Abstract
AMPK is a highly conserved master regulator of metabolism, which restores energy balance during metabolic stress both at the cellular and physiological levels. The identification of numerous AMPK targets has helped explain how AMPK restores energy homeostasis. Recent advancements, however, demonstrate that regulation of AMPK is also affected by novel contexts, such as subcellular localization and phosphorylation by non-canonical upstream kinases. Notably, the therapeutic potential of AMPK is widely recognized and heavily pursued for treatment of metabolic diseases such as diabetes, but also obesity, inflammation and cancer. Moreover, the recently solved crystal structure of AMPK has shed light both into how nucleotides activate AMPK but, importantly, also into the sites bound by small molecule activators, thus providing a path for improved drugs.
Keywords: AMPK, trimeric complex, energy restoration, metabolic adaptation, therapeutic activation
Introduction
Maintenance of energy homeostasis and the execution of adaptive responses during periods of low nutrients are critical functions of all cells. The main sensor of cellular energy status in effectively all eukaryotic cells is the AMP-activated protein kinase (AMPK), which is highly conserved across all eukaryotic species. In general, AMPK is activated in response to energy stress by sensing increases in AMP:ATP and ADP:ATP ratios and restores energy balance by inhibiting anabolic processes that consume ATP, while promoting catabolic processes that generate ATP. Moreover, the activity of AMPK is extensively regulated by multiple upstream signals, thus making AMPK a central node exploited by cells to coordinate their metabolism with specific energy demands. Importantly, the primordial role of AMPK as an energy sensor has been co-opted in higher eukaryotes, such as mammals, to also coordinate growth and metabolism both in specialized tissues and at the whole body level. Therefore, this ability of AMPK to reprogram metabolism is heavily pursued as a therapeutic avenue for the treatment of several metabolic diseases, especially diabetes, but also for obesity, inflammation, and cancer. Here we review recent advances in our understanding of AMPK structure, its complex upstream regulation and downstream effects, and provide perspective on novel approaches for the therapeutic activation of AMPK.
The AMPK trimeric complex
AMPK exists as a trimeric complex consisting of a catalytic subunit (α-subunit) and two regulatory subunits (β- and γ-subunits). In mammals, the α-subunit is encoded by two isoforms, and the β- and γ-subunits are encoded by two and three isoforms, respectively (Fig 1A). The expression levels of these AMPK isoforms vary across tissues, leading to diverse subunit combinations in different cell types. Although the distinct complexes are for the most part functionally redundant, they can exhibit somewhat different biochemical properties (Ross et al., 2016). While AMPKα1, AMPKβ1, and AMPKγ1 are ubiquitously expressed, the other isoforms show a more restricted expression pattern. AMPK α2 is expressed at high levels in skeletal and cardiac muscle, where it is the dominant α-subunit, but is also expressed in the liver and at lower levels in other tissues. Similarly, AMPKβ2 is the dominant β-subunit in skeletal and cardiac muscle, but it is found at lower levels in many other tissues. The expression of AMPKγ2 and AMPKγ3 appears to be restricted to skeletal and cardiac tissue.
Figure 1. Domains and structure of the AMPK complex.
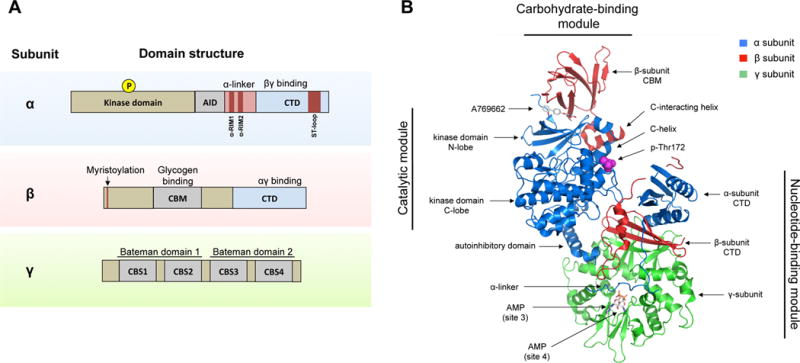
A) AMPK exists as a trimeric complex consisting of a catalytic subunit (α) and two regulatory subunits (β and γ). The main protein domains are shown. Abbreviations: AID (autoinhibitory domain), CTD (C-terminus domain), NTD (N-terminus domain), CBM (carbohydrate-binding module), CBS (cystathionine β-synthase repeats), RIM (regulatory-subunit-interacting motif), ST-loop (serine/threonine enriched loop).
B) The crystal structure of the AMPK α2β1γ1 trimeric complex is shown. Major structural domains are indicated. The structure shows the activator A769662 bound to a pocket formed by the interface between the kinase domain and the CBM. Also shown are two AMP molecules bound to site 3 and site 4, respectively, and phospho-Thr172. The ST-loop in the α-subunit and the myristoylation site in the β-subunit are not resolved in this structure. Structure sourced and adapted from PDB file 4CFF, using PyMOL software.
The N-terminus of the α-subunit is comprised of the kinase domain (KD). Phosphorylation of a conserved threonine in the activation loop (by convention referred to as Thr172) of the kinase domain is required for full activation of AMPK. The C-terminus of the α-subunit binds the β- and γ-subunits and contains important regulatory domains, namely, a so-called autoinhibitory domain (AID), the α-linker (which interacts with the γ-subunit through two regulatory-subunit interacting motifs, or α-RIM motifs) and a serine/threonine rich domain referred to as the “ST-loop”. The β-subunit contains two conserved domains: a carbohydrate-binding module (CBM) (also called the glycogen-binding domain), which allows AMPK to sense glycogen, and a C-terminus domain that binds the α- and γ-subunits. The β-subunit also contains a myristoylation site at its N-terminus, which facilitates targeting of AMPK to cellular membranes. The γ-subunit is comprised of two Bateman domains, each of which contains two cystathionine β-synthase repeats (CBS). The four CBS repeats form the four sites in AMPK that can potentially bind AMP, ADP or ATP in a competitive manner. In mammals, site 1 and site 3 exchange adenine nucleotides competitively, whereas site 2 is missing the key aspartate required for nucleotide binding, which renders it non-functional (Xiao et al., 2011). Site 4 can also bind AMP and ATP, but has higher affinity for AMP (Calabrese et al., 2014; Chen et al., 2012). The ability of the γ-subunit to bind AMP, ADP and ATP confers AMPK with its exquisite ability to sense the energy state of the cell.
AMPK structure
Our molecular understanding of the regulation and function of AMPK has significantly been advanced by the elucidation of the crystal structures of a variety of AMPK holoenzymes (Calabrese et al., 2014; Chen et al., 2012; 2013; Li et al., 2015a; Xiao et al., 2011; 2013; Xin et al., 2013). Although the reports differ in certain details, together they provide a detailed view of the architecture of the AMPK complex. The structure of the AMPK trimeric complex consists of three major segments or “modules”: the catalytic module, the carbohydrate-binding module (CBM) and the nucleotide-binding module (also called “regulatory fragment”) (Fig 1B). The activation loop of the α-subunit resides at the interface between the catalytic and nucleotide-binding modules, in close proximity to the C-terminus of the β-subunit and the CBS repeats of the γ-subunit. This structural arrangement ensures that phosphorylation and dephosphorylation of Thr172 is sensitive to conformational rearrangements induced by nucleotide binding. The catalytic domain exhibits a typical eukaryotic serine/threonine kinase domain structure with a small N-lobe and a large C-lobe. The CBM directly contacts the N-lobe of the kinase domain and the interface between these two modules forms a discrete pocket that was identified as the binding site for many direct AMPK-activating compounds. It is speculated that natural metabolites might bind this site to regulate AMPK, however, no such metabolite has been yet identified. The nucleotide-binding module is made up mostly by the γ-subunit, which forms a flattened disk with the CBS repeats symmetrically arranged around the disk, one in each quadrant.
