Abstract
We describe imaging the luminance of green fluorescent protein (GFP)-expressing bacteria from outside intact infected animals. This simple, nonintrusive technique can show in great detail the spatial–temporal behavior of the infectious process. The bacteria, expressing the GFP, are sufficiently bright as to be clearly visible from outside the infected animal and recorded with simple equipment. Introduced bacteria were observed in several mouse organs including the peritoneal cavity, stomach, small intestine, and colon. Instantaneous real-time images of the infectious process were acquired by using a color charge-coupled device video camera by simply illuminating mice at 490 nm. Most techniques for imaging the interior of intact animals may require the administration of exogenous substrates, anesthesia, or contrasting substances and require very long data collection times. In contrast, the whole-body fluorescence imaging described here is fast and requires no extraneous agents. The progress of Escherichia coli-GFP through the mouse gastrointestinal tract after gavage was followed in real-time by whole-body imaging. Bacteria, seen first in the stomach, migrated into the small intestine and subsequently into the colon, an observation confirmed by intravital direct imaging. An i.p. infection was established by i.p. injection of E. coli-GFP. The development of infection over 6 h and its regression after kanamycin treatment were visualized by whole-body imaging. This imaging technology affords a powerful approach to visualizing the infection process, determining the tissue specificity of infection, and the spatial migration of the infectious agents.
Keywords: green fluorescent protein, external optical imaging, Escherichia coli, antibiotic response, mice
Many biological research techniques depend on detecting uniquely distinguishable cells, e.g., cells exhibiting unique or aberrant morphologies. Far greater sensitivity is achieved by adding exogenous markers, especially those that are optically visible. The techniques of early embryologists marking developmental pathways with India ink injections have evolved into modern methods that include genetically transducing cells with reporters such as dye-activating enzymes. However, such methods still could not detect small, rare targets and usually could not be visualized in living systems. A qualitative advance in sensitivity is afforded by the new class of cell marking reagents exemplified by green fluorescent protein (GFP) and its derivatives. Genetically based, these can be permanent and nontoxic, and are very strongly fluorescent. Most importantly, because their presence seems to have little effect on the marked cell, observations in living tissue are now possible. We show here that GFP enables previously impossible, noninvasive tracking of bodily disease agents in space and time.
GFP has been used as a reporter gene for numerous biological processes (1). Its freedom from required substrates or cofactors allows it to be expressed and visualized in living cells with no apparent biological damage (1). The extreme sensitivity afforded by GFP-expressing cells has been a powerful tool in studying metastasis. We developed the necessary GFP-expressing human cancer cells and showed that these gave rise to metastases that can be visualized at the single-cell level in freshly dissected tissue or in intravital examination (1, 2). Studies with GFP-expressing cancer cells of the lung (3, 4), ovary (2), prostate (5), and pancreas (6) have revealed the earliest metastatic events and afford rapid, accurate determination of the metastatic potential of these tumor types.
In the course of experiments with implanted GFP-labeled tumors, we noted that the fluorescence was sufficiently strong as to be visible from outside the animal. This led to a simple but effective technique for imaging whole, intact mice so that internal GFP-fluorescent tumors were clearly visible. These whole-body images afforded unique, real-time views of tumor growth and metastasis of GFP-expressing cancer cells (7). The whole-body imaging technique requires only simple (490-nm from a xenon or mercury lamp) illumination of mice bearing GFP-expressing tumors and image capture with a charge-coupled device color video camera (7). Tumor growth and metastasis were imaged in the colon, brain, liver, skeleton, and other organs (7). The nonintrusiveness of the technique is worth noting. In contrast to other imaging techniques, no substrate injection, radioactivity, contrast agent, or anesthesia is required (8). Also, whereas most imaging technologies require lengthy exposures of immobilized animals (8–18), the whole-body fluorescence images of GFP-expressing tumors are acquired essentially instantaneously (7).
