Summary
Dual leucine zipper kinase (DLK) has been implicated in cell death signaling secondary to axonal damage in retinal ganglion cells (RGCs) and other neurons. To better understand the pathway through which DLK acts, we developed enhanced functional genomic screens in primary RGCs, including use of arrayed, whole-genome, small interfering RNA libraries. Explaining why DLK inhibition is only partially protective, we identify leucine zipper kinase (LZK) as cooperating with DLK to activate downstream signaling and cell death in RGCs, including in a mouse model of optic nerve injury, and show that the same pathway is active in human stem cell-derived RGCs. Moreover, we identify four transcription factors (JUN), activating transcription factor 2 (ATF2), myocyte-specific enhancer factor 2A (MEF2A), and SRY-Box 11 (SOX11)) as being the major downstream mediators through which DLK/LZK activation leads to RGC cell death. Increased understanding of the DLK pathway has implications for understanding and treating neurodegenerative diseases.
eTOC
Welsbie et al. use high-throughput whole genome siRNA-based screening in primary retinal ganglion cells to identify novel pathway members of DLK-mediated axon injury signaling, including the related kinase LZK and the transcription factors, MEF2A and SOX11.
Introduction
Neuronal cell death in response to axon injury is a key feature of many neurodegenerative diseases. For example, axonal injury triggers the death of projection neurons called retinal ganglion cells (RGCs), resulting in vision loss in a group of diseases known as optic neuropathies. The most common of the optic neuropathies, glaucoma, is expected to blind nearly 11 million people worldwide by 2020 (Quigley and Broman, 2006). While a component of glaucomatous injury is intraocular pressure-related, lowering pressure can be challenging in some patients, and in others, RGC damage continues despite pressure lowering. Thus, the development of effective neuroprotective strategies could complement pressure lowering by mitigating the RGC response to injury. This makes the RGC both a useful model to study neurodegenerative cell death signaling and, in and of itself, a highly disease-relevant neuron.
In response to axon injury, neurons execute an active genetic program that leads to axonal degeneration and, eventually, cell death. In axon degeneration signaling, this requires Sterile Alpha and TIR Motif Containing 1 (SARM1) and mitogen-activated protein kinase (MAPK) activation (Gerdts et al., 2013; Osterloh et al., 2012; Yang et al., 2015). To identify the signaling components responsible for cell death, using an unbiased approach, we developed a method for delivering small interfering RNA (siRNA) to primary mouse RGCs and performed a high-throughput, functional genomic screen of kinases involved in RGC cell death (Welsbie et al., 2013). This and other work identified dual leucine zipper kinase (DLK) as being a key mediator of RGC cell death in response to axon injury (Fernandes et al., 2014; Watkins et al., 2013; Welsbie et al., 2013). Moreover, DLK is involved in the signaling associated with axon degeneration and regeneration (Miller et al., 2009; Watkins et al., 2013; Yang et al., 2015). This, along with its druggability, makes DLK an attractive neuroprotective target for chronic neurodegenerative diseases like glaucoma that are associated with low-level, repeated injury.
To further define the molecular network involved in RGC cell death signaling, we have developed a series of novel high-throughput functional genomic screens. Here we show that DLK cooperates with a related kinase, leucine zipper kinase (LZK), to trigger activation of downstream kinases including mitogen activated protein kinases 4 and 7 (MKK4/7) and JUN N-terminal kinases 1-3 (JNK1-3). We identify the four downstream transcription factors JUN, activating transcription factor 2 (ATF2), myocyte-specific enhancer factor 2A (MEF2A) and SRY-Box 11 (SOX11) as being required for full DLK/LZK-mediated cell death. Additionally, supporting the potential development of DLK/LZK inhibition as a viable therapeutic strategy for human neurodegenerative disease, we demonstrate that treatment of human stem cell-derived RGCs with a protein kinase inhibitor (PKI) with activity against DLK and LZK potently promotes human RGC survival. Finally, we identify an already FDA-approved PKI, sunitinib, as also being protective of human RGCs.
Results
Sensitized kinome screen identifies LZK as a mediator of RGC cell death
In the process of exploring various small molecule- and genomic-based strategies for DLK inhibition, we repeatedly observed that PKIs with activity against DLK were more efficacious than siRNA-mediated knockdown of DLK in their ability to promote survival of primary mouse RGCs (Welsbie et al., 2013). To exclude the possibility that the difference in activity was due to residual levels of DLK activity remaining after siRNA-mediated knockdown, we tested the effect of Cre recombinase-mediated DLK knockout (Figure 1A). Primary RGCs were immunopanned from floxed Dlk mice (Miller et al., 2009), transduced with adenovirus expressing Cre or a GFP control, and then treated with maximally-protective doses of the DLK inhibitor, tozasertib, or a vehicle control. After two days, to allow time for turnover of extant DLK, immunopanning injury was exacerbated by the addition of colchicine, which destabilizes microtubules and engages axon injury signaling cascades, including DLK (Bounoutas et al., 2011; Miller et al., 2009; Valakh et al., 2013, 2015). After an additional two days, viability was measured with CellTiter-Glo, an ATP-based luminescence assay that highly correlates with survival measured by conventional stains of viability and automated image analysis (Figure S1A). Similar to the results with siRNA-mediated knockdown, genetic deletion of Dlk was protective, but markedly less so than pharmacologic inhibition. This suggested that PKIs like tozasertib might be inhibiting one or more additional kinase targets that cooperate with DLK to promote cell death and that simultaneous inhibition of both was required to promote maximal RGC survival.
Figure 1. Sensitized siRNA screening of the kinome identifies LZK as a mediator of RGC cell death in vitro.
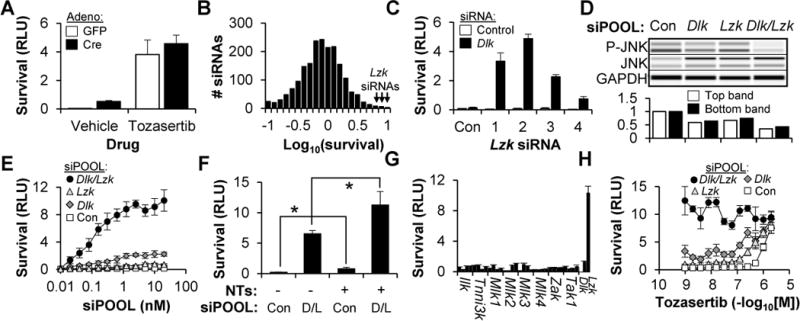
(A) Survival (±SD) of Dlkfl/fl RGCs, transduced with adenovirus and cultured with or without tozasertib (1 μM), two days after colchicine (1 μM) addition.
(B) Histogram showing the normalized survival for all 1,869 siRNAs in the kinome library (transfected in the presence of Dlk siRNA).
(C) Survival (±SD) of WT RGCs transfected with control or Dlk siRNA, and one of four independent Lzk siRNAs or the non-targeting control (Con), two days after colchicine (1 μM) addition.
(D) Capillary-based immunoassay (top) and quantification (bottom) of JNK phosphorylation in WT RGCs, one day after transfection with siPOOLs.
(E) Survival (±SD) of WT RGCs transfected with siPOOLs, two days after colchicine (1 μM) addition.
(F) Survival (±SD) of WT RGCs transfected with siPOOLs, in the presence or absence of neurotrophins (NTs, 50 ng/mL BDNF, 5 ng/mL GDNF, 5 ng/mL CNTF), two days after colchicine (1 μM) addition. *P<0.05, Mann-Whitney U test. D/L, Dlk/Lzk
(G) Survival (±SD) of WT RGCs, transfected with Dlk siRNA and either control siRNAs or one of four independent siRNAs targeting the other members of the MLK family of kinases, two days after colchicine (1 μM) addition.
(H) Survival (±SD) of WT RGCs, transfected with siPOOLs and cultured in tozasertib, two days after colchicine (1 μM) addition.
In order to identify the other putative kinase target(s), we modified our original siRNA screen (Welsbie et al., 2013) to include Dlk siRNA in every well, thereby sensitizing it to those kinase siRNAs that synergized with DLK knockdown to further increase RGC survival (Figure 1B). Using this approach, we re-screened a library of 1,869 siRNAs, targeting 623 kinases, for the ability to increase RGC survival after a colchicine challenge. The top gene nominated by the screen was Lzk (Map3k13), hitherto unknown to have any role in disease or neuronal survival. Given the abundance of false-positive hits from siRNA screens (Bushman et al., 2009; Schultz et al., 2011), we sought to validate the findings by using the “redundancy and rescue” approach (Echeverri et al., 2006). First, RGCs were treated with four new Lzk siRNAs, with sequences not tested in the initial screen, and their effect on LZK levels (Figure S1B) and cell survival (Figure 1C) were assessed. While none of the four siRNAs had survival-promoting activity by themselves, explaining why our first kinome screen did not identify LZK, in the setting of DLK knockdown, all four robustly increased RGC survival. As another layer of validation, we turned to siPOOLs. These are pools of approximately 30 siRNAs targeting a common mRNA, thereby diluting out off-target effects while maintaining on-target silencing (Hannus et al., 2014). Primary RGCs were transfected with control siPOOLs or siPOOLs targeting Dlk and/or Lzk and assayed for protein expression using a capillary-based immunoassay (Protein Simple, San Jose, CA). As expected, each siPOOL knocked down its cognate target (Figure S1C) and simultaneous knockdown of both DLK and LZK increased the suppression of JNK signaling (Figures 1D). In the survival assay, Lzk siPOOLs alone had little effect on survival, but synergized with Dlk siPOOLs to greatly increase survival (Figure 1E). By comparison, simultaneous addition of the neurotrophins brain-derived neurotrophic factor (BDNF), glia-derived neurotrophic factor (GDNF) and ciliary-derived neurotrophic factor (CNTF) led to a far less robust increase in RGC survival, but did modestly increase the effect when combined with DLK/LZK knockdown (Figure 1F). Conventional stains of viability and automated fluorescent microscopy confirmed that the survival increase was not an artifact of the luminescence assay (Figure S1D) and that the effect was sustained (Figure S1E). As one further confirmation, we showed that the effect of LZK on cell death could be reconstituted after siRNA-mediated dual DLK/LZK knockdown by expressing mouse siRNA-resistant human LZK cDNA (Figure S1F–G). Additionally, we found that DLK and LZK can have an additive role in promoting neurite outgrowth, an in vitro surrogate for axon regeneration (Figure S1H–I).
As a complementary approach to explore LZK’s effect on RGCs, we expressed a point mutant of human LZK (K195A) that lacks kinase activity and should function as a dominant-negative (Ikeda et al., 2001a). Unexpectedly, even without inhibiting DLK, the K195A LZK mutant had robust survival promoting activity that was dependent on its dimerization domain (Figure S1J). Since this suggested that the dominant-negative might also be interfering with DLK signaling, possibly by heterodimer formation, we next asked whether DLK and LZK interacted either directly or indirectly. Endogenous DLK was immunoprecipitated from primary RGCs and immunoassayed for LZK, or vice versa (Figure S1K). In both cases, DLK and LZK co-immunoprecipitated, suggesting that they indeed participate in a common endogenous complex. Taken together, these results suggest that LZK signaling can partially compensate for DLK in activating RGC cell death and that simultaneous inhibition is required to arrest the cell death program.