Mechanistically, these crystallographic studies reveal the molecular details of how adenine nucleotides and small molecule activators activate AMPK. In the case of nucleotides, the crystal structures show that when AMP is bound to site 3, the γ-subunit forms stable interactions with a few amino acids within the α-linker’s α-RIM1 and α-RIM2, which interact with the unoccupied site 2 and the AMP molecule bound at site 3, respectively (Chen et al., 2013; Xiao et al., 2013; Xin et al., 2013). The binding of the α-RIM motifs to the γ-subunit restricts the flexibility of the α-linker, resulting in tighter association of the catalytic and nucleotide-binding modules, which physically protects Thr172 from dephosphorylation. Interestingly, the same effect is proposed to occur when ADP binds site 3, raising the possibility that in some contexts ADP might be the relevant AMPK activating signal (Xiao et al., 2011). Moreover, the binding of the α-RIM motifs to the γ-subunit shifts the AID in the α-subunit away from the kinase domain when AMP is bound, thus releasing the AID’s negative effects on the kinase domain (Calabrese et al., 2014; Chen et al., 2013; Li et al., 2015a). This rearrangement of the AID domain may represent the molecular basis for the allosteric activation effect of AMP. According to this model, the AID can shift between kinase domain-bound (inactive AMPK) and nucleotide-module-bound states (active AMPK) depending on the nucleotide binding status. In summary, the published crystal structures concur that binding of AMP, especially at site 3, induces a conformational change that is transmitted to the kinase domain by changes in the interaction of the α-RIM motifs and the AID with the nucleotide-binding module. These structural changes, which are opposed by ATP, result in the allosteric activation of AMPK and a compaction of the interface between the catalytic module and the nucleotide-binding module, which protects Thr172 from dephosphorylation. However, it is not clear whether these structural rearrangements also promote Thr172 phosphorylation.
On the other hand, activating compounds, such as A769662, activate AMPK by a different mechanism. Binding of these compounds, together with phosphorylation of serine 108 in the β-subunit, stabilizes the CBM and strengthens the interaction of the CBM with the kinase domain (Calabrese et al., 2014; Li et al., 2015a; Xiao et al., 2013). Specifically, binding of activating compounds induces the formation of an α-helix in the β-subunit, termed the C-interacting helix, which interacts with the so-called C-helix of the kinase domain (a conserved helix across multiple kinases which is important for ATP binding). This conformational change results in a shift towards a closed, active conformation of the kinase domain, protection from Thr172 dephosphorylation, and increased substrate affinity. Interestingly, glycogen inhibits the CBM-KD interaction and this may be the mechanism by which glycogen inhibits AMPK (McBride et al., 2009).
Nucleotide-dependent and nucleotide-independent regulation of AMPK
The regulation of AMPK as a sensor of changes in intracellular levels of AMP, ADP and ATP, such as in the case of energy stress, is significantly understood and is well established as the canonical regulation of AMPK (Figure 2). Accordingly, AMPK becomes fully activated through a three-pronged mechanism. First, binding of AMP or ADP to the γ-subunit promotes Thr172 phosphorylation in the activation loop in the kinase domain by upstream kinases. The main upstream kinase responsible for Thr172 phosphorylation in response to energy stress is the serine/threonine kinase LKB1 (liver-kinase-B1) (Hawley et al., 2003; Shaw et al., 2004; Woods et al., 2003). Phosphorylation of Thr172 in the α-subunit is the principal event required for full activation of AMPK. This phosphorylation event can increase AMPK activity up to 100-fold in vitro, although fold activation in intact cells is usually more modest (Gowans et al., 2013; Oakhill et al., 2011; Suter et al., 2006). Second, binding of AMP or ADP to the γ-subunit induces a conformational change that protects against Thr172 dephosphorylation by protein phosphatases (Gowans et al., 2013; Xiao et al., 2011). Notably, the phosphatases that normally dephosphorylate AMPK under physiological conditions remain largely unknown, though recent reports implicate roles for different phosphatases (Garcia-Haro et al., 2010; Joseph et al., 2015). Lastly, binding of AMP, but not ADP, results in up to 10 fold allosteric activation of AMPK (Gowans et al., 2013). Of note, ATP inhibits all three mechanisms.
Figure 2. Upstream regulation of AMPK and metabolic consequences of AMPK activation.
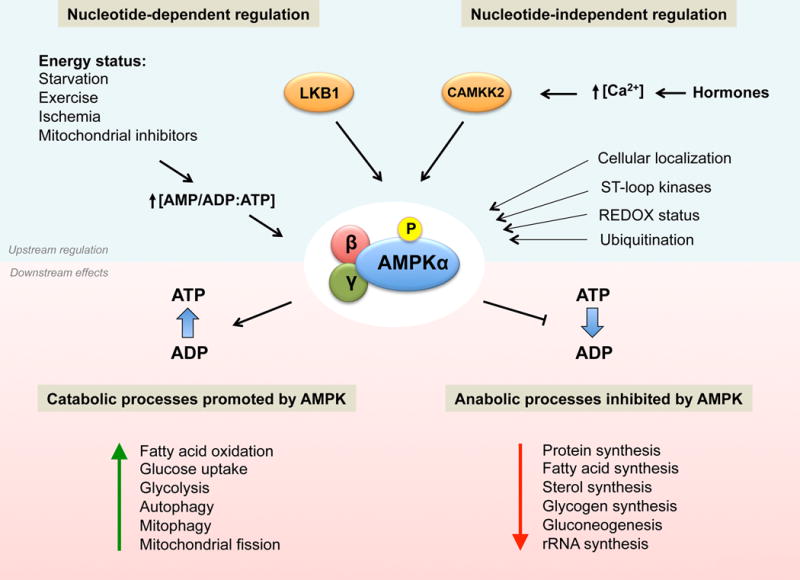
Phosphorylation by upstream kinases is the main AMPK activating event. The regulation of AMPK phosphorylation can be nucleotide-dependent (canonical regulation), resulting from changes in AMP:ATP or ADP:ATP ratios that are induced by a variety of energy stresses. Several important alternative modes of AMPK regulation have been described, which can be classified as nucleotide-independent regulation. In response to energy stress AMPK restores ATP levels by acutely inhibiting ATP-consuming biosynthetic pathways while simultaneously activating catabolic pathways that regenerate ATP through the breakdown of macromolecules. Some of the metabolic processes that are affected by AMPK activation are shown.
In addition to changes in adenine nucleotide levels, it has become increasingly clear that there are other important, non-canonical modes of AMPK regulation (Figure 2). The best characterized nucleotide-independent regulation of AMPK is phosphorylation of Thr172 by CAMKK2 (calcium/calmodulin-dependent kinase kinase 2, also known as CAMKKβ), the other major upstream kinase of AMPK (Hawley et al., 2005; Hurley et al., 2005; Woods et al., 2005). CAMKK2 is activated by increases in intracellular Ca2+ levels. Indeed, Ca2+-mediated CAMKK2 activation of AMPK is a frequent mechanism by which metabolically relevant hormones induce transient activation of AMPK. Thus, although CAMKK2 does not itself sense cellular energy status, it is nonetheless critical for the regulation of many aspects of whole body metabolism by AMPK.
