Abstract
A characteristic feature of the order Rhodobacterales is the presence of a large number of photoautotrophic and photoheterotrophic species containing bacteriochlorophyll. Interestingly, these phototrophic species are phylogenetically mixed with chemotrophs. To better understand the origin of such variability, we sequenced the genomes of three closely related haloalkaliphilic species, differing in their phototrophic capacity and oxygen preference: the photoheterotrophic and facultatively anaerobic bacterium Rhodobaca barguzinensis, aerobic photoheterotroph Roseinatronobacter thiooxidans, and aerobic heterotrophic bacterium Natronohydrobacter thiooxidans. These three haloalcaliphilic species are phylogenetically related and share many common characteristics with the Rhodobacter species, forming together the Rhodobacter-Rhodobaca (RR) group. A comparative genomic analysis showed close homology of photosynthetic proteins and similarity in photosynthesis gene organization among the investigated phototrophic RR species. On the other hand, Rhodobaca barguzinensis and Roseinatronobacter thiooxidans lack an inorganic carbon fixation pathway and outer light-harvesting genes. This documents the reduction of their photosynthetic machinery towards a mostly photoheterotrophic lifestyle. Moreover, both phototrophic species contain 5-aminolevulinate synthase (encoded by the hemA gene) incorporated into their photosynthesis gene clusters, which seems to be a common feature of all aerobic anoxygenic phototrophic Alphaproteobacteria. Interestingly, the chrR-rpoE (sigma24) operon, which is part of singlet oxygen defense in phototrophic species, was found in the heterotrophic strain Natronohydrobacter thiooxidans. This suggests that this organism evolved from a photoheterotrophic ancestor through the loss of its photosynthesis genes. The overall evolution of phototrophy among the haloalkaliphilic members of the RR group is discussed.
Keywords: Anoxygenic photosynthesis, bacteriochlorophyll, horizontal gene transfer, photosynthesis gene cluster, Rhodobacteraceae
Introduction
The order Rhodobacterales (Alphaproteobacteria) encompasses a highly diverse ensemble of species (Simon et al. 2017). They largely differ in their phenotype, metabolic traits, and ecological niches they inhabit. Its members conduct many fundamental metabolic processes such as aerobic respiration, anaerobic fermentation, sulfur oxidation, autotrophic carbon fixation, nitrogen fixation, or hydrogen production in various combinations (Garrity et al. 2005; Androga et al. 2012). One of the most interesting features of Rhodobacterales is the presence of many species that perform anoxygenic photosynthesis. Members of the Rhodobacter (Rba.) and Rhodovulum (Rdv.) genera represent classical examples of purple nonsulfur bacteria—organisms capable of photoautotrophic growth under anaerobic conditions utilizing the Calvin cycle, or heterotrophic growth in the presence of oxygen (Androga et al. 2012). On the other hand, there exist many marine photoheterotrophic species belonging to the so called Roseobacter group, which grow and conduct photosynthesis in the presence of oxygen (Wagner-Döbler and Biebl 2006; Moran et al. 2007; Brinkhoff et al. 2008), do not contain RuBisCO and use light only as an additional energy source (Koblížek et al. 2013).
In the last 20 years, seven haloalkaliphilic photoheterotrophic species were isolated from several saline and soda lakes in Africa, America, Asia, and Antarctica. Remarkably, these organisms are phylogenetically closer to freshwater Rhodobacter species than to marine photoheterotrophs. Rhodobaca (Rca.) bogoriensis was isolated from soda lakes in the East African Rift Valley (Milford et al. 2000), and is capable of both aerobic and anaerobic growth on organic substrates, however it does not contain Calvin cycle enzymes and thus cannot grow photoautotrophically. A closely related organism with the same physiology, Rca. barguzinensis, was later isolated from a small soda lake in the Barguzin valley in south-eastern Siberia (Boldareva et al. 2008). In contrast, the closely related aerobic anoxygenic phototrophic (AAP) bacterium Roseinatronobacter (Rna.) thiooxidans, isolated from a soda lake in the Kunkurskaya steppe (Chita region, south-easter Siberia) grows solely under aerobic conditions (Sorokin et al. 2000b; Stadnichuk et al. 2009). Later, a very similar species, Rna. monicus was isolated from the hypersaline Soda Mono lake in California, USA (Boldareva et al. 2006). Closely related to the above mentioned species are two Antarctic AAP isolates Roseibaca ekhonensis and Roseicitreum antarcticum, which were collected from Lake Ekho (Labrenz et al. 2009), and sandy intertidal sediments (Yu et al. 2011), respectively. The last described AAP bacterium in this group is Roseibacula alcaliphilum, which was isolated from Lake Doroninskoe, East Transbaikal region, Russia (Boldareva and Gorlenko 2014). This photoheterotrophic group is complemented by some nonphototrophic species such as the haloalkaliphilic heterotrophic bacterium Natronohydrobacter (Nhb.) thiooxidans isolated from a soda lake in Kenya.
Based on the close phylogenetic proximity of photoautrophic, photoheterotrophic, and lithoautotrophic species, Keppen et al. (2013) suggested that the photoheterotrophic species such as Rhodobaca and Roseinatronobacter represent the intermediate species on the regressive evolutionary pathway leading from photoautotrophic purple nonsulfur bacteria to heterotrophic species. A similar scenario has also been proposed for photoheterotrophic and heterotrophic species in the marine Roseobacter group (Koblížek et al. 2013). However, the existence of photoheterotrophic species can be explained using an alternative scenario where the originally heterotrophic species acquired their photosynthesis genes via horizontal gene transfer (HGT). A typical feature of purple phototrophic bacteria is that most of their genes involved in the light-phase of photosynthesis are organized in photosynthesis gene clusters (PGCs) (Zsebo and Hearst 1984; Zheng et al. 2011). It has been speculated that clustering of photosynthesis genes in PGCs can facilitate HGT. In line with this hypothesis is the report that the whole PGC was likely transferred between Proteobacteria and Gemmatimonadetes (Zeng et al. 2014). Rhodobacterales species often carry various extra chromosomal elements (Petersen et al. 2013), transposable elements (Vollmers et al. 2013), or gene transfer agents (Luo and Moran 2014), which can facilitate gene transfer. Moreover, PGC-containing plasmids have been identified in some Roseobacter-group representatives (Petersen et al. 2013). HGT has also been proposed for puf genes encoding the protein subunits of the bacterial reaction centers within the Roseobacter group (Koblížek et al. 2015).