LZK and DLK are the key targets of neuroprotective PKIs
LZK is a 959 amino acid member of the mixed-lineage kinase (MLK) family of kinases (Sakuma et al., 1997). To determine whether other MLKs also had a role in RGC cell death, but were simply missed by the initial kinome screens, we tested four additional, independent siRNA sequences for each member of the MLK family. Perhaps surprisingly, LZK was the only MLK whose knockdown synergized with DLK knockdown to promote primary RGC survival (Figure 1G). Based on these results, we hypothesized that LZK might be the as-yet-unidentified “missing target” of our neuroprotective PKIs. In fact, while LZK shares only 60% overall homology with DLK, the kinase domain, the target of small molecule inhibitors, is 86% conserved. Moreover, kinase profiling has demonstrated that each one of the neuroprotective PKIs (i.e. tozasertib, foretinib, crizotinib, lestaurtinib and KW2449) has biochemical activity against both purified DLK and LZK (Davis et al., 2011). We reasoned that if LZK and DLK were the relevant targets of the neuroprotective PKIs, individual knockdown of either kinase should sensitize the cells to subthreshold concentrations of tozasertib. Consistent with this hypothesis, knockdown of either DLK or LZK left-shifted the dose-response curve to tozasertib (Figure 1H). Additionally, simultaneous knockdown of both DLK and LZK fully recapitulated the protective effect of the drug and further addition of tozasertib did not produce a significant increase in survival. Although we cannot rule out minor contributions from other kinases, the major survival-promoting effect of neuroprotective PKIs appears to be mediated by the simultaneous inhibition of DLK and LZK.
Development of an improved method for quantifying RGC survival in vivo
To test the role of LZK in a mouse model of RGC axon injury, we developed a highly-quantitative method for measuring RGC survival. Most previous approaches utilized flatmounted retinas in which RGCs are labeled by immunofluorescent staining for RGC antigens like β-III-tubulin/TUBB3 (Welsbie et al., 2013), BRN3A/B (Nadal-Nicolás et al., 2009), NeuN (Buckingham et al., 2008), γ-synuclein in situ hybridization (Soto et al., 2011), or injection of retrograde tracers (Vidal-Sanz et al., 1988). Unfortunately, these methods can be difficult to quantify, have limited specificity, are labor-intensive, and/or are incompatible with optic nerve injury models that downregulate RGC markers (Mead et al., 2014; Soto et al., 2008). We adapted a flow cytometry (FC)-based approach (Chang et al., 2012) to use two intracellular antigens, β-III-tubulin (TUBB3) and γ-synuclein (SNCG), that are expressed by RGCs (Figure S2A) but not downregulated after injury. Retinas from wildtype (WT), uninjured mice were isolated, dissociated, stained and analyzed by FC. As expected, 0.8–1.0% of total retinal cells were double-positive for TUBB3/SNCG, consistent with past estimates of RGC frequency (Williams et al., 1996). Moreover, the intersection of SNCG and TUBB3 staining by FC was both highly specific and sensitive for RGCs immunopanned from neonatal retina (Figure S2B) or dissociated in adult retina (Figure S2C). SNCG and TUBB3 staining was preserved after optic nerve injury (Figure S2D), allowing for quantification in the mouse optic nerve crush (ONC) model (Figure S2E). Finally, we showed that the FC technique identifies a similar reduction in RGC survival two weeks after ONC when compared to traditional imaging of immunostained flatmounted retinas (71% vs 75%, respectively; Figure S2F).
Targeted disruption of LZK confirms that LZK plays a role in RGC survival in vitro and in vivo
In light of the perinatal lethality associated with homozygous Dlk disruption (Hirai et al., 2006), we took a “knockout first” approach in order to generate mice with targeted deletion of Lzk. An Lzk null allele was engineered by targeting a promoterless splice acceptor/β-galactosidase cassette (Skarnes et al., 2011) to the second exon of Lzk in C57BL/6 mice and then further converted into a conditional deletion by crossing to FLP-expressing mice (Figure 2A). Interestingly, unlike the situation with Dlk, mice homozygous for the null Lzk allele were both viable and fertile, exhibiting no obvious phenotype. We then isolated RGCs from either WT or Lzk null mice and assayed for LZK expression (Figure 2B). As was the case with DLK, LZK is robustly upregulated 24 hours after immunopanning (which necessarily axotomizes the cells), but only in WT mice, confirming the specificity of our antibody and the validity of our knockout strategy.
Figure 2. RGCs with targeted deletion of Dlk and Lzk are resistant to axon injury-induced cell death in vitro and in vivo.
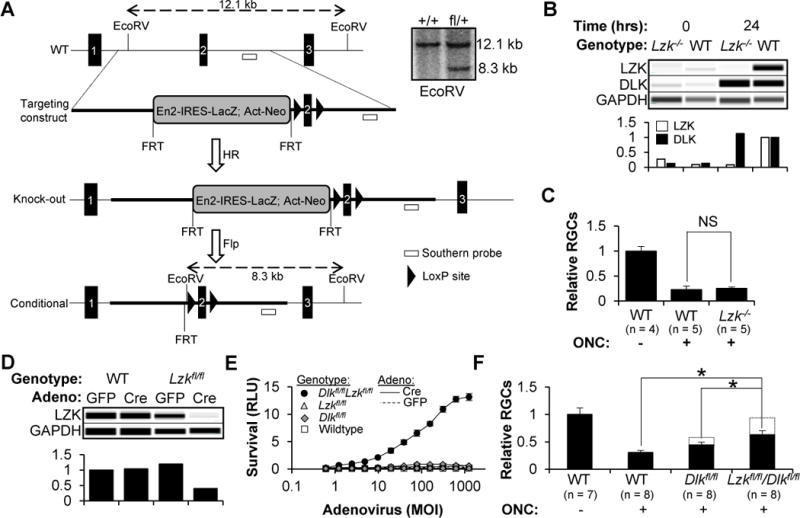
(A) Approach used to generate constitutive and conditional Lzk knockout mice. Inset shows a Southern blot confirming the presence of a single targeting construct in the heterozygous animals.
(B) Capillary-based immunoassay (top) and quantification (bottom) of DLK and LZK in RGCs, 0 or 24 hours after immunopanning injury.
(C) FC-based quantification of surviving RGCs, normalized to the uninjured control (±SD), two weeks after ONC or sham surgery. NS, non-significant, Mann-Whitney U test.
(D) Capillary-based immunoassay (top) and quantification (bottom) of LZK in RGCs one day after transduction with adenovirus.
(E) Survival (±SD) of RGCs, transduced with adenovirus, two days after a colchicine challenge. Dlkfl/fl RGCs transduced with Cre have a 2–5-fold increase in survival over the controls.
(F) FC-based quantification of surviving RGCs, normalized to the uninjured control (±SD), two weeks after ONC or sham surgery. All eyes were injected with 109 vg AAV2-Cre two weeks prior to the surgery. *P<0.05, Mann-Whitney U test.
To evaluate whether knockout of LZK was protective in vivo, Lzk null or WT control mice were subjected to ONC or sham control. Two weeks later, allowing time for the wave of RGC cell death that follows axon injury, retinas were isolated and the number of surviving RGCs was quantified by FC. Lzk knockout did not promote RGC survival in vivo (Figure 2C), which was not surprising given the minimal effect of Lzk siRNAs in vitro. We next crossed floxed Lzk mice with floxed Dlk mice (Miller et al., 2009) to generate four strains of animals: Dlk+/+Lzk+/+, Dlkfl/flLzk+/+, Dlk+/+Lzkfl/fl and Dlkfl/flLzkfl/fl. After confirming that LZK levels were reduced after Cre-mediated recombination (Figure 2D), RGCs were isolated from each strain and transduced with Cre or GFP control adenovirus. After two days, the cells were challenged with colchicine and survival was measured an additional two days later (Figure 2E). While genetic disruption of Dlk had a modest effect on survival (consistent with the two-to-four-fold increase seen previously), simultaneous knockout of Dlk and Lzk synergistically increased RGC survival (up to 100-fold), validating the siRNA findings. In order to extend the findings in vivo, floxed Dlk, floxed Dlk/Lzk and WT mice received an intravitreal injection of adeno-associated virus 2 (AAV2) expressing Cre. After two weeks to allow time for recombination to occur, ONC or sham surgery was performed and the number of surviving RGCs was assayed by FC after an additional two weeks (Figure 2F). While targeted deletion of Dlk was partially protective, simultaneous targeting of both Dlk and Lzk led to increased survival, nearing uninjured levels when accounting for the percentage of retina that was transduced by the virus (dotted lines).
DLK/LZK inhibition with an FDA-approved PKI protects human stem cell-derived RGCs
In order to test whether the DLK/LZK pathway had a similar role in human RGCs (hRGCs), we turned to human stem cell-derived RGCs. We previously described a method for differentiating and purifying hRGCs that utilizes an embryonic stem cell (hESC) reporter line that was generated using clustered regularly interspaced short palindromic repeats (CRISPR) technology to knock a P2A-mCherry cassette into the BRN3B (POU4F2) locus (Sluch et al., 2015). RGCs differentiated from this line demonstrate morphological, ultrastructural, gene expression, and electrophysiological properties consistent with hRGCs. For the studies described here, we used a modified version of this approach, based upon an improved cell line, which incorporates a P2A-tdTomato-P2A-Thy1.2 reporter cassette (Sluch et al., manuscript submitted). After anti-Thy1.2 immunopurification, hRGCs were challenged with colchicine as a pharmacologic model of axonal injury (Miller et al., 2009) and survival was measured 48 hours later using CellTiter-Glo (Fig. 3A) or conventional stains of viability (data not shown). As with primary mouse RGCs, colchicine killed the vast majority of hRGCs and the effect was reversed with the dual DLK/LZK-inhibitor tozasertib and a structurally-distinct, high-potency DLK (Cohen et al., 2013) that similarly inhibits LZK (data not shown). With the goal of potentially developing DLK/LZK inhibition as a treatment approach for optic neuropathies and other forms of neurodegenerative disease, we tested the FDA-approved drug sunitinib in the hRGC axonal injury assay. Sunitinib, which is approved for treatment of renal cell carcinoma and gastrointestinal stromal tumor, is a broad spectrum PKI with activity against both DLK and LZK (Davis et al., 2011; Karaman et al., 2008; Rock et al., 2007). As shown in Fig. 3B, sunitinib potently and in a dose-dependent manner promotes the survival of hRGCs challenged by colchicine-induced axonal injury.
Figure 3. Pharmacologic inhibition of DLK and LZK, including by sunitinib, an FDA-approved small-molecule inhibitor, promotes the survival of human ESC-derived RGCs.
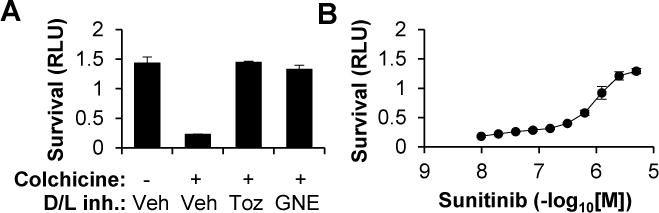
(G) Survival (±SD) of hESC-derived RGCs two days after a challenge with vehicle or colchicine (1 μM) in the presence of the DLK/LZK inhibitors tozasertib (1 μM), Genentech inhibitor 123 (0.1 μM) or a vehicle control.
(H) Survival (±SD) of hESC-derived RGCs two days after a challenge with colchicine (1 μM) in the presence of increasing doses of sunitinib.