Besides phosphorylation of Thr172, phosphorylation of the ST-loop has emerged as an important site for the regulation and inhibition of AMPK by other kinases (Hardie, 2014). Examples of this include phosphorylation of serine 485 (Ser485) in AMPKα1 and the equivalent serine 491 (Ser491) in AMPKα2 by PKA (cyclic-AMP-dependent protein kinase), which was proposed to be important for counteracting AMPK activity during gluconeogenic periods (Hurley et al., 2006), and by AKT (insulin-activated protein kinase), which was proposed to be a mechanism by which insulin inhibits AMPK (Horman et al., 2008). Similarly, S6K (p70S6 kinase) has been reported to inhibit AMPK activity by phosphorylating Ser491 in AMPKα2, and this phosphorylation was proposed as the mechanism by which leptin inhibits AMPK in the hypothalamus (Dagon et al., 2012). Other kinases reported to inhibit AMPK by phosphorylating various residues in the ST-loop are GSK3 (glycogen synthesis kinase 3) (Suzuki et al., 2013), PKD1 (protein kinase D) (Coughlan et al., 2016) and PKC (protein kinase C) (Heathcote et al., 2016), though it remains to be determined which of these kinases are relevant in different tissues in vivo. Although the mechanism of AMPK inhibition is not entirely clear, it seems that phosphorylation of the ST-loop reduces net phosphorylation of Thr172, by either physically interfering with its phosphorylation or by promoting its dephosphorylation (Hawley et al., 2014). Collectively, these phosphorylation events on the ST-loop may represent an important mechanism to keep AMPK activity low during periods when anabolic metabolism is required. Indeed, phosphorylation of AMPK by oncogenically deregulated AKT, a common feature in tumor cells, could be a mechanism used by tumors to downregulate AMPK activity, whose activity would otherwise oppose their proliferative metabolism (Hawley et al., 2014). This requirement for some tumors to reduce AMPK activity is further demonstrated by the identification of yet another mechanism to downregulate AMPK, namely, through its ubiquitination and degradation. Thus, for instance, MAGE (melanoma antigen genes)-A3 and MAGE-A6 proteins, which are testis-specific genes that are abnormally expressed in a variety of tumors, potently promote tumor growth by, in part, mediating the ubiquitin-dependent degradation of AMPKα1 by the TRIM28 ubiquitin ligase (Pineda et al., 2015). In addition, UBE20 (ubiquitin-conjugating enzyme E20) was similarly reported to promote tumor growth by targeting AMPKα2 for ubiquitination and degradation (Vila et al., 2017). Moreover, another ubiquitin ligase, called WWP1, was reported to degrade AMPKα2 under high-glucose conditions in muscle cells (Lee et al., 2013), suggesting that ubiquitin-dependent degradation of AMPK may be a more broad mechanism for the regulation of AMPK than is currently recognized.
Another context that is increasingly appreciated as important for the regulation of AMPK activity is its localization to cellular membranes. Indeed, myristoylation of the β-subunit can promote localization of AMPK to membranes (Liang et al., 2015; Oakhill et al., 2010). Interestingly, LKB1 is farnesylated and can also localize to membranes, which raised the question of whether co-localization of AMPK and LKB1 into two-dimensional surfaces of cellular membranes could promote activation of AMPK by LKB1. Strong support for this hypothesis has come from the analysis of knock-in mice that express an LKB1 mutant that cannot be farnesylated (LKB1C433S) and does not, therefore, localize to membranes (Houde et al., 2014). Although LKB1C433S was fully capable of activating AMPK in vitro, tissues and cells from LKB1C433S mice exhibited a significant reduction of both basal and induced AMPK activation (Houde et al., 2014), suggesting that the reduced AMPK activation was due to defective membrane localization of LKB1C433S in cells. Notably, which cellular membranes were important for LKB1-AMPK activation was left unanswered.
Interestingly, recent studies suggest that the lysosome might be one relevant membrane surface where LKB1 phosphorylates AMPK (Zhang et al., 2014; 2013). In brief, these studies describe a complex mechanism in which, under conditions of low nutrients, AMPK and LKB1 form a complex with the scaffolding protein AXIN (Zhang et al., 2013) and localize to the lysosomes via binding to the lysosomal protein LAMTOR1 (Zhang et al., 2014), a component of the mTORC1-activating RAGULATOR complex. Moreover, LAMTOR1 knockout cells and tissues had defective AMPK activation, suggesting that recruitment of AMPK to the lysosome might be necessary for proper activation (Zhang et al., 2014). Although this was not tested, it is tempting to speculate that the lipid modifications in AMPK and LKB1 might be necessary for their lysosomal localization. Intriguing as this might be, a systematic analysis of the intracellular location of AMPK activity found it to be enriched not only in lysosomal membranes, but also in Golgi, endoplasmic reticulum, mitochondrial and plasma membranes, suggesting AMPK membrane localization is broadly exploited in cells (Miyamoto et al., 2015). Consequently, even though most AMPK is cytosolic, a critical question for now is whether discrete subcellular pools of AMPK (such as membrane-bound AMPK) respond to specific stress stimuli, and perhaps in turn may target subsets of downstream targets.
Finally, there is increasing evidence suggesting that AMPK may also be a redox-sensing protein. Reactive oxygen species (ROS) are naturally produced my many metabolic reactions, most notably by the production of ATP in mitochondria, and management of their levels is important for cellular homeostasis. Although ROS can indirectly activate AMPK through increases in AMP (Hawley et al., 2010), a few studies have demonstrated that ROS can modulate AMPK activity by direct posttranslational modification (Shao et al., 2014; Zmijewski et al., 2010). The nature and the effects of such modifications, however, seem to be context dependent. In HEK293 and lung cells, H2O2 was shown to induce oxidation and S-glutathionylation of cysteines 299 and 304 in the α-subunit, resulting in activation of AMPK (Zmijewski et al., 2010). In cardiomyocytes, on the other hand, increased H2O2 levels and ischemia induced oxidation of the highly conserved cysteines 130 and 174 in the α-subunit, which resulted in inhibition of AMPK through aggregation of AMPK molecules and blockage of phosphorylation by upstream kinases (Shao et al., 2014). Importantly, this study also demonstrated that Trx1 (thioredoxin1), a major reducing enzyme, prevented AMPK oxidation and was necessary for maintenance of AMPK activity. Collectively, even though a comprehensive picture is yet to emerge, the evidence points to AMPK as a nexus between cellular metabolism and cellular redox status.
Metabolic consequences of AMPK activation
Our understanding of the mechanisms by which AMPK regulates metabolism has been greatly expanded over the last few years by the identification of numerous AMPK substrates, aided substantially by the decoding of the AMPK substrate motif (Gwinn et al., 2008; Hardie et al., 2016). Moreover, novel proteomic approaches have considerably expanded the list and the scope of putative AMPK substrates (Banko et al., 2011; Hoffman et al., 2015; Schaffer et al., 2015), although rigorous analysis of the functional roles of these novel substrates will require further investigation. As explained above, AMPK restores ATP levels during metabolic stress by acutely inhibiting ATP-consuming biosynthetic pathways while simultaneously activating pathways that regenerate ATP through the breakdown of macromolecules (Figure 2). In addition, AMPK phosphorylates several transcription factors (or co-factors) that are themselves master regulators of biosynthetic pathways and metabolism (Mouchiroud et al., 2014). In this way, AMPK can acutely restore energy balance but also reprogram cell metabolism transcriptionally in response to prolonged energetic decreases. Some of the best established AMPK substrates are depicted in Figure 3. We discuss some of the metabolic effects of AMPK in the following section.
Figure 3. Substrates of AMPK regulate multiple metabolic processes in cells.