To elucidate the most likely evolutionary pathway, we decided to sequence genomes of three closely related (on average 98.7% 16S rRNA pairwise similarity) haloalkaliphilic organisms differing in their metabolic capacities and oxygen preference. Photoheterotrophic Rca. barguzinensis strain alga05 exhibits both aerobic and anaerobic growth. It lacks RuBisCO and nitrogenase, but it has the capacity to utilize thiosulfate and sulfide as electron donors (Boldareva et al. 2008). The second organism, Rna. thiooxidans strain ALG1, is a photoheterotrophic bacterium combining aerobic respiration and photophosphorylation, but it can also oxidize thiosulfate and sulfide during aerobic lithoheterotrophic growth (Sorokin et al. 2000b). The last organism, Nhb. thiooxidans strain AH01, is a strictly aerobic, nonphototrophic bacterium. It is capable of growing lithoheterotrophically with acetate and thiosulfate, sulfide, polysulfide, and elemental sulfur which are oxidized to sulfate, and in addition it can oxidize H2 (Sorokin et al. 2000a). We mostly focused on the presence of photosynthesis genes, their organization and phylogeny in order to test the two proposed evolutionary scenarios.
Materials and Methods
Bacterial Strains and Cultivation Conditions
Rna. thiooxidans strain ALG1, Rca. barguzinensis strain alga05 and Nhb. thiooxidans strain AH01 were obtained from the UNIQEM Culture Collection of the Winogradsky Institute of Microbiology, Russian Academy of Sciences. The phototrophic strains were grown under aerobic conditions in the presence of light at 28 °C in 250 ml Erlenmeyer flasks with 100 ml of medium under shaking (150 rpm). The media used was as described in Sorokin et al. (2000b) and Boldareva et al. (2008). The chemotrophic strain AH01 was grown in a 5 l gas bottle with 1 l of acetate-containing medium and with a 20% H2/80% air gas phase (Sorokin et al. 2000a). The purity of the cultures was controlled by repeated streaking of the individual colonies formed on the medium solidified with 2% (wt/vol) agar and verified by microscopy.
Genome Sequencing and Data Analyses
About 45 ml of dense culture was harvested by centrifugation at 13,000 × g for 5 min. The genomic DNA was extracted and purified using a commercial kit (Tiangen Biotech, Beijing, China). DNA quantity and quality were determined using a NanoDrop ND-1000. The identity of the cultures was verified by amplification and sequencing their 16S rRNA genes. The whole-genome shotgun sequencing was performed using the Illumina HiSeq 2000 platform and a Roche 454 GS FLX+ system at Macrogen (Seoul, South Korea). Procedures for DNA shearing, library preparation and quality control, sample loading, and sequencer operation were performed according to Macrogen’s standard protocols (https://dna.macrogen.com).
Raw reads produced from both types of whole-genome shotgun sequencing were trimmed and de novo assembled into contigs using CLC Genomics Workbench 9.0 software (QIAGEN Aarhus, Denmark). To decrease the number of final contigs, they were further processed using the “Genome Finishing Module” of the CLC software.
Annotation of contigs was performed with the RAST server (Aziz et al. 2008). The obtained genomic data was compared with other Rhodobacterales species [Rba. sphaeroides 2.4.1, Rba. capsulatus SB 1003, Rdv. sulfidophilum DSM 2351, Paracoccus (Pco.) denitrificans PD 1222] obtained from the NCBI GenBank.
To detect clustered regularly interspaced short palindromic repeats (CRISPR) in studied genomes, we used the web-based tool CRISPRFinder (Grissa et al. 2007).
Orthologous gene cluster analysis was performed using the OrthoVenn web server (Wang et al. 2015). Pairwise sequence similarities between all input protein sequences were calculated with an e-value cut-off of 1e−5. An inflation value (−I) of 1.5 was used to define orthologous cluster structure.
Phylogenetic Analyses
16S rRNA gene sequences obtained from GenBank™ were aligned using ClustalX version 2.1. Ambiguously aligned regions and gaps were excluded from further analyses using Gblocks (Talavera and Castresana 2007). Phylogenetic trees based on 16S rRNA gene sequences were constructed using PhyML/MEGA 6.0 software using a maximum likelihood (ML) algorithm with a HKY85 nucleotide substitution model and a bootstrap of 1,000×. For the BchIDHLNB concatenated tree, amino acid sequences were retrieved from GenBank™ and aligned using ClustalX version 2.1. The sites containing gaps and ambiguously aligned regions were stripped from the final alignment using Gblocks. Amino acid sequence alignments of each peptide were concatenated with Geneious version 8.1.2 (Biomatters Ltd.). The 5-aminolevulinate synthase (ALAS) phylogenetic tree involves amino acid sequences derived by BLASTing from ALAS sequences of Dinoroseobacter (Drb.) shibae (Dshi_3546, Dshi_1182, and Dshi_3190). Phylogenetic trees were inferred by MEGA 6.0 software using ML algorithm with LG model. 16S rRNA sequence pairwise similarities were calculated using GGDC tool under the recommended settings (Meier-Kolthoff et al. 2013).
Sequence Accession Numbers
The Rca. barguzinensis strain alga05, Rna. thiooxidans strain ALG1 and Nhb. thiooxidans strain AH01 genome sequences have been deposited at NCBI GenBank under the accession numbers: GCA_001870665.1, GCA_001870675.1, and GCA_001884735.1.
Results
The sequencing and assembly of the genomes of the three studied organisms resulted in draft assemblies consisting of 5–52 contigs (table 1). The smallest genome (∼3.5 Mb) was found in Rna. thioxidans ALG1, while Rca. barguzinensis alga05 and Nhb. thiooxidans AH01 had somewhat larger genomes (∼3.9 and 4.1 Mb). The three strains share 2,240 orthologs which account for ∼80% of their genes (supplementary fig. 1, Supplementary Material online).
Table 1.
