Abstract
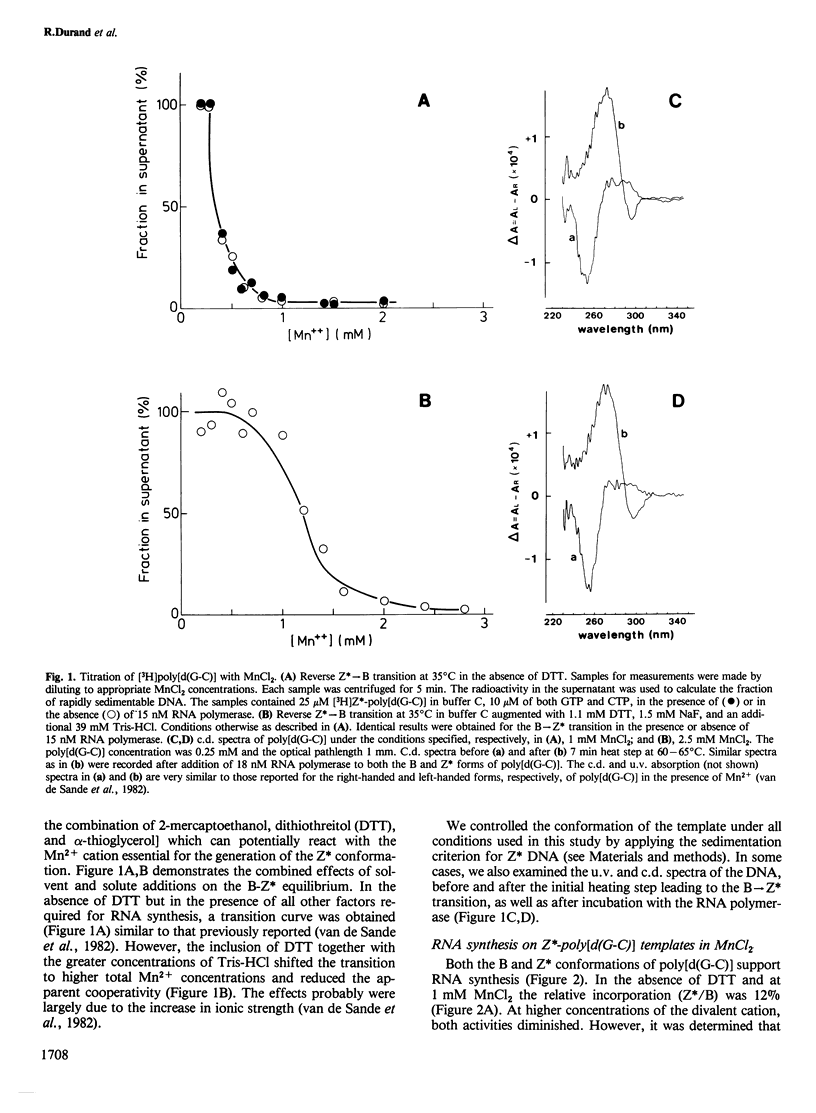
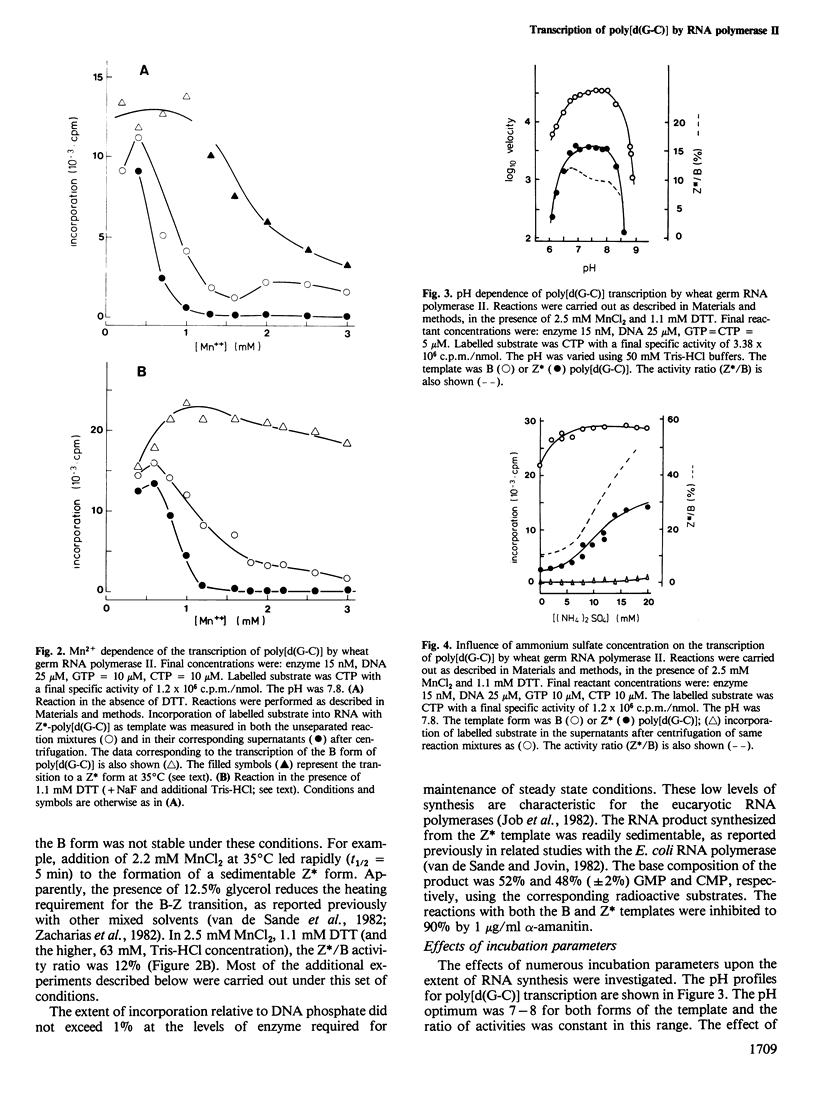
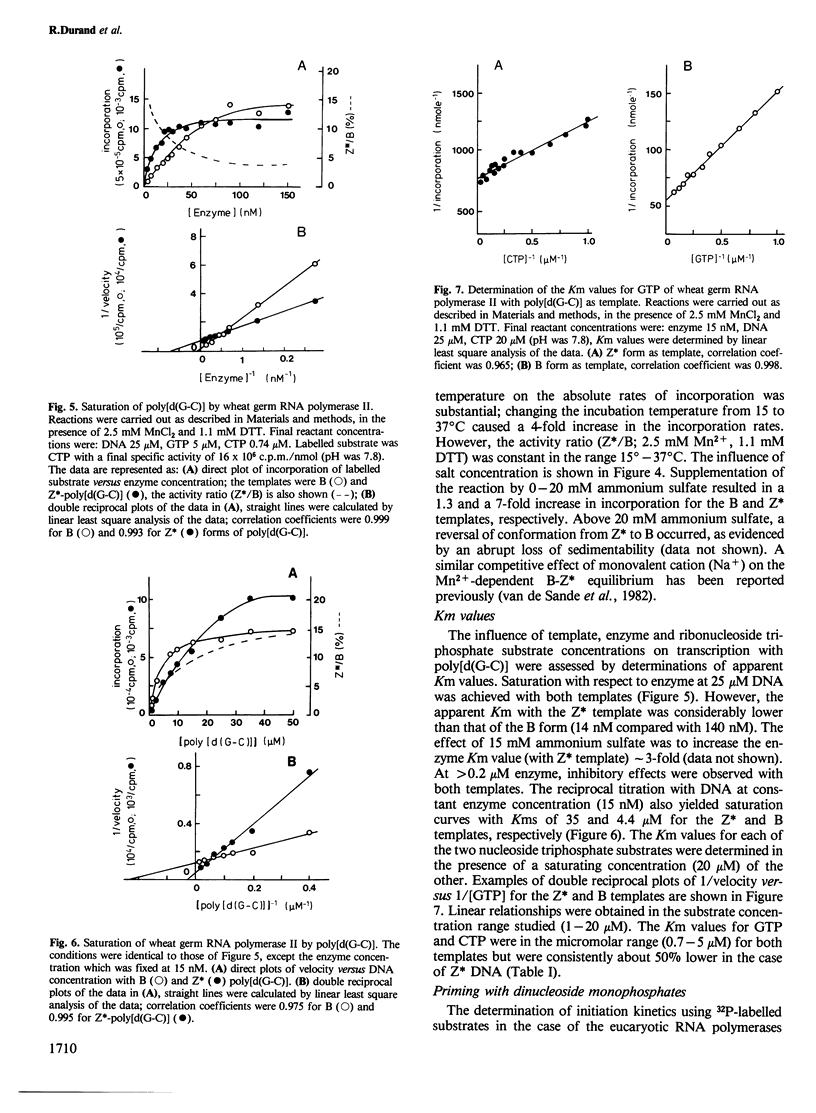
The template properties of left-handed synthetic polymers, the Z* form of poly[d(G-C)] and the Z form of poly[d(G-m5C)], have been investigated using an eucaryotic RNA polymerase, the class II enzyme from wheat germ. Results from a comparative kinetic study of transcription using the polynucleotide substrates in the B and Z conformations are reported. Optimal conditions for enzyme activity compatible with the preservation of the desired template conformation were determined. On the basis of several criteria, both physical (c.d. spectra of the polymers, sedimentability of the Z* form) and biochemical, it was demonstrated that the left-handed conformations of poly[d(G-C)] and poly[d(G-m5C)] serve as templates for wheat germ RNA polymerase II. The level of incorporation was less than that exhibited by the B form of poly[d(G-C)], the relative activity being a function of the precise experimental conditions. Activity ratios (Z*/B or Z/B) ranged from 0.1 to 0.5. The effect of various incubation parameters, including pH, salt concentration, temperature, and the presence of dinucleoside monophosphate primers were investigated. The Km values for nucleoside triphosphate substrates were slightly smaller for the Z* form of poly[d(G-C)] than for the B conformation. Titration of DNA (Z* or B) with enzyme and reciprocal experiments suggested that the reduced activity of left-handed templates might derive from the availability of fewer and/or lower affinity sites for initiation and/or translocation on these templates. Specific antibodies raised against left-handed DNA strongly inhibited the observed transcription of Z* and Z DNAs by wheat germ RNA polymerase II.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anthony D. D., Goldthwait D. A., Wu C. W. Studies with the ribonucleic acid polymerase. II. Kinetic aspects of initiation and polymerization. Biochemistry. 1969 Jan;8(1):246–256. doi: 10.1021/bi00829a035. [DOI] [PubMed] [Google Scholar]