LZK and DLK trigger RGC cell death through a JNK- and MKK-dependent kinase cascade
In C. elegans, the LZK homolog DLK-1 has an intrinsic calcium-sensing motif that, in response to axonal injury, binds calcium and leads to kinase activation (Yan and Jin, 2012). Given that the motif is conserved in mammalian LZK, we asked whether the same mechanism was active in RGCs. WT RGCs were immunopanned and tested in the human LZK reconstitution assay described above, comparing WT and mutant LZK (Figure 4A). A deletion of the homologous calcium-sensing domain (Δ6), but not a mutation which renders the kinase domain inactive, had activity comparable to WT LZK, demonstrating that the calcium-sensing motif is not necessary for LZK’s ability to promote cell death, at least in the setting of overexpression, but that active kinase signaling is required.
Figure 4. LZK kinase signaling triggers RGC cell death via the MKK4/7 and JNK1-3 kinase cascade.
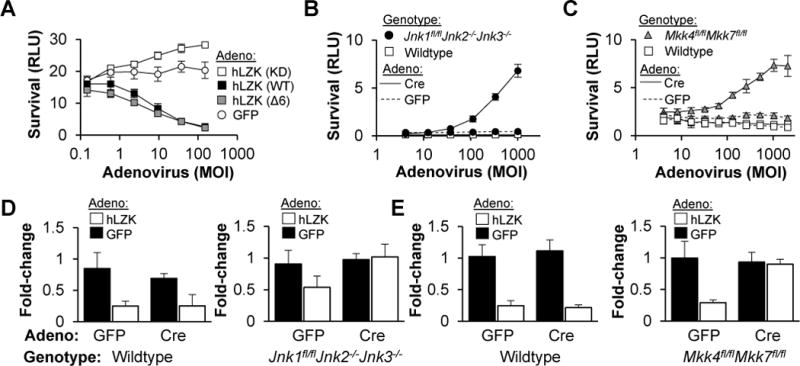
(A) Survival (±SD) of WT RGCs transfected with Dlk/Lzk siPOOL, two days after reconstitution of LZK signaling with adenovirus expressing mouse siRNA-resistant, human LZK cDNA or a GFP control (“LZK reconstitution assay”).
(B–C) Survival (±SD) of RGCs, transduced with adenovirus, two days after colchicine (1 μM) addition.
(D–E) Fold-change in survival (±SD) of RGCs, pretreated with adenovirus to eliminate JNK (D) or MKK4/7 (E) signaling, in response to adenovirus (MOI 400) to reconstitute LZK signaling.
DLK/LZK, and homologs in lower organisms, have been shown to signal through two downstream MAPK pathways, p38 and JNK (Bounoutas et al., 2011; Fan et al., 1996; Tulgren et al., 2011). While we have consistently failed to observe survival-promoting activity with p38 inhibitors (data not shown), both kinase screens identified knockdown of MKK4 and MKK7, both JNK kinases, as survival-promoting. In order to confirm a role for MKK4/7 and JNK1-3 in RGC cell death, we isolated primary RGCs from Mkk4/7 (Mazzitelli et al., 2011)t and Jnk1-3 (Xu et al., 2011) conditional knockout mice, and transduced them with Cre or GFP adenovirus. As expected, deletion of Jnk1, Jnk2 and Jnk3 (Figure 4B) or Mkk4 and Mkk7 (Figure 4C) increased RGC survival, more than individual knockout of Mkk4 or Mkk7 (data not shown). We then combined the human LZK reconstitution assay with knockout of each tier to examine whether LZK had cell death-promoting activity that signaled outside of the MKK4/7, JNK1-3 axis. RGCs were isolated from WT, floxed Mkk4/Mkk7 or floxed Jnk1, Jnk2/3-null mice, transfected with Dlk and Lzk siPOOLs and transduced with Cre-or GFP-expressing adenovirus. After two days, RGCs were challenged with GFP or human LZK-expressing adenovirus to reconstitute LZK signaling, and then survival was measured after an additional two days. LZK reconstitution promoted cell death in WT cells, but this activity was attenuated when only Jnk1 was present, and completely absent when all three Jnk genes were deleted (Figure 4D). Simultaneous knockout of Mkk4 and Mkk7 also rendered RGCs insensitive to LZK reconstitution (Figure 4E). On the other hand, individual knockout of Mkk4 or Mkk7 was only modestly protective (data not shown). A similar pattern of results was seen when the pathway was reconstituted by expressing siRNA-insensitive rat Dlk cDNA (data not shown). Together, these results suggest a linear cascade in which DLK and LZK activate MKK4 and MKK7, which in turn, leads to JNK1-3 activation and, ultimately, cell death.
Arrayed, whole-genome siRNA screen in primary RGCs
In order to identify non-kinase genes that are involved in RGC cell death signaling, we expanded our kinase screen and assayed an arrayed, whole-genome library of 52,725 siRNAs, providing three-fold coverage of 17,575 genes, for each siRNA’s ability to promote RGC cell survival (Figure 5A). Since seed-based off-target effects can dominate the results of siRNA screens looking at complex phenotypes like survival (Jackson et al., 2003, 2006), we used bioinformatic signal correction approaches to take advantage of the extensive and redundant dataset provided by the whole genome screen, which included nearly every possible seed combination multiple times. In one approach, we tracked the survival effect of every seed sequence within the library and used this information to generate a correction factor to subtract out off-target contributions, resulting in “corrected” on-target cell survival-promoting activities (Marine et al., 2012). Using this common-seed method, we generated a rank order of the genes whose median siRNA had the most survival-promoting activity (Figure S3A). In a second approach, we predicted the potential off-target mRNAs that are being silenced by the bioactive seed sequences and then used a Haystack analysis to identify commonly-targeted genes by similarly-behaving seeds in order to deconvolute the off-target effects and identify the genes whose off-target silencing affects survival (Buehler et al., 2012). This latter approach nominated a distinct but overlapping set of genes (Figure S3B). Validating the approach, kinases known to be involved in RGC cell death, including Map2k4/Mkk4, Map2k7/Mkk7, and Map3k12/Dlk, were amongst the top hits. We then used independent siRNAs to test the other genes nominated by the on-target (Figure S3C) and off-target analyses (Figure S3D) and were able to replicate the survival-promoting activity (Figure 5B) associated with knockdown (Figure S4A) of three genes: Atf2, Mef2a and Bbc3/Puma. Finally, we tested siPOOLs for their ability to knockdown ATF2, MEF2A and PUMA levels (Figure S4B) and confirmed that knockdown of each one improved RGC survival in vitro (Figure 5C).
Figure 5. Whole-genome siRNA screen identifies ATF2, PUMA and MEF2A as mediators of RGC cell death.
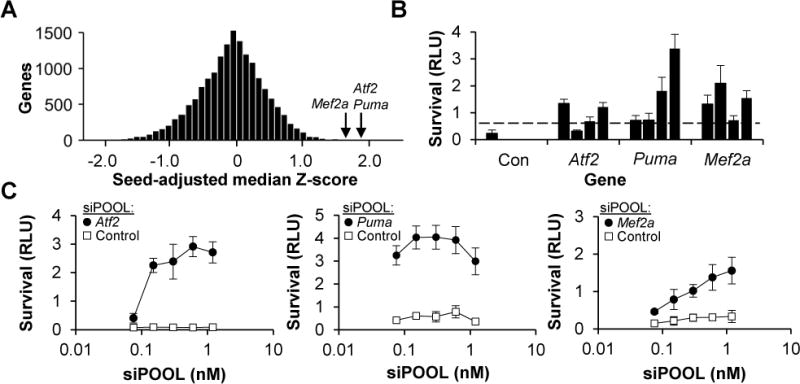
(A) Histogram showing the normalized, seed-adjusted survival for the median survival-promoting siRNA targeting each of the 17,575 genes in the whole-genome library.
(B) Survival (±SD) of WT RGCs, transfected with one of four independent siRNAs targeting Atf2, Puma or Mef2a or the non-targeting control, two days after colchicine addition. Dashed line shows the threshold of survival greater than 3SD from the negative control.
(C) Survival (±SD) of WT RGCs, transfected with siPOOLs, two days after colchicine (1 μM) addition.
In peripheral sensory neurons, neurotrophin deprivation activates the Puma promoter through the transcription factors, FOXO3A and JUN, thereby integrating the activity of the pro-survival AKT and pro-death JNK pathways (Ambacher et al., 2012; Simon et al., 2016). The resulting increase in PUMA levels leads to cell death and axon degeneration. That we identified knockdown of PUMA as being survival-promoting suggested that the pathway might be similar in injured CNS neurons like RGCs. Indeed, RGC axon injury in vivo leads to an upregulation of Puma transcripts that is both JUN- (Fernandes et al., 2013) and DLK-dependent (Watkins et al., 2013). However, in the primary RGC survival assay, neither PUMA knockdown nor JUN knockdown was able to fully rescue cell death (data not shown) and, in vivo, targeted deletion of either Jun or Puma only serves to delay, but not prevent, cell death following optic nerve injury (Fernandes et al., 2012; Harder and Libby, 2011, 2013; Yoshida et al., 2002). Since our screen identified ATF2, a known heterodimeric partner of JUN, and since axon injury was known to trigger phosphorylation of ATF2 (Levkovitch-Verbin et al., 2005), we queried whether ATF2 inhibition was required to prevent RGC cell death. To test this, we obtained floxed Atf2 mice (Shah et al., 2010), in which Cre-dependent recombination triggers an in-frame deletion of portions of the DNA-binding and dimerization domains (Figure S4C). While targeted disruption of Atf2 increased RGC survival, albeit modestly, in vitro (Figure S4D) and in the mouse model of ONC (Figure S4E), combined knockdown of ATF2 and JUN was still unable to fully rescue cell death (data not shown). This suggested the possibility that in the absence of ATF2/JUN, unlike the case with sensory neurons, RGC cell death signaling might additionally proceed through other transcription factors. Based on our siRNA results, we hypothesized that MEF2A might be one of these other factors.
MEF2A is a DLK-regulated mediator of RGC cell death
To validate the siRNA findings and test whether the transcriptional-regulatory function of MEF2A was critical for RGC cell death, we utilized conditional Mef2a knockout mice in which the second coding exon of Mef2a is floxed and Cre-dependent recombination produces an in-frame deletion that disrupts the DNA-binding MADS-box and MEF2 domains (Akhtar et al., 2012). As expected, RGCs isolated from the floxed mice displayed a shift in the size of MEF2A in response to Cre expression (Figure 6A). To test the effect on survival, RGCs were isolated from WT or floxed Mef2a mice, transduced with adenovirus expressing Cre or a GFP control, and then viability was measured two days after a colchicine challenge (Figure 6B). While increasing amounts of Cre-expressing adenovirus had no effect on WT RGCs, it led to a modest, but dose-dependent, increase in survival of floxed Mef2a RGCs. We then considered the possibilities that dominant-negative forms of MEF2A were interfering with the signaling of other highly-related MEF2 family members, including MEF2C and MEF2D. However, arguing against this possibility, RGCs isolated from mice with combined conditional deletions of Mef2a, Mef2c and Mef2d (Akhtar et al., 2012) had no reversal of the phenotype associated with Mef2a knockout (Figure 6C). To confirm these findings in vivo, WT or floxed Mef2a mice were intravitreally injected with AAV2 expressing Cre and then, after two weeks, subjected to ONC or sham surgery. After an additional two weeks, retinas were harvested and assayed for RGC survival using the TUBB3/SNCG FC technique described above (Figure 6D). Consistent with the in vitro findings, targeted disruption of the MEF2A transcriptional regulatory domains increased RGC survival.