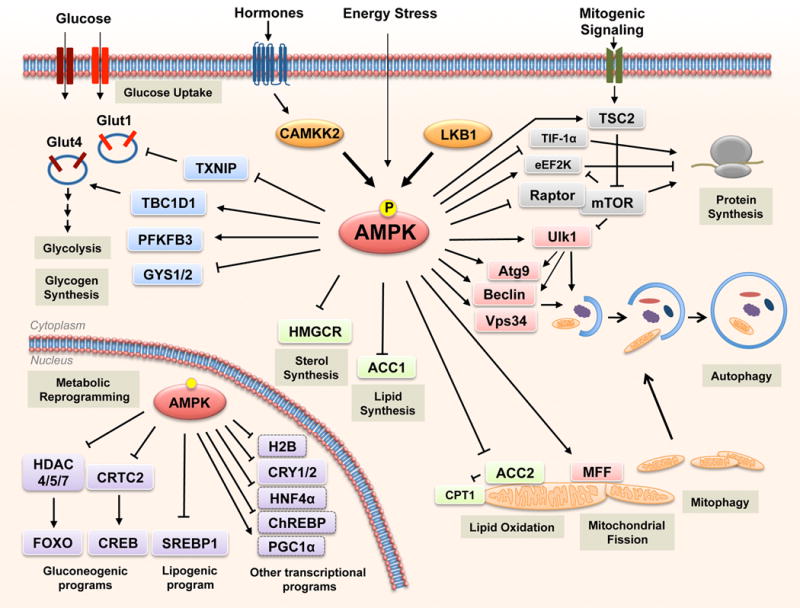
AMPK is phosphorylated and activated by LKB1 and CAMKKβ in response to stimuli that increase AMP/ADP levels (energy stress) or Ca2+ flux, respectively. Once active, AMPK induces metabolic changes through the phosphorylation of substrates. Some of the best established metabolic processes regulated by AMPK are shown, together with the relevant substrates.
Glucose and lipid metabolism
Glucose and lipids are major sources for the supply and storage of energy in cells. AMPK increases ATP levels by promoting their breakdown and inhibiting their synthesis and storage. AMPK promotes glucose uptake by phosphorylating TBC1D1 (TBC domain family, member 1) and TXNIP (thioredoxin-interacting protein), which control the translocation and cell-surface levels of glucose transporters GLUT4 and GLUT1, respectively (Hardie, 2013; Wu et al., 2013), though through distinct mechanisms. AMPK also acutely regulates glycolysis in some tissue types by phosphorylating PFKFB3 (6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3), while inhibiting storage of glucose in some tissues by inhibiting multiple isoforms of GYS (glycogen synthase) (Hardie, 2013). AMPK controls overall cellular lipid metabolism through direct phosphorylation of ACC1 (acetyl-CoA carboxylase 1) and ACC2, suppressing fatty acid synthesis and simultaneously promoting fatty acid oxidation by relieving the suppression of CPT1 (carnitine palmitoyltransferase 1) by malonyl-CoA locally produced at the mitochondria outer membrane by ACC2, which has a mitochondrial targeting sequence in its amino terminus. AMPK also phosphorylates and inhibits HMGCR (3-hydroxy-3-methyl-glutaryl-coA reductase), which, collectively with its effects on ACC1 and ACC2, leads to a preprogramming of lipid and sterol synthesis in the cell. AMPK also promotes lipid absorption and release by phosphorylating lipases such as HSL (hormone-sensitive lipase) and ATGL (adipocyte-triglyceride lipase) (Ahmadian et al., 2011; Kim et al., 2016).
AMPK inhibits the transcriptional induction of gluconeogenesis, the process of de novo synthesis of glucose, via phosphorylation and nuclear exclusion of CRTC2 (cyclic-AMP-regulated transcriptional co-activator 2) and class II HDACs (histone deacetylases), which are necessary co-factors for the transcription of gluconeogenic genes (Koo et al., 2005; Mihaylova et al., 2011), In addition, AMPK phosphorylates and inhibits transcription factors that activate glycolytic and lipogenic transcriptional programs, most notably SREBP1 (sterol regulatory element binding protein 1), a master transcriptional regulator of lipid synthesis (Li et al., 2011), but also HNF4α (hepatocyte nuclear factor-4α) and ChREBP (carbohydrate-responsive element binding protein) (Hong et al., 2003; Kawaguchi et al., 2002). Thus, acute AMPK activation favors glucose uptake to promote ATP restoration, while sustained AMPK activation reprograms cells to limit glucose and lipid synthesis and favor oxidation of fatty acids as an energy source.
mTOR and protein metabolism
The ability of AMPK to inhibit protein synthesis is mediated in large part by direct inhibition of the mTORC1 complex (mechanistic target of rapamycin complex 1). mTOR is a central integrator of nutrient and growth factor signals that activates many biosynthetic pathways, especially protein translation, and stimulates cellular growth. At many levels, AMPK and mTORC1 function antithetically in the regulation of cellular metabolism. AMPK inhibits mTORC1 activity by a two-pronged mechanism, through phosphorylation and activation of TSC2 (tuberous sclerosis complex 2) (Inoki et al., 2003), a negative regulator of mTORC1, and phosphorylation and inhibition of Raptor (regulatory-associated protein of mTOR), a subunit of the mTORC1 complex (Gwinn et al., 2008). Besides inhibition of mTOR, AMPK has been reported to limit protein synthesis by blocking ribosomal RNA synthesis through phosphorylation and inhibition of TIF-IA (transcription initiation factor IA), a transcription factor for RNA-polymerase I (Hoppe et al., 2009). AMPK also inhibits protein elongation, through phosphorylation and activation of eEF2K (eukaryotic elongation factor 2 kinase), an inhibitor of elongation (Leprivier et al., 2013). Importantly, mTORC1 is also a dominant regulator of eEF2K (Faller et al., 2015), providing an example of the many downstream targets of AMPK that are also directly phosphorylated by mTORC1 or S6K1 to antagonistically regulate their function from AMPK phosphorylation. In this fashion, AMPK and mTOR control anabolism and catabolism by traveling around the cell flipping on and off a limited set of master metabolic switches.
Autophagy and mitophagy
Autophagy is a cellular process in which proteins, organelles and other macromolecules are delivered to the lysosomes for degradation. It is a process used by cells both for normal turnover and for the generation of nutrients in response to energy shortages. AMPK potently promotes autophagy through several mechanisms. AMPK phosphorylates and activates ULK1 (unc-51-like autophagy-activating kinase 1), which triggers the initiation of the autophagic cascade (Egan et al., 2011; Kim et al., 2011; Mack et al., 2012). Importantly, mTOR strongly suppresses autophagy, in part by directly phosphorylating and inhibiting ULK1 (Kim et al., 2011). Accordingly, AMPK promotes autophagy not only by direct activation of ULK1 but also by negatively regulating mTORC1 and blocking its inhibitory effect on ULK1. Thus, ULK1 is yet another node at which AMPK and mTOR regulate a specific metabolic process in opposing fashion. AMPK also stimulates autophagy initiation by differential regulation of VPS34 (vacuolar protein sorting 34) containing complexes (Kim et al., 2013), which are important for the initiation and formation of autophagosomes. AMPK was reported to directly phosphorylate and inhibit VPS34 in non-autophagic complexes that do not contain autophagy adaptor proteins, while enhancing VPS34 activity in pro-autophagic complexes that contain Beclin-1 by directly phosphorylating Beclin-1 (Kim et al., 2013). In this way, AMPK presumably suppresses nonessential vesicle trafficking in favor of membrane trafficking into the autophagy pathway during nutrient starvation. Given that both AMPK and ULK1 have been reported to directly phosphorylate distinct sites in both Beclin-1 and Vps34, much remains to be clarified about the temporal and spatial control of autophagy initiation in response to different stresses. In addition, AMPK and ULK1 have also both been reported to phosphorylate and control the localization of Atg9, a transmembrane protein involved in early autophagosome formation (Mack et al., 2012; Weerasekara et al., 2014; Zhou et al., 2017).