- Arndt-Jovin D. J., Robert-Nicoud M., Zarling D. A., Greider C., Weimer E., Jovin T. M. Left-handed Z-DNA in bands of acid-fixed polytene chromosomes. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4344–4348. doi: 10.1073/pnas.80.14.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behe M., Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B--Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc Natl Acad Sci U S A. 1981 Mar;78(3):1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behe M., Zimmerman S., Felsenfeld G. Changes in the helical repeat of poly(dG-m5dC) . poly(dG-m5dC) and poly(dG-dC) . poly(dG-dC) associated with the B-Z transition. Nature. 1981 Sep 17;293(5829):233–235. doi: 10.1038/293233a0. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bull P., Garrido J. Structure of yeast RNA polymerases determined by electron microscopy. Arch Biochem Biophys. 1982 Nov;219(1):163–166. doi: 10.1016/0003-9861(82)90145-x. [DOI] [PubMed] [Google Scholar]
- Dezélée S., Sentenac A. Role of DNA-RNA hybrids in eukaryotes. Purification and properties of yeast RNA polymerase B. Eur J Biochem. 1973 Apr 2;34(1):41–52. doi: 10.1111/j.1432-1033.1973.tb02726.x. [DOI] [PubMed] [Google Scholar]
- Downey K. M., Jurmark B. S., So A. G. Determination of nucleotide sequences at promoter regions by the use of dinucleotides. Biochemistry. 1971 Dec 21;10(26):4970–4975. doi: 10.1021/bi00802a021. [DOI] [PubMed] [Google Scholar]
- Downey K. M., So A. G. Studies on the kinetics of ribonucleic acid chain initiation and elongation. Biochemistry. 1970 Jun 9;9(12):2520–2525. doi: 10.1021/bi00814a019. [DOI] [PubMed] [Google Scholar]
- Durand R., Job C., Teissère M., Job D. Non-processive transcription of poly[d(A-T)] by wheat germ RNA polymerase II. FEBS Lett. 1982 Dec 27;150(2):477–481. doi: 10.1016/0014-5793(82)80793-x. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Burgess R. R. In vitro transcription by wheat germ ribonucleic acid polymerase II: effects of heparin and role of template integrity. Biochemistry. 