Figure 6. RGCs with a targeted disruption of the transcriptional regulatory domain of MEF2A are partially resistant to axon injury-induced cell death in vivo.
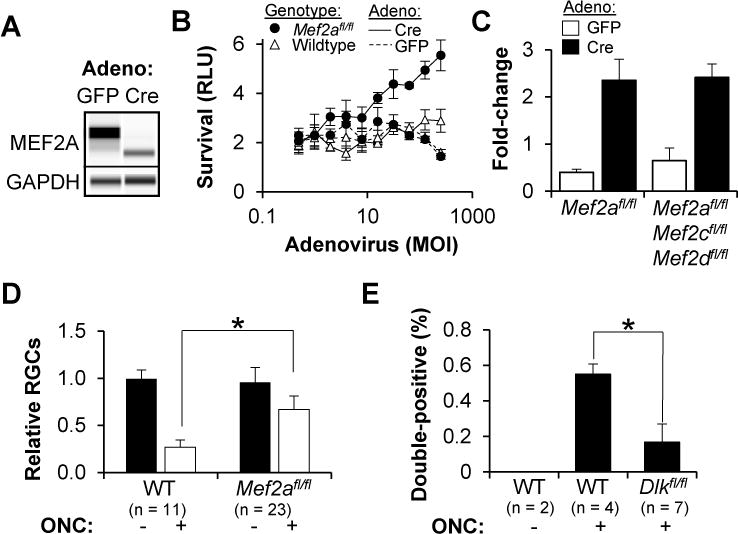
(A) Capillary-based immunoassay of MEF2A in RGCs, two days after transduction with adenovirus.
(B) Survival (±SD) of RGCs, transduced with adenovirus, two days after colchicine (1 μM) addition.
(C) Fold-change in survival (±SD) of RGCs from adenoviral transduction (MOI 400), two days after colchicine (1 μM) addition.
(D) FC-based quantification of surviving RGCs, normalized to the uninjured control (±SD), two weeks after ONC or sham surgery. All eyes were injected with 109 vg AAV2-Cre two weeks prior to the surgery. *P<0.05, Mann-Whitney U test.
(E) FC-based quantification of TUBB3/phospho-S408 MEF2A double-positive cells, expressed as a percentage of total retina (±SD), two days after ONC or sham surgery. *P<0.05, Mann-Whitney U test.
Since we had demonstrated that MEF2A played a role in RGC cell death, we considered the obvious possibility that it was somehow regulated by DLK/LZK, or downstream JNK, signaling. First, we sampled several antibodies against known phosphosites and found that S408, whose phosphorylation is associated with inactive or transcriptionally-repressive forms of MEF2A (Flavell et al., 2006; Shalizi et al., 2006), could be reliably quantified in vivo using an FC-based approach. WT or floxed Dlk mice were intravitreally injected with AAV2 expressing Cre and then subjected to ONC or sham surgery. Two days later, the number of double-positive TUBB3/phosphorylated-MEF2A cells was quantified by FC. Indeed, axon injury increased phosphorylation of MEF2A S408 in RGCs in vivo, in a manner that was highly-dependent on the presence of DLK (Figures 6E, S5A and S5B). Collectively, these results highlight the ability of a whole-genome siRNA screen to be able to find novel and unexpected mediators of DLK-dependent cell death.
Sensitized, whole-genome siRNA screen identifies SOX11 as a downstream mediator of RGC cell death
The finding that LZK knockdown by itself did little to improve RGC survival, but greatly increased the effect of DLK knockdown, afforded an opportunity to perform a “sensitized” whole-genome siRNA screen for additional DLK pathway members. Each well was transfected with Lzk siPOOL (to sensitize the cells to DLK inhibition) and one of 16,698 siRNA minipools (comprised of four siRNAs each), in duplicate, challenged with colchicine two days later, and then assayed for viability after another two days. Using the common-seed method, we generated a list of the most survival-promoting siRNA minipools (Figure S6A). Validating the approach, the top hit was the minipool targeting Dlk (Figure 7A). Moreover, other confirmed hits, including Atf2, Jun, Puma, Mkk4, Mkk7 and Jnk1-3, were all in the top 1% of genes (Figure S6B). Since none of the novel hits were confirmed when we tried to validate them with independent siRNAs (Figure S6C), we turned to Haystack analysis to deconvolute the off-target effects. This analysis identified validated genes like Puma and Mkk4 (Figure S6D). Additionally, it identified Sox11, which was confirmed with independent siRNAs (Figure S6E).
Figure 7. Sensitized, whole-genome siRNA screen identifies SOX11 as a downstream mediator of RGC cell death.
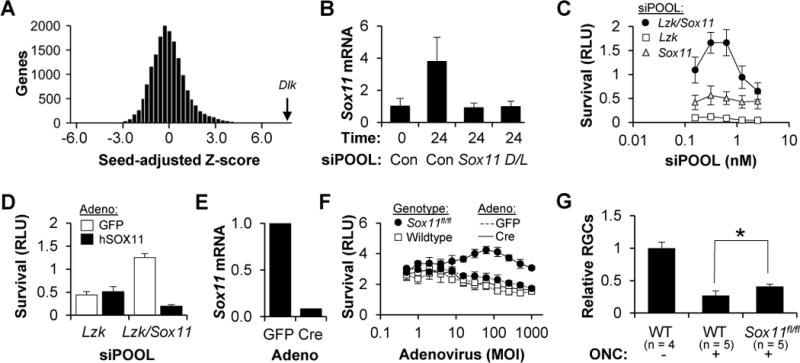
(A) Histogram showing the normalized, seed-adjusted survival for the siRNA minipool targeting each of the 16,698 genes in the whole-genome library (transfected in the presence of Lzk siPOOL).
(B) QPCR assay for Sox11 mRNA, normalized to GAPDH levels (±SD), in WT RGCs transfected with siPOOLs, at the indicated time following immunopanning injury.
(C) Survival (±SD) of WT RGCs, transfected with siPOOL, two days after colchicine (1 μM) addition.
(D) Survival (±SD) of WT RGCs transfected with siPOOLs, two days after reconstitution of SOX11 signaling with an adenovirus expressing mouse siRNA-resistant human SOX11 cDNA or a GFP control.
(E) QPCR assay of Sox11 mRNA, normalized to GAPDH levels, in Sox11fl/fl RGCs transduced with adenovirus.
(F) Survival (±SD) of RGCs, transduced with adenovirus, two days after a colchicine (1 μM) addition.
(G) FC-based quantification of surviving RGCs, normalized to the uninjured control (±SD), two weeks after ONC or sham surgery. All eyes were injected with 109 vg AAV2-Cre two weeks prior to the surgery. *P<0.05, Mann-Whitney U test.
SOX11 is one of the most DLK-dependent, upregulated genes in the retina following ONC (Watkins et al., 2013). However, along with SOX4, SOX11 is required for RGC differentiation and survival in development; thus its role in RGC cell death was unexpected (Jiang et al., 2013). In order to validate this finding, we first obtained an siPOOL targeting Sox11. After injury, consistent with the findings in vivo, primary RGCs upregulate Sox11 mRNA in a DLK/LZK-dependent and Sox11 siPOOL-sensitive manner (Figure 7B). Moreover, Sox11 siPOOLs had a mild survival-promoting effect that synergized with LZK knockdown (Figure 7C) and could be reversed with adenoviral expression of a mouse siPOOL-resistant, human SOX11 cDNA (Figure 7D). No effect was seen when RGCs were transfected with siPOOLs targeting Sox4 (data not shown). For more definitive validation, we examined conditional Sox11 mice (Wang et al., 2013) and found that RGCs isolated from floxed Sox11 mice had reduced cell death when transduced with Cre-, but not GFP-expressing, adenovirus (Figure 7E). Finally, using the mouse ONC model we demonstrated that targeted deletion of Sox11 produced a statistically significant, albeit modest, increase in RGC survival in vivo (Figure 7F).
DLK/LZK-dependent cell death requires SOX11, MEF2A, JUN and ATF2
Since targeted in vivo disruption of Sox11, Mef2a, Jun and Atf2, one factor at a time, produced significant but modest improvements in RGC survival, we asked whether simultaneous inhibition of all four would lead to more robust neuroprotection. WT primary RGCs were transfected with siPOOLs against Atf2, Jun, Sox11, Mef2a, Dlk or Lzk, alone or in various combinations and assayed for survival (Figure 8A) and neurite outgrowth (Figure S7). Consistent with the hypothesis, combined knockdown of all four transcription factors was highly survival-promoting and inhibitory to neurite outgrowth, rivaling the effect of combined DLZ/LZK knockdown. To test whether these factors were downstream of DLK/LZK, we turned to the LZK-reconstitution assay and found that combined knockdown of all four completely abrogated DLK/LZK-mediated cell death (Figure 8B). While highly suggestive, this result could have been confounded by siRNA off-target effects. Since breeding mice with conditional knockout alleles for all four transcription factors would be prohibitively time-consuming, we developed a rapid CRISPR-based method to knockout genes, including in combination, in primary RGCs. RGCs were isolated from mice that express Streptococcus pyogenes Cas9 (SpCas9) from the Rosa26 locus (Platt et al., 2014) and transfected with T7-transcribed synthetic guide RNAs (sgRNAs). As a proof-of-principle, tracrRNA and six Dlk-targeting, chimeric sgRNAs were synthesized and cotransfected, along with Lzk siPOOL, into WT or SpCas9-expressing RGCs and assayed for their ability to inhibit cell death. Those sgRNAs that target Dlk, but not the control tracrRNA, were able to increase survival in the presence of SpCas9 (Figure 8C). Similar results were obtained in the reciprocal experiment with cotransfection of Lzk-targeting sgRNAs and Dlk siPOOL (Figure 8D) or with cotransfection of both Dlk- and Lzk-targeting sgRNAs (Figure 8E). We then leveraged the fact that SpCas9-directed mutagenesis disrupts an endogenous BglII site in one of the Dlk targets and all of the most likely off-targets, to demonstrate that the system produces highly-specific knockout (Figure S8A). Moreover, subcloning of the target site in the Dlk locus showed the typical spectrum of small indels (Figure S8B). Consistent with the siPOOL results, combined Dlk/Lzk targeting was far more robust than the addition of neurotrophins although the combination led to a modest increase in survival (Figure S8C), a finding which may be relevant to future therapeutic strategies centered around DLK/LZK pathway inhibition. We then designed sgRNAs targeting Jun, Atf2, Mef2a and Sox11 and showed that all four had modest survival promoting activity when tested individually (Figure 8F). However, when combined, the four synergized to increase primary RGC survival to a level comparable to combined knockout of Dlk and Lzk. A similar degree of synergy was seen with an independent set of sgRNAs, arguing against an off-target mediated phenotype (Figure S8D). Finally, CRISPR-based knockout of the four transcription factors rendered RGCs completely resistant to the effects of LZK pathway activation (Figure 8G). Taken together, these data reinforce the siRNA-based results and establish that SOX11, ATF2, JUN and MEF2A cooperate to promote RGC cell death following DLK/LZK activation (Figure 8H).
Figure 8. DLK/LZK-dependent cell death is mediated by a set of four transcription factors: JUN, ATF2, SOX11 and MEF2A.