AMPK has also recently been shown to promote autophagy through transcriptional mechanisms, via regulation of Tfeb (transcription factor EB), a master transcriptional regulator of lysosomal genes and autophagy. Although no direct link between AMPK and Tfeb has been reported, AMPK can activate Tfeb through inhibition of mTORC1, thus blocking the ability of mTOR to phosphorylate and translocate Tfeb out of the nucleus (Young et al., 2016). Furthermore, through phosphorylation and activation of the transcription factor FOXO3a (Forkhead box O3) (Greer et al., 2007), AMPK has been reported to increase the levels of CARM1 (co-activator-associated arginine methyltransferase 1), an important cofactor for Tfeb transcription (Shin et al., 2016).
In addition to general autophagy, several lines of evidence indicate that AMPK promotes mitophagy, the process of degradation of defective mitochondria. Indeed, activation of ULK1 by AMPK was shown to be required for proper removal of damaged mitochondria via mitophagy, though the details of how ULK1 regulates mitophagy are not fully resolved (Egan et al., 2011). A necessary step preceding removal of damaged mitochondria is the fragmentation of mitochondria in response to mitochondrial insults, in order to separate and target damaged mitochondrial fragments to turnover via the mitophagy pathway. This highly conserved process is known as mitochondrial fission. Recently, a novel mechanism was elucidated by which AMPK promotes mitochondrial fission (Toyama et al., 2016). In this study, AMPK was demonstrated to induce mitochondrial fission during energy stress through direct phosphorylation of MFF (mitochondrial fission factor), which then serves as a receptor for DRP1 (dynamin-related protein 1), the enzyme that catalyzes mitochondrial fission (Toyama et al., 2016). Once at the mitochondria, DRP1 splits damaged mitochondria into smaller fragments that are presumably more efficiently cleared by autophagosomes. Furthermore, AMPK activates PGC1α (peroxisome proliferator-activated receptor gamma, coactivator 1α), a master regulator of mitochondrial biogenesis, reportedly via direct phosphorylation of PGC1α (Jäger et al., 2007) but also by promoting NAD+-dependent activation of PGC1α by Sirt1 (sirtuin 1) (Cantó et al., 2009). Interestingly, Tfeb, similar to its family member Tfe3, was recently reported to drive mitochondrial biogenesis as well (Mansueto et al., 2017; Wada et al., 2016), which offers the possibility that activation of Tfeb, or Tfe3, might be yet another mechanism by which AMPK can promote the regeneration of mitochondria. In all, AMPK coordinates mitochondrial fission and mitophagy in the acute response to mitochondrial insults, and after sustained energy stress, AMPK promotes transcriptional induction of mitochondrial biogenesis. In this fashion, AMPK serves as a central mediator of mitochondrial quality, ensuring metabolic efficiency in cells and tissues.
AMPK activity in other biological contexts
A great deal of evidence points to a role for AMPK as an inhibitor of cell growth and proliferation. Of considerable interest, however, are recent studies that further document this ability of AMPK to inhibit cell proliferation through phosphorylation of novel substrates in important mitogenic pathways. Specifically, AMPK was reported to inhibit Hedgehog signaling through phosphorylation and destabilization of the transcription factor GLI1 (Glioma-associated oncogene 1) (Li et al., 2015b). Similarly, AMPK was shown to inhibit the Hippo-YAP pathway during energy stress via phosphorylation and stabilization of AMOTL1 (angiomotin-like 1) (DeRan et al., 2014), a negative regulator of YAP (Yes-associated protein), and by phosphorylation and inhibition of YAP itself (Mo et al., 2015; Wang et al., 2015). Furthermore, AMPK was reported to phosphorylate and inhibit MDMX (mouse double minute X), a negative regulator of the tumor suppressor p53, resulting in p53 activation and subsequent cell cycle arrest (He et al., 2014). All of these pathways have well-established roles in cell growth and proliferation and are frequently deregulated in tumor cells. Their regulation by AMPK may indicate novel mechanisms by which AMPK serves as a “metabolic checkpoint” in cells.
Novel biological contexts in which AMPK function might be relevant are constantly being reported. One such notable context is management of reactive oxygen species. AMPK is not only a redox sensitive protein, as described above, but is also involved in the response to oxidative stress. Indeed, AMPK was reported to phosphorylate Nrf2 (nuclear factor erythroid 2-related factor 2), a master transcriptional regulator of antioxidant gene programs, which resulted in nuclear accumulation of Nrf2 and subsequent expression of antioxidant genes (Joo et al., 2016). Moreover, AMPK can indirectly mitigate ROS by maintaining high levels NADPH and GSH (which are cellular antioxidants) through inhibition of fatty acid synthesis, a process that consumes NADPH, and by promotion of fatty acid oxidation, a process that generates NADPH (Jeon et al., 2012). Intriguingly, this mechanism of indirect regulation of key metabolites by AMPK was recently described for yet another novel biological context. In a recent study, AMPK was shown to promote the epigenetic remodeling of the Prdm16 promoter by maintaining high levels of α-ketoglutarate in brown adipocytes (Yang et al., 2016). Briefly, high α-ketoglutarate levels (maintained by AMPKα1) were shown to be required for demethylation of the Prdm16 promoter by TET (ten-eleven translocation hydroxylases) enzymes, which then led to expression of PRDM16 (PR domain containing 16 protein), the key transcription factor for brown adipogenesis.
Finally, AMPK has been implicated in the regulation of circadian rhythms. The core circadian clock is set by the activity of transcription factors CLOCK and BMAL1 and their cyclical repression by chryptochrome (CRY1, CRY2) and period proteins (PER1, PER2, PER3). AMPK directly regulates the circadian core clock by phosphorylating CRY1 and CRY2, which targets them for degradation (Lamia et al., 2009). AMPK also indirectly promotes degradation of PER proteins (Um et al., 2007). Thus, the ability of AMPK to regulate the circadian clock may be important for the entrainment of circadian rhythms in tissues. Indeed, AMPK-dependent degradation of clock core components may have evolved as a light-independent clock-setting signal that allows coupling of circadian rhythms to energy availability (Jordan and Lamia, 2013). Given the newly appreciated role of CRY1 and CRY2 to act as repressors of specific nuclear receptors (NRs), AMPK dependent degradation of CRY proteins may also serve to promote specific NR-dependent gene expression programs (Lamia et al., 2011).
Therapeutic activation of AMPK
Given the therapeutically beneficial effects associated with AMPK activation, it is not surprising that many AMPK activating compounds have been identified (Hardie, 2013). How much of the effect of these compounds can be ascribed to AMPK depends not only on drug specificity but also on the particular mechanism of AMPK activation. The mechanisms by which these compounds activate AMPK can be broadly divided into three classes: 1) agents that increase intracellular AMP and ADP, thus indirectly activating AMPK; 2) compounds that mimic AMP and are thus able to bind AMPK at the γ-subunit; 3) compounds that activate AMPK by selectively binding the interface between the CBM and the kinase domain (Fig 4).
Figure 4. AMPK activating compounds.
