Abstract
In mammals, homologous chromosomes rarely pair outside of meiosis. An exception is the X-chromosome, which transiently pairs during X-chromosome inactivation (XCI). How two chromosomes find each other in 3D space is not known. Here, we reveal a required interaction between the X-inactivation center (Xic) and the telomere in mouse embryonic stem cells. The sub-telomeric, pseudoautosomal region (PAR) of both sex chromosomes (X,Y) also undergoes pairing. PAR transcribes a class of telomeric RNA, dubbed “PAR-TERRA”, which accounts for a vast majority of all TERRA transcripts. PAR-TERRA binds throughout the genome, including PAR and Xic. PAR-TERRA anchors the Xic to PAR, creating a “tetrad” of pairwise homologous interactions (Xic:Xic, PAR:PAR, Xic:PAR). Xic pairing occurs within the tetrad. Depleting PAR-TERRA abrogates pairing and blocks initiation of XCI, whereas autosomal PAR-TERRA induces ectopic pairing. We proposed a Constrained Diffusion Model in which PAR-TERRA creates an interaction hub to guide Xic homology searching during XCI.
INTRODUCTION
The mammalian genome is ubiquitously transcribed and the ends of telomeres are no exception. The telomere produces a heterogeneous population of long noncoding RNAs known as “TERRA” 1–3. TERRA contains the telomeric repeat sequence, UUAGGG, and sequences unique to sub-telomeric regions of each chromosome. TERRA biology has been of major interest, as it has been linked to many human diseases and forms an integral part of telomeric architecture 4,5. Although TERRA research has focused exclusively on telomeres, only a fraction of detectable TERRA transcripts resides at telomeres 6 and TERRA has a propensity to localize near the inactive X-chromosome (Xi) of female cells 3,7. While TERRA also concentrates next to the Y-chromosome 3, a functional link to sex chromosome biology has remained elusive. Until now, efforts to understand functional linkages have been hampered by difficulties in depleting TERRA, an incomplete understanding of TERRA’s transcriptional origin, and limited knowledge of genomic targets.
Here we interrogate TERRA’s potential link to X-chromosome inactivation (XCI). XCI inactivates one X-chromosome in mammalian females to balance gene expression with XY males. XCI is driven by the X-inactivation center (Xic) via noncoding genes, Xite, Tsix, and Xist 8–12. During early development, a counting mechanism determines X-chromosome number and initiates XCI in cells with two or more X-chromosomes 13. An allelic choice mechanism then randomly selects one active and one inactive X (Xa, Xi). On the Xi-elect, the Xist gene initiates chromosome-wide silencing, whereas Tsix antagonizes Xist action on the Xa-elect. A switch from biallelic to monoallelic Tsix expression marks the onset of XCI and establishes allelic choice 14,15. Interestingly, Tsix’s transition to a monoallelic state coincides with a homologous pairing event between two Xic’s 16–20. The required 15-kb region within Tsix and Xite (“pairing center” 17,18) is thought to provide a platform on which biallelically bound transcription factors could be partitioned to one allele during pairing and, in this way, ensure that only one Tsix allele continues to be expressed 18,19,21. Abolishing pairing results in loss of mutually exclusive choice of Xa and Xi, whereas ectopic pairing prevents initiation of XCI by preventing trans-interactions between two Xic alleles 16,18,22. Thus, pairing appears to be critical to counting and allelic choice.
X:X pairing is one of the only known examples of homologous pairing in the mammalian soma. Given emergence of other examples 23–26, somatic pairing may be more prevalent than currently suspected. X:X pairing is an excellent model system to investigate pairing mechanisms, as it can be recapitulated by a genetically tractable system — mouse embryonic stem (ES) cells. In female ES cells, X:X pairing is observed between days 1–4 of differentiation 18,27. Contact occurs for <30 minutes 17,20. Recent evidence suggests that Tsix and Xite RNAs co-transcriptionally recruit the chromosomal architectural protein, CTCF, which in turn serves as interchromosomal “glue” 17,28. Currently unknown is how the two pairing centers find each other in 3D space. Here we investigate this question and uncover a central role for the pseudoautosomal region (PAR), through an RNA tether that we call PAR-TERRA.
RESULTS
Telomeric pairing of sex chromosomes in ES cells
In a previous study, we noted that a significant number of female ES cells show a coalescence of X-telomeric ends early in cell differentiation 3. Because Xic:Xic pairing is one of the earliest measurable events in differentiating female cells, this older observation led us to question whether telomeric agglomeration maybe related to Xic pairing. We performed 3D DNA FISH assays and measured inter-allelic distances for the Xic (Xist/Tsix), the telomere, and various unrelated X-linked locus (Fig. 1a–b, S1a–c). For all pairing assays, we simultaneously tracked at least two X-linked loci (labelled with different fluorophores) in order to avoid scoring sister chromatids and ensure inclusion only of distinct alleles. To track X-linked telomeres, we labelled distal Chromosome X (ChrX) BAC probes: RP23-461E16 containing Arhgap6, and RP24-500I4 mapping within the pseudoautosomal region (PAR). The PAR is a subtelomeric region shared by Chr X and Y 29,30. To isolate PAR-specific probes, we generated unique PCR fragments from RP24-500I4 to create a cocktail containing P3, P4, P5, P6, and P8 that specifically identifies the ends of both sex chromosomes (P34568; Fig. S1d–f). We plotted inter-allelic distances as a fraction of nuclear diameters (ND) between 0.0 and 1.0, and applied the Kolmogorov-Smirnov (KS) test to assess statistical significance (Fig. S1g). An increase in the number of nuclei with inter-allelic distances of <0.1 ND (<1 μm) is broadly defined as “paired” 17–20,27.
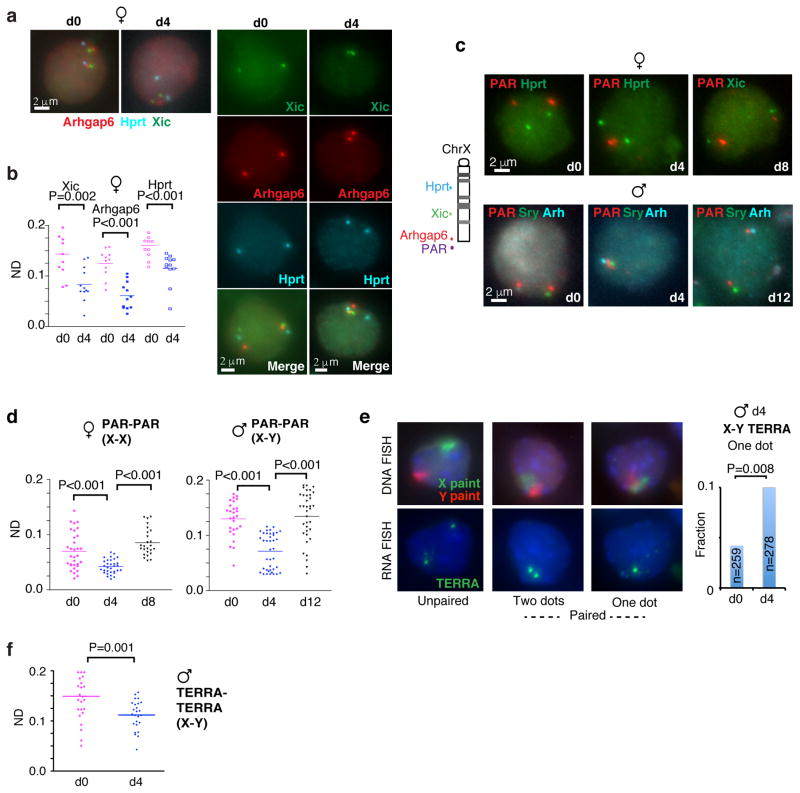
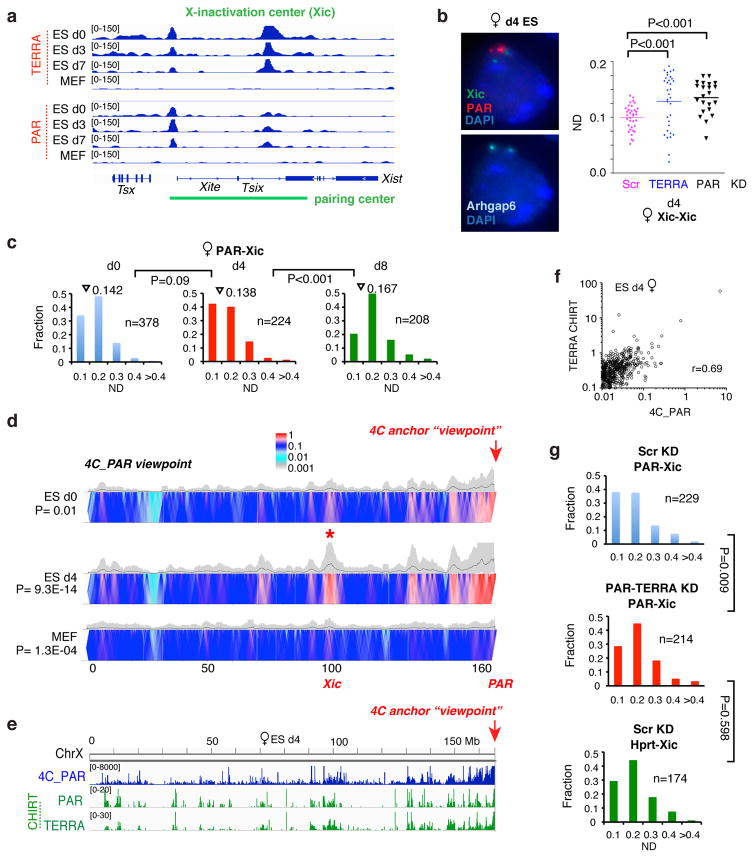
Figure 1. Homologous sex chromosome pairing via the pseudoautosomal region (PAR) in differentiating XX and XY ES cells.
a. DNA FISH pairing assays using Xic, Arhgap6 (BAC RP23-461E16), and Hprt probes in female ES cells undergoing differentiation on d0 and d4. FISH signals are pseudocolored as indicated.
b. Dotplot of inter-allelic distances for the top decile of nuclei (with smallest distances) shown in Fig. 1a and S1g. ND, normalized distance = distance/nuclear diameter. P values determined using two-tailed student t-test.
c. Multi-color DNA FISH shows PAR:PAR pairing in both male and female ES cells. P34568 sub-probe sets derived from RP24-500I4 were used to detact the PAR. Additional X-linked (Hprt, Xic, Arhgap6 [Arh]) and Y-linked (Sry) were used as controls both to rule out full-length pairing and to ensure efficient hybridization to both homologues. We scored only those nuclei with discernible signals for both PAR and control locus. Multiple biological replicates showed similar results.
d. PAR-to-PAR distances for the top decile during female and male ES cell differentiation for the experiment in c. P values determined using two-tailed student t-test.
e. Left panel: Serial RNA-DNA FISH using X and Y painting probes to detect ChrX and ChrY (DNA FISH) and using a TERRA probe to detect the distal telomeric ends (RNA FISH) in d4 male ES cells. Right panel: Histogram shows that, on d4 of differentiation, X- and Y-PAR signals are frequently merged (one dot, or unresolvable dots considered true pairing events). P value determined using Fisher exact test.
f. Dotplot of the top decile of inter-TERRA distances in d0 versus d4 male ES cells. P value was determined using two-tailed student t-test.