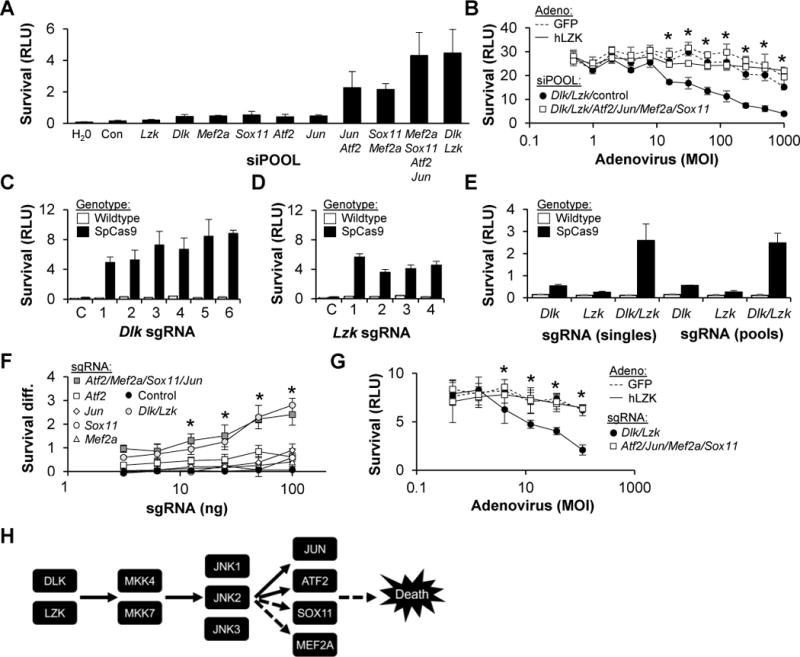
(A) Survival (±SD) of WT RGCs transfected with siPOOL, two days after colchicine (1 μM) addition.
(B) Survival (±SD) of WT RGCs transfected with siPOOL, two days after adenoviral transduction to reconstitute LZK signaling. *P<0.05 Mann-Whitney U test comparing absence/presence of transcription factor siPOOLs.
(C–E) Survival (±SD) of RGCs, transfected with Lzk siPOOL and either tracrRNA or sgRNAs targeting Dlk (C), Dlk siPOOL and either tracrRNA or sgRNAs targeting Lzk (D) or pools versus individual sgRNAs targeting Dlk and Lzk (E).
(F) Difference in survival (SpCas9-WT; ±SD) conferred by transfecting sgRNA targeting each of the four transcription factors, alone or in combination, and compared to negative control tracrRNA or positive control sgRNAs targeting Dlk/Lzk. *P<0.05 Mann-Whitney U test compared to control; no significant difference between Dlk/Lzk and transcription factor sgRNAs.
(G) Survival (±SD) of SpCas9 RGCs, transfected with sgRNA, two days after adenoviral transduction to activate LZK signaling. *P<0.05 Mann-Whitney U test comparing Dlk/Lzk and transcription factor sgRNAs.
(H) Proposed pathway for RGC cell death following axon injury. Dashed lines indicate that SOX11 and MEF2A are downstream of DLK/LZK, but not necessarily JNK1-3, and could be indirectly activated.
Discussion
In response to axon injury, a variety of neurons, including RGCs, respond by initiating an active genetic program of cell death. It is becoming increasingly appreciated that DLK plays a role as a key upstream trigger of this cascade (Shin et al., 2012; Watkins et al., 2013; Welsbie et al., 2013; Xiong et al., 2010). In an effort to better define the DLK injury-signaling pathway, we have employed a variety of kinome-wide and whole genome screens in conjunction with a disease-relevant primary neuron, the RGC. Using this unbiased and comprehensive functional genomic approach, we made the unexpected discoveries that cell death signaling can partially circumvent DLK and proceed through the related kinase, LZK, and that two transcription factors, SOX11 and MEF2A, previously only known to play a role in promoting neuronal survival, cooperate with ATF2 and JUN to execute the DLK/LZK-mediated cell death program.
LZK was initially identified based on its homology to other MLKs (Sakuma et al., 1997) and, similar to the case with DLK, shown to have biochemical activity as an MKK7 kinase (Ikeda et al., 2001b). Subsequently, it was found that developing motor neurons use MKK4, not MKK7, for DLK-dependent cell death (Itoh et al., 2014). Using genetic deletion and rescue experiments, we have shown that LZK and DLK signaling in RGCs utilizes both MKK4 and MKK7, although further work will be required to dissect out the specific role for each. LZK can act as an intracellular mediator of PirB signaling, thought to be important in myelin’s ability to retard axon growth (Dickson et al., 2010). Interestingly, in primary RGCs, we have found that LZK has the opposite function, promoting neurite outgrowth after axotomy. Our finding is consistent with the recent report by Chen et al. who demonstrated that LZK can stimulate axon growth of cultured cerebellar granule neurons (CGNs)(Chen et al., 2016). However, there also appear to be interesting differences between the activity of LZK in RGCs versus CGNs. As just noted above, LZK signaling in RGCs appears to go through both MKK4 and MKK7. This is distinct from CGNs, where LZK signaling seems to be mediated exclusively by MKK4, similar to that of DLK in motor neurons (Itoh et al., 2014). That LZK inhibition alone decreases outgrowth, as well as our finding that the effect of DLK and LZK inhibition on neurite growth is additive, suggests that DLK and LZK have non-redundant roles in RGCs. Again, the mechanism may be different from the case in CGNs, as in that case the effect of DLK and LZK on axon growth is not additive (Chen et al., 2016).
In contrast to the case with neurite outgrowth, in RGC cell death signaling DLK and LZK appear to be able to compensate for each other, at least partially, likely reflecting their participation in common biochemical complexes. The fact that LZK was not previously discovered to have a role in promoting neuronal cell death is perhaps surprising, given its structural and biochemical similarity to DLK, but it highlights the power and utility of unbiased phenotypic screens such as those we employed here. From a therapeutic perspective, these results highlight the need to develop small molecule inhibitors that target both DLK and LZK in order to treat neurodegenerative diseases related to axonopathy. However, as more is learned about the distinct functional roles of DLK and LZK, it may turn out to be desirable to develop drugs that can separately and independently modulate DLK and LZK activity.
After axonal injury and/or cytoskeletal disruption activates DLK/LZK signaling, our data suggests that the four transcription factors SOX11, JUN, ATF2 and MEF2A are engaged and act as downstream mediators of cell death and, possibly, axon regeneration. Our SOX11 finding is consistent with the work of Norsworthy et al., appearing in this issue, who using a totally independent approach identified SOX11 as playing a role in the death of RGCs following optic nerve crush. Highlighting the complexity of the response to axon injury, they found that SOX11 is preferentially involved in the death of a specific subclass of RGCs, alpha cells. Additionally, again consistent with our results, their data also implicates SOX11 in axon regeneration following crush. The finding of JUN and ATF2 involvement is not unexpected, as these factors are well known to be JNK-regulated through phosphorylation following neuronal injury. More surprising are our findings related to MEF2A, since based on prior literature one would have expected the opposite result. MEF2A undergoes axon injury-triggered, DLK-dependent phosphorylation at S408, albeit not necessarily directly catalyzed by JNK. Typically, phosphorylation of MEF2A S408 leads to the sumoylation at K403 and the formation of a transcriptional repressor. This repressor has been shown to regulate synaptic differentiation (Shalizi et al., 2006). Hitherto, MEF2A was known only to promote survival, with multiple groups showing MEF2A inactivation, either by phosphorylation or cleavage, triggering neuronal cell death (Gong et al., 2003; Okamoto et al., 2002). We hypothesize that in RGCs, a transcriptional repressor form of MEF2A serves to downregulate the expression of pro-survival factors and tip the balance in favor of cell death. Importantly, when multiple MEF2 paralogs are disrupted, the survival increase is not attenuated, suggesting that the repressor must be doing more than simply interfering with the already-known, pro-survival effect of MEF2 signaling. Future profiling of the transcriptional changes mediated by MEF2A and, for that matter, JUN, ATF2 and SOX11, will help to clarify the relevant targets and understand how the cell death and regeneration signals bifurcate. We also recognize that our primary RGC assay is a mixture of RGC subtypes (Duan et al., 2015) and that some of the transcription factor synergy could reflect differing requirements in the different subtypes, as for example demonstrated in the results of Norsworthy et al.
Our evolving understanding of the role of DLK and LZK, and their downstream signaling and effector molecules, is providing new insights into the mechanisms by which axon injury is detected, how the injury signal is relayed to the cell body, and how this information is then interpreted so as to result in either cell death and/or a regenerative response. These new insights, in addition to increasing our understanding of basic neuronal processes, is opening possibilities for novel neuroprotective strategies for treatment of the optic neuropathies and other forms of neurodegenerative disease. Furthering this case for the potential clinical translatability of DLK/LZK inhibition, our data with human stem cell-derived RGCs shows that small molecule protein kinase inhibitors with activity against DLK and LZK, including an already FDA approved drug, sunitinib, are also active in promoting the survival of human neurons.
Star Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Donald J. Zack (donzack@gmail.com).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All animal use was in accordance with the Association for Research in Vision and Ophthalmology (ARVO) Statement for the Use of Animals, following animal protocols approved by the Institutional Animal Care and Use Committee at Johns Hopkins University. Male/female, 2–4 month-old, male/female, C57BL/6 mice (genotype indicated in the figure) were used for optic nerve injury experiments. SpCas9 knockin mice (Rosa26 locus) were obtained from Jackson labs (stock 026179). Mice with the following alleles: Dlkfl/fl (Miller et al., 2009, RRID: MGI:5440728), Mkk4fl/flMkk7fl/fl (Mazzitelli et al. 2011), Jnk1fl/flJnk2−/−Jnk3−/− (Xu et al., 2011), Mef2afl/fl and Mef2afl/flMef2cfl/flMef2dfl/fl (Akhtar et al., 2012), Sox11fl/fl (Wang et al., 2013; RRID: MGI:5510984) and Atf2fl/fl (Shah et al., 2010) were generously provided by the respective investigators. In cases where a single, pure-breeding genotype received different treatments, littermates were randomly assigned to experimental groups. Animals were housed in the Johns Hopkins School of Medicine Animal facility, with full veterinary supervision, with 14-hour light/8-hour dark cycles, at 72 degrees C, 30–70% relative humidity and continuous food and water supply.
Creation of Lzk knockout mice
Lzk knockout mice were generated at ingenious targeting laboratory by electroporating the targeting construct shown in Fig. 1H (from the KOMP repository) into C57BL/6J ES cells and selecting for integration with G418. After confirming targeting with PCR and Southern blotting, ES cells were injected into C57BL/6 blastocysts to produce chimeras. Those with germline transmission were either intercrossed to generate the Lzk null line or crossed with FLP-expressing, C57BL/6 mice to excise the targeting construct and generate heterozygous floxed Lzk mice. These were subsequently intercrossed to generate the homozygous conditional Lzk knockout line. Subsequently, floxed Dlk mice were crossed with floxed Lzk mice to generate the combined conditional knockout.