General Features of the Studied Genomes as Compared with the Published Genomes of the Closest Relatives
| Rca. barguzinensis alga05 | Rna. thiooxidans ALG1 | Nhb. thiooxidans AH01 | Rba. sphaeroides 2.4.1 | Rdv. sulfidophilum DSM 2351 | Rba. capsulatus SB 1003 | Pco. denitrificans PD 1222 | |
|---|---|---|---|---|---|---|---|
| Total bases | 3,893,644 | 3,504,907 | 4,075,255 | 4,628,173 | 4,732,772 | 3,871,920 | 5,236,194 |
| No. of contigs | 5 | 52 | 39 | Closed | Closed | Closed | Closed |
| GC content [%] | 59 | 60 | 63 | 69 | 67 | 67 | 67 |
| No. of RNAs | 53 | 42 | 43 | 60 | 59 | 66 | 61 |
| chromosomes | n.a. | n.a. | n.a. | 2 | 1 | 1 | 2 |
| plasmids | n.a. | n.a. | n.a. | 5 | 3 | 1 | 1 |
| ORFs | 3,805 | 3,385 | 3,885 | 4,347 | 4,398 | 3,685 | 5,121 |
| proteins | 2,760 (73%) | 2,502 (74%) | 2,834 (73%) | 3,076 (71%) | 2,837 (65%) | 3,100 (84%) | 4,032 (79%) |
| hyp.proteins | 1, 045 (27%) | 883 (26%) | 1,051 (27%) | 1,271 (29%) | 1,561 (35%) | 610 (16%) | 1,089 (21%) |
| PGC length [kbp] | 45.4 | 45.1 | – | 44.7 | 42.1 | 44.3 | – |
| No. of CRISPR spacers | 4 | 38 | 29 | 0 | 11 | 59 | 9 |
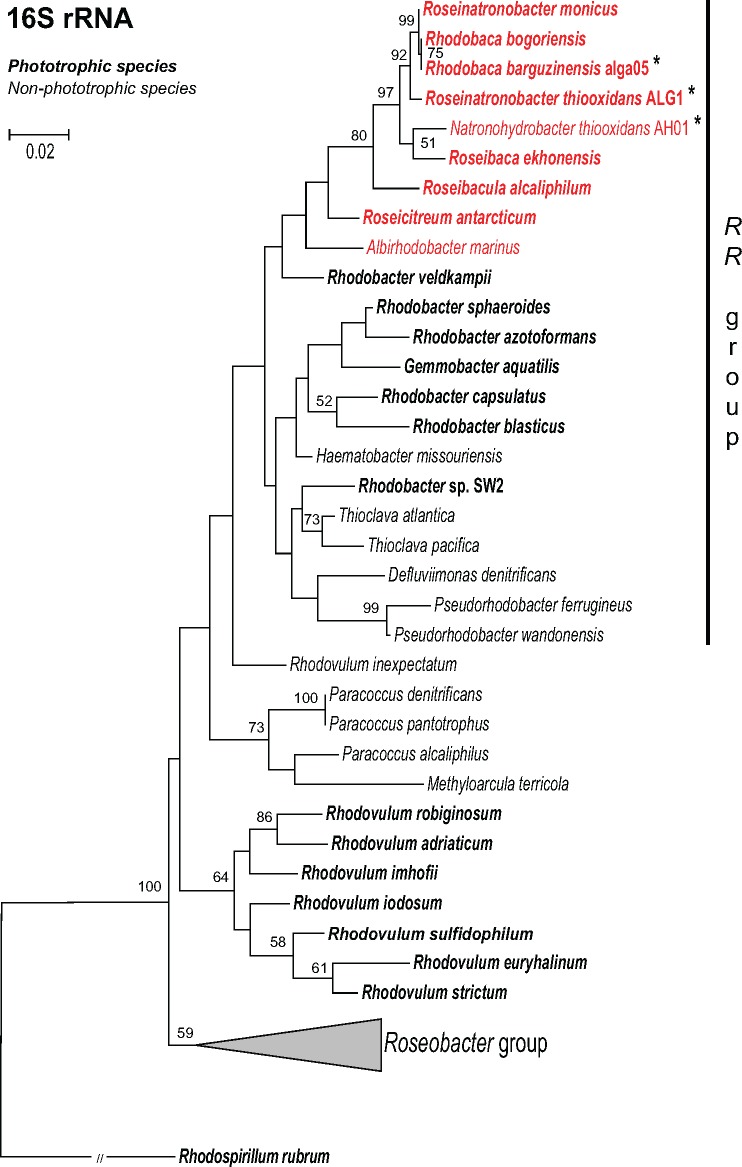
The 16S rRNA sequences of the three sequenced strains were compared with other members of order Rhodobacterales. The obtained phylogenetic tree documents that the sequenced haloalkaliphilic strains are related not only to Roseibaca, Roseicitreum, and Roseibacula species but more distantly also to Rhodobacter species Rba. sphaeroides, Rba. capsulatus, Rba. sp. SW2, and Rba. veldkampii (fig. 1). We denote this group Rhodobacter-Rhodobaca group, or, in shorthand RR group. Freshwater photoautotrophic bacterium Rdv. sulfidophilum, and Paracoccus species, were closely related to the RR-group. The marine species belonging to the so-called Roseobacter group were phylogenetically more distant (fig. 1).
Fig. 1.—
16S rRNA phylogenetic tree showing the position of studied strains (marked with an asterisk) within the Rhodobacterales clade. Halophilic strains are highlighted in red. The phylogenetic tree was calculated using maximum likelihood algorithm with HKY85 nucleotide substitution model and bootstrap 1,000×. Rhodospirillum rubrum was used as an outgroup organism. Scale bar represents changes per position. Bootstrap values >50% are shown.
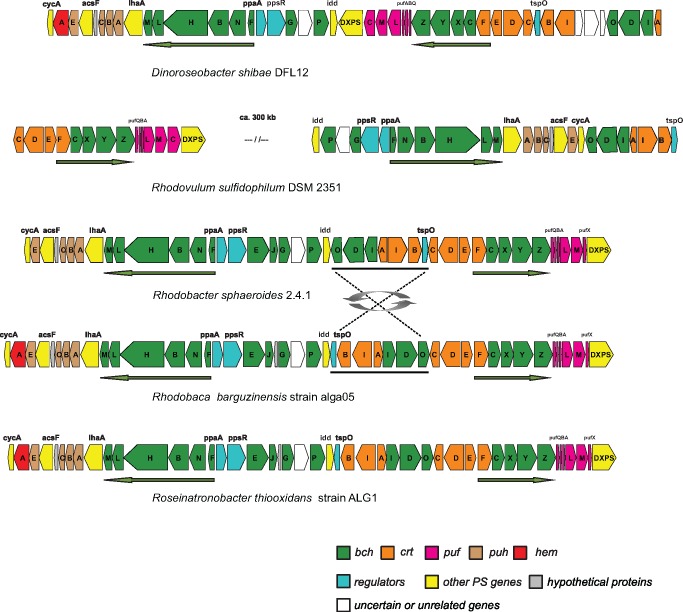
As expected, a complete PGC was found in alga05 and ALG1 genomes. Its organization was identical in both strains, and it was very similar to the organization of the PGC in Rhodobacter species (fig. 2). The only difference was the presence of the hemA gene in the haloalkaliphilic strains and the transposition of tspO-crtBIA-bchIDO genes. The PGC in Drb. shibae and Rvu. sulfidophilum has a clearly different organization. Moreover, in Rvu. sulfidophilum, it is split in two parts placed ∼300 kb apart. A typical feature for all phototrophic RR-species is the divergent orientation (← →) of superoperons bchFNBHLM-IhaA-puh and crt-bchCXYZ-puf, while in other Rhodobacterales groups they have convergent or colinear orientation (Zheng et al. 2011).
Fig. 2.—
Comparison of gene organization in PGCs of the studied and reference genomes. Arrows show directions of superoperons bchFNBHLM-IhaA-puh and superoperons crt-bchCXYZ-puf. Note: Rba. sp. SW2 has the same PGC organization as Rba. sphaeroides. PGC in Rba. capsulatus differs from Rba. sphaeroides only by the absence of the acsF gene.