We observed a significant reduction in average inter-chromosomal distances between days 0 and 4 (d0, d4) not only at the Xic’s, but also at the distal ends of ChrX. A left shift in the full distribution and an overall increase in the number of allelic pairs in the 0.0–0.1 ND bin indicated allelic clustering of Xic’s and telomeres. By contrast, few Hprt alleles fell in the 0.0–0.1 ND range. Because pairing is transient (t1/2 <30 minutes) and only a few percent of asynchronously differentiating ES cells are paired in a single snapshot 17, we focused on nuclei in the first decile – the top 10% with the shortest inter-allelic distances (Fig. 1b). Among this fraction, the transition from d0 to d4 resulted in a decrease in allelic Xic and Arhgap6 distances suggestive of pairing. At d4, a large number of nuclei showed inter-Arhgap6 distances of <0.1 ND. The same was true of inter-Xic distances, but not for negative control X-linked control loci (Fig. 1b,S1a–c,g). Clustering was most dramatic when viewed from the PAR probe, RP24-500I4 (Fig. 1c–d,S1a,f,h). At d4, ~14% of PAR:PAR measurements fell within the 0.0–0.1 ND bin and the top 10% of nuclei showed a major reduction of inter-allelic distances (Fig. S1h). Thus, like the Xic’s, telomeric ends of ChrX undergo transient allelic pairing.
Given that ChrY also has PAR, we asked whether PAR pairing occurs in differentiating male ES cells as well. Srikingly, inter-PAR distances also decreased (Fig. 1c–d,S1h), indicating that the X- and Y-PAR indeed pair. In agreement, X- and Y-painting probes also showed transient clustering of Chr X and Y, but they came together without merging (Fig. 1e). By performing RNA FISH on the same nuclei to detect TERRA 3, we observed a merging of two TERRA signals (“two dots”) into one (“one dot”; dots unresolvable by light microscopy)(Fig. 1f), suggesting that X-Y pairing is confined to the PAR. Among the 1st decile, the number of nuclei with X-Y distances of <0.1 ND also increased significantly (Fig. 1h). We conclude that sex chromosomes (X,Y) undergo transient PAR:PAR pairing during the same time window of Xic:Xic pairing.
Identification of sex-linked subtelomeric transcripts: PAR-TERRA
Our previous work had revealed a large telomeric RNA cluster next to sex chromosomes 3. By RNA/DNA fluorescence in situ hybridization (FISH) using TERRA oligo probes, we confirmed two prominent TERRA foci at distal ChrX telomeres, colocalizing with PAR DNA, in female ES cells (Fig. S1f). Additionally, we noticed that RNA signals generated by PAR hybridization greatly resembled TERRA signals (Fig. 2a–e), suggesting that PAR is transcribed and its transcript accumulates. This intriguing relationship led us to explore a possible structural and functional linkage between PAR and TERRA. Although previous cytological analysis showed TERRA localization to the ends of most, if not all chromosomes 1,7, where TERRA originates is not fully known. TERRA may be transcribed by all telomeres and retained in cis, or be transcribed by select loci and disseminated in trans to other sites. One study suggested TERRA originates predominantly in Chr18 31. Because sub-telomeric sequences of most mouse chromosomes have not been fully assembled, additional origins could have escaped detection. Because PAR is contiguous to the telomere and resembles TERRA signals, we asked if PAR and TERRA may be one long sex-linked telomeric transcript.
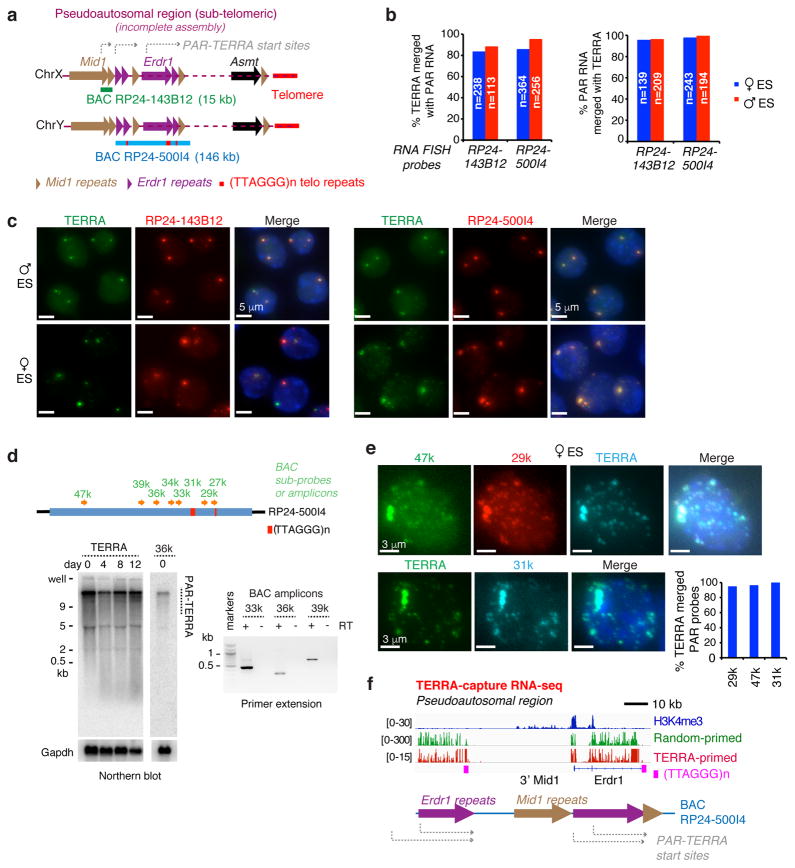
Figure 2. PAR transcripts produced by the sex chromosomes.
a. The pseudoautosomal region (PAR) at the distal ends of Chr X and Y. Dotted purple lines indicate that this region is incompletely sequenced and assembled in the current genome assemblies (mm9, mm10). Mid1 and Erdr1 have repeated fragments within PAR (brown and purple triangles). The telomeric repeats (red bars) are present within PAR. PAR BAC clones: 15 kb BAC RP24-143B12 and the ~146 kb RP24-500I4.
b. Quantitation of percent overlapping TERRA and PAR RNA signals for the experiments in c. Multiple biological replicates showed similar results.
c. Two color RNA FISH detecting TERRA (Alexa488, green) and PAR transcripts (BAC probes, Cy3, red) in ES cells.
d. Top panel: Map of sub-BAC probes and PCR amplicons. Left panel: Northern blot analysis of PAR-TERRA in ES cells using TERRA-specific or PAR-specific oligo probes. Left panel: Northern blot analysis of PAR-TERRA RNA using either TERRA or PAR-36K oligo probes in ES cells on different differentiation days. GAPDH, loading control. Right panel: Primer extension using an antisense TERRA oligo probe with PCR amplification using PAR-specific primer pairs located at 33, 36, and 39 k (kb) from the end of BAC RP24-500I4. +, with RT; −, without RT.
e. RNA FISH indicating colocalization of TERRA and PAR signals at both large and small foci in ES cells. Three-color RNA FISH (upper panel): TERRA oligo probe (cyan blue); PAR-specific probes, 47k (green) and 29k (red). Two-color RNA FISH (lower panel): TERRA oligo probe (green); PAR specific probe, 31k (cyan blue). DAPI (blue) for nuclear stain. Right graph, quantitation of colocalization.
f. IGV screenshots of TERRA-capture RNA-seq experiments show deduced PAR-TERRA transcription start sites. PAR transcripts are linked to telomeric repeat RNA. RT was conducted with TERRA-specific primers versus random hexamers, as indicated.
The PAR comprises the coding genes, Mid1, Erdr1, and Asmt, and a number of highly repetitive sequences, including internal telomeric (TTAGGG)n repeats and various intronic/exonic repeats of Mid1 and Erdr1 (Fig. 2a). Because of its repetitive nature, the current genome assembly remains incomplete in this region. RNA FISH using PAR BAC probes, RP24-143B12 and RP24-500I4, showed large foci that nearly perfectly overlapped TERRA signals in both male and female ES cells (Fig. 2a–c). Northern blot analyses using various PAR probes (Fig. S2a, 2d, top and left panels) showed that each detected a heterogeneous population dominated by a high-molecular-weight band of ≫9kb, in a pattern that resembled TERRA’s 1,31. PAR patterns revealed by probes 27k, 29k, 34k, 36k, and 47k (all unique to PAR), were similar and remained consistent during cell differentiation (Fig. 2d-left panel, S2a). RT primer extension using an antisense TERRA oligo yielded positive PCR amplification with PAR-specific primer pairs at 33, 36, and 39 kb from the distal end of BAC RP24-500I4 (Fig. 2d-right panel). Notably, RNA FISH showed that PAR and TERRA signals colocalized with each other not only at sex chromosomes, but also within finer speckles throughout the nucleus (>90%, Fig. 2e). Thus, PAR and TERRA show striking resemblance as viewed by orthogonal assays.
To investigate the possibility of PAR being continuous with TERRA, we conducted TERRA-capture RNA-seq. We first captured TERRA transcripts from total ES cell RNA using biotinylated oligos complementary to (UUAGGG)n repeats. These TERRA-containing PAR transcripts were enriched 737x and 151x over 18S and GAPDH RNAs, respectively. We then sequenced cDNA generated using TERRA-specific primers or random-hexamers, with similar results using either (Fig. 2f; S2b–d). Unique sequences contiguous to (UUAGGG)n repeats enabled unambiguous alignment to the chromosome of origin. Intriguingly, X- and Y-PAR sequences accounted for the overwhelming majority of reads (>99%) from subtelomeric regions (Fig. S2c,d). TERRA transcripts were also produced from subtelomeric regions of autosomes, but they accounted for <1% of total TERRA. These data argue that PAR and TERRA occur continuously and that sex chromosome produce a large quantity of TERRA.