The relative ease and efficacy with which implanted fluorescent tumors can be imaged externally suggested extending the technique to other foreign agents such as infecting bacteria. Escherichia coli was transfected with a high-expression plasmid containing the GFP gene. The GFP-expressing E. coli (E. coli-GFP) was administered to mice by various routes and the fate of the bacteria was readily visualized in real time by whole-body imaging. This technique makes possible the rapid in vivo screening and evaluation of antibiotics, the role of virulence genes, and other determinants of infection, pathology, and immunity.
Materials and Methods
GFP Vector.
A variant of the Renilla mulleri (RMV)-GFP (M.Z., M. Xu., and R.M.H., unpublished data) was used. RMV-GFP was cloned into the BamHI and NotI sites of the pUC19 derivative pPD16.43 (CLONTECH) with GFP expressed from the lac promoter. The vector was termed pRMV-GFP.
E. coli-GFP.
pRMV-GFP was transfected into E. coli JM 109 competent cells (Stratagene) by standard methods. Transformed E. coli JM109 were selected by ampicillin resistance on agar plates. High-expression E. coli-GFP clones were selected by fluorescence microscopy.
Mice.
nu/nu/CD-1 mice (4-week-old females) were used for infection studies. All animal studies were conducted in accordance with the principles and procedures outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals under assurance number A3873-1.
E. coli-GFP Infection of Stomach, Small Intestine, and Colon in Mice.
Mice were gavaged with 1 ml of an E. coli-GFP suspension (1 × 1011 cells per ml) with a 20-gauge barrel tip feeding needle (Fine Science Tools, Belmont, CA) and latex-free syringe (Becton Dickinson).
E. coli-GFP Direct Colon Infection.
A solution (1 ml) containing 1 × 1011 E. coli-GFP per mouse was administered into the colon by enema using a 20-gauge barrel-tip feeding needle (Fine Science Tools) and latex-free syringe (Becton Dickinson).
E. coli-GFP Peritoneal Infection.
The mice in each group were given an i.p injection of 109 to 1010 E. coli-GFP. A 1-ml 29G1 latex-free syringe (Becton Dickinson) was used.
Antibiotic Treatment.
E. coli-GFP-infected mice were given an i.p. injection of 2 mg of kanamycin (Fisher Scientific) in 100 μl. Mice in the control group were given an i.p. injection of 100 μl of PBS instead of antibiotic.
Whole-Body and Intravital Imaging of E. coli-GFP (7).
Imaging was carried out in a light box illuminated by blue light fiber optics (Lightools Research, Encinitas, CA). Images were captured by using a Hamamatsu C5810 three-chip cooled color charge-coupled device camera (Hamamatsu Photonics Systems, Bridgewater, NJ). Images of 1024 × 724 pixels were captured either directly on an IBM PC or continuously through video output on a high-resolution Sony VCR (model SLV–R1000; Sony, Tokyo). Images were processed for contrast and brightness and analyzed with the use of IMAGE PRO PLUS 3.1 software (Media Cybernetics, Silver Spring, MD).
Results
External Whole-Body Imaging of Gastrointestinal Infection with E. coli-GFP.
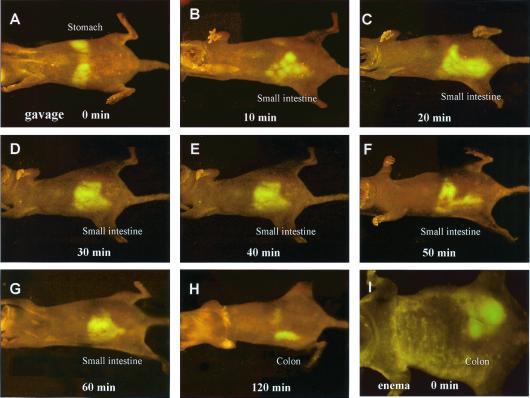
E. coli-GFP introduced to the mouse gastrointestinal tract by gavage became visible in the stomach in whole-body images almost immediately (Fig. 1A). The stomach emptied within 10 min after gavage and the E. coli-GFP next appeared in the small intestine (Fig. 1 B–G). The bacterial population in the small intestine appeared to peak at 40 min after gavage (Fig. 1E) and disappeared by 120 min (Fig. 1H). After 120 min, E. coli-GFP appeared in the colon (Fig. 1H). Direct colonic inoculation with E. coli-GFP was also visualized after intra-anal enema delivery (Fig. 1I).