The major difference between the investigated species and Rhodobacter species was the complete absence of genes for carbon and nitrogen fixation. We detected a RuBisCO-like protein in the genome of the AH01 strain, but due to low homology (36% amino-acid identity) and absence of the other necessary enzymes it does not seem to be involved in carbon fixation. All three studied strains also lack the pucBAC operon encoding the outer light harvesting complexes (table 2), which is consistent with the lack of LH2 bands in their absorption spectra (Sorokin et al. 2000a).
Table 2.
The Presence of Genes Related to Photosynthesis in the Studied and Reference Genomes
| DSM 2351 | 2.4.1 | SB 1003 | alga05 | ALG1 | AH01 | PD 1222 | |
|---|---|---|---|---|---|---|---|
| Carbon fixation | |||||||
| RubisCO (large subunit) | ○ | ○ | ○ | – | – | –/? | ○ |
| puf genes | |||||||
| pufC | • | – | – | – | – | – | – |
| pufX | – | • | • | • | • | – | – |
| puh genes | |||||||
| puhABC, puhE | • | • | • | • | • | – | – |
| puc genes | |||||||
| pucBAC | ○ | ○ | ○ | – | – | – | – |
| Other PS genes | |||||||
| cycA | • | • | ○ | • | • | ○ | ○ |
| lhaA | • | • | • | • | • | – | – |
| DXPS | •○ | •○ | •○ | •○ | •○ | ○ | ○ |
| Porphyrin biosynthesis | |||||||
| hemA | ○ | ○○ | ○ | •○ | •○ | ○ | ○ |
| hemF* | ○ | ○ | – | ○ | ○ | ○ | ○ |
| hemN | ○ | ○ | ○ | ○ | ○ | ○ | ○ |
| hemY* | ○ | ○ | ○ | ○ | ○ | ○ | ○ |
| hemG | – | – | – | – | – | – | – |
| acsF* | • | • | – | • | • | – | – |
| bchE | ○ | • | • | • | • | – | – |
| Regulatory proteins | |||||||
| ppaA/aerR | • | • | • | • | • | – | – |
| appA | – | ○ | – | – | – | – | – |
| ppsR/crtJ | • | • | • | • | • | – | – |
| hvrB | – | – | • | – | – | ○ | – |
| tspO | • | • | • | • | • | – | – |
| pufQ | • | • | • | • | • | – | – |
| Transcription factors | |||||||
| Sigma factors | ○ | ○ | ○ | ○ | ○ | ○ | ○ |
| RpoE, RpoHI, RpoHII | |||||||
| Anti-sigma factor ChrR | ○ | ○ | – | ○ | ○ | ○ | – |
Note.—The PS genes present inside the PGC are marked “•”; the PS genes located outside the PGC are marked “○”; “• ○” means two forms of gene are present; “–” means gene is absent in the genome. Genes coding for the oxygen-dependent form of the enzyme are marked with asterisk. hemF and hemN are aerobic and anaerobic forms of Coproporphyrinogen III oxidase, hemY and hemG are aerobic and anaerobic forms Protoporphyrinogen IX oxidase. acsF and bchE are aerobic and anaerobic forms of Mg-protoporphyrin IX monomethylester oxidative cyclase.‘-/?’ = RubisCO-like protein identified in the genome of AH01.
Tetrapyrrole Biosynthesis Genes
Bacteriochlorophylls are the main light-harvesting pigments in anoxygenic phototrophs (Willows and Kriegel 2009). The genes responsible for their biosynthesis have been routinely used as suitable phylogenetic markers for the investigation of the origin of photosynthesis in bacteria (Raymond et al. 2002; Koblížek et al. 2013).
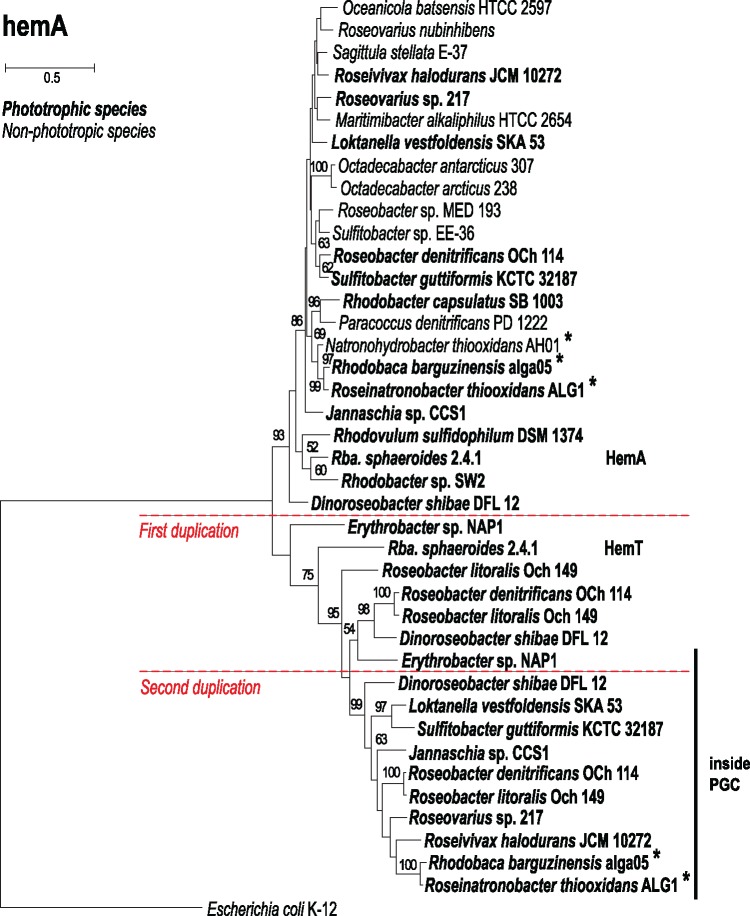
The biosynthesis of all tetrapyrrols, including BChl a, starts with the synthesis of 5-aminolevulinate (Willows and Kriegel 2009; Dailey et al. 2017). This compound is synthesized in all Proteobacteria via the Shemin pathway by 5-aminolevulinate synthase (ALAS). In Rba. sphaeroides 2.4.1., there exist two homologous isoenzymes coded by the hemA and hemT genes (Fanica-Gaignier and Clement-Metral 1973; Bolt et al. 1999). Similarly, there were both ALAS genes in the phototrophic strains alga05 and ALG1 (table 2). Only one gene was found (hemA form) in the heterotrophic strain AH01, in photoautotrophic bacteria Rba. capsulatus, Rba. sp. SW2, as well as in Rvu. sulfidophilum. To better understand the origin of the multiple ALAS forms, we constructed a maximum-likelihood tree with all ALAS genes found in various members of Rhodobacterales. The constructed tree clearly splits into two major groups (fig. 3). The first group encompasses hemA-like genes from both heterotrophic and phototrophic species, including the hemA gene from Rba. sphaeroides. It also contains hemA genes from Rba. capsulatus, Rba. sp. SW2, and Rvu. sulfidophilum. The second group contains hemT related genes, mostly from phototrophic Rhodobacterales. Both, alga05 and ALG1 contain one ALAS gene from the hemA group, and one from the hemT group. Noteworthly, the latter is a part of the PGC (fig. 2), which seems to be a universal characteristics of all AAP Rhodobacterales. Interestingly, a homologue of this gene was also found in the PGC of AAP bacterium Erythrobacter sp. NAP1 (order Sphingomanadales), though, here the gene was likely acquired through the HGT.