Because of the repetitive nature of PAR, the exact transcription start site of sex-linked TERRA is presently impossible to pinpoint. However, approximate start sites could be deduced by combining Northern analysis and RNA FISH using chromosome-walking probes, TERRA-capture RNA-seq analyses, and H3K4me3 ChIP-seq profiles. Together, these data suggest multiple potential start sites within the Erdr1 repeats and as far proximal as a central intron of Mid1 (grey broken arrows; Fig. 2a,f). Paired-end RNA-seq showed that these transcripts include long telomeric repeats of 300 to >3000 nt (Fig. S2e,f). In another BAC clone, RP24-338D22, 3kb-long telomeric repeats were found within PAR (Fig. S2e). Tophat alignment indicated continuity between unique PAR-sequences and telomeric repeats (Fig. 2f, S2f), consistent with continuity between PAR and TERRA. However, because telomeric repeat sequences [(TTAGGG)n] are present throughout sub-telomeric regions, it is difficult to determine whether the PAR transcripts extend to the very ends of telomeres.
In sum, these data demonstrate that (i) PAR RNA is a large, heterogeneous transcript containing telomeric repeats, (ii) PAR and TERRA RNA have nearly identical subnuclear localization, and (iii) the overwhelming majority of telomeric transcripts is derived from sex chromosomes in mouse ES cells. We name the sex-linked telomeric transcript, “PAR-TERRA,” to denote its PAR origin and inclusion of (UUAGGG)n repeats.
PAR-TERRA targets chromatin sites both in cis and in trans
To map PAR-TERRA chromatin binding sites, we performed CHIRT6, a hybrid ChIRP 32 and CHART 33 method optimized for PAR-TERRA. Because capture probes could potentially interact with DNA rather than RNA, we included an RNaseH elution step (Fig. S3a). Several capture probe sets were designed: (i) TERRA antisense (TERRA-AS), to capture transcripts containing UUAGGG, (ii) PAR, to capture PAR-containing transcripts (Fig. 3a), and (iii) TERRA sense (TERRA-S, the reverse complement), to control for strand-specificity and further rule out DNA capture. Quantitative RT-PCR indicated that PAR-TERRA transcripts were specifically enriched relative to other nuclear RNAs using TERRA-AS probes (Fig. S3b). PAR DNA was also enriched relative to Hprt in both PAR and TERRA-AS capture (Fig. 3b). Enrichment was dependent on RNaseH and abolished by RNase A (Fig. 3c), indicating that the pulldown was mediated by interaction between DNA capture probes and RNA targets. To exclude artifacts due to probe hybridization to genomic DNA rather than the intended RNA target, we sequenced two critical controls: (i) an RNaseH-control in which RNaseH was omitted in the elution step, which should preclude elution of RNA-dependent interactions; and (ii) a TERRA-S control, which would not hybridize to TERRA RNA but could pull down genomic DNA.
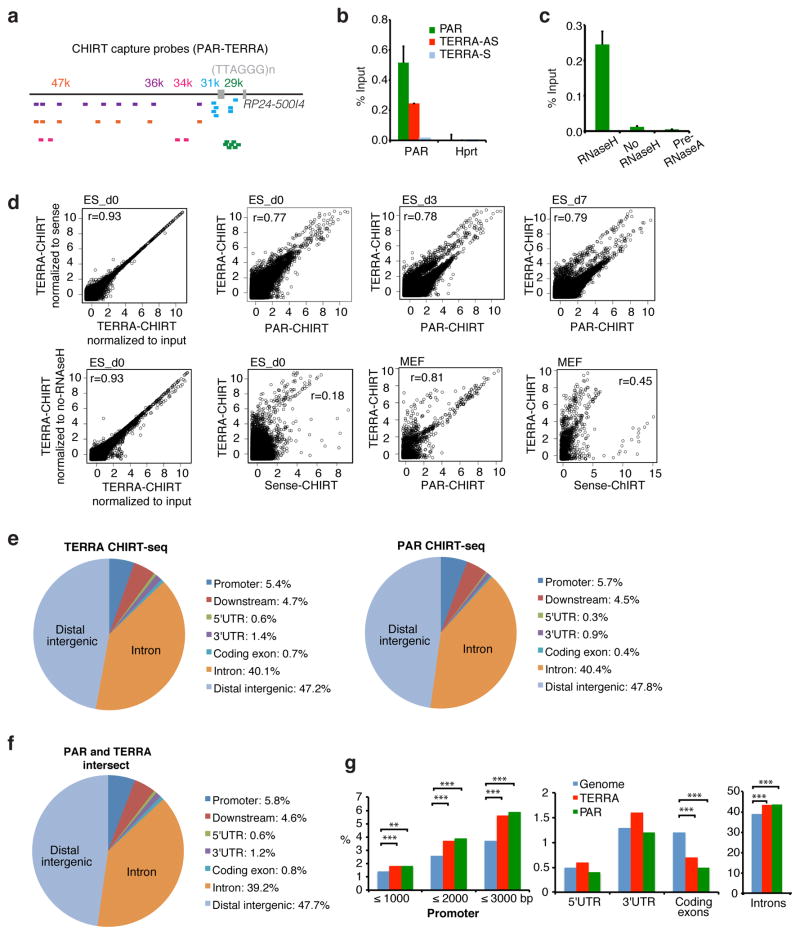
Figure 3. Mapping genomic PAR-TERRA binding sites by CHIRT-seq.
a. PAR CHIRT using five capture probe sets: 29k, 31k, 34k, 36k, and 47k. Each probe cocktail is shown as bars with matching colors.
b. Quantitative PCR showing the enrichment of PAR DNA in TERRA-AS CHIRT and PAR CHIRT, but not TERRA-S CHIRT in ES cells.
c. Enrichment of PAR DNA following TERRA CHIRT was observed only when eluted with RNaseH. Enrichment was abolished by RNase A pre-treatment.
d. Scatterplot analysis comparing Log2 coverages of TERRA and PAR CHIRT. Pearson’s r shown. CHIRT results were normalized to input unless otherwise indicated.
e. Pie charts show relative representation of various genomic regions in PAR CHIRT in d0 female ES cells, as compared to TERRA CHIRT6.
f. Peaks in common between PAR and TERRA CHIRT.
g. CEAS analysis shows significant over-representation of noncoding sequences in PAR and TERRA CHIRT in d0 ES cells. ***, P<0.001 (one-sided binomial test). The genome reference was obtained from the CHIRT-seq input.
We performed CHIRT paired-end sequencing of d0, d3, and d7 female ES samples and mouse embryonic fibroblasts (MEF). PAR-specific binding sites were called using the peak caller MACS and compared to TERRA sites 6 with normalization to (i) input library, (ii) TERRA-S library, or (iii) no-RNaseH library, with each normalization method yielding similar results in two biological replicates (Fig. 3d;S3c,d). In total, 2000–5000 significant peaks were called using PAR and TERRA probes in ES cells (d0, d3, d7), whereas very few were called for the control TERRA-S probe (Table 1). In MEFs, the number of PAR and TERRA peaks were reduced to 400–500, correlating with a 6-fold reduction in MEFs relative to ES cells (Fig. S3e). There was considerable overlap of PAR with TERRA CHIRT profiles, with high Pearson’s r values in correlation plots for d0, d3, d7 ES cells, and MEFs, but not for comparisons to sense-CHIRT controls (Fig. 3d; Table 1). There was also high correlation between cells of various differentiation states (Fig. S4). In MEFs, consistent with a dramatic reduction in PAR-TERRA expression, the number of peaks shared between PAR and TERRA decreased. Interestingly, in all samples, there existed a set of binding sites unique to TERRA, possibly reflecting binding sites for autosomal TERRA (Table 1). These data supported two types of telomeric transcripts — PAR-TERRA and TERRA — with PAR-TERRA being the dominant species in ES cells.
Table 1.
PAR-TERRA binding sites in mouse ES cells and MEFs.
| Numbers of PAR and TERRA binding sites | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ES_d0 | ES_d3 | ES_d7 | MEF | |||||||
| TERRA | PAR | Sense | TERRA | PAR | TERRA | PAR | TERRA | PAR | Sense | |
| Normalized to input | 4054 | 2304 | 78 | 2125 | 5373 | 1884 | 3199 | 500 | 440 | 544 |
| Normalized to sense | 4076 | - | - | - | - | - | - | 492 | - | - |
| Normalized to no-RNase H | 3880 | - | - | - | - | - | - | 492 | - | - |
| Peaks shared between PAR and TERRA | ||||||||||
| Peaks | ES_d0 | ES_d3 | ES_d7 | MEF | ||||||
| PAR,TERRA intersect | 3219 | 1751 | 1643 | 76 | ||||||
| Unique to TERRA | 267 | 61 | 238 | 431 | ||||||
Top: Table of CHIRT results indicating the number of PAR and TERRA binding sites in ES cells on different days of differentiation and in MEFs. Different normalization methods produced similar results.
Bottom: Numbers of peaks common to PAR and TERRA CHIRT (“intersect”) and those unique to TERRA CHIRT.
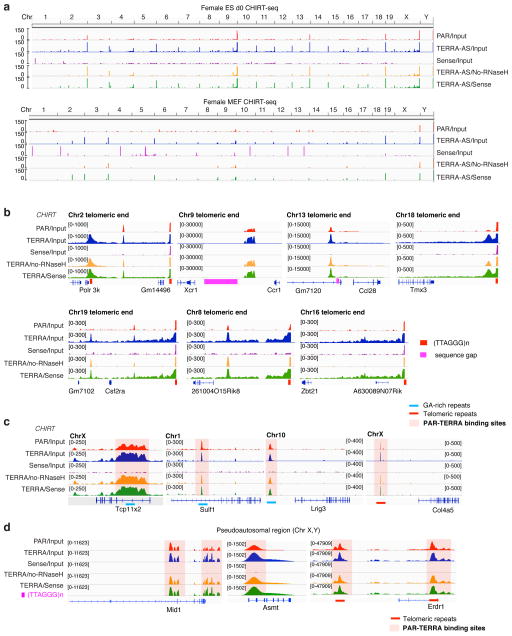
CEAS analysis indicated that, like TERRA 6, PAR was enriched for binding in noncoding space (Fig. 3e–g). Strongest enrichment occurred at subtelomeric and telomeric regions (Fig. 4a,b). At Chr 2, 9, 13, and 18, for example, strong PAR and TERRA peaks dominated the subtelomeric landscape (Fig. 4b). Moreover, PAR and TERRA binding occurred together at non-telomeric repeat regions (Fig. 4c,3f). Binding was also observed at internal (TTAGGG)n repeats present throughout the genome. The highly similar binding profiles of PAR and TERRA further support the idea of a continuous transcript. Together, these data indicate that, like TERRA 6, X-linked telomeric transcripts, PAR-TERRA, targets chromatin sites both in cis and in trans.