Figure 1.
Whole-body imaging of E. coli-GFP infection in various organs. (A) E. coli-GFP infection in the stomach immediately after gavage of 1011 E. coli-GFP. (B) E. coli-GFP infection in the small intestine 10 min after gavage. (C) E. coli-GFP infection in the small intestine 20 min after gavage. (D) E. coli-GFP infection in the small intestine 30 min after gavage. (E) E. coli-GFP infection in the small intestine 40 min after gavage. (F) E. coli-GFP infection in the small intestine 50 min after gavage. (G) E. coli-GFP infection in the small intestine 60 min after gavage. (H) E. coli-GFP infection in the colon 120 min after gavage. (I) E. coli-GFP infection in the colon immediately after enema of 1011 E. coli-GFP.
Comparison of Whole-Body and Intravital Imaging of Gastrointestinal E. coli-GFP.
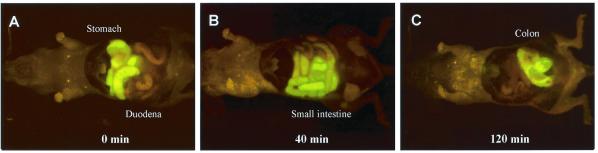
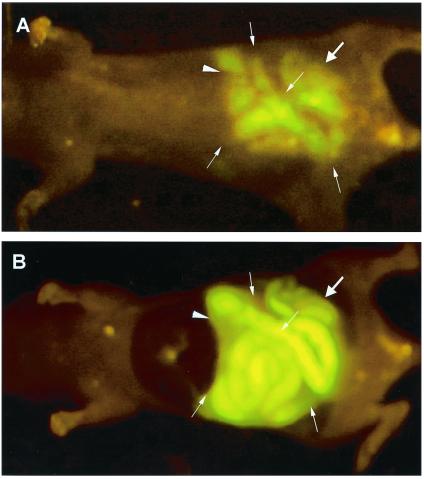
At appropriate times after gavage, the abdominal cavity was opened and intravital images made of the E. coli-GFP fluorescence. The stomach (Fig. 2A), small intestine (Fig. 2B), and colon (Fig. 2C) were brightly fluorescent with E. coli-GFP as seen by intravital imaging. Multiple gavage with E. coli-GFP allowed simultaneous inoculation of the stomach, small intestine, and colon, which were imaged by whole-body (Fig. 3A) and intravital techniques (Fig. 3B). Comparison of whole-body and intravital images of E. coli-GFP in the stomach, small intestine, and colon showed a high degree of correspondence (Figs. 1–3).
Figure 2.
Intravital imaging of E. coli-GFP infection in the stomach, small intestine, and colon after gavage. (A) E. coli-GFP infection in the stomach and the duodenum immediately after gavage of 1011 E. coli-GFP. (B) E. coli-GFP infection in the small intestine 40 min after gavage. (C) E. coli-GFP infection in the colon 120 min after gavage.
Figure 3.
Whole-body and intravital imaging of E. coli-GFP infection in the stomach, small intestine, and colon after gavage. (A) Whole-body image of E. coli-GFP infection in the stomach (arrowhead), the small intestine (fine arrows), and the colon (thick arrow) after multiple gavage of aliquots 3 × 1011 E. coli-GFP. (B) Intravital image of E. coli-GFP infection in the stomach (arrowhead), the small intestine (fine arrows), and the colon (thick arrow) after multiple gavage of aliquots of 3 × 1011 E. coli-GFP.
External Whole-Body Imaging of an i.p. Infection and Its Response to an Antibiotic.