Fig. 3.—
Phylogenetic analysis based on alignment of amino acid sequences of ALAS (HemA or HemT). Maximum likelihood (ML) tree with bootstrap 500× was constructed for studied strains and other representative species from the Rhodobacterales clade. Escherichia coli K-12 was used as an outgroup organisms. Scale bars represent changes per position. Bootstrap values >50% are shown. Horizontal bar marks sequences found inside the PGC. Studied strains are marked with an asterisk. Hypothetical gene duplication events are marked with horizontal dashed lines.
The tetrapyrrole pathway then proceeds from 5-aminolevulinate to protoporphyrin IX by action of six different enzymes. The last two are oxidases, which exist in oxygen-dependent and oxygen-independent forms. The coproporphyrinogen III oxidase is present in both forms (hemF, hemN) in all studied strains except Rba. capsulatus, which contains only the oxygen-independent form of this enzyme. In the case of protoporphyrinogen IX oxidase, all the investigated strains contained only the oxygen-dependent form encoded by the hemY gene (table 2).
Bacteriochlorophyll Biosynthesis
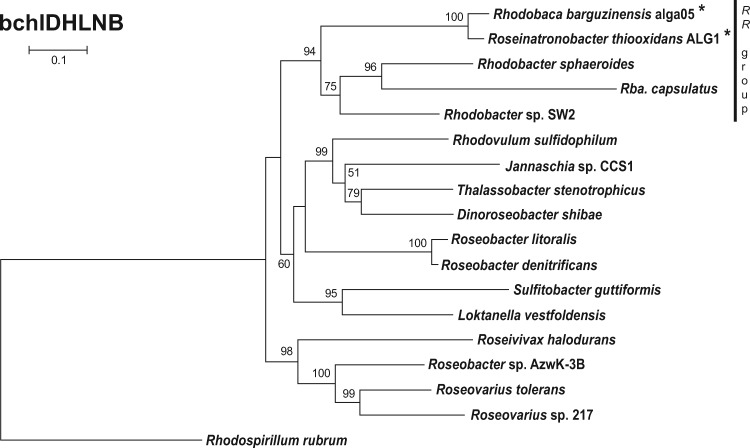
The BChl a pathway on its own starts with magnesium chelatase. This enzyme catalyzes the insertion of Mg into protoporphyrin IX. It is composed of three subunits encoded by the bchI, bchD, and bchH genes. Another enzyme of the BChl a biosynthesis pathway is light-independent protochlorophyllide reductase (subunits encoded by the bchL, bchN, and bchB genes). This protein is required for the light-independent reduction of protochlorophyllide to chlorophyllide a (Willows and Kriegel 2009). To investigate the phylogenetic relationship of the photosynthesis genes between studied and reference strains, we calculated the tree using the concatenated amino acid sequences of these two enzymes. The inferred phylogeny documents a very close relationship of the two photoheterotrophic strains, alga05 and ALG1, and all the RR-species (fig. 4).
Fig. 4.—
Phylogenetic tree based on concatenated alignments of amino acid sequences of magnesium chelatase and light-independent protochlorophyllide reductase (BchIDHLNB; 2317 common amino acid positions). Maximum-likelihood (ML) tree with bootstrap 1,000× was constructed for studied strains and other representative species from the Rhodobacterales clade. Rhodospirillum rubrum was used as an outgroup organism. Scale bars represent changes per position. Bootstrap values >50% are shown. Studied strains are marked with an asterisk.
Another important step of the BChl a biosynthetic pathway is the cyclization of its fifth ring by Mg-protoporphyrin IX monomethyl ester oxidative cyclase (Willows and Kriegel 2009). This enzyme has two forms: an oxygen-dependent and oxygen-independent form encoded by acsF and bchE gene, respectively. Rba. sphaeroides, Rba. sp. SW2, Rvu. sulfidophilum, alga05 and ALG1 contain both forms of this enzyme, while Rba. capsulatus contains only the oxygen independent form. Heterotrophic strains AH01 and Pco. denitrificans obviously do not contain these genes. Interestingly these two forms of the cyclase have a very different phylogeny. The oxygen-dependent form of the cyclase (acsF gene product) was used previously as a convenient gene marker for the detection of AAPs (Boldareva et al. 2013; Zeng et al. 2014). Indeed, all RR-species formed a distinct group, clearly separated from the marine species as well as Rvu. sulfidophilum (supplementary fig. 2, Supplementary Material online). The situation was very different for the oxygen-independent form (bchE gene product), where in Rba. spp., ALG1, and alga05, the bchE gene is part of the PGC, while in Rvu. sulfidophilum and in many marine AAP species it is outside the PGC.
Reaction Center Proteins
One part of the PGC comprises of puf (photosynthesis unit forming), an operon encoding the proteins for the inner antenna (pufBA) and the photosynthesis reaction center subunits L and M (pufLM). In all Rhodobacter species (Rba. sphaeroides, Rba. capsulatus, and Rba. sp. SW2) these genes are accompanied by a pufX gene encoding the light-harvesting associated protein PufX, with the typical pufQBALMX operon organization. This gene was also present in the two phototrophic strains alga05 and ALG1. In contrast, Rdv. sulfidophilum contains the pufC gene encoding the cytochrome c subunit of the complex (table 2).
Since the pufM gene represents a convenient genetic marker of anoxygenic phototrophs with type 2 reaction centers, it is frequently used to infer phylogeny of new phototrophic organisms. The computed PufM phylogenetic tree of phototrophic Rhodobacterales shows a major split into two branches. The first encompasses Rhodobacter species, the phototrophic species alga05 and ALG1, and a few members of the marine Roseobacter group. The second group contains Rvu. sulfidophilum, Roseobacter (Rsb.) denitrificans, Drb. shibae and most of the marine AAP species. This phylogeny further confirms the close relationship of the studied haloalkalophiles with Rhodobacter species, and their distinction from Rhodovulum and Roseobacter species (supplementary fig. 3, Supplementary Material online).