1979 Oct 16;18(21):4581–4588. doi: 10.1021/bi00588a019. [DOI] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6465–6469. doi: 10.1073/pnas.79.21.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrisak J. J., Burgess R. R. A new method for the large-scale purification of wheat germ DNA-dependent RNA polymerase II. Biochemistry. 1975 Oct 21;14(21):4639–4645. doi: 10.1021/bi00692a012. [DOI] [PubMed] [Google Scholar]
- Job D., Durand R., Teissere M. Enzymatic properties and cooperative effects in the kinetics of wheat-germ RNA polymerases. A comparative study of the three nuclear enzyme classes. Eur J Biochem. 1982 Nov;128(1):35–39. doi: 10.1111/j.1432-1033.1982.tb06927.x. [DOI] [PubMed] [Google Scholar]
- Jovin T. M., van de Sande J. H., Zarling D. A., Arndt-Jovin D. J., Eckstein F., Füldner H. H., Greider C., Grieger I., Hamori E., Kalisch B. Generation of left-handed Z-DNA in solution and visualization in polytene chromosomes by immunofluorescence. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):143–154. doi: 10.1101/sqb.1983.047.01.019. [DOI] [PubMed] [Google Scholar]
- Klevan L., Schumaker V. N. Stabilization of Z-DNA by polyarginine near physiological ionic strength. Nucleic Acids Res. 1982 Nov 11;10(21):6809–6817. doi: 10.1093/nar/10.21.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. A. The structure and mechanism of action of bacterial DNA-dependent RNA polymerase. Prog Biophys Mol Biol. 1981;38(3):165–210. doi: 10.1016/0079-6107(81)90013-4. [DOI] [PubMed] [Google Scholar]
- Lafer E. M., Möller A., Nordheim A., Stollar B. D., Rich A. Antibodies specific for left-handed Z-DNA. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3546–3550. doi: 10.1073/pnas.78.6.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescure B., Williamson V., Sentenac A. Efficient and selective initiation by yeast RNA polymerase B in a dinucleotide-primed reaction. Nucleic Acids Res. 1981 Jan 10;9(1):31–45. doi: 10.1093/nar/9.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfoy B., Rousseau N., Leng M. Interaction between antibodies to Z-form deoxyribonucleic acid and double-stranded polynucleotides. Biochemistry. 1982 Oct 26;21(22):5463–5467. doi: 10.1021/bi00265a013. [DOI] [PubMed] [Google Scholar]