Primary RGCs
Retinas were isolated from postnatal day 0–3 mice and dissociated with papain. Microglia were immunodepleted with CELLection Dynabeads (Invitrogen) conjugated to anti-CD11b (BD Pharmingen, 554859). The suspension of retinal cells was immunopanned on plates pre-conjugated with anti-Thy1.2 antibodies (BioRad, MCA02R) and goat anti-mouse IgM (Jackson Immunoresearch, 115-001-020) at room temperature (RT). After washing, retinal ganglion cells (RGCs) were released from the plate with trypsin (Sigma T9201), counted, and seeded at a density of 5,000–10,000 per well in 96-well plates, 2000–3500 per well in 384-well plates, 500–1000 per well in 1536-well plates or 150,000–300,000 per well in 4-well plates (Nunclon plates, Poly-D-lysine and laminin coated). Growth media was composed of Neurobasal (Life Technologies) supplemented with NS21, Sato, L-glutamine, penicillin/streptomycin, N-acetyl-cysteine, insulin, sodium pyruvate, triiodothyronine (T3), forskolin, BDNF, CNTF, and GDNF (Chen et al., 2008). Transfection of small RNAs (siRNAs, siPOOLs or sgRNAs) was performed at the time of isolation, using NeuroMag magnetic nanoparticle (OZ Biosciences, Marseille). Adenovirus and colchicine were added as indicated. Survival was measured [in relative light units (RLU)] at the times indicated using CellTiter-Glo (Promega, Madison). Four replicates were typically used for each data point in non-screen experiments with graphs plotting mean and error bars showing one standard deviation from the mean.
Human stem cell-derived RGCs
H7 hESCs (normal karyotype, XX) were modified to express mouse Thy1.2 and tdTomato from the BRN3B locus (Sluch et al., manuscript submitted). Cells were then dissociated to single cells and plated on Matrigel coated plates at a density of 50K/cm2 in mTeSR1 with 5 μM blebbistatin. One day after plating, mTeSR1 was completely exchanged for N2/B27 media [1:1 mix of DMEM/F12 and Neurobasal with 1× GlutaMAX Supplement, 1× antibiotic-antimycotic, 1% N2 Supplement, and 2% B27 Supplement (all from Life Technologies)] to start differentiation. Cells were fed with a full exchange of N2B27 media every other day. Differentiation was carried out at 37°C in 5% CO2/20% O2. On day 35, cells were dissociated and purified using the anti-Thy1.2 MACS system (Miltenyi Biotec), per the manufacturer’s protocol, and plated in 96-well plates at a density of 2500 cells per well. After five days, cells were challenged with 1 μM colchicine in the presence or absence of the DLK inhibitors. Survival was measured 48 hours later using CellTiter-Glo (Promega).
METHOD DETAILS
Reagents
Small molecules used were tozasertib (Selleckchem, S1048), sunitinib (LC labs, S-8803) and colchicine (Sigma, C3915). The Genentech compound (“123” in Cohen et al., 2013) was synthesized by D. Ferraris at the Brain Science Institute, Johns Hopkins University. All siPOOLs (mouse Atf2, Jun, Mef2a, Sox11, Dlk, Lzk, Puma and a non-targeting control) were purchased from siTOOLs Biotech. Active siRNAs in the secondary screening targeted Puma (Qiagen), Atf2 (Qiagen), Mef2a (Qiagen, Lzk (Dharmacon) and Sox11 (Dharmacon). Antibodies for Western blotting, flow cytometry and immunostaining were as follows: DLK (Novus Biologicals NBP2-17218, RRID: AB_2650533), LZK (Sigma HPA016497, RRID: AB_10670027), P-JNK Thr183/Tyr185 (Cell Signaling Technology 4671, RRID: AB_331342), JNK (Cell Signaling Technology 9252, RRID: AB_2250373), JUN (Cell Signaling Technology 9165, RRID: AB_2130165), P-JUN Ser63 (Cell Signaling Technology 9261, RRID: AB_2130162), GAPDH (Abcam ab9485, RRID: AB_307275), ATF2 C-19 (Santa Cruz Biotechnology sc-187, RRID: AB_630885), MEF-2A H-300 (Santa Cruz Biotechnology sc-10794, RRID: AB_670016), MEF2A P-S408 (Abcam ab51151, RRID: AB_881494), TUBB3 (Biolegend 801201, RRID: AB_2313773), Biotinylated NEUN, clone A60 (EMD Millipore MAB377B, RRID: AB_2313773), SNCG rabbit polyclonal (generous gift from N. Marsh-Armstrong), PerCP-Vio700-Thy1.2 (Miltenyi 130-102-204, RRID: AB_2650534)
Production of sgRNAs
For CRISPR studies, sgRNAs against mouse Dlk, Lzk, Atf2, Mef2a, Sox11, and Jun were produced using Gibson Assembly of a custom T7 gRNA vector (AvrII-digested) and gene specific oligo duplexes. Resulting constructs were amplified in 10-beta cells (New England Biosciences C3019), NotI-linearized and used for in vitro transcription with HiScribe T7 Quick High Yield RNA Synthesis Kit (New England Biolabs E2050S). RNA was purified with MEGAclear™ Transcription Clean-Up Kit (Ambion AM1908) and quantified using a Nanodrop (Thermo Fisher Scientific).
Genotyping Lzk knockout mice
Genotyping primers for Lzk null mice are LZKko_fwd: CCAGTGGGTTAAATGCACTTGCTG, LZKko_lacZ_rev: TGGCCTGTCCCTCTCACCTTCTAC and LZKko_wt_rev: CTGAGATCTCGGGAGACTTCCTC; LZKko_fwd/LZKko_lacZ_rev yields a band at 932 bp if the KO construct is present while LZKko_fwd/LZKko_wt_rev gives a band at 883 bp if the KO construct is absent (i.e. wildtype). Genotyping primers for floxed Lzk mice are: LZKcKO_fwd AGGCCAGAGACATAAAGGTGG and LZKcKO_rev CTGAGATCTCGGGAGACTTCCTC; a band at 464 bp indicates LoxP sites are present while one at 340 bp indicates wildtype.
Transfection with siRNAs and sgRNAs
RGCs were transfected with siRNAs (0.5–50 nM) or sgRNAs (1–30 nM) by complexing RNA with NeuroMag (OZ Biosciences) in Optimem (Life Technologies) before adding to wells. RGCs were then reverse transfected overnight on a stationary magnet (OZ Biosciences). In the case of transfecting with multiple RNAs, different siRNAs or siRNA and sgRNA were complexed together in the same mixture of Optimem/Neuromag. For experiments in which the amount of siRNA/siPOOL is not indicated in the legend, 0.5–1 nM total was used. Protospacer sequences: Atf2 – gtcgactcggggtgaggtaa, Mef2a – gttgagcactacagacctca, Jun – gctctcggactggaggaacg, Sox11 – gcgagaagatcccgttcatc.
Viability/Neurite length
Cells were assayed for survival by adding a 50% volume of CellTiter-Glo (Promega G8462) or by staining with calcein AM, ethidium homodimer and Hoechst 33342. For the former, luminescence was measured with a plate reader (BMG Labtech) while, for the latter, images were taken using Cellomics ArrayScan VTI HCS Reader (Thermo Fisher) and analyzed with Cellomics Neuronal Profiling software. This allowed for a calculation of the number of viable cells as well as neurite length.
Adenovirus
Ad5-CMV-eGFP and Ad5-CMV-Cre-eGFP adenoviruses were purchased from the Vector Development Lab at Baylor College of Medicine. Viruses expressing human SOX11 and LZK cDNA were made by subcloning into pAd-V5-DEST using a gateway-compatible entry vector. The templates were an expression plasmid for human LZK, generously provided by L. Holzman and human cDNA (for SOX11). Deletion and point mutants were cloned via site-directed mutagenesis (Quikchange, Agilent). Adenoviruses were made using the Virapower Adenoviral Expression System (ThermoFisher) and concentrated using Adenopure kits from Puresyn, Inc.
Sensitized kinome screen
Primary RGCs were isolated as described above, seeded into 384 well plates with 2500 cells per well and transfected, in triplicate, with 20 nM siRNA from the Sigma mouse kinome library (3 siRNAs per gene) and 20 nM Dharmacon Dlk siRNA. After 48 hours, cells were challenged with 1 μM colchicine. CellTiter-Glo was added at 96 hours to assay viability. The activity of each well was normalized for the plate’s activity and the genes were ranked by the activity of the median siRNA.
Whole-genome screen
Primary RGCs were isolated as described above, seeded into 1536 well plates with 500 cells per well and transfected with 40 nM siRNA from the Ambion Silencer mouse whole-genome siRNA library. After 72 hours, viability was measured with CellTiter-Glo. Survival from each well was normalized to negative controls present on each plate and then analyzed using common-seed (Marine et al., 2012) or Haystack analyses (Buehler et al., 2012). Automated liquid handing was used at all steps to minimize variability.
Sensitized whole-genome screen
Primary RGCs were isolated as described above, seeded into 384 well plates with 1000 cells per well and transfected, in duplicate, with 1.25 nM siRNA minipools (set of four siRNAs) from the Dharmacon siGENOME mouse whole-genome and 1.25 nM Lzk siPOOL. After 48 hours, cells were challenged with 1 μM colchicine. CellTiter-Glo was added at 96 hours to assay viability. Survival from each well was normalized to negative controls present on each plate and then analyzed using common-seed (Marine et al., 2012) or Haystack analyses (Buehler et al., 2012). Automated liquid handing was used at all steps to minimize variability.
Intravitreal injection/optic nerve crush
Wild type and transgenic mice at were anesthetized using a ketamine/xylazine cocktail injected intraperitoneally and eyes were locally anesthetized using 0.5% proparacaine hydrochloride. Mice were intravitreally injected with 1 μl of 109 DNA-containing particles of capsid-mutant (Y444, 500, 730F) AAV2 (gift from W. Hauswirth laboratory), expressing Cre recombinase or GFP from the chicken β-actin promoter, using a 1.5 cm 33-gauge Hamilton needle (Hamilton Company, Reno, NV). Two weeks following AAV2 injection, mice were injured by optic nerve crush as follows: Mice were anesthetized as done previously and a small incision was made in the superior posterior area of the conjunctiva while the eye muscles were gently moved to reveal the optic nerve. The nerve was crushed for 5 seconds using Dumont N7 fine self-closing forceps at approximately 1 mm behind the globe (being careful not to damage retinal blood vessels). Following optic nerve crush, eyes were treated topically with a Bacitracin ophthalmic ointment. At the indicated timepoints after nerve crush, mice were euthanized and eyes were enucleated for analyses. Investigators were blinded to the genotype when performing the intravitreal injections and both genotype and past intravitreal injection when performing the optic nerve crush. No animals that completed the experimental protocol were excluded from the analyses. Based on past effect size with DLK inhibition (Welsbie et al., 2013) in the optic nerve crush assay, using an alpha of 0.05 and beta of 0.2, we estimated a minimum of four animals per group. Anticipating smaller effect sizes for other genes, larger sample sizes were used in some cases.