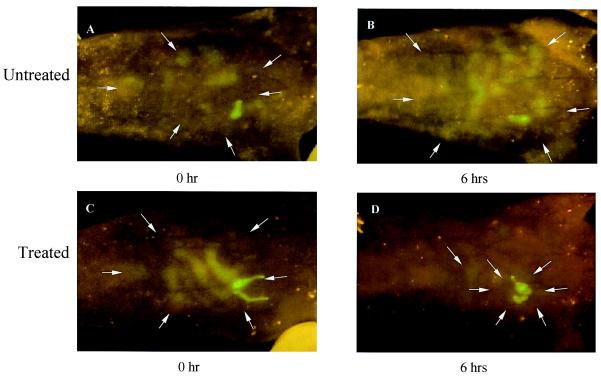
An authentic infection was established in a mouse by i.p. inoculation with E. coli-GFP. Immediately after injection, the fluorescent bacteria were seen localized around the injection site by external whole-body imaging (Fig. 4 A and C). Six hours later, the E. coli-GFP were seen to spread throughout the peritoneum (Fig. 4B), coinciding with the death of the animal. Intravital imaging of E. coli-GFP in the open peritoneal cavity at 6 h (Fig. 5) showed a bacterial distribution similar to that seen by external whole-body imaging. Intraperitoneally infected animals were next treated with kanamycin after inoculation. Whole-body imaging showed a marked reduction of the bacterial population over the next 6 h (Fig. 4 C and D). Previous attempts to image infection in intact animals used bacteria expressing luciferase (14), which, because of the much lower luminosity, is a far more difficult and intrusive procedure.
Figure 4.
Whole-body imaging of E. coli-GFP peritoneal cavity infection and antibiotic response. (A and C) E. coli-GFP infection in the peritoneal cavity immediately after i.p. injection of 109 E. coli-GFP. (B) Untreated mouse 6 h after i.p. injection. Animal died at this time point. (D) Kanamycin-treated mouse 6 h after i.p. injection. Animal survived. Arrows indicate the fluorescent images.
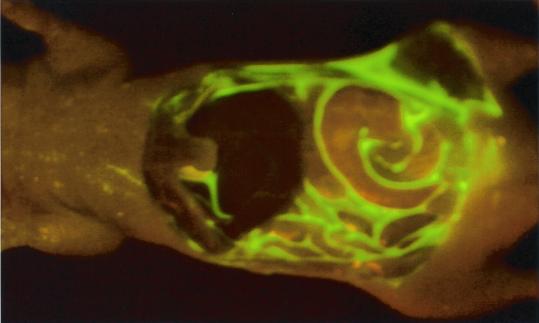
Figure 5.
Intravital imaging of E. coli-GFP peritoneal cavity infection. E. coli-GFP infection in the peritoneal cavity immediately after i.p. injection of 109 E. coli-GFP. The wall of the abdominal cavity was removed.
Discussion
The advent of GFP as a fluorescent cell marker has brought unprecedented sensitivity and discrimination to the study of cell behavior. These qualities were uniquely applicable to our ongoing studies of tumor metastases. GFP-labeled tumors were developed and implanted orthotopically, where they express their proper metastatic behavior. The strong GFP fluorescence allowed detection of micrometastases down to the single-cell level. In the course of these studies, we noted that the strong tumor luminescence was visible from outside the intact, living animal. The ability to noninvasively image tumors enabled an entire program of research.
We also applied the GFP whole-body imaging technique to visualize gene expression in several organs of the mouse (19). Mice were labeled in the brain, liver, pancreas, prostate, or bone by using adenoviral-GFP (19). This technique may allow the noninvasive visualizing of the expression and modulation of specific genes that have been coupled to GFP.
Recently, we have shown whole-body images of angiogenesis in GFP-expressing tumors in mice, including orthotopically implanted tumors (20). In this technique the nonfluorescent tumor induced blood vessels are readily apparent in stark contrast to the GFP-fluorescent tumors. The whole-body images, enable quantitation of tumor vascularization noninvasively in real time.
GFP-expressing bacteria have been previously used in a number of studies that did not involve intact, living animals (21–29). An example of such studies was the visualization of the in vitro infection of muscle tissue by the pathogenic E. coli O157H GFP (28). Another approach examined the mouse gastrointestinal tract after gavage infection by removal and fixation of the gastrointestinal tissue (29). Fish infected with GFP-transduced Edwardsiella tarda were imaged for infection after removal of their organs (22). Genes associated with virulence and other infectious processes were evaluated by linkage to GFP expression (22–25).