Figure 4. CHIRT-seq: PAR-TERRA RNA binds in cis and in trans throughout the genome.
a. CHIRT-seq tracks representing PAR-TERRA enrichment at chromosomal ends in female ES cells and MEFs. PAR data are compared to TERRA data 6, and are normalized to input (TERRA/input, PAR/Input), no-RNase H control (TERRA/no RNase H), or the sense control (TERRA/sense).
b. PAR-TERRA enrichment in subtelomeric regions of multiple autosomes in female ES cells. Red bars, TTAGGG repeats. Pink bars, sequence gaps.
c. PAR-TERRA binds to internal chromosomal regions as well.
d. PAR-TERRA binds to pseudoautosomal regions of ChrX and ChrY.
PAR-TERRA directs telomeric sex chromosomal pairing
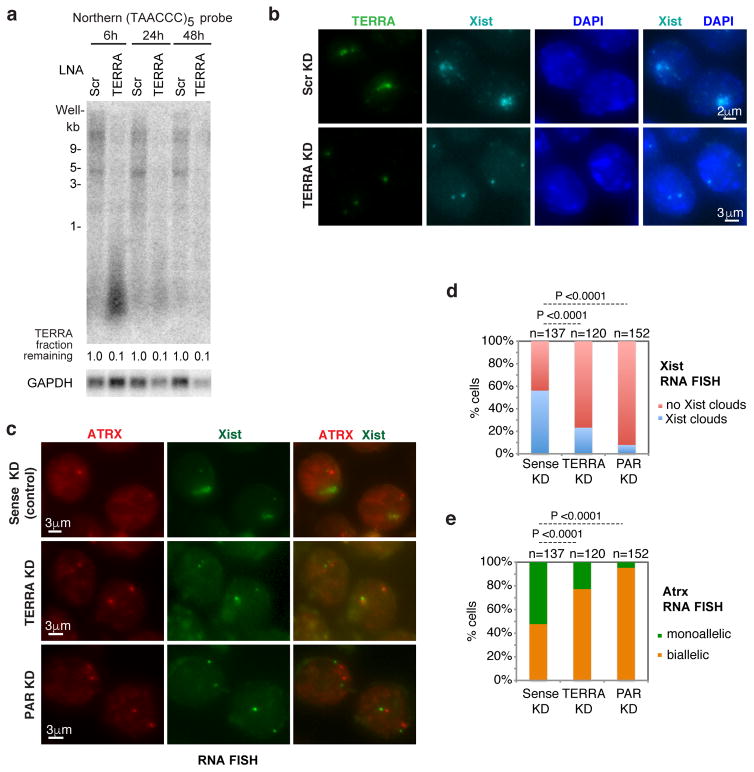
We asked whether subtelomeric transcription may aid in pairing. Interestingly, PAR-TERRA showed greatest density at discrete positions within Mid1, Erdr1, and Asmt (Fig. 4a,d). We depleted PAR-TERRA using locked nucleic acids gapmers (LNA) directed against either PAR or TERRA sequences (Fig. 5a). At 1–6 hours post-treatment, only 20–30% of PAR-TERRA remained by Northern analysis (Fig. 5a). RNA FISH confirmed disappearance of PAR-TERRA foci (Fig. 5b). Depleting PAR resulted in a concomitant depletion of TERRA and vice versa, consistent with the idea that the two transcripts are linked.
Figure 5. PAR-TERRA mediates trans-PAR pairing of sex chromosomes.
a. PAR-TERRA depletion by LNA-mediated knockdown. Northern blot analysis after knockdown at various timepoints (1h, 3h and 6h) in female ES cells. Control, scrambled LNA gapmer (Scr KD). PAR KD, LNA against PAR-31K sequence, TERRA KD, LNA against UUAGGG repeats. Fraction remaining is quantitated by densitometry. GAPDH mRNA, loading control.
b. TERRA RNA signals are greatly reduced after PAR or TERRA knockdown. RNA FISH detecting TERRA (green, (TAACCC)7-Alexa488) after 6 hours of knockdown in female ES cells. Blue, DAPI-stained nuclei.
c. PAR-TERRA knockdown disrupts inter-PAR pairing in female ES cells. Full distributions of inter-PAR distances in d4 ES cells after PAR-TERRA versus control (Scr) knockdown for 6 hours. P values determined by the KS test. Mean values indicated by triangles.
d. Dotplot of inter-PAR distances for the top decile of nuclei shown in c. P values determined using two-tailed student t-test.
e. Analysis of ES cells carrying a PAR-TERRA transgene (Tg, RP24-500I4 BAC). Top panel: RNA FISH using indicated probes show Tg PAR-TERRA expression (arrows). Middle panel: DNA FISH distinguishes location of Tg (P1 vecto-PAR colocalization, asterisks) from endogenous PAR. Lower panel: Three colored-DNA FISH for P1, Arhgap6, and Hprt.
f. Dot plot shows Tg-Arhgap6 pairing in PAR-TERRA Tg cells. Top decile shown in dotplot. P values determined using a two-tailed student t-test.
g. A cartoon rendering the dynamics of PAR:PAR pairing in wildtype ES cells (left panel) and Tg-PAR pairing in Tg ES cells (right panel).
We then assessed pairing in d4 female ES cells 6h after knockdown (KD). Indeed, PAR pairing was significantly disrupted relative to scrambled (Scr) KD controls (Fig. 5c–d). This was also the case in male ES cells (Fig. S5). Thus, PAR-TERRA is required for PAR:PAR pairing. To ask if PAR-TERRA is sufficient for pairing, we created ES cells carrying an autosomal PAR-TERRA BAC transgene (RP24-500I4; Fig. 1a). FISH densitometry showed that the transgene copy number did not exceed the endogenous PAR (Fig. S6a,b). RNA FISH demonstrated transgene expression of PAR-TERRA, and linked P1 vector sequences (Fig. 5e). To assess whether the transgene could pair with PAR, we performed DNA FISH using a P1 and an Arhgap6 probe. Significantly, transgenic PAR-TERRA induced ectopic pairing with the distal end of ChrX (Fig. 5e,f). No pairing was observed with the Hprt control (Fig. 5e, lowest panels; 5f). By performing 3-color DNA FISH for PAR, Arhgap6-Mid1 (ChrX_end) and Sry (Y-linked)(Fig. S6c–d), we observed a simultaneous disruption of X-Y PAR-pairing (Fig. S6e), suggesting a competitive interaction between PAR sequences in vivo. Thus, PAR is both necessary and sufficient to direct interchromosomal pairing between sex chromosome telomeres (Fig. 5g).
PAR-TERRA tethers the Xic to PAR
We next investigated the relationship between PAR and Xic pairing. Interestingly, time-course CHIRT analysis revealed hotspots of PAR-TERRA binding in the 15-kb X-pairing center of Tsix and Xite 17,18 (Fig. 6a). Binding was specific to ES cells (which are pairing-competent), and not seen in MEFs (which are not pairing-competent). To ask if PAR-TERRA affects Xic:Xic pairing, we depleted PAR-TERRA, confirmed loss of PAR pairing, and performed Xic pairing assays (Fig. 6b). Significantly, depleting PAR-TERRA impacted inter-Xic pairing in d4 female ES cells, with a substantial decrease in the number of nuclei exhibiting inter-allelic distances of <0.1 ND (Fig. 6b,c). Therefore, both PAR:PAR and Xic:Xic pairing require PAR-TERRA.
Figure 6. Intra-chromosomal interactions between PAR and Xic occur in a PAR-TERRA-dependent manner.
a. Time-course analysis (d0, d3, d7 ES cells; MEFs) using PAR-TERRA CHIRT-seq reveals binding sites at the Xic pairing center.
b. PAR-TERRA knockdown disrupts inter-Xic pairing in d4 female ES cells at 6h post-transfection. Left panel: DNA FISH using probes to the Xic, PAR, and Arhgap6 in d4 female ES cells. Arhgap6 signals served as hybridization control and ensured scoring only those nuclei with two discernible signals for each probe. Right panel: Dotplot of inter-allelic distances shown for the top decile of nuclei. P values were determined using two-tailed student t-test.
c. Frequency of PAR-Xic association in differentiating female ES cells (d0, d4, d8). Full distributions of PAR-Xic distances are shown. n, sample size. Triangles, mean values. P values determined by the KS test.
d. Heatmap of 4C analysis with a viewpoint at PAR/Erdr1 in d0 and d4 female mESCs and in MEFs. The heatmap represents the log mean coverage for a given window size (100k-5Mb sliding windows). Two biological replicates show the similar pattern of interactions. Significance of interaction between Xic and PAR was determined by fitting the observed empirical distribution of normalized contact data with Weibull distribution and calculating p-values. The “*” indicates the PAR:Xic interaction in d4 ES cells.
e. Strong correlation between contact frequencies (blue track, 4C) and PAR-TERRA RNA binding (green tracks, CHIRT) in female ES cells undergoing XCI (d4).
f. Scatterplot: 4C analysis of post-XCI female MEFs also showed strong correlation between PAR-TERRA binding (CHIRT) and PAR interaction frequency (4C). Pearson’s r = 0.69. Each dot represents the mean coverage of 100 kb bin size for 4C (X-axis) and CHIRT-seq (Y-axis) in the scatterplot.
g. Frequency of PAR-Xic and Hprt-Xic associations in differentiating d4 female ES cells after Scr or PAR-TERRA knockdown for 6 hours. Full distributions for each pairwise distance measurements are shown. n, sample size. P values determined by the KS test.
The relationship between PAR and Xic intrigued us, as meiotic chromosome pairing appears to be driven by a telomeric bouquet 34–38. This conglomeration of telomeres has been proposed to facilitate initial synapsis at one chromosomal end and enable synaptic extension through a “zippering” mechanism to the other end. However, because somatic X-X pairing does not involve the full chromosome 17,18,27, a telomere-initiated zippering mechanism seems unlikely to drive Xic:Xic pairing. In principle, a long-range looping mechanism could bring the Xic to the PAR which, when itself paired, would constrain the random walk for homology searching between two Xic alleles. To test this idea, we measured PAR-Xic distances and observed a significant decrease on d4, but not on d8 (Fig. 6c). To confirm using an unbiased sampling method, we employed 4C technology (Circularized Chromosome Conformation Capture) to capture all pair-wise interactions between the PAR and 150 Mb of X-linked sequence. By anchoring a viewpoint primer in PAR/Erdr1, we observed a hotspot of interaction (*, Fig. 6d) between Xic and PAR on d4. We conclude that PAR interacts with the Xic at long range during the timeframe of Xic:Xic pairing.
Comparing the PAR 4C map with PAR-TERRA CHIRT map revealed an excellent correlation between PAR-interacting loci (4C) and PAR-TERRA binding sites (Fig. 6e,f; Pearson’s r=0.69). This correlation suggested that PAR-Xic DNA interactions may be influenced by PAR-TERRA RNA binding. To test this possibility, we depleted PAR-TERRA RNA using LNA’s and observed that the heterotypic Xic:PAR interactions were abolished, whereas no effects were seen on the relative localization of the Xic and Hprt (Fig. 6g). Taken together, our data indicate that all three pairwise interactions — Xic:Xic, PAR:PAR, and Xic:PAR — are regulated by PAR-TERRA.