Regulatory Proteins
All photosynthetic organisms control expression of their photosynthesis genes in response to changes in the environment. All seven studied strains contain these common regulators: AhcY, FnrL, HvrA, PrrA/RegA, PrrB/RegB, and PrrC/SenC. All studied phototrophic strains accommodate ppaA/aerR, ppsR/crtJ, tspO, and pufQ genes inside their PGCs. Rba. sphaeroides is the only strain with appA gene, which is located outside the PGC. Interestingly, Rba. sp. SW2 also does not possess this gene in the genome (data not shown). Two investigated strains, that is, phototrophic Rba. capsulatus and chemotrophic strain AH01 possess the gene coding for the HvrB protein. The phototrophic strain contains this gene inside its PGC (table 2).
One of the most important physiological processes in phototrophic species is the protection of cells against reactive oxygen species. Singlet oxygen is generated under aerobic conditions by light-excited BChl a (Borland et al. 1989). In Rba. sphaeroides three alternative sigma factors are involved in response to the singlet oxygen, the sigma-24 factor RpoE, and the sigma-32 factors RpoHI and RpoHII. While RpoH is part of a more general oxidative stress response, RpoE and ChrR have been shown to be triggered specifically by the oxygen singlet. In these organisms, RpoE and ChrR are encoded in an operon and together with a second operon transcribed from the same promoter in the opposite direction form a conserved gene cluster. This gene cluster has also been found in all investigated phototrophic species, but it was absent in most of the heterotrophic species. Surprisingly, the rpoE-chrR cluster was also present in the studied heterotrophic strain AH01, as well as in several marine heterotrophic bacteria Maritimibacter alkaliphilus, Octadecabacter antarcticus, Octadecabacter arcticus, Oceanicola batsensis, and Sagittula stellata which should not be prone to singlet oxygen formation (fig. 5).
Fig. 5.—

Phylogenetic tree of sigma-24 factors in Rhodobacterales. Species with a photosynthesis gene cluster are in bold. Sigma-24 factors in an operon with chrR form a distinct phylogenetic group, marked by a vertical bar. The tree has been constructed using the Neighbour-joining (NJ) method with pairwise deletion of gaps and 500 bootstraps.
Discussion
Two evolutionary scenarios explaining the presence of phototrophic and heterotrophic strains among haloalkaliphilic species in the RR-group were considered in this study (1) the “regressive evolution” scenario, where the ancestors of the haloalkaliphilic RR-group were photoautotrophic organisms, which later lost part (or all) of their photosynthesis gene, or (2) the ancestors of the RR-group were heterotrophs, which adopted their photosynthesis genes via HGT of photosynthesis genes. Our data indicates that phototrophy is ancestral in the RR-group. There are two main lines of evidence. First, all the studied phototrophic organisms belonging to the RR-group shared an almost identical organization of their PGCs (fig. 2). A unique feature for all phototrophic RR-species is the divergent orientation of superoperons bchFNBHLM-IhaA-puh and crt-bchCXYZ-puf. This orientation has not been found in any other Rhodobacterales species (Zheng et al. 2011), which represents the main evidence against the hypothesis that the RR-species adopted the PGC through HTG from other Rhodobacterales genera. Another distinctive feature present in all studied phototrophic members of the RR-group is the presence of the pufX gene, and the absence of the pufC gene (table 2). The gene pufX is also present in several marine AAP species, but they have likely received it via the HGT as discussed before (Koblížek et al. 2015). Second, the performed phylogenetic analyses on the photosynthesis genes bchIDHLNB, acsF and pufM (fig. 4, supplementary figs. 2 and 3, Supplementary Material online) document that the RR-group always cluster together, with an exception of the pufM tree where the RR-group is mixed with several marine AAP species (supplementary fig. 3, Supplementary Material online).
An important finding is the presence of genes connected with the singlet oxygen defense system in chemotrophic strain AH01. This strain contains the same singlet-oxygen-stress-response mechanism (encoded in rpoE-chrR gene operon) as Rba. sphaeroides (Anthony et al. 2004), as well as in ALG1 and alga05 strains. When bound to its anti-sigma factor ChrR, RpoE is inactive in the cell. The presence of singlet oxygen leads to a dissociation of the heterodimer, thus the activation of RpoE and subsequently its more than 180 target genes (Anthony et al. 2004). A similar response has been shown for the AAP species Rsb. litoralis (Berghoff et al. 2011) and Drb. shibae (Tomasch et al. 2011). As the main source of singlet oxygen in bacteria is BChl a (Borland et al. 1989), all the phototrophic Rhodobacterales contain the RpoE–ChrR system to protect their cellular machinery against damage. However, chemotrophic organisms (which lack BChl a) should not be prone to the formation of singlet oxygen, and the presence of RpoE–ChrR system is unnecessary. This suggests that the ancestor of AH01 was originally a phototrophic bacterium, which later lost its photosynthetic genes, but yet retained the RpoE–ChrR system.
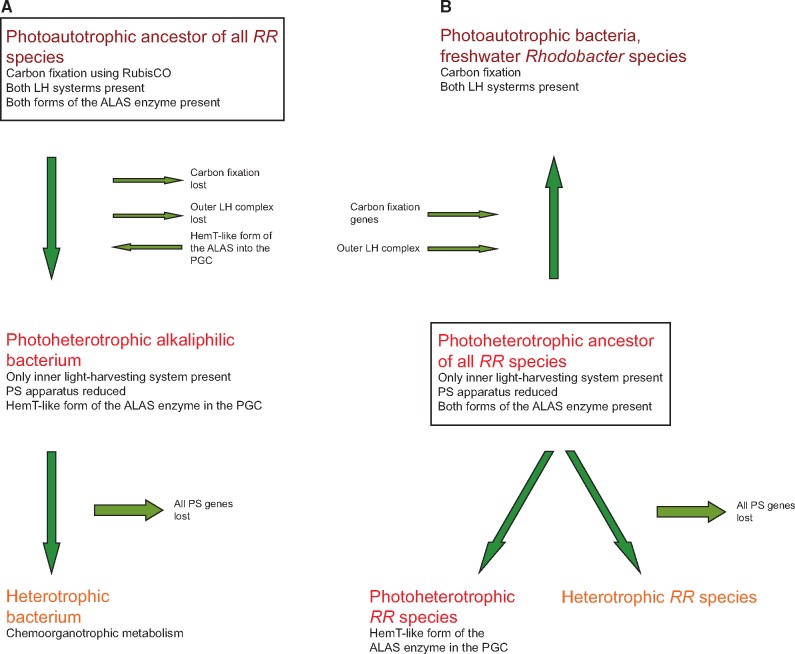
From the presented pieces of evidence, we conclude that the ancestors of the haloalkaliphilic members of the RR-group were phototrophic organisms. A more difficult question to answer is whether these ancestral species were photoautotrophic and contained RubisCO. There are two possible scenarios (fig. 6). The first [compatible with Keppen et al. (2013)] assumes that the ancestors of the RR-group were photoautotrophs similar to most of the modern Rhodobacter species. Then, during evolution, some RR-lineages lost a part or all of their photosynthesis genes producing photoheterotrophic, lithoheterotrophic, or chemoheterotrophic species. In the second scenario, the ancestors of the RR-group were photoheterotrophs. Here, some lineages (e.g., Rhodobacter species) gained RubisCO genes and became photoautotrophs, whereas some lineages lost all their photosynthesis genes and became heterotrophs (fig. 6). These two scenarios are difficult to reconcile based on the available data, but based on the performed phylogenetic analyses we prefer the photoautotrophic ancestor scenario. Indeed, the photoautotrophic Rhodobacter species (especially Rba. veldkampii) are located closer to the root of the RR-group in