Flow cytometry
Retinas were carefully extracted and dissociated in 0.07% papain in DPBS for 40 min at 37 °C. Followi ng dissociation, papain was neutralized with 10% Lo Ovo in DPBS at room temperature (RT). Cells were then washed with PBS and pelleted by centrifugation at 800 RPM for 7 min. Supernatant was removed and cells were resuspended in blocking buffer (HBSS (Life Technologies) with 1% BSA and 2% donkey serum). Cells were then fixed by adding an equal volume of acetone at −20°C for 10 min. Following fixation, cells w ere diluted in blocking buffer and pelleted at 800 RPM for 7 min. Cells were resuspended in blocking buffer with antibodies labeled against antibodies against TUBB3 (TUJ1) conjugated with Alexa Fluor 488 (BD Pharmingen 560381, 1:100 dilution, mouse, RRID: AB_1645344) and either SNCG (gift from N. Marsh-Armstrong laboratory, 1: 1000 dilution, rabbit) or P-S408 MEF2A (Abcam ab51151, 1:250 dilution, rabbit) for 25 min at RT. Cells were then washed in blocking buffer and pelleted by centrifugation at 800 RPM for 7 min to remove excess primary antibody. Pellets were resuspended in blocking buffer containing fluorescently-labeled secondary antibodies (Alexa Fluor donkey anti-rabbit, Life Technologies, 1:1000 dilution) for 25 min at RT. Cells were washed in blocking buffer and pelleted 800 RPM for 7 min following antibody incubation. Finally, cells were resuspended in PBS, filtered to remove debris and analyzed with an Accuri C6 flow cytometer (Becton, Dickinson and Company, Franklin Lakes, USA). Sample blinding was not employed for these experiments because quantification was performed with whole retina samples that were processed and analyzed by flow cytometry using standard methods for all samples.
qPCR
RNA was collected from RGCs using the RNeasy Micro Kit (Qiagen 74004). cDNA was made using iScript cDNA synthesis kit (Bio Rad 1708891). qPCRs were run on a CFX384 Touch™ Real-Time PCR Detection System (Bio Rad) using SsoAdvanced Universal SYBR Green Supermix (Bio Rad 1725271). Primers for PUMA qPCR were PUMA_fwd ATGGCGGACCTCAAC and PUMA_rev AGTCCCATGAAGAGATTGTACATGAC. Primers for Sox11 qPCR were Sox11_fwd ACCCGGACTGGTGCAAGAC and Sox11_rev CGACTGCTCCATGATCTTCCT.
Immunoprecipitation (IP)
RGCs were purified and plated as described above. 24 h after isolation/plating, cells were washed once with 4°C PBS, and lysis buffer was added directly to wel ls. Approximately 3 million RGCs were plated and lysed for each immunoprecipitation. Lysis buffer consisted of 50 mM Tris, 150 mM NaCl, 5 mM EDTA, 0.1% Triton X-100, with protease inhibitors (Roche 4693159001) and phosphatase inhibitors (Roche 4906845001) added immediately before use. Lysate was transferred to microfuge tubes and spun down at 4°C for 10 mins at 10,000g. 1 ml of supernatant per IP was removed to new tubes and either DLK, LZK, or control (rabbit IgG) antibodies were added (1 μg antibody per 1 ml lysate). Lysate with antibody was incubated overnight at 4°C. Protein A beads (Santa Cruz Biotechnology sc-2001) were then added and allowed to complex with antibody/lysate for an additional 2 hours at 4 °C. Beads were then spun down and washed 4 times with lysis buffer. Finally, 2X Laemmli buffer with β-mercaptoethanol (BIO-RAD Laboratories 1610737; Sigma-Aldrich 63689) was added to the beads before boiling them for 5 mins. Beads were spun down, and supernatant was removed and analyzed using a capillary-based immunoassay.
Capillary-based immunoassay
Protein levels in RGCs were measured using the Wes™ automated Western blotting system (ProteinSimple, San Jose, CA). Wes-specific reagents, including their mouse or rabbit secondary antibodies, were used according to the manufacturer’s recommendations. “Virtual blot” electrophoretic images were generated using ProteinSimple‘s Compass Software. As such, images from lanes on the same run were reordered as needed.
Flatmount immunostaining
Eyes were enucleated and fixed in 4% PFA for 1 hr. Following fixation, intact retinas were extracted and washed in PBS with 0.3% Triton X-100 (PBST). Retinas were blocked in 10% horse serum in PBST for 2 hours followed by primary antibody/blocking solution incubation for 3–5 days at 4°C. Following primary antibody incubation, retinas were washed in PBS and incubated in secondary antibody in PBST overnight at 4°C. Retinas were then carefully flattened and mou nted on slides in Fluoro-gel mounting media (EMS 17985-11) and imaged using a Zeiss 510 confocal microscope. Investigators were blinded as to the treatment when processing and imaging the tissue.
Cryosection immunostaining
Eyes were enucleated and fixed in 4% PFA for 1 hr. Eyes were then transferred to a 20–30% sucrose solution in PBS overnight and then embedded in cryosection molds using OCT solution (Tissue-Tek 4583). Eyes were cryosectioned at a thickness of 8 μm and placed on slides. Slides were washed in PBS and blocked in 10% horse serum in PBST. Slides were incubated in primary antibody/blocking solution overnight, washed in PBS, and incubated in secondary antibody in block overnight. After a final wash, retinal sections were mounted and counterstained using 4′, 6-diamidino-2-phenylindole (DAPI) to identify cell nuclei and imaged using a Zeiss 510 confocal microscope. Investigators were blinded as to the treatment when processing and imaging the tissue.
QUANTIFICATION AND STATISTICAL ANALYSIS
Details of the statistical tests can be found in the figure legends. A Mann-Whitney U test was used to determine statistical significance and a P<0.05 was used as the threshold for significance. In all cases, means are plotted with error bars representing standard deviation. For animal experiments, n represents the number of eyes per group.
Supplementary Material
Highlights.
Arrayed, whole genome siRNA screening identifies RGC cell death mediators
LZK cooperates with DLK to promote RGC cell death in response to axon injury
FDA-approved inhibitor of DLK/LZK prevents human RGC cell death
DLK/LZK-mediated cell death involves SOX11, JUN, ATF2 and MEF2A
Short paragraph for email alerts
DLK has been shown to partially mediate RGC cell death in response to axonal injury, a key feature of glaucoma. Using a functional genomic approach, Welsbie et al. identify LZK as cooperating with DLK to signal through a set of four downstream transcription factors in order to promote cell death.
Acknowledgments
We thank J. Maruotti for assistance with the FC assay, N. Marsh-Armstrong and C. Davis for SNCG antibody, N. Hong for subcloning, D. Ferraris, B. Slusher and T. Tsukamoto for synthesis of the Genentech DLK inhibitor, L. Holzman and W.W. Hauswirth for reagents, P. Ormanoglu, C. Klumpp-Thomas and M. Lal for help with siRNA screens and E. Olson, L. Monteggia, C. Tournier and R. Davis for providing mice. We also thankfully acknowledge funding from NIH (including core grant 5P30EY001765), Research to Prevent Blindness, American Glaucoma Society, E. Matilda Ziegler Foundation, Brightfocus Foundation, Maryland Stem Cell Research Fund, and the Guerrieri Family Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, Supervision and Funding Acquisition, DSW and DJZ; Investigation, KLM, AP, PZ, ZY, JK, BK, EH, PZ, YD, YG and JF; Methodology, DSW, DJZ, KLM, VR, XC, JK, ZY, CAB, JF and VMS; Formal Analysis, CAB, EB and SEM; Resources, LL, ZAR and IDCF.
References
- Akhtar MW, Kim MS, Adachi M, Morris MJ, Qi X, Richardson JA, Bassel-Duby R, Olson EN, Kavalali ET, Monteggia LM. In vivo analysis of MEF2 transcription factors in synapse regulation and neuronal survival. PLoS One. 2012;7:e34863. doi: 10.1371/journal.pone.0034863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambacher KK, Pitzul KB, Karajgikar M, Hamilton A, Ferguson SS, Cregan SP. The JNK-and AKT/GSK3β-signaling pathways converge to regulate Puma induction and neuronal apoptosis induced by trophic factor deprivation. PLoS One. 2012;7:e46885. doi: 10.1371/journal.pone.0046885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounoutas A, Kratz J, Emtage L, Ma C, Nguyen KC, Chalfie M. Microtubule depolymerization in Caenorhabditis elegans touch receptor neurons reduces gene expression through a p38 MAPK pathway. Proc Natl Acad Sci U S A. 2011;108:3982–3987. doi: 10.1073/pnas.1101360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham BP, Inman DM, Lambert W, Oglesby E, Calkins DJ, Steele MR, Vetter ML, Marsh-Armstrong N, Horner PJ. Progressive ganglion cell degeneration precedes neuronal loss in a mouse model of glaucoma. J Neurosci. 2008;28:2735–2744. doi: 10.1523/JNEUROSCI.4443-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehler E, Khan AA, Marine S, Rajaram M, Bahl A, Burchard J, Ferrer M. siRNA off-target effects in genome-wide screens identify signaling pathway members. Sci Rep. 2012;2:428. doi: 10.1038/srep00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD, Malani N, Fernandes J, D’Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, König R, et al. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5:e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang ZY, Lu DW, Yeh MK, Chiang CH. A novel high-content flow cytometric method for assessing the viability and damage of rat retinal ganglion cells. PLoS One. 2012;7:e33983. doi: 10.1371/journal.pone.0033983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Geoffroy CG, Wong HN, Tress O, Nguyen MT, Holzman LB, Jin Y, Zheng B. Leucine Zipper-bearing Kinase promotes axon growth in mammalian central nervous system neurons. Sci Rep. 2016;6:31482. doi: 10.1038/srep31482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen F, Huestis M, Ly C, Patel S, Siu M, Zhao X. Substituted dipyridylamines and uses thereof. WO2013174780 A1 Patent application. 2013
- Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- Dickson HM, Zurawski J, Zhang H, Turner DL, Vojtek AB. POSH is an intracellular signal transducer for the axon outgrowth inhibitor Nogo66. J Neurosci. 2010;30:13319–13325. doi: 10.1523/JNEUROSCI.1324-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Qiao M, Bei F, Kim IJ, He Z, Sanes JR. Subtype-specific regeneration of retinal ganglion cells following axotomy: effects of osteopontin and mTOR signaling. Neuron. 2015;85:1244–1256. doi: 10.1016/j.neuron.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri CJ, Beachy Pa, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N, et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- Fan G, Merritt SE, Kortenjann M, Shaw PE, Holzman LB. Dual leucine zipper-bearing kinase (DLK) activates p46SAPK and p38mapk but not ERK2. J Biol Chem. 1996;271:24788–24793. doi: 10.1074/jbc.271.40.24788. [DOI] [PubMed] [Google Scholar]
- Fernandes KA, Harder JM, Fornarola LB, Freeman RS, Clark AF, Pang IH, John SWM, Libby RT. JNK2 and JNK3 are major regulators of axonal injury-induced retinal ganglion cell death. Neurobiol Dis. 2012;46:393–401. doi: 10.1016/j.nbd.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes KA, Harder JM, Kim J, Libby RT. JUN regulates early transcriptional responses to axonal injury in retinal ganglion cells. Exp Eye Res. 2013;112:106–117. doi: 10.1016/j.exer.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes KA, Harder JM, John SW, Shrager P, Libby RT. DLK-dependent signaling is important for somal but not axonal degeneration of retinal ganglion cells following axonal injury. Neurobiol Dis. 2014;69:108–116. doi: 10.1016/j.nbd.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Gerdts J, Summers DW, Sasaki Y, DiAntonio A, Milbrandt J. Sarm1-mediated axon degeneration requires both SAM and TIR interactions. J Neurosci. 2013;33:13569–13580. doi: 10.1523/JNEUROSCI.1197-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Tang X, Wiedmann M, Wang X, Peng J, Zheng D, Blair LAC, Marshall J, Mao Z. Cdk5-mediated inhibition of the protective effects of transcription factor MEF2 in neurotoxicity-induced apoptosis. Neuron. 2003;38:33–46. doi: 10.1016/s0896-6273(03)00191-0. [DOI] [PubMed] [Google Scholar]