The tetrad
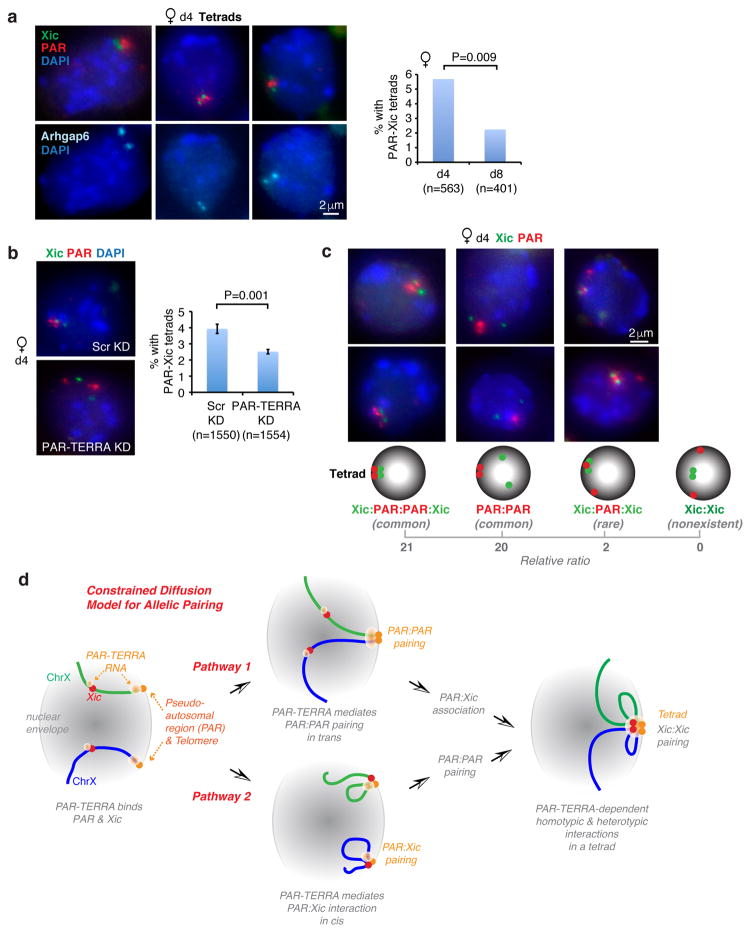
A scenario involving coincident pairwise interactions would predict existence of a “tetrad” formed by homotypic (Xic:Xic, PAR:PAR) and heterotypic (Xic:PAR) interactions. In d4 female ES cells, 5.7% of nuclei harbored a tetrad, as defined by distances of all pairwise interactions occurring within <1.5μm, and almost all Xic:Xic pairs were found within such tetrads (Fig. 7a). These tetrads almost always occurred at the nuclear edge, consistent with chromosomal ends being anchored to the nuclear envelope 34,38. By d8, the number of tetrads decreased to 2.2%. To determine whether PAR-TERRA mediates tetrad formation, we depleted PAR-TERRA and found a significant loss of tetrads in d4 female cells (Fig. 7b). Thus, tetrads form during homologous sex chromosome pairing in a PAR-TERRA-dependent manner.
Figure 7. The tetrad as a hub for pairing interactions.
a. DNA FISH using probes to the Xic, PAR, and Arhgap6 in d4 female ES cells reveals a high frequency of tetrads. Mean percentages are plotted in graph, with sample sizes (n) and statistical analysis. P value determined using Fisher’s exact test.
b. DNA FISH shows that PAR-TERRA knockdown disrupts tetrad formation at 6h post-transfection in d4 femal ES cells. Three biological replicates are averaged. N, sample sizes. P value determined using Fisher’s exact test.
c. Cartoons show possible pairing species and the prevalence (relative ratio) of each in d4 female ES cells. DNA FISH images for two representative examples of each species are shown above the cartoon. Xic:Xic pairs without PAR interaction are not observed.
d. Constrained Diffusion Model for homology-searching. PAR-TERRA RNA binds to the pseudoautosomal region and the Xic in ES cells. In Pathway 1, PAR-TERRA then directs PAR:PAR interactions, followed by PAR:Xic interactions, which then facilitate Xic:Xic homology searching and pairing. In Pathway 2, PAR-TERRA directs PAR:Xic interactions in cis, followed by PAR:PAR pairing, which in turn facilitates Xic:Xic homology searching and pairing. The two pathways are not mutually exclusive. PAR-TERRA RNA is required for all pairwise interactions.
Can PAR:PAR pairing could occur without Xic:Xic pairing and, conversely, can Xic:Xic pairing could occur without PAR:PAR pairing? In d4 ES cells, tetrads accounted for approximately half of all nuclei with PAR:PAR pairing (Fig. 7c). In the remaining half, nuclei showing PAR:PAR pairing without Xic:Xic pairing were the predominant species. Never observed were nuclei with Xic:Xic pairing without a simultaneous PAR interaction. Therefore, while Xic:PAR interactions and PAR:PAR interactions can occur in isolation, Xic:Xic interactions do not (Fig. 7c). In rare nuclei, Xic:Xic pairing occurred when only one PAR allele was in contact – with the single PAR allele sandwiched between two Xic’s (Xic:PAR:Xic; Fig. 7c), suggesting that a single PAR could occasionally nucleate Xic:Xic pairing. Taken together, these data lead to a model in which Xic:Xic pairing is facilitated by PAR:PAR interactions. We conclude that X:X pairing occurs in a step-wise fashion, with PAR-TERRA tethering PAR to Xic in all pairwise interactions and thereby creating an interaction “hub” (Fig. 7d).
Finally, we asked if PAR-TERRA’s effects had consequences for XCI. Indeed, RNA FISH showed that depleting PAR-TERRA precluded formation of Xist clouds in differentiating female ES cells. Fewer TERRA KD cells demonstrated large foci of Xist RNA than Scr KD cells on d8 (P= 0.01; 29.1% [Scr KD, n=168] vs 17.8% [n=219, TERRA KD]; Fig. 8b). The failure of XCI was supported by a larger fraction of d8 nuclei retaining a biallelic pattern of X-linked gene expression (Fig. 8c–e). Depleting either PAR or TERRA RNA resulted in the same effect on Xist upregulation (Fig. 8d) and Atrx silencing (Fig. 8e), with the knockdown of PAR having an especially strong effect. By contrast, treatment with either Scr or TERRA-sense control LNA’s did not adversely affect XCI. PAR KD also did not affect expression of autosomal TERRA (Fig. S7). We conclude that PAR-TERRA RNA is required for homologous X-X pairing and for proper XCI17,18,27.
Figure 8. PAR-TERRA is required for proper initiation of XCI.
a. TERRA depletion by LNA-mediated knockdown. Northern blot analysis after knockdown at various timepoints (6h, 24h and 48h). LNA transfection started on differentiation d4 in female ES cells. Control, scrambled LNA gapmer (Scr KD). TERRA KD, LNA against UUAGGG repeats. Fraction remaining is quantitated by densitometry. GAPDH mRNA, loading control.
b. Xist RNA FISH after LNA knockdown for 48 hours. LNA transfection started on differentiation d4 in female ES cells and RNA FISH performed on d6 cells. %nuclei with Xist clouds: For Scr KD: d5, 3.6% (n=166); d6, 6.9% (n=173); d8, 29.1% (n=168). For TERRA KD: d5, 2.0% (n=196); d6, 3.5% (n=169); d8, 17.8% (n=219). The difference between Scr and TERRA KD on d8 is significant (P=0.01), as determined by Fisher exact test.
c. RNA FISH shows that depletion of either PAR or TERRA results in blunted Xist upregulation and failure of XCI. LNA was introduced on differentiation d4 in female ES cells, and Xist and ATRX RNA FISH was performed on d8 cells. Control KD, TERRA-sense LNA gapmer. TERRA KD, LNA against UUAGGG repeats. PAR KD, LNA against PAR-31K sequence.
d. Bar graph shows percentage of cells with Xist clouds on d8 of female ES differentiation for the experiment in c. There appeared to be robust Xist upregulation after treatment with sense LNA. The opposite effects achieved by sense versus TERRA LNA achieved opposite effects supports specificity of the TERRA knockdown and the effect of TERRA on Xist upregulation. P values were determined by Fisher exact test. n, sample size.
e. Bar graph shows percentage of cells with monoallelic and biallelic Atrx expression on d8 for the experiment in c. P values were determined by Fisher exact test. n, sample size.
DISCUSSION
Here we have (i) identified the subtelomeric region (PAR) of sex chromosomes as a major source of TERRA transcripts in ES cells, (ii) demonstrated that PAR-TERRA binds in cis and in trans throughout the genome, (iii) uncovered PAR-TERRA as a mediator of homologous X-chromosome pairing, and (iv) defined PAR:PAR interactions as a key to nucleating Xic:Xic pairing for subsequent initiation of XCI.
We propose a Constrained Diffusion Model (Fig. 7d) in which telomeric pairing guides Xic:Xic pairing and PAR-TERRA RNA serves as a tether to bring the PAR and Xic into contact within a tetrad. This interaction hub would serve to constrain the space for Xic:Xic interactions, thereby reducing the effective volume within which Xic alleles must engage in a random walk during homology searching. Our data show that all pairing occurs within the tetrad (Fig. 7). PAR transcription is key in establishing all pairwise interactions, both homotypic (PAR:PAR) and heterotypic (PAR:Xic) interactions. Two pathways can be envisioned. In Pathway 1, PAR-TERRA directs PAR:PAR interactions. Independent binding of PAR-TERRA to the Xic then enables PAR to interact with Xic, resulting in formation of PAR:Xic interactions, which then in turn facilitate Xic:Xic homology searching and in trans-pairing. Alternatively (Pathway 2), PAR-TERRA directs PAR:Xic interactions to occur first in cis. PAR:PAR pairing would occur independently, and this in turn would facilitate Xic:Xic homology searching and trans-pairing. The models are not mutually exclusive. Nevertheless, additional factors must work with PAR-TERRA to control the timing of pairing, given that PAR-TERRA expression is constitutive. Our study suggests that a key difference between meiotic and somatic pairing is that, while both appear to be telomere-mediated, the latter does not apparently involve a whole-chromosome zippering mechanism.
The purpose of somatic X-X pairing has been actively debated. Our present data provide clear evidence for a role of PAR-TERRA and PAR/Xic pairing during the initiation of XCI, and lend further support to the idea that allelic pairing mediates counting and allelic choice of one Xa and one Xi 17,18. A recent study suggested that XCI can also occur in the absence of X-X pairing 39, as heterokaryons made by fusing XX and XY cells occasionally resulted in XCI in the XY nucleus (where pairing was presumptively not possible). However, heterokaryons can continue to divide after fusion, in which case breakdown of the nuclear envelope could enable physical contact between X’s and mixing of chromosomes. Alternatively, Xist dysregulation could occur in heterokaryons, akin to “chaotic XCI” 40,41.