our 16S rRNA tree, whereas the photoheterotrophic species branch off later (fig. 1). The weakness of this argument is the very low statistical support of the 16S phylogenetic tree. However, a similar conclusion can also be made based on the ALAS phylogeny. The ALAS (hemA/hemT gene) tree shows a major split, documenting the existence of two different forms of the gene in Rhodobacterales (hemA-like and hemT-like form). These two forms probably originate from a gene duplication event. Since the hemA and hemT genes of Rba. sphaeroides lay close to the split, it seems that the gene duplication occurred in the ancestors of Rhodobacter-like species and all the subsequent photoheterotrophic ancestors retained the two forms of the gene, which gradually diverged. Photoheterotrophic species always contain both forms, which indicate that they only evolved after Rba. sphaeroides. Later, the AAP species incorporated the hemT-like form of ALAS into the PGC. The presence of ALAS in the PGC seems to be a common characteristic of all the AAP species belonging to Alphaproteobacteria (Zheng et al. 2011). This arrangement is probably advantageous for convenient regulation of BChl a synthesis in AAP species, which need to tightly control the initial step of the tetrapyrrol pathway with the final part of the BChl a pathway. Indeed, this has been documented in Drb. shibae, where the inducible ALAS gene located in the PGC (hemT-like form) was under strong light regulation together with all bch genes (Tomasch et al. 2011). Interestingly, all these ALAS genes present in the PGC cluster together form a distinct subclade of the entire hemT-like group (fig. 3). This indicates that the hemA split occurred first in photoautotrophic Rhodobacter-like species, which probably needed to differentially regulate the tetrapyrrol biosynthesis pathway for heme (hemA form) and BChl a synthesis (hemT form). Later, after the evolution of the AAP species, the hemT form of the gene was moved under the common regulation of the PGC cluster. Moreover, in some species (Drb. shibae and Roseobacter species), the hemT form of ALAS underwent a second duplication, indicating the need for even more delicate regulation of the gene.
Fig. 6.—
Proposed schemes of the evolution of phototrophy among the members of the RR-group: (A) Regressive evolution model compatible with Keppen et al. (2013). (B) Mixed model assuming a photoheterotrophic ancestor of all modern RR-species—heterotrophs, photoheterotrophs, and photoautotrophs.
In conclusion, the presented data indicate that the ancestors of the haloalkaliphilic members of the RR-group were phototrophic species. The heterotrophic species have evolved through the regressive loss of their photosynthetic apparatus. In addition, we have demonstrated that the important step enabling the evolution of photoheterotrophic species was the duplication of the hemA gene, and its later incorporation into the PGC of the AAP species.
Many interesting questions were left to be answered such as if there is an alternative pathway of descent from photoautotrophic species through the facultatively autotrophic aerobic or facultatively anaerobic lithotrophs, such as Paracoccus (loss of the PGC), to aerobic chemolithoheterotrophs (loss of RubisCO) and finally organoheterotrophs? What was the role of the hydrogenase in this alternative scenario? Did Rba. capsulatus evolve regressively from a Rba. sphaeroides-like species through the loss of genes (acsF, hemF, hemT, and anti-sigma factor), or does it represent a former (more ancient) photoautotrophic species? We believe, that the constantly expanding genomic data, will allow us to address these questions in the near future.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors are grateful to Dr Katya Boldareva-Nuyanzhina for providing her strains Rca. barguzinensis and Rna. thiooxidans. We also thank Jason Dean BSc. for the language correction. This research has been supported by the GAČR project P501/12/G055, the DAAD project 57155424, the EC-funded project Algatech Plus (LO1416), and the RFBR grant 16-04-00035. J.T. was supported by the Deutsche Forschungsgemeinschaft (DFG) within the Transregio 51 “Roseobacter”. D.Y.S. was supported by the Gravitation Program (SIAM, grant 24002002) from the Dutch Ministry of Education and Science.
Literature Cited
- Androga DD, Özgür E, Eroglu I, Yücel M, Gündüz U.. 2012. Photofermentative hydrogen production in outdoor conditions. Croatia: INTECH Open Access Publisher. [Google Scholar]
- Anthony JR, Newman JD, Donohue TJ.. 2004. Interactions between the Rhodobacter sphaeroides ECF sigma factor, σ E, and its anti-sigma factor, ChrR. J Mol Biol. 341(2):345–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auch AF, von Jan M, Klenk HP, Göker M.. 2010. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2(1):117–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, et al. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9(1):1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghoff BA, et al. 2011. Anoxygenic photosynthesis and photooxidative stress: a particular challenge for Roseobacter. Environ Microbiol 13(3):775–791. [DOI] [PubMed] [Google Scholar]
- Boldareva EN, et al. 2006. The new alkaliphilic bacteriochlorophyll a-containing bacterium Roseinatronobacter monicus sp. nov. from the hypersaline Soda Mono Lake (California, United States). Microbiology 76:82–92. [PubMed] [Google Scholar]
- Boldareva EN, et al. 2008. Rhodobaca barguzinensis sp. nov., a new alkaliphilic purple nonsulfur bacterium isolated from a soda lake of the Barguzin Valley (Buryat Republic, Eastern Siberia). Microbiology 77(2):206–218. [PubMed] [Google Scholar]
- Boldareva EN, Bláhová Z, Sobotka R, Koblížek M.. 2013. Distribution and origin of oxygen-dependent and oxygen-independent forms of Mg-protoporphyrin monomethylester cyclase among phototrophic proteobacteria. Appl Environ Microbiol. 79(8):2596–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldareva EN, Gorlenko VM.. 2014. Roseibacula alcaliphilum gen. nov. sp. nov., a new alkaliphilic aerobic anoxygenic phototrophic bacterium from a meromictic soda lake Doroninskoe (East Siberia, Russia). Microbiology 83:381–390. [PubMed] [Google Scholar]
- Bolt EL, et al. 1999. Characterization of the Rhodobacter sphaeroides 5-aminolaevulinic acid synthase isoenzymes HemA and HemT, isolated from recombinant Escherichia coli. Eur J Biochem. 265(1):290–299. [DOI] [PubMed] [Google Scholar]
