].Kane JF, Hartley DL. Properties of recombinant protein containing inclusion bodies in Escherichia coli. Bioprocess Technol 1991; 12:121-45; PMID:1369303 [PubMed] [Google Scholar]
- [20].Baig F, Fernando LP, Salazar MA, Powell RR, Bruce TF, Harcum SW. Dynamic transcriptional response of Escherichia coli to inclusion body formation. Biotechnol Bioeng 2014; 111:980-99; PMID:24338599; https://doi.org/ 10.1002/bit.25169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].de Marco A. Recombinant polypeptide production in Escherichia coli: towards a rational approach to improve the yields of functional proteins. Microbial Cell Fact 2013; 12:101; https://doi.org/ 10.1186/1475-2859-12-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tagkopoulos I. Microbial factories under control: auto-regulatory control through engineered stress-induced feedback. Bioengineered 2013; 4:5-8; PMID:22922761; https://doi.org/ 10.4161/bioe.21935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Marschall L, Sagmeister P, Herwig Ch. Tunable recombinant protein expression in Escherichia coli: enabler for continuous processing? Appl Microb Biotechnol 2016; 100:5719-28; https://doi.org/ 10.1007/s00253-016-7550-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sambrook J, Russell DW. 2001. Molecular cloning: A laboratory manual (3rd Ed.). Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press. [Google Scholar]