Our study shows that male cells also undergo PAR:PAR pairing. If pairing were for the express purpose of initiating XCI correctly, why does PAR pairing also occur in male cells? While male cells do not initiate XCI, they nevertheless must count X-chromosomes. In male cells, PAR:PAR pairing would not result in a productive Xic homology search due to the absence of a second X. Accordingly, XCI does not initiate.
An important sidebar to our discovery is that TERRA is predominantly expressed from sex chromosomes, with PAR-TERRA providing >99% of all (UUAGGG)n-containing transcripts in mouse ES cells. There may be differences between species and cell types 42,43,31. Still, one wonders why sex chromosomes produce so much TERRA. Facilitation of homologous pairing might be only one of its sex-linked functions. Notably, somatic pairing also occurs at a number of autosomal loci, including Oct4, cytokine genes, and imprinted loci 23,25,26,44. It would be of interest to determine whether subtelomeric pairing and transcription also play a role in those contexts. Finally, our study shows that PAR-TERRA targets loci on autosomes as well. Indeed, it is now known that TERRA in general can operate in trans at non-telomeric loci 6. Henceforth, a major goal will be to understand whether sex-linked PAR-TERRA can influence genome-wide activity and what those activities might be.
MATERIALS AND METHODS
FISH & pairing assays
Male (J1) and female (16.7) ES cells were cytospun onto glass slides and permeabilized with CSK buffer containing 0.5% Triton X-100, and fixed in 4 % paraformaldehyde. DNA oligos probes for RNA FISH were ordered from Integrated DNA Technologies. For TERRA: (TAACCC)7-Alexa488-3′ and 5′-Cy5-(TAACCC)7. For I4 oligos: I4-47k 5′-Alexa488-TGC ACT GAC GTC CTG TGG CCA CTG GGT GGC GCC AGA GCAT; I4-29k: 5′-Cy3-TAA TCT GAA TAT CTG GGC CTC CGT GTG CAG ACC TGA GGT T; I4 31k: 5′-Cy5-GTC TCT GTG TCT GTC TCT CTG TCT CTG TCG CTA ACT CTA T. Other FISH probes: RP23-461E16 BAC DNA for Arhgap6 DNA FISH; RP24-500I4 or RP24-143B12 for PAR DNA or RNA FISH; pSx9-3 for Xist DNA or RNA FISH; pSacBII P1-puromycin vector DNA for P1 DNA FISH. RP23-450B21 for ATRX DNA and RNA FISH; RP23-69B11 for Ngfrap1 DNA FISH. RP24-283L17 for ChrX-end (located between Arhgap6 and Mid1) DNA FISH. RP24-332J21 for SRY on Chromosome Y. DNA oligo probes for RNA-FISH were used at 0.5 pmol/μl in hybridization buffer (50% formamide, 2’SSC, 2 mg/ml BSA, 10% Dextran Sulfate-500K). BAC DNA probes and PCR-PAR probes were nick-translated with fluorophore-dUTP and used 1 ng/μl for RNA-FISH and 50 ng/μl for DNA FISH in hybridization buffer. For RNA FISH, hybridization occurred 42°C overnight and slides were washed with 2’SSC/50% formamide for 5 min three times at 44°C, then with 2’SSC for 5 min twice at 44°C. For DNA FISH, slides were treated with 0.4 mg/ml RNase A in PBS at 37°C for 1 hr, washed with PBS, incubated with 0.1 N HCl for 10 min. Slides were washed in PBST (0.2% Tween 20 in 1 X PBS) at RT for 5 mins, and then denatured in 70% formamide/2X SSC at 80°C for 15 mins. Slides were dehydrated with EtOH, air dried, and hybridized to probe at 37°C overnight. Washing is identical to RNA FISH except for an additional wash in 0.1’SSC for 5 min at 44°C. For metaphase spread, cells were incubated with 50 ng/ml colcemid for 2 hr, harvested, washed with PBS, incubated in cold 0.056M KCl on ice for 30 min, fixed in methanol/acetic acid (3:1), spread onto glass slides, air dried, and fixed in 4% formaldehyde.
For 3D pairing assay, digital images were captured using a Nikon workstation and processed using Volocity. Z-sections were captured at 0.2 μm intervals and distances were measured. Only nuclei with two resolvable X signals were scored (single dots were excluded). ‘Normalized distance’ (ND) is defined as x/d, where d is the nuclear diameter, defined as 2(A/π)0.5. PCR-PAR PCR primer pairs were used as follows: P3-F: CTCAGAGCCCAGTGTCAATCAC, P3-R: CACGACCGCTTAGAAGAACCGG; P4-F: GAGACGGCCTACCATGTGCTTC, P4-R: GTGAGTGCTGTGAACTCGGCTG; P5-F: CAGGGCCTGATTTGGCTTGAAAC, P5-R: GAAGAGTAGTCTGACCTCATCTC; P6-F: CAGGGCATGATATCCTCTTTGG, P6-R: CATTCAATGGTGTTGATGATGGTAC; P8-F: GGTTAGAATACAGCGCGGACATTCA, P8-R: GTGAATCTCCGAGGCAACTGTC.
TERRA-capture RNA-seq
20 μg of Trizol purified total RNA was treated with 4U of TURBO DNase at 37°C for 10 min in 100 μl with RNase inhibitor, 10nM of ribonucleoside vanadyl complex (VRC). 5 mM EDTA was added to disrupt VRC and RNA was extracted again by Trizol. 20 μg of RNA was mixed with 10 pmol of biotin-labeled oligo probes ((TAACCC)3TA-/3′BioTEG), denatured at 70°C in 100 μl of 6XSSC hybridization buffer for 10 min, transferred to 44°C heat block, and incubated for 30 min. RNA was then captured by 100 μl of MyOneC1 beads at 37°C for 15 min, washed with 2XSSC/0.1% NP40 at 37°C for 4 times (5 min each time), washed twice with 1XSSC/0.1% NP40 at 37°C, and rinsed once with 1X SSC at RT. TERRA-enriched RNA was eluted in 30 μl of DEPC-treated water at 70°C for 5 min and reversed transcribed with random or (TAACCC)4 primers using Superscript III (Invitrogen) at 55°C for 60 min. 1.25 mM of dUTP was supplied during second-strand synthesis. cDNA was then briefly sonicated by covaris E220 in microTUBEs (Duty factor=2%, peak intensity power=140, cycle per burst=200, time=15 second). End repair, dA-tailing, adaptor ligation, and USER enzyme digestion were performed using NEBNext Ultra Directional RNA-seq Library Prep protocol, except some modifications on size selection. Larger fragments (>500 bp) were selected by 0.6x AMpure XP beads and pair-end sequenced using Illumina MiSeq instrument (300 bp reads). Reads were aligned to GRCm38/mm10 using Tophat2 after adaptor removal (Trim Galore). After PCR duplicate removal, reads were normalized by frequency per million (FPM) mapped reads and FPM values are shown for subtelomerically mapped reads. Note: Subtelomeric regions (PAR) of ChrX and Y are not fully sequenced or assembled in mm10.
CHIRT-seq analysis
CHIRT combines ChIRP and CHART protocols 32,33 as follows: We used a minimum number of capture probes to reduce off-target effects; we increased the shearing size to 0.5 – 3 kb to preserve lncRNA integrity; because we observed that RNaseH is not active in SDS, we used NP40 instead of SDS or N-lauroyl sarcosine during DNA elution to preserve RNaseH activity. In brief, 15 millions of exponentially growing ES cells were washed with PBS and resuspended in 10 ml of PBS. Cells with then fixed by adding 10 ml of 2% of glutaraldehyde at room temperature for 10 min and crosslinking was quenched with 0.125 M glycine for 5 min. Cells were spun at 2000g for 5 min at 4°C, washed with cold PBS, respun, snap-frozen in liquid nitrogen, and stored at −80°C. For differentiating ES cells (d3, d7), embryoid bodies were trypsinized, filtered with cell strainers (40 μm), prior to the above steps. For nuclei isolation during CHIRT, cells were thawed, resuspened in 1 ml of swelling buffer (0.1 M Tris pH 7.0 10 mM KOAc, 15 mM MgOAc, 1% NP40, 1mM DTT, 1mM PMSF, 100 U/ml Superase-In[Ambion]) for 10 min on ice, dounced and pelleted at 2500 g for 5 min. Nuclei were further lyzed in 50 mM Tris pH7.0, 10 mM EDTA, 1 % SDS, 1mM DTT, 1mM PMSF, protease inhibitor, 100 U/ml Superase-In on ice for 10 min, and sonicated (Bioruptor) for a 0.5–3 kb size range. Lysates were spun down at 13,000 rpm for 5 min to remove debris, snap-frozen in liquid nitrogen, and stored in −80°C. Streptavidin-magnetic C1 (Life Technologies) beads were blocked with 500 ng/ul yeast total RNA, and 1mg/ml BSA for 1 hr at 37°C, and respuspended in 1X hybridization buffer (1 volume of lysis buffer plus 2 volume of 2X hybridization buffer). Lysates were diluted in two volume of 2X hybridization buffer (lysate: 2Xhyb = 1:2) (750 mM NaCl, 1% SDS, 50 mM Tris pH 7.0, 1 mM EDTA, 15% Formamide, 1 mM DTT, PMSF, protease inhibitor, and 100 U/ml Superase-in), precleaned with Streptavidin-magnetic C1 beads at 37°C for 1 hr (100 μl of beads for 1 ml lysates), and incubated with pooled probes (100 pmol for 3 ml of diluted cell lysates) at 37°C for 3 hr. Three hundred microliters washed/blocked C1 beads were added per 100 pmol of probes, and the reaction was mixed for another 1 hr at 37°C. DNA probes for CHIRT were ordered from Integrated DNA Technologies and labeled with 3′ biotin-TEG. PAR DNA probes are as follows: 36K: GAGCGCCTCAGTGTGCAAATCT, 47K: ACTGGGTGGCGCCAGAGCAT, 29K: CTCCGTGTGCAGACCTGAGGTT, 34K: CCCTACCTACCCTCCAGAGA, 31K: TCTCTGTCTCTGTCGCTAAC. TERRA-AS probe, TAACCCTAACCCTAACCCTA. TERRA-sense probe, TTAGGGTTAGGGTTAGGGTT. Beads:biotin-probes:RNA:chromatin adducts were captured by magnets, washed five times at 37°C for 5 min with wash buffer (2X SSC, 0.5% SDS, 1 mM DTT, 1 mM PMSF), and washed twice for 5 min at room temperature with 0.1% NP40 buffer (150 mM NaCl, 50 mM Tris pH8.0, 3 mM MgCl2, 10 mM DTT, 0.1% NP40). DNA was eluted twice for 20 min in 450 μl of 0.1% NP40 buffer with 200 U/ml RNase H (NEB) at room temperature. DNA for no RNase H controls was eluted in 0.1% NP40 buffer without RNaseH. Eluted DNA was treated with RNase A (1 mg/ml) at 37°C for 1 hr, then treated with proteinase K (1 mg/ml), and mixed with SDS to 0.5% concentration at 55°C for 16 hr. DNA was extracted with phenol/chloroform using phase lock gel tubes. For pre-RNaseA treatment control, cell lysates were treated with RNase A at 37°C overnight before hybridization. For RNA elution after hybridization, beads:biotin-probes:RNA:chromatin adducts were washed 5 time in wash buffer, then treated with proteinase K in PK buffer (100 mM NaCl, Tris pH 7.0, 1 mM EDTA, 0.5% SDS) at 55°C for 30 min. Beads suspension was boiled at 90°C for 5 min, and then RNA was extracted using TRIzol (Invitrogen). Primer pairs for qPCR are: PAR-DNA-F: TGGAGGTTAAACGATTATTTATCTGC, PAR-DNA-R: ACGAGTTTCCAAGGTGCTG; Hprt-F: CTGCTACTTCAACTCCTGGTGTGC, Hprt-R: AGGCGAATTGGGATGTAGCTCAG.