- Borland CF, Cogdell RJ, Land EJ, Truscott TG.. 1989. Bacteriochlorophyll a triplet state and its interactions with bacterial carotenoids and oxygen. J Photochem Photobiol B 3(2):237–245. [Google Scholar]
- Brinkhoff T, Giebel HA, Simon M.. 2008. Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch Microbiol. 189(6):531–539. [DOI] [PubMed] [Google Scholar]
- Brocks JJ, et al. 2005. Biomarker evidence for green and purple sulphur bacteria in a stratified Palaeoproterozoic sea. Nature 437(7060) 866–870. [DOI] [PubMed] [Google Scholar]
- Canfield DE, et al. 2008. Ferruginous conditions dominated later Neoproterozoic deep-water chemistry. Science 321(5891):949–952. [DOI] [PubMed] [Google Scholar]
- Dailey HA, et al. 2017. Prokaryotic heme biosynthesis: multiple pathways to a common essential product. Microbiol Mol Biol Rev. 81(1): e00048–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanica-Gaignier M, Clement-Metral J.. 1973. 5-Aminolevulinic acid synthetase of Rhodopseudomonas spheroides Y. Eur J Biochem. 40(1): 13–18. [DOI] [PubMed] [Google Scholar]
- Fitch WM. 1970. Distinguishing homologous from analogous proteins. Syst Zool. 19:99–113. [PubMed] [Google Scholar]
- Garrity GM, Bell JA, Lilburn T.. 2005. Order III. Rhodobacterales ord. nov In: Vos P, et al. , editors. Bergey’s Manual® of Systematic Bacteriology. New York: Springer US; p. 270–323. [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C.. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35(Web Server):52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppen OI, Krasil’nikova EN, Lebedeva NV, Ivanovskii RN.. 2013. Comparative study of metabolism of the purple photosynthetic bacteria grown in the light and in the dark under anaerobic and aerobic conditions. Microbiology 82(5):547–553. [PubMed] [Google Scholar]
- Koblížek M, Moulisová V, Muroňová M, Oborník M.. 2015. Horizontal transfers of two types of puf operons among phototrophic members of the Roseobacter clade. Folia Microbiol. 60(1):37–43. [DOI] [PubMed] [Google Scholar]
- Koblížek M, Zeng Y, Horák A, Oborník M.. 2013. Regressive evolution of photosynthesis in the Roseobacter clade. Adv Bot Res. 66:385–405. [Google Scholar]
- Labrenz M, Lawson PA, Tindall BJ, Hirsch P.. 2009. Roseibaca ekhonensis gen. nov., sp. nov., an alkalitolerant and aerobic bacteriochlorophyll a-producing alphaproteobacterium from hypersaline Ekho Lake. Int J Syst Evol Microbiol. 59 (Pt 8):1935–1940. [DOI] [PubMed] [Google Scholar]
- Liotenberg S, et al. 2008. Organization and expression of photosynthesis genes and operons in anoxygenic photosynthetic proteobacteria. Environ Microbiol. 10(9):2267–2276. [DOI] [PubMed] [Google Scholar]
- Luo H, Moran MA.. 2014. Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev. 78(4):573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Göker M, Spröer C, Klenk HP.. 2013. When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol. 195(6):413–418. [DOI] [PubMed] [Google Scholar]
- Milford AD, Achenbach LA, Jung DO, Madigan MT.. 2000. Rhodobaca bogoriensis gen. nov. and sp. nov., an alkaliphilic purple nonsulfur bacterium from African Rift Valley soda lakes. Arch Microbiol. 174(1–2):18–27. [DOI] [PubMed] [Google Scholar]
- Moran MA, et al. 2007. Ecological genomics of marine Roseobacters. Appl Environ Microbiol. 73(14):4559–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Frank O, Göker M, Pradella S.. 2013. Extrachromosomal, extraordinary and essential—the plasmids of the Roseobacter clade. Appl Microbiol Biotechnol. 97(7):2805–2815. [DOI] [PubMed] [Google Scholar]
- Raymond J, Zhaxybayeva O, Gogarten JP, Gerdes SY, Blankenship RE.. 2002. Whole-genome analysis of photosynthetic prokaryotes. Science 298(5598):1616–1620. [DOI] [PubMed] [Google Scholar]
- Simon M, et al. 2017. Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 11(6):1483–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin DY, Tourova TP, Kuenen JG.. 2000a. A new facultatively autotrophic hydrogen- and sulphur-oxidizing bacterium from alkaline environment. Extremophiles 4(4):237–245. [DOI] [PubMed] [Google Scholar]
- Sorokin DY, Tourova TP, Kuznetsov BB, Bryantseva IA, Gorlenko VM.. 2000b. Roseinatronobacter thiooxidans gen. nov., sp. nov., a new alkaliphilic aerobic bacteriochlorophyll-a-containing bacteria from a soda lake. Microbiology 69(1):89–97. [PubMed] [Google Scholar]
- Stadnichuk IN, et al. 2009. Photosynthetic activity and components of the electron transport chain in the aerobic bacteriochlorophyll a-containing bacterium Roseinatronobacter thiooxidans. Microbiology 78(1):7–15. [PubMed] [Google Scholar]
- Talavera G, Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577. [DOI] [PubMed] [Google Scholar]
- Tomasch J, Gohl R, Bunk B, Diez MS, Wagner-Döbler I.. 2011. Transcriptional response of the photoheterotrophic marine bacterium Dinoroseobacter shibae to changing light regimes. ISME J. 5(12):1957–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers J, et al. 2013. Poles apart: Arctic and Antarctic Octadecabacter strains share high genome plasticity and a new type of xanthorhodopsin. PLoS One 8(5):e63422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner-Döbler I, Biebl H.. 2006. Environmental biology of the marine Roseobacter lineage. Annu Rev Microbiol. 60:255–280. [DOI] [PubMed] [Google Scholar]
- Wang Y, Coleman-Derr D, Chen G, Gu YQ.. 2015. OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 43(W1):78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willows RD, Kriegel AM.. 2009. Biosynthesis of bacteriochlorophylls in purple bacteria In: Hunter CN, Daldal F, Thurnauer MC, Beatty JT, editors. The purple phototrophic bacteria. Netherlands: Springer; p. 57–79. [Google Scholar]
- Yu Y, Yan SL, Li HR, Zhang XH.. 2011. Roseicitreum antarcticum gen. nov., sp. nov., an aerobic bacteriochlorophyll a-containing alphaproteobacterium isolated from Antarctic sandy intertidal sediment. Int J Syst Evol Microbiol. 61(Pt 9):2173–2179. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Feng F, Medová H, Dean J, Koblížek M.. 2014. Functional type 2 photosynthetic reaction centers found in the rare bacterial phylum Gemmatimonadetes. Proc Natl Acad Sci USA. 111(21):7795–7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, et al. 2011. Diverse arrangement of photosynthetic gene clusters in aerobic anoxygenic phototrophic bacteria. PLoS One 6(9):e25050.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsebo KM, Hearst JE.. 1984. Genetic-physical mapping of a photosynthetic gene cluster from R. capsulata. Cell 37(3):937–947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.