A very different technique for generating and imaging interior luminescence uses bioluminescent bacteria. The light source is the luciferin–luciferase reaction, the quantum yield of which appears to be far lower than for an equivalent bacterial population labeled with GFP. Although the infection could be whole-body imaged, the signal was relatively weak. Consequently, imaging required long collection times during which the animal had to be immobilized and anesthetized. The signal from the much brighter GFP-labeled bacteria allowed instantaneous image capture with high organ resolution, even in a simple light box, with freely moving animals in a lighted room. No substrates, radioactivity, contrast agent, anesthesia, or other perturbation is required, just illumination with blue light.
The technique of whole-body imaging of E. coli-GFP infection in mice reported here is a significant advance that enables real-time infection studies in a mammal without perturbing the animal. The whole-body imaging capability could be used to screen and study the efficacy of new antibiotics on drug-resistant, GFP-labeled bacteria. It will be possible to see the bacterial behavior in various organs of the mouse. Virulence genes can also be studied with regard to how they influence infection in various organs by whole-body imaging in real time. The whole-body imaging technique should allow greatly increased precision and detail in examining bacterial–host interactions.
Abbreviations
- GFP
green fluorescent protein
- RMV
a variant of Renilla mulleri
References
- 1.Hoffman R M. Methods Enzymol. 1999;302:20–31. doi: 10.1016/s0076-6879(99)02005-4. [DOI] [PubMed] [Google Scholar]
- 2.Chishima T, Miyagi Y, Wang X, Yamaoka H, Shimada H, Moossa A R, Hoffman R M. Cancer Res. 1997;57:2042–2047. [PubMed] [Google Scholar]
- 3.Rashidi B, Yang M, Jiang P, Baranov E, An Z, Wang X, Moossa A R, Hoffman R M. Clin Exp Metastasis. 2000;18:57–60. doi: 10.1023/a:1026596131504. [DOI] [PubMed] [Google Scholar]
- 4.Yang M, Hasegawa S, Jiang P, Wang X, Tan Y, Chishima T, Shimada H, Moossa A R, Hoffman R M. Cancer Res. 1998;58:4217–4221. [PubMed] [Google Scholar]
- 5.Yang M, Jiang P, Sun F X, Hasegawa S, Baranov E, Chishima T, Shimada H, Moossa A R, Hoffman R M. Cancer Res. 1999;59:781–786. [PubMed] [Google Scholar]
- 6.Bouvet M, Yang M, Nardin S, Wang X, Jiang P, Baranov E, Moossa A R, Hoffman R M. Clin Exp Metastasis. 2000;18:213–218. doi: 10.1023/a:1006767405609. [DOI] [PubMed] [Google Scholar]
- 7.Yang M, Baranov E, Jiang P, Sun F-X, Li X-M, Li L, Hasegawa S, Bouvet M, Al-Tuwaijri M, Chishima T, et al. Proc Natl Acad Sci USA. 2000;97:1206–1211. doi: 10.1073/pnas.97.3.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Budinger T F, Benaron D A, Koretsky A P. Ann Rev Biomed Eng. 1999;1:611–648. doi: 10.1146/annurev.bioeng.1.1.611. [DOI] [PubMed] [Google Scholar]
- 9.Herschman H R, MacLaren D C, Iyer M, Namavari M, Bobinski K, Green L A, Wu L, Berk AJ, Toyokuni T, Barrio J R, et al. J Neurosci Res. 2000;59:699–705. doi: 10.1002/(SICI)1097-4547(20000315)59:6<699::AID-JNR1>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 10.Louie A Y, Huber M M, Ahrens E T, Rothbacher U, Moats R, Jacobs R E, Fraser S E, Meade T J. Nat Biotechnol. 2000;18:321–325. doi: 10.1038/73780. [DOI] [PubMed] [Google Scholar]