Before library construction, equal amounts of lambda DNA (0.015 pg of PCR products, ~250bp) were spiked into each CHIRT sample as normalization control. Spike-in DNA was amplified using Lambda 5-F, 5′-GCA TAT GTT GTG TTT TAC AG-3′, and Lambda 5-R, 5′-GCA ACA AAT TGA TAA GCA-3′. 30 million 50-bp paired-end reads were obtained for each library. Following adaptor trimming and PCR duplicate removal, reads aligned to either GRCm38/mm10 or NCBI37/mm9 using Novoalign (v3.00.02) (http://www.novocraft.com/products/novoalign/). >70% of reads uniquely mapped to the mouse genome. Coverage files were generated using R and SPP software 45 with smoothing using 500 bp bins with a 100 bp step size to generate control-subtracted, normalized read densities. Data were visualized using IGV software to display all tracks with a mean windowing function and scales indicated in each figure. Other methods to generate normalized coverage files, including the generation of conservative enrichment and maximum likelihood estimates, resulted in similar distribution patterns. Scatter plots for correlation analysis used input-normalized coverage produced by SPP, windowed by 3 kb bins and filtered for enriched bins with an averaged density of at least 4. Peaks were called by MACS (1.4.2)46 using normalization to indicated controls (input, sense, no RNaseH or pre-RNaseA), and filtered by peak length greater than 1 kb, and >10 fold enrichment. Metagene profiles were produced by software CEAS (0.9.9.7) 47 using 2-fold enriched over input wig files and bed files produced by MACS peak-calling. At least two biological replicates of each CHIRT experiment were examined and they showed similar results.
PAR-TERRA knockdown analysis
2 X 106 ES cells (16.7, cas/mus hybrid) were grown in antibiotic free media and differentiated into embryonic bodies for 4 days. Cells then were trypsinzed and transfected with LNA gapmers (Exiqon) at 2 μM in 100 μl nucleuofector solution using A30 program (Lonza) for pairing assay. For Xist RNA-FISH, cells were transfected with 10nM of LNA using Lipofeatamine 2000 or RNAiMax, and collected at 6, 24, 48, 96 hours after transfection. Gapmer LNAs with a phosphothiolated backbone were: Scr, 5′-CAC GTC TAT ACA CCA C-3′; PAR, 5′-TCT CTG TCT CTG TCG C-3′; TERRA, 5′-TAA CCC TAA CCC TAA C-3′; sense, 5′-TTA GGG TTA GGG TTA G-3′. Multiple biological replicates were examined for each knockdown experiment. Degree of knockdown was determined by quantitative Northern analysis with probes as follows: TERRA: TAACCCTAACCCTAACCCTAACCCTAACCC; GAPDH: GTAGACCCACGACATACTCAGCACCGGCCTCACCCCATT; I4-29k: TAATCTGAATATCTGGGCCTCCGTGTGCAGACCTGAGGTT; I4-27k: TTGGGGGCGTGTCTCAGAGCAGGAGGGGTGTGGTCTGGCA; I4-31k: GTCTCTGTGTCTGTCTCTCTGTCTCTGTCGCTAACTCTAT; I4-34k: AAAGCCACCAGGCCTCTAATCCCTACCTACCCTCCAGAGA; I4-42k: CCTGGAGAAATCAAGTCTGCGAAGATCCAAAAATTAAAAT; I4-47k: TGCACTGACGTCCTGTGGCCACTGGGTGGCGCCAGAGCAT. All oligo probes were end labeled using T4 polynucleotide kinase. Total RNA was extracted using TRIzol followed by acid phenol extraction. Total RNA (5 μg) was loaded and hybridization was carried out at 42°C overnight using ULTRAhyb-Oligo hybridization buffer (Ambion).
Quantitative RT-PCR
Total RNA was extracted using TRIzol followed by acid phenol extraction, and cDNA was synthesized using random primers. Wls-F, 5′-CCAGTCTAATGGTGACCTGGG-3′; Wls-R, 5′-TGAGAGTCAGCATGCACCAG-3′. GAPDH-F, 5′-CGTCCCGTAGACAAAATGGT-3′; GAPDH-R, 5′-TTGATGGCAACAATCTCCAC-3′.
Generation of PAR-TERRA transgenic ES cells
Male ES cells (J1) were transfected by lipofectamine 2000 with NcoI-cut BAC DNA (RP24-500I4) along with the pSacBII P1-puromycin resistance vector DNA (Molar ratio of I4:P1=4:1). Media was changed after 6 hours post-transfection. Puro-resistant clones were picked and maintained under puromycin selection. Transgene integration was confirmed by DNA FISH and RNA-FISH.
4C Library construction
6–8 million cells were trypsinized and crosslinked in 40 mL of media with 2% formaldehyde for 10 minutes at room temperature. The formaldehyde was quenched by adding glycine to a final concentration of 125 mM; cells were then washed twice in 1X PBS, frozen in liquid nitrogen and stored at −80°C. To prepare 3C libraries, we used a method similar to the “in situ Hi-C” method48. Cells were lysed by resuspending them in 500 μl permabilization buffer (10 mM Tris-HCl pH=8.0, 10 mM NaCl, 0.2% Igepal CA-630) supplemented with 50 μl 100X EDTA-free protease inhibitor cocktail for 30 minutes on ice with occasional agitation to keep them from settling. Cells were spun at 1350g for 5 minutes at room temperature, then resupsended in 358 μl 1.25X DpnII buffer + 11 μl 10% SDS. Cells were rotated for 1 hour at 37°C, then 75 μl 10% Triton X-100 was added and cells were rotated at 37 °C for an additional hour to quench the SDS. To digest the chromatin, we added 5000 units of DpnII (NEB) and rotated the lysate at 37°C overnight. To perform in-situ ligation, we added 820 μl 10X T4 DNA Ligase Buffer (NEB), 82 μl 10 mg/ml BSA (NEB), 7.61 ml water and 50 μl T4 DNA Ligase (Enzymatics) and rotated for 4 hours at room temperature. To purify the DNA, 2 mg Proteinase K was added to the ligation reactions and left at 65°C overnight, then the DNA was cleaned by phenol:cholorform:isoamyl alcohol extraction followed by ethanol precipitation.
Next, we converted the 3C libraries into 4C libraries, using a protocol similar to the modified-4C protocol49, but with some modifications. First, we digested 3 μg of each 3C library with 4 units of FatI (NEB) for 4 hours at 55 degrees. We then dephosphorylated the ends of the FatI digested molecules by adding 20 units of Calf Intestinal Phosphotase (NEB) and heating at 37°C for 1 hour. The DNA was then purified using the PCR Purification Kit (Qiagen). To ligate the universal adaptor to the 3C library, we incubated the 3C library with 200 picomoles universal adaptor and 2 μl T4 DNA Ligase (Enzymatics) in 1X T4 DNA Ligase Buffer (NEB) in a volume of 50 μl for 1 hour at 16°C, then 2 hours at 16°C. We then purified the DNA and removed excess adaptor by adding 90 μl Ampure XP SPRI beads and performing a standard SPRI cleanup. Next, we used ligation-mediated PCR (LM-PCR) to selectively amplify molecules containing the viewpoint within the PAR region. We performed LM-PCR using 1.5 μM biotinylated viewpoint primer, 1.5 μM Illumina Index Primer in a 100 μl reaction with Phusion HF buffer as the master mix for 12 cycles. We then performed a SPRI cleanup with a 1:1 bead:PCR reaction ratio to remove unincorporated primers. To enrich for library molecules containing the viewpoint primer, we washed 10 μl MyOne C1 Streptavidin Beads (ThermoFisher) 3X in 2X Binding and Wash buffer (10 mM Tris-HCl pH-8.0, 2M NaCl, 1 mM EDTA, 0.1% Tween-20), then incubated the washed beads with the purified PCR products for 15 minutes with rotation. We then washed 2X with 1X Binding and Wash Buffer (10 mM Tris-HCl pH-8.0, 1M NaCl, 1 mM EDTA, 0.1% Tween-20), the 1X in TE+Tween (10 mM Tris-HCl pH-8.0 mM EDTA, 0.1% Tween-20). To increase yield and incorporate full-length Illumina primer sequences, we added the beads to 100 μl PCR reactions, using 2 μl Illumina Universal primer and 2 μl Illumina Index primer and Phusion HF master mix for 13 cycles. We then purified and size selected using a 0.85:1 ratio of SPRI beads to PCR mix. Library sizes were determined using the High Sensitivity Bioanalyzer (Agilent), and quantifications were determined using Library Quantification Kit (KAPA Biosystems). 2x76 bp paired-end reads were obtained using an Illumina MiSeq. 4C-PAR Dpn1view-point primer: /5Biosg/ccctacacgacgctcttccgatctNNNNCGAGAACGGGTGTCTCGGGGTT.
4C data analysis
Our design of the 4C adaptors allows us to filter out several sources of noise in 4C experiments, most notably spurious amplification of off-target sequences and PCR duplicates. Our paired-end sequencing strategy allows us to sequence the viewpoint primer in read 1, and then identify potential interaction reads by sequence has ligated onto the viewpoint in read 2. This allows us to identify library molecules that actually contain the viewpoint, as opposed to library molecules that arise from off-target priming or spurious amplification of from the universal adaptor sequences, thus allowing us to eliminate several sources of noise from our data. To achieve this, we first filtered read pairs so that we include only read pairs with the viewpoint sequence present at the beginning of the read 1. We included a 6 nt random barcode sequence at the beginning of read 2; we stripped this sequence from the beginning of read 2 and appended it to the name of the read in the fastq file. We also removed the FatI sequence next to the barcode. Finally, we trimmed read 2 for sequence past DpnII sites, as we are interested in the molecules that ligated onto the DpnII site near the viewpoint primer. All filtering steps were conducted using cutadapt 1.7.1 using its built-in paired-end filtering options.