- Hannus M, Beitzinger M, Engelmann JC, Weickert MT, Spang R, Hannus S, Meister G. siPools: highly complex but accurately defined siRNA pools eliminate off-target effects. Nucleic Acids Res. 2014;42:8049–8061. doi: 10.1093/nar/gku480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder JM, Libby RT. BBC3 (PUMA) regulates developmental apoptosis but not axonal injury induced death in the retina. Mol Neurodegener. 2011;6:50. doi: 10.1186/1750-1326-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder JM, Libby RT. Deficiency in Bim, Bid and Bbc3 (Puma) do not prevent axonal injury induced death. Cell Death Differ. 2013;20:182. doi: 10.1038/cdd.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai S, Cui DF, Miyata T, Ogawa M, Kiyonari H, Suda Y, Aizawa S, Banba Y, Ohno S. The c-Jun N-terminal kinase activator dual leucine zipper kinase regulates axon growth and neuronal migration in the developing cerebral cortex. J Neurosci. 2006;26:11992–12002. doi: 10.1523/JNEUROSCI.2272-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A, Masaki M, Kozutsumi Y, Oka S, Kawasaki T. Identification and characterization of functional domains in a mixed lineage kinase LZK. FEBS Lett. 2001a;488:190–195. doi: 10.1016/s0014-5793(00)02432-7. [DOI] [PubMed] [Google Scholar]
- Ikeda A, Hasegawa K, Masaki M, Moriguchi T, Nishida E, Kozutsumi Y, Oka S, Kawasaki T. Mixed lineage kinase LZK forms a functional signaling complex with JIP-1, a scaffold protein of the c-Jun NH(2)-terminal kinase pathway. J Biochem. 2001b;130:773–781. doi: 10.1093/oxfordjournals.jbchem.a003048. [DOI] [PubMed] [Google Scholar]
- Itoh T, Horiuchi M, Ikeda RH, Xu J, Bannerman P, Pleasure D, Penninger JM, Tournier C, Itoh A. ZPK/DLK and MKK4 form the critical gateway to axotomy-induced motoneuron death in neonates. J Neurosci. 2014;34:10729–10742. doi: 10.1523/JNEUROSCI.0539-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Ding Q, Xie X, Libby RT, Lefebvre V, Gan L. Transcription factors SOX4 and SOX11 function redundantly to regulate the development of mouse retinal ganglion cells. J Biol Chem. 2013;288:18429–18438. doi: 10.1074/jbc.M113.478503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- Levkovitch-Verbin H, Quigley HA, Martin KRG, Harizman N, Valenta DF, Pease ME, Melamed S. The transcription factor c-jun is activated in retinal ganglion cells in experimental rat glaucoma. Exp Eye Res. 2005;80:663–670. doi: 10.1016/j.exer.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Marine S, Bahl A, Ferrer M, Buehler E. Common seed analysis to identify off-target effects in siRNA screens. J Biomol Screen. 2012;17:370–378. doi: 10.1177/1087057111427348. [DOI] [PubMed] [Google Scholar]
- Mazzitelli S, Xu P, Ferrer I, Davis RJ, Tournier C. The loss of c-Jun N-terminal protein kinase activity prevents the amyloidogenic cleavage of amyloid precursor protein and the formation of amyloid plaques in vivo. J Neurosci. 2011;31:16969–16976. doi: 10.1523/JNEUROSCI.4491-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead B, Thompson A, Scheven BA, Logan A, Berry M, Leadbeater W. Comparative evaluation of methods for estimating retinal ganglion cell loss in retinal sections and wholemounts. PLoS One. 2014;9:e110612. doi: 10.1371/journal.pone.0110612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BR, Press C, Daniels RW, Sasaki Y, Milbrandt J, DiAntonio A. A dual leucine kinase-dependent axon self-destruction program promotes Wallerian degeneration. Nat Neurosci. 2009;12:387–389. doi: 10.1038/nn.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal-Nicolás FM, Jiménez-López M, Sobrado-Calvo P, Nieto-López L, Cánovas-Martínez I, Salinas-Navarro M, Vidal-Sanz M, Agudo M. Brn3a as a marker of retinal ganglion cells: qualitative and quantitative time course studies in naive and optic nerve-injured retinas. Invest Ophthalmol Vis Sci. 2009;50:3860–3868. doi: 10.1167/iovs.08-3267. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Li Z, Ju C, Scholzke MN, Mathews E, Cui J, Salvesen GS, Bossy-Wetzel E, Lipton SA. Dominant-interfering forms of MEF2 generated by caspase cleavage contribute to NMDA-induced neuronal apoptosis. Proc Natl Acad Sci U S A. 2002;99:3974–3979. doi: 10.1073/pnas.022036399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterloh JM, Yang J, Rooney TM, Fox AN, Adalbert R, Powell EH, Sheehan AE, Avery MA, Hackett R, Logan MA, et al. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science. 2012;337:481–484. doi: 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock EP, Goodman V, Jiang JX, Mahjoob K, Verbois SL, Morse D, Dagher R, Justice R, Pazdur R. Food and Drug Administration drug approval summary: Sunitinib malate for the treatment of gastrointestinal stromal tumor and advanced renal cell carcinoma. Oncologist. 2007;12:107–113. doi: 10.1634/theoncologist.12-1-107. [DOI] [PubMed] [Google Scholar]
- Sakuma H, Ikeda A, Oka S, Kozutsumi Y, Zanetta JP, Kawasaki T. Molecular cloning and functional expression of a cDNA encoding a new member of mixed lineage protein kinase from human brain. J Biol Chem. 1997;272:28622–28629. doi: 10.1074/jbc.272.45.28622. [DOI] [PubMed] [Google Scholar]
- Schultz N, Marenstein DR, De Angelis Da, Wang WQ, Nelander S, Jacobsen A, Marks DS, Massagué J, Sander C. Off-target effects dominate a large-scale RNAi screen for modulators of the TGF-β pathway and reveal microRNA regulation of TGFBR2. Silence. 2011;2:3. doi: 10.1186/1758-907X-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M, Bhoumik A, Goel V, Dewing A, Breitwieser W, Kluger H, Krajewski S, Krajewska M, Dehart J, Lau E, et al. A role for ATF2 in regulating MITF and melanoma development. PLoS Genet. 2010;6:e1001258. doi: 10.1371/journal.pgen.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi A, Gaudillière B, Yuan Z, Stegmüller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- Shin JE, Cho Y, Beirowski B, Milbrandt J, Cavalli V, DiAntonio A. Dual leucine zipper kinase is required for retrograde injury signaling and axonal regeneration. Neuron. 2012;74:1015–1022. doi: 10.1016/j.neuron.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DJ, Pitts J, Hertz NT, Yang J, Yamagishi Y, Olsen O, Tešić Mark M, Molina H, Tessier-Lavigne M. Axon Degeneration Gated by Retrograde Activation of Somatic Pro-apoptotic Signaling. Cell. 2016;164:1031–1045. doi: 10.1016/j.cell.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluch VM, Davis CO, Ranganathan V, Kerr JM, Krick K, Martin R, Berlinicke CA, Marsh-Armstrong N, Diamond JS, Mao HQ, et al. Differentiation of human ESCs to retinal ganglion cells using a CRISPR engineered reporter cell line. Sci Rep. 2015;5:16595. doi: 10.1038/srep16595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto I, Oglesby E, Buckingham BP, Son JL, Roberson EDO, Steele MR, Inman DM, Vetter ML, Horner PJ, Marsh-Armstrong N. Retinal ganglion cells downregulate gene expression and lose their axons within the optic nerve head in a mouse glaucoma model. J Neurosci. 2008;28:548–561. doi: 10.1523/JNEUROSCI.3714-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto I, Pease ME, Son JL, Shi X, Quigley HA, Marsh-Armstrong N. Retinal ganglion cell loss in a rat ocular hypertension model is sectorial and involves early optic nerve axon loss. Invest Ophthalmol Vis Sci. 2011;52:434–441. doi: 10.1167/iovs.10-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulgren ED, Baker ST, Rapp L, Gurney AM, Grill B. PPM-1, a PP2Cα/β phosphatase, regulates axon termination and synapse formation in Caenorhabditis elegans. Genetics. 2011;189:1297–1307. doi: 10.1534/genetics.111.134791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valakh V, Walker LJ, Skeath JB, DiAntonio A. Loss of the spectraplakin short stop activates the DLK injury response pathway in Drosophila. J Neurosci. 2013;33:17863–17873. doi: 10.1523/JNEUROSCI.2196-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valakh V, Frey E, Babetto E, Walker LJ, DiAntonio A. Cytoskeletal disruption activates the DLK/JNK pathway, which promotes axonal regeneration and mimics a preconditioning injury. Neurobiol Dis. 2015;77:13–25. doi: 10.1016/j.nbd.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal-Sanz M, Villegas-Pérez MP, Bray GM, Aguayo AJ. Persistent retrograde labeling of adult rat retinal ganglion cells with the carbocyanine dye diI. Exp Neurol. 1988;102:92–101. doi: 10.1016/0014-4886(88)90081-7. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lin L, Lai H, Parada LF, Lei L. Transcription factor Sox11 is essential for both embryonic and adult neurogenesis. Dev Dyn. 2013;242:638–653. doi: 10.1002/dvdy.23962. [DOI] [PubMed] [Google Scholar]
- Watkins TA, Wang B, Huntwork-Rodriguez S, Yang J, Jiang Z, Eastham-Anderson J, Modrusan Z, Kaminker JS, Tessier-Lavigne M, Lewcock JW. DLK initiates a transcriptional program that couples apoptotic and regenerative responses to axonal injury. Proc Natl Acad Sci U S A. 2013;110:4039–4044. doi: 10.1073/pnas.1211074110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsbie DS, Yang Z, Ge Y, Mitchell KL, Zhou X, Martin SE, Berlinicke CA, Hackler L, Fuller J, Fu J, et al. Functional genomic screening identifies dual leucine zipper kinase as a key mediator of retinal ganglion cell death. Proc Natl Acad Sci U S A. 2013;110:4045–4050. doi: 10.1073/pnas.1211284110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW, Strom RC, Rice DS, Goldowitz D. Genetic and environmental control of variation in retinal ganglion cell number in mice. J Neurosci. 1996;16:7193–7205. doi: 10.1523/JNEUROSCI.16-22-07193.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Wang X, Ewanek R, Bhat P, Diantonio A, Collins CA. Protein turnover of the Wallenda/DLK kinase regulates a retrograde response to axonal injury. J Cell Biol. 2010;191:211–223. doi: 10.1083/jcb.201006039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Das M, Reilly J, Davis RJ. JNK regulates FoxO-dependent autophagy in neurons. Genes Dev. 2011;25:310–322. doi: 10.1101/gad.1984311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Jin Y. Regulation of DLK-1 kinase activity by calcium-mediated dissociation from an inhibitory isoform. Neuron. 2012;76:534–548. doi: 10.1016/j.neuron.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wu Z, Renier N, Simon DJ, Uryu K, Park DS, Greer PA, Tournier C, Davis RJ, Tessier-Lavigne M. Pathological axonal death through a Mapk cascade that triggers a local energy deficit. Cell. 2015;160:161–176. doi: 10.1016/j.cell.2014.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Behrens A, Le-Niculescu H, Wagner EF, Harada T, Imaki J, Ohno S, Karin M. Amino-terminal phosphorylation of c-Jun regulates apoptosis in the retinal ganglion cells by optic nerve transection. Invest Ophthalmol Vis Sci. 2002;43:1631–1635. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