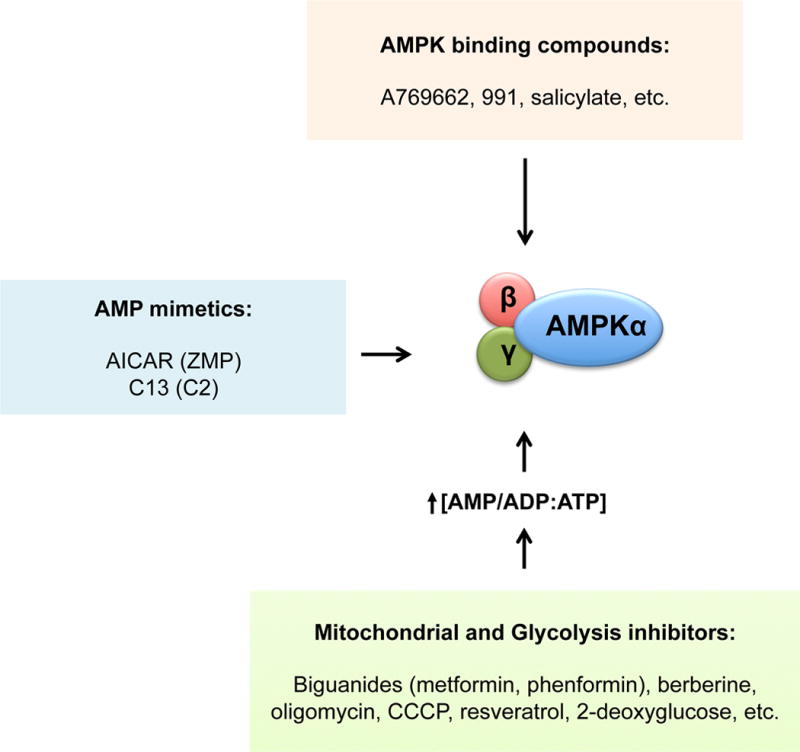
Compounds that activate AMPK can be divided into three categories, depending on the mechanism used to activate AMPK. Examples of some of these compounds are shown.
The majority of drugs that activate AMPK by increasing cellular AMP/ADP levels do so by inhibiting mitochondrial respiration and its production of ATP. Biguanides, which include metformin and its more potent analog phenformin, are modest inhibitors of Complex I in the respiratory chain and thus activate AMPK via this mechanism. Metformin is the most widely prescribed drug for type II diabetes and a large proportion of its blood glucose lowering properties are thought to be mediated by AMPK activation (Cao et al., 2014; Duca et al., 2015; Fullerton et al., 2013; He and Wondisford, 2015; Shaw et al., 2005), although AMPK-independent effectors are likely also involved in metformin action (Madiraju et al., 2015; Miller et al., 2013). Mitochondrial poisons (such as oligomycin, CCCP, rotenone, etc.) and a large number of plant-derived drugs (such as resveratrol, berberine, galegine, etc.) have also been shown to activate AMPK by inhibiting different components of the respiratory chain (Hardie, 2013; Hawley et al., 2010). Glycolysis inhibitors, such as 2-deoxyglucose (a non-hydrolysable form of glucose), also elevate AMP levels to activate AMPK, by suppressing glycolysis in cells that are reliant on it for ATP-generation. In a more novel approach, inhibition of AMP deaminase (AMPD) has been tested as yet another way to increase intracellular AMP. AMPD converts AMP into IMP, so its inhibition also increases AMP pools, which in turn activate AMPK (Plaideau et al., 2014). Indeed, one report suggested that metformin may control AMP levels via effects on AMPD (Ouyang et al., 2011).
A second class of compounds activates AMPK by functioning as AMP mimetics. These agents are administered as pro-drugs, which are taken up by cells and then metabolized to the actual AMP analog. The best-known member of this class of compounds is 5-aminoimidazole-4-carboxamide-1-β-D-ribofuranoside (AICAR). AICAR is taken up by adenosine transporters in cells and then converted to ZMP, which mimics all of the effects of AMP on AMPK. A new AMPK-activating AMP analog was recently reported, 5-(5- hydroxyl-isoxazol-3-yl)-furan-2-phosphonic acid compound 2 (C2) (Gómez-Galeno et al., 2010), which is administered as the pro-drug C13. C2 is more potent than AMP or ZMP, but unlike AMP or ZMP, it only activates AMPK complexes containing AMPKα1 (Hunter et al., 2014). Also, although ZMP is expected to affect many AMP-dependent processes independently of AMPK, C2 is reported to have high specificity for AMPK (Hunter et al., 2014). Interestingly, multiple recent reports have exploited modulation of purine biosynthesis as a mechanism to alter ZMP or AMP levels to activate AMPK, including the widely used chemotherapeutics pemetrexed and methotrexate (Asby et al., 2015; Pirkmajer et al., 2015; Racanelli et al., 2009).
The third class of compounds consists of novel small molecules that directly bind and activate AMPK. The first member of this class was the thienopyridone A769662 (Cool et al., 2006). Multiple additional related compounds have since been reported, such as 991 (also known as ex229), MT-63-78, and GSK621 (Li et al., 2015a; Sujobert et al., 2015; Xiao et al., 2013). Like AMP, A769662 was shown to allosterically activate AMPK and to protect from Thr172 dephosphorylation (Göransson et al., 2007; Sanders et al., 2007). However, contrary to AMP, A769662 was demonstrated to specifically require the CBM domain in AMPKβ1 and phosphorylation of AMPKβ1 at serine 108 (Sanders et al., 2007). Phosphorylation of AMPKβ1 at serine 108 appears to be mostly due to autophosphorylation, although phosphorylation by other kinases cannot be excluded (Sanders et al., 2007; Scott et al., 2014). The crystal structure of the actual single binding site for A769662 and 991 has now been resolved (Xiao et al., 2013). As described above, the CBM domain of the β-subunit directly contacts the kinase domain of the α-subunit, forming a pocket between the two domains that is bound by 991 and A769662, where these small molecules form hydrophobic interactions with residues from both domains. A769662 and AMP potently synergize to activate AMPK, remarkably, even in the absence of Thr172 phosphorylation (Scott et al., 2014). Of note, salicylate, the breakdown product of aspirin and salsalate, was also found to directly activate AMPK (Hawley et al., 2012). Surprisingly, the CBM-kinase domain pocket is very likely to be the same binding site for salicylate since, like A769662, activation of AMPK by salicylate requires the CBM domain of AMPKβ1 and its phosphorylation at Ser108 (Hawley et al., 2012). Consistent with its ability to activate AMPK, mice treated with salicylate were shown to have higher rates of fatty acid oxidation and reduced levels of circulating fatty acids in an AMPK-dependent manner (Hawley et al., 2012). Indeed, some of the beneficial effects of salicylate in humans might be mediated by its ability to activate AMPK, especially its metabolic, anti-tumorigenic and anti-inflammatory effects (Ford et al., 2015; Fullerton et al., 2015).
Concluding remarks
AMPK is a highly conserved master regulator of metabolism, both at the cellular and organismal levels, whose function is extremely relevant not only for normal physiology, but also for the understanding of many metabolic diseases. The examination of novel mechanisms of AMPK regulation and the identification of additional AMPK substrates continue to enhance our understanding of not only AMPK biology, but also of how cells manage their energy demands generally. Importantly, many AMPK activating compounds, in addition to the clinically approved metformin and salsalate, have enormous therapeutic potential, in particular for the treatment of metabolic disorders such as diabetes. However, these drugs are being experimentally tested for a variety of other applications, including the treatment of obesity, muscle dysfunction, and cancer, and there will undoubtedly be much to learn from these trials.