After these filtering steps, read2 was aligned to the mm9 reference genome using the same single-end alignment strategy we have previously used to align Hi-C reads allele-specifically50, which is an extension of our pipeline for aligning ChIP-seq reads allele-specifically28,51,52. Next, we employed are random-barcode strategy to eliminate PCR duplicates. Properly identifying and removing PCR duplicates in 4C experiments is usually not possible using standard methods because the positions of 4C reads are “anchored” by the positions of the 1st and second restriction sites. Thus, genuine interactions will result in multiple reads that align to exactly the same coordinates and have the same sequence; these reads are indistinguishable from PCR duplicates. But since we have included a random barcode sequence, we can properly identify duplicates. If multiple reads at the same position arise from duplication during PCR, then all reads will have the same barcode, whereas if the arise from a ligation event present multiple times in the population, then the reads will have distinct barcodes. Thus, we collapsed the counts of all reads aligning to with the same barcode to the same position to 1, eliminating PCR duplicates from the data. Read counts were normalized at each position by read depth and represent read counts as reads/million reads (RPM). Only uniquely mapped reads were used in the downstream analysis. Reads counts were normalized in 10Kb genomic bins by 4cseq_pipeline 53. Statistical significance of detected interaction domains was estimated as P-values based on fitting the observed empirical distribution of normalized contact data with Weibull distribution.
Growth conditions and ES cell differentiation
Cells were all tested and confirmed that they were free from mycoplasma contamination. Mouse embryonic stem cells were cultured in regular ES medium (500 ml DMEM with addition of 1 ml of β-mercaptoethanol, 6 ml of MEM NEAA, 25 ml of 7.5% NaHCO3, 6 ml of GlutaMAX-1, 15 ml of 1M HEPES, 90 ml of FBS, 300 μl of LIF, 6 ml of PEN/STREP) with feeders. ES cell differentiation was iniated by removing LIF and cultured in low adherent plates to form embryonic bodies in suspension (day 0 to day4). At differentiation day 5–8, embryonic bodies were plated onto gelatinized plates to allow them to attach and outgrow.
Data availability
CHIRT-seq, TERRA-captured RNA seq, and 4C-seq data have been deposited in GEO (GSE69887).
Supplementary Material
Acknowledgments
We are grateful to Y. Jeon and H. Sunwoo for technical advice. This work was funded by a grant from the NIH (R01-GM58839) to J.T.L. J.T.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
H.P.C and J.T.L designed the experiments and analyzed data; H.P.C performed experiments, including FISH, CHIRT-seq, TERRA-capture RNA-seq and LNA knockdown. J.E.F. performed the 4C experiment. S.F.P established 4C protocol. H.P.C. and H.J.O. optimized CHIRT protocols together. F. J., R. S., B.K.S and H.P.C performed bioinformatics analyses. H.P.C and J.T.L wrote the manuscript.
References
- 1.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric Repeat Containing RNA and RNA Surveillance Factors at Mammalian Chromosome Ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 2.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nature Cell Biology. 2007;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 3.Zhang LF, et al. Telomeric RNAs Mark Sex Chromosomes in Stem Cells. Genetics. 2009;182:685. doi: 10.1534/genetics.109.103093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azzalin CM, Lingner J. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015;25:29–36. doi: 10.1016/j.tcb.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Maicher A, Kastner L, Dees M, Luke B. Deregulated telomere transcription causes replication-dependent telomere shortening and promotes cellular senescence. Nucleic Acids Res. 2012;40:6649–59. doi: 10.1093/nar/gks358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu H-P, et al. TERRA RNA antagonizes ATRX and protects telomeres. Cell. 2017 doi: 10.1016/j.cell.2017.06.017. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–36. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 8.Starmer J, Magnuson T. A new model for random X chromosome inactivation. Development. 2009;136:1–10. doi: 10.1242/dev.025908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Disteche CM. Dosage compensation of the sex chromosomes. Annu Rev Genet. 2012;46:537–60. doi: 10.1146/annurev-genet-110711-155454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JT. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat Rev Mol Cell Biol. 2011;12:815–26. doi: 10.1038/nrm3231. [DOI] [PubMed] [Google Scholar]
- 11.Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet. 2011;12:542–53. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 12.van Bemmel JG, Mira-Bontenbal H, Gribnau J. Cis- and trans-regulation in X inactivation. Chromosoma. 2016;125:41–50. doi: 10.1007/s00412-015-0525-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun S, et al. Jpx RNA activates Xist by evicting CTCF. Cell. 2013;153:1537–51. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–4. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 15.Brown CJ, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–42. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 16.Carrel L. Molecular biology. “X”-rated chromosomal rendezvous. Science. 2006;311:1107–9. doi: 10.1126/science.1124662. [DOI] [PubMed] [Google Scholar]
- 17.Xu N, Donohoe ME, Silva SS, Lee JT. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat Genet. 2007;39:1390–6. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- 18.Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–52. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- 19.Donohoe ME, Silva SS, Pinter SF, Xu N, Lee JT. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460:128–32. doi: 10.1038/nature08098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masui O, et al. Live-cell chromosome dynamics and outcome of X chromosome pairing events during ES cell differentiation. Cell. 2011;145:447–58. doi: 10.1016/j.cell.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scialdone A, Nicodemi M. Mechanics and dynamics of X-chromosome pairing at X inactivation. PLoS Comput Biol. 2008;4:e1000244. doi: 10.1371/journal.pcbi.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J. Regulation of X-chromosome counting by Tsix and Xite sequences. Science. 2005;309:768. doi: 10.1126/science.1113673. [DOI] [PubMed] [Google Scholar]
- 23.LaSalle JM, Lalande M. Homologous association of oppositely imprinted chromosomal domains. Science. 1996;272:725–8. doi: 10.1126/science.272.5262.725. [DOI] [PubMed] [Google Scholar]
- 24.Brandt VL, Hewitt SL, Skok JA. It takes two: communication between homologous alleles preserves genomic stability during V(D)J recombination. Nucleus. 2010;1:23–9. doi: 10.4161/nucl.1.1.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogan MS, Parfitt DE, Zepeda-Mendoza CJ, Shen MM, Spector DL. Transient pairing of homologous Oct4 alleles accompanies the onset of embryonic stem cell differentiation. Cell Stem Cell. 2015;16:275–88. doi: 10.1016/j.stem.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zorca CE, et al. Myosin VI regulates gene pairing and transcriptional pause release in T cells. Proc Natl Acad Sci U S A. 2015;112:E1587–93. doi: 10.1073/pnas.1502461112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bacher CP, et al. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8:293–9. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- 28.Kung JT, et al. Locus-specific targeting to the X chromosome revealed by the RNA interactome of CTCF. Mol Cell. 2015;57:361–75. doi: 10.1016/j.molcel.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berletch JB, Yang F, Disteche CM. Escape from X inactivation in mice and humans. Genome Biol. 2010;11:213. doi: 10.1186/gb-2010-11-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–4. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 31.de Silanes IL, et al. Identification of TERRA locus unveils a telomere protection role through association to nearly all chromosomes. Nat Commun. 2014;5:4723. doi: 10.1038/ncomms5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon MD, et al. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci U S A. 2011;108:20497–502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiang Y, Miller DE, Ross EJ, Sanchez Alvarado A, Hawley RS. Synaptonemal complex extension from clustered telomeres mediates full-length chromosome pairing in Schmidtea mediterranea. Proc Natl Acad Sci U S A. 2014;111:E5159–68. doi: 10.1073/pnas.1420287111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maguire MP. The mechanism of meiotic homologue pairing. J Theor Biol. 1984;106:605–15. doi: 10.1016/0022-5193(84)90010-9. [DOI] [PubMed] [Google Scholar]
- 36.Rockmill B, Roeder GS. Telomere-mediated chromosome pairing during meiosis in budding yeast. Genes Dev. 1998;12:2574–86. doi: 10.1101/gad.12.16.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reig-Viader R, et al. Telomeric repeat-containing RNA and telomerase in human fetal oocytes. Hum Reprod. 2013;28:414–22. doi: 10.1093/humrep/des363. [DOI] [PubMed] [Google Scholar]
- 38.Zickler D, Kleckner N. Recombination, Pairing, and Synapsis of Homologs during Meiosis. Cold Spring Harb Perspect Biol. 2015;7 doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barakat TS, et al. The trans-activator RNF12 and cis-acting elements effectuate X chromosome inactivation independent of X-pairing. Molecular Cell. 2014;53:965–978. doi: 10.1016/j.molcel.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 40.Lee JT. Homozygous Tsix mutant mice reveal a sex-ratio distortion and revert to random X-inactivation. Nat Genet. 2002;32:195–200. doi: 10.1038/ng939. [DOI] [PubMed] [Google Scholar]
- 41.Lee JT. Regulation of X-Chromosome Counting by Tsix and Xite Sequences. Science. 2005;309:768–771. doi: 10.1126/science.1113673. [DOI] [PubMed] [Google Scholar]
- 42.Feretzaki M, Lingner J. A practical qPCR approach to detect TERRA, the elusive telomeric repeat-containing RNA. Methods. 2017;114:39–45. doi: 10.1016/j.ymeth.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 43.Farnung BO, Giulotto E, Azzalin CM. Promoting transcription of chromosome ends. Transcription. 2010;1:140–143. doi: 10.4161/trns.1.3.13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–45. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 45.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–9. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin H, Liu T, Manrai AK, Liu XS. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–6. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 48.Nagano T, et al. Comparison of Hi-C results using in-solution versus in-nucleus ligation. Genome Biol. 2015;16:175. doi: 10.1186/s13059-015-0753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Apostolou E, et al. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell. 2013;12:699–712. doi: 10.1016/j.stem.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minajigi A, et al. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015;349 doi: 10.1126/science.aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinter SF, et al. Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res. 2012;22:1864–76. doi: 10.1101/gr.133751.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yildirim E, Sadreyev RI, Pinter SF, Lee JT. X-chromosome hyperactivation in mammals via nonlinear relationships between chromatin states and transcription. Nat Struct Mol Biol. 2011;19:56–61. doi: 10.1038/nsmb.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van de Werken HJ, et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat Methods. 2012;9:969–72. doi: 10.1038/nmeth.2173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CHIRT-seq, TERRA-captured RNA seq, and 4C-seq data have been deposited in GEO (GSE69887).