- Miller F. D., Rattner J. B., van de Sande J. H. Nucleosome-core assembly on B and Z forms of poly[d(G-m5C)]. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):571–575. doi: 10.1101/sqb.1983.047.01.067. [DOI] [PubMed] [Google Scholar]
- Nickol J., Behe M., Felsenfeld G. Effect of the B--Z transition in poly(dG-m5dC) . poly(dG-m5dC) on nucleosome formation. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1771–1775. doi: 10.1073/pnas.79.6.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordheim A., Tesser P., Azorin F., Kwon Y. H., Möller A., Rich A. Isolation of Drosophila proteins that bind selectively to left-handed Z-DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7729–7733. doi: 10.1073/pnas.79.24.7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oen H., Wu C. W. DNA-dependent single-step addition reactions catalyzed by Escherichia coli RNA polymerase. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1778–1782. doi: 10.1073/pnas.75.4.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oen H., Wu C. W., Haas R., Cole P. E. T7 deoxyribonucleic acid directed, rapid-turnover, single-step addition reactions catalyzed by Escherichia coli ribonucleic acid polymerase. Biochemistry. 1979 Sep 18;18(19):4148–4155. doi: 10.1021/bi00586a015. [DOI] [PubMed] [Google Scholar]
- Pohl F. M., Jovin T. M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J Mol Biol. 1972 Jun 28;67(3):375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- Pohl F. M., Thomae R., DiCapua E. Antibodies to Z-DNA interact with form V DNA. Nature. 1982 Dec 9;300(5892):545–546. doi: 10.1038/300545a0. [DOI] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Shaner S. L., Melançon P., Lee K. S., Burgess R. R., Record M. T., Jr Ion effects on the aggregation and DNA-binding reactions of Escherichia coli RNA polymerase. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):463–472. doi: 10.1101/sqb.1983.047.01.055. [DOI] [PubMed] [Google Scholar]