Acknowledgments
We thank Kristina Hellberg for her assistance with PyMOL software. D.G. is funded by a postdoctoral fellowship to the Salk Institute Glenn Center for Aging Research. R.J.S. holds the William R. Brody Chair and is a professor in the Molecular and Cell Biology Department at the Salk Institute of Biological Studies. The work from our laboratory described in this review was supported by grants from the National Institutes of Health (R01DK080425, R01CA172229, P01CA120964), and The Leona M. and Harry B. Helmsley Charitable Trust (grant #2012-PGMED002). We apologize to the many authors whose works could not be cited due to space limitations, including many insightful discoveries into new targets of AMPK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmadian M, Abbott MJ, Tang T, Hudak CSS, Kim Y, Bruss M, Hellerstein MK, Lee HY, Samuel VT, Shulman GI, et al. Desnutrin/ATGL Is Regulated by AMPK and Is Required for a Brown Adipose Phenotype. Cell Metabolism. 2011;13:739–748. doi: 10.1016/j.cmet.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asby DJ, Cuda F, Beyaert M, Houghton FD, Cagampang FR, Tavassoli A. AMPK Activation via Modulation of De Novo Purine Biosynthesis with an Inhibitor of ATIC Homodimerization. Chem Biol. 2015;22:838–848. doi: 10.1016/j.chembiol.2015.06.008. [DOI] [PubMed] [Google Scholar]
- Banko MR, Allen JJ, Schaffer BE, Wilker EW, Tsou P, White JL, Villén J, Wang B, Kim SR, Sakamoto K, et al. Chemical Genetic Screen for AMPKα2 Substrates Uncovers a Network of Proteins Involved in Mitosis. Molecular Cell. 2011;44:878–892. doi: 10.1016/j.molcel.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese MF, Rajamohan F, Harris MS, Caspers NL, Magyar R, Withka JM, Wang H, Borzilleri KA, Sahasrabudhe PV, Hoth LR, et al. Structural Basis for AMPK Activation: Natural and Synthetic Ligands Regulate Kinase Activity from Opposite Poles by Different Molecular Mechanisms. Structure/Folding and Design. 2014;22:1161–1172. doi: 10.1016/j.str.2014.06.009. [DOI] [PubMed] [Google Scholar]
- Cantó C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Meng S, Chang E, Beckwith-Fickas K, Xiong L, Cole RN, Radovick S, Wondisford FE, He L. Low Concentrations of Metformin Suppress Glucose Production in Hepatocytes through AMP-activated Protein Kinase (AMPK) J Biol Chem. 2014;289:20435–20446. doi: 10.1074/jbc.M114.567271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang J, Zhang YY, Yan SF, Neumann D, Schlattner U, Wang ZX, Wu JW. AMP-activated protein kinase undergoes nucleotide-dependent conformational changes. Nature Structural & Molecular Biology. 2012;19:716–718. doi: 10.1038/nsmb.2319. [DOI] [PubMed] [Google Scholar]
- Chen L, Xin FJ, Wang J, Hu J, Zhang YY, Wan S, Cao LS, Lu C, Li P, Yan SF, et al. Conserved regulatory elements in AMPK. Nature. 2013;498:E8–E10. doi: 10.1038/nature12189. [DOI] [PubMed] [Google Scholar]
- Cool B, Zinker B, Chiou W, Kifle L, Cao N, Perham M, Dickinson R, Adler A, Gagne G, Iyengar R, et al. Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell Metabolism. 2006;3:403–416. doi: 10.1016/j.cmet.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Coughlan KA, Valentine RJ, Sudit BS, Allen K, Dagon Y, Kahn BB, Ruderman NB, Saha AK. PKD1 Inhibits AMPKα2 through Phosphorylation of Serine 491 and Impairs Insulin Signaling in Skeletal Muscle Cells. J Biol Chem. 2016;291:5664–5675. doi: 10.1074/jbc.M115.696849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagon Y, Hur E, Zheng Bin, Wellenstein K, Cantley LC, Kahn BB. p70S6 Kinase Phosphorylates AMPK on Serine 491 to Mediate Leptin’s Effect on Food Intake. Cell Metabolism. 2012;16:104–112. doi: 10.1016/j.cmet.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRan M, Yang J, Shen CH, Peters EC, Fitamant J, Chan P, Hsieh M, Zhu S, Asara JM, Zheng Bin, et al. Energy Stress Regulates Hippo-YAP Signaling Involving AMPK-Mediated Regulation of Angiomotin-like 1 Protein. CellReports. 2014;9:495–503. doi: 10.1016/j.celrep.2014.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca FA, Côté CD, Rasmussen BA, Zadeh-Tahmasebi M, Rutter GA, Filippi BM, Lam TKT. Metformin activates a duodenal Ampk-dependent pathway to lower hepatic glucose production in rats. Nature Medicine. 2015;21:506–511. doi: 10.1038/nm.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-Activated Protein Kinase Connects Energy Sensing to Mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faller WJ, Jackson TJ, Knight JRP, Ridgway RA, Jamieson T, Karim SA, Jones C, Radulescu S, Huels DJ, Myant KB, et al. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature. 2015;517:497–500. doi: 10.1038/nature13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford RJ, Fullerton MD, Pinkosky SL, Day EA, Scott JW, Oakhill JS, Bujak AL, Smith BK, Crane JD, Blümer RM, et al. Metformin and salicylate synergistically activate liver AMPK, inhibit lipogenesis and improve insulin sensitivity. Biochem J. 2015;468:125–132. doi: 10.1042/BJ20150125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton MD, Ford RJ, McGregor CP, LeBlond ND, Snider SA, Stypa SA, Day EA, Lhoták Š, Schertzer JD, Austin RC, et al. Salicylate improves macrophage cholesterol homeostasis via activation of Ampk. J Lipid Res. 2015;56:1025–1033. doi: 10.1194/jlr.M058875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton MD, Galic S, Marcinko K, Sikkema S, Pulinilkunnil T, Chen ZP, O’;Neill HM, Ford RJ, Palanivel R, O’Brien M, et al. Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nature Medicine. 2013;19:1649–1654. doi: 10.1038/nm.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Haro L, Garcia-Gimeno MA, Neumann D, Beullens M, Bollen M, Sanz P. The PP1-R6 protein phosphatase holoenzyme is involved in the glucose-induced dephosphorylation and inactivation of AMP-activated protein kinase, a key regulator of insulin secretion, in MIN6 beta cells. Faseb J. 2010;24:5080–5091. doi: 10.1096/fj.10-166306. [DOI] [PubMed] [Google Scholar]
- Gowans GJ, Hawley SA, Ross FA, Hardie DG. AMP Is a True Physiological Regulator of AMP-Activated Protein Kinase by Both Allosteric Activation and Enhancing Net Phosphorylation. Cell Metabolism. 2013;18:556–566. doi: 10.1016/j.cmet.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Galeno JE, Dang Q, Nguyen TH, Boyer SH, Grote MP, Sun Z, Chen M, Craigo WA, van Poelje PD, MacKenna DA, et al. A Potent and Selective AMPK Activator That Inhibits de Novo Lipogenesis. ACS Med Chem Lett. 2010;1:478–482. doi: 10.1021/ml100143q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göransson O, McBride A, Hawley SA, Ross FA, Shpiro N, Foretz M, Viollet B, Hardie DG, Sakamoto K. Mechanism of Action of A-769662, a Valuable Tool for Activation of AMP-activated Protein Kinase. J Biol Chem. 2007;282:32549–32560. doi: 10.1074/jbc.M706536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK Phosphorylation of Raptor Mediates a Metabolic Checkpoint. Molecular Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. AMPK: A Target for Drugs and Natural Products With Effects on Both Diabetes and Cancer. Diabetes. 2013;62:2164–2172. doi: 10.2337/db13-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. AMPK-Sensing Energy while Talking to Other Signaling Pathways. Cell Metabolism. 2014;20:939–952. doi: 10.1016/j.cmet.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Schaffer BE, Brunet A. AMPK: An Energy-Sensing Pathway with Multiple Inputs and Outputs. Trends in Cell Biology. 