- 11.Weissleder R, Moore A, Mahmood U, Bhorade R, Benveniste H, Chiocca E A, Basilion J P. Nat Med. 2000;6:351–354. doi: 10.1038/73219. [DOI] [PubMed] [Google Scholar]
- 12.Gambhir S S, Barrio J R, Phelps M E, Iyer M, Namavari M, Satyamurthy N, Wu L, Green L A, Bauer E, MacLaren D C, et al. Proc Natl Acad Sci USA. 1999;96:2333–2338. doi: 10.1073/pnas.96.5.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tjuvajev J G, Finn R, Watanabe K, Joshi R, Oku T, Kennedy J, Beattie B, Koutcher J, Larson S, Blasberg R G. Cancer Res. 1996;56:4087–4095. [PubMed] [Google Scholar]
- 14.Contag P R, Olomu I N, Stevenson D K, Contag C H. Nat Med. 1998;4:245–247. doi: 10.1038/nm0298-245. [DOI] [PubMed] [Google Scholar]
- 15.Alfano R R, Demos S G, Gayen S K. Ann NY Acad Sci. 1997;820:248–270. doi: 10.1111/j.1749-6632.1997.tb46200.x. [DOI] [PubMed] [Google Scholar]
- 16.Masters B R, So P T, Gratton E. Ann NY Acad Sci. 1998;838:58–67. doi: 10.1111/j.1749-6632.1998.tb08187.x. [DOI] [PubMed] [Google Scholar]
- 17.Wu J, Perelman L, Dasari R, Feld M. Proc Natl Acad Sci USA. 1997;94:8783–8788. doi: 10.1073/pnas.94.16.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alfano R R, Demos S G, Galland P, Gayen S K, Guo Y, Ho P P, Liang X, Liu F, Wang L, Wang Q Z, Wang W B. Ann NY Acad Sci. 1998;838:14–28. doi: 10.1111/j.1749-6632.1998.tb08184.x. [DOI] [PubMed] [Google Scholar]
- 19.Yang M, Baranov E, Moossa A R, Penman S, Hoffman R M. Proc Natl Acad Sci USA. 2000;97:12278–12282. doi: 10.1073/pnas.97.22.12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang M, Baranov E, Li X-M, Wang J-W, Jiang P, Li L, Moossa A R, Penman S, Hoffman R M. Proc Natl Acad Sci USA. 2001;98:2616–2621. doi: 10.1073/pnas.051626698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu H, Song Z, Hentzer M, Andersen J B, Heydorn A, Mathee K, Moser C, Eberl L, Molin S, Hoiby N, Givskov M. Microbiology. 2000;146:2481–2493. doi: 10.1099/00221287-146-10-2481. [DOI] [PubMed] [Google Scholar]
- 22.Ling S H, Wang X H, Xie L, Lim T M, Leung K Y. Microbiology. 2000;146:7–19. doi: 10.1099/00221287-146-1-7. [DOI] [PubMed] [Google Scholar]
- 23.Badger J L, Wass C A, Kim K S. Mol Microbiol. 2000;36:174–182. doi: 10.1046/j.1365-2958.2000.01840.x. [DOI] [PubMed] [Google Scholar]
- 24.Kohler R, Bubert A, Goebel W, Steinert M, Hacker J, Bubert B. Mol Gen Genet. 2000;262:1060–1069. doi: 10.1007/pl00008649. [DOI] [PubMed] [Google Scholar]
- 25.Valdivia R H, Hromockyj A E, Monack D, Ramakrishnan L, Falkow S. Gene. 1996;173:47–52. doi: 10.1016/0378-1119(95)00706-7. [DOI] [PubMed] [Google Scholar]
- 26.Valdivia R H, Falkow S. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 27.Scott K P, Mercer D K, Richardson A J, Melville C M, Glover L A, Flint H J. FEMS Microbiol Lett. 2000;182:23–27. doi: 10.1111/j.1574-6968.2000.tb08867.x. [DOI] [PubMed] [Google Scholar]
- 28.Prachaiyo P, McLandsborough L A. J Food Prot. 2000;63:427–433. doi: 10.4315/0362-028x-63.4.427. [DOI] [PubMed] [Google Scholar]
- 29.Geoffroy M C, Guyard C, Quatannens B, Pavan S, Lang M, Mercenier A. Appl Environ Microbiol. 2000;66:383–391. doi: 10.1128/aem.66.1.383-391.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]