- Shimamoto N., Wu C. W. Mechanism of ribonucleic acid chain initiation. 1. A non-steady-state study of ribonucleic acid synthesis without enzyme turnover. Biochemistry. 1980 Mar 4;19(5):842–848. doi: 10.1021/bi00546a003. [DOI] [PubMed] [Google Scholar]
- Singleton C. K., Klysik J., Stirdivant S. M., Wells R. D. Left-handed Z-DNA is induced by supercoiling in physiological ionic conditions. Nature. 1982 Sep 23;299(5881):312–316. doi: 10.1038/299312a0. [DOI] [PubMed] [Google Scholar]
- Sylvester J. E., Cashel M. Stable RNA-DNA-RNA polymerase complexes can accompany formation of a single phosphodiester bond. Biochemistry. 1980 Mar 18;19(6):1069–1074. doi: 10.1021/bi00547a004. [DOI] [PubMed] [Google Scholar]
- Wang A. J., Quigley G. J., Kolpak F. J., van der Marel G., van Boom J. H., Rich A. Left-handed double helical DNA: variations in the backbone conformation. Science. 1981 Jan 9;211(4478):171–176. doi: 10.1126/science.7444458. [DOI] [PubMed] [Google Scholar]
- Zacharias W., Larson J. E., Klysik J., Stirdivant S. M., Wells R. D. Conditions which cause the right-handed to left-handed DNA conformational transitions. Evidence for several types of left-handed DNA structures in solution. J Biol Chem. 1982 Mar 25;257(6):2775–2782. [PubMed] [Google Scholar]
- van de Sande J. H., Jovin T. M. Z* DNA, the left-handed helical form of poly[d(G-C)] in MgCl2-ethanol, is biologically active. EMBO J. 1982;1(1):115–120. doi: 10.1002/j.1460-2075.1982.tb01133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Sande J. H., McIntosh L. P., Jovin T. M. Mn2+ and other transition metals at low concentration induce the right-to-left helical transformation of poly[d(G-C)]. EMBO J. 1982;1(7):777–782. doi: 10.1002/j.1460-2075.1982.tb01247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]