2016;26:190–201. doi: 10.1016/j.tcb.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Mäkelä TP, Alessi DR, Hardie DG. Complexes between the LKB1 tumor suppressor, STRADα/β and MO25α/β are upstream kinases in the AMP-activated protein kinase cascade. Journal of Biology. 2003;2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Fullerton MD, Ross FA, Schertzer JD, Chevtzoff C, Walker KJ, Peggie MW, Zibrova D, Green KA, Mustard KJ, et al. The ancient drug salicylate directly activates AMP-activated protein kinase. Science. 2012;336:918–922. doi: 10.1126/science.1215327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Pan DA, Mustard KJ, Ross L, Bain J, Edelman AM, Frenguelli BG, Hardie DG. Calmodulin-dependent protein kinase kinase-β is an alternative upstream kinase for AMP-activated protein kinase. Cell Metabolism. 2005;2:9–19. doi: 10.1016/j.cmet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Hawley SA, Ross FA, Chevtzoff C, Green KA, Evans A, Fogarty S, Towler MC, Brown LJ, Ogunbayo OA, Evans AM, et al. Use of Cells Expressing γ Subunit Variants to Identify Diverse Mechanisms of AMPK Activation. Cell Metabolism. 2010;11:554–565. doi: 10.1016/j.cmet.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Ross FA, Gowans GJ, Tibarewal P, Leslie NR, Hardie DG. Phosphorylation by Akt within the ST loop of AMPK-α1 down-regulates its activation in tumour cells. Biochem J. 2014;459:275–287. doi: 10.1042/BJ20131344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Zhang YW, Lee JH, Zeng SX, Wang YV, Luo Z, Dong XC, Viollet B, Wahl GM, Lu H. AMP-activated protein kinase induces p53 by phosphorylating MDMX and inhibiting its activity. Molecular and Cellular Biology. 2014;34:148–157. doi: 10.1128/MCB.00670-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Wondisford FE. Metformin action: concentrations matter. Cell Metabolism. 2015;21:159–162. doi: 10.1016/j.cmet.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Heathcote HR, Mancini SJ, Strembitska A, Jamal K, Reihill JA, Palmer TM, Gould GW, Salt IP. Protein kinase C phosphorylates AMP-activated protein kinase α1 Ser487. Biochem J. 2016;473:4681–4697. doi: 10.1042/BCJ20160211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman NJ, Parker BL, Chaudhuri R, Fisher-Wellman KH, Kleinert M, Humphrey SJ, Yang P, Holliday M, Trefely S, Fazakerley DJ, et al. Global Phosphoproteomic Analysis of Human Skeletal Muscle Reveals a Network of Exercise- Regulated Kinases and AMPK Substrates. Cell Metabolism. 2015;22:922–935. doi: 10.1016/j.cmet.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YH, Varanasi US, Yang W, Leff T. AMP-activated protein kinase regulates HNF4alpha transcriptional activity by inhibiting dimer formation and decreasing protein stability. J Biol Chem. 2003;278:27495–27501. doi: 10.1074/jbc.M304112200. [DOI] [PubMed] [Google Scholar]
- Hoppe S, Bierhoff H, Cado I, Weber A, Tiebe M, Grummt I, Voit R. AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proceedings of the National Academy of Sciences. 2009;106:17781–17786. doi: 10.1073/pnas.0909873106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horman S, Morel N, Vertommen D, Hussain N, Neumann D, Beauloye C, Najjar El N, Forcet C, Viollet B, Walsh MP, et al. AMP-activated protein kinase phosphorylates and desensitizes smooth muscle myosin light chain kinase. J Biol Chem. 2008;283:18505–18512. doi: 10.1074/jbc.M802053200. [DOI] [PubMed] [Google Scholar]
- Houde VP, Ritorto MS, Gourlay R, Varghese J, Davies P, Shpiro N, Sakamoto K, Alessi DR. Investigation of LKB1 Ser 431phosphorylation and Cys 433farnesylation using mouse knockin analysis reveals an unexpected role of prenylation in regulating AMPK activity. Biochem J. 2014;458:41–56. doi: 10.1042/BJ20131324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter RW, Foretz M, Bultot L, Fullerton MD, Deak M, Ross FA, Hawley SA, Shpiro N, Viollet B, Barron D, et al. Mechanism of Action of Compound-13: An α1-Selective Small Molecule Activator of AMPK. Chbiol. 2014;21:866–879. doi: 10.1016/j.chembiol.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley RL, Anderson KA, Franzone JM, Kemp BE, Means AR, Witters LA. The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J Biol Chem. 2005;280:29060–29066. doi: 10.1074/jbc.M503824200. [DOI] [PubMed] [Google Scholar]
- Hurley RL, Barré LK, Wood SD, Anderson KA, Kemp BE, Means AR, Witters LA. Regulation of AMP-activated Protein Kinase by Multisite Phosphorylation in Response to Agents That Elevate Cellular cAMP. J Biol Chem. 2006;281:36662–36672. doi: 10.1074/jbc.M606676200. [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Jäger S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Pnas. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661–665. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo MS, Kim WD, Lee KY, Kim JH, Koo JH, Kim SG. AMPK Facilitates Nuclear Accumulation of Nrf2 by Phosphorylating at Serine 550. Molecular and Cellular Biology. 2016;36:1931–1942. doi: 10.1128/MCB.00118-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan SD, Lamia KA. AMPK at the crossroads of circadian clocks and metabolism. Molecular and Cellular Endocrinology. 2013;366:163–169. doi: 10.1016/j.mce.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph BK, Liu HY, Francisco J, Pandya D, Donigan M, Gallo-Ebert C, Giordano C, Bata A, Nickels JT. Inhibition of AMP Kinase by the Protein Phosphatase 2A Heterotrimer, PP2APpp2r2d. J Biol Chem. 2015;290:10588–10598. doi: 10.1074/jbc.M114.626259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T, Osatomi K, Yamashita H, Kabashima T, Uyeda K. Mechanism for fatty acid “sparing” effect on glucose-induced transcription: regulation of carbohydrate-responsive element-binding protein by AMP-activated protein kinase. J Biol Chem. 2002;277:3829–3835. doi: 10.1074/jbc.M107895200. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL. Differential Regulation of Distinct Vps34 Complexes by AMPK in Nutrient Stress and Autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Tang T, Abbott M, Viscarra JA, Wang Y, Sul HS. AMPK Phosphorylates Desnutrin/ATGL and Hormone-Sensitive Lipase To Regulate Lipolysis and Fatty Acid Oxidation within Adipose Tissue. Molecular and Cellular Biology. 2016;36:1961–1976. doi: 10.1128/MCB.00244-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Papp SJ, Yu RT, Barish GD, Uhlenhaut NH, Jonker JW, Downes M, Evans RM. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480:552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JO, Lee SK, Kim N, Kim JH, You GY, Moon JW, Jie S, Kim SJ, Lee YW, Kang HJ, et al. E3 ubiquitin ligase, WWP1, interacts with AMPKα2 and down-regulates its expression in skeletal muscle C2C12 cells. J Biol Chem. 2013;288:4673–4680. doi: 10.1074/jbc.M112.406009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprivier G, Remke M, Rotblat B, Dubuc A, Mateo ARF, Kool M, Agnihotri S, El-Naggar A, Yu Bin, Somasekharan SP, et al. The eEF2 Kinase Confers Resistance to Nutrient Deprivation by Blocking Translation Elongation. Cell. 2013;153:1064–1079. doi: 10.1016/j.cell.2013.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang L, Zhou XE, Ke J, de Waal PW, Gu X, Tan MHE, Wang D, Wu D, Xu HE, et al. Structural basis of AMPK regulation by adenine nucleotides and glycogen. Cell Res. 2015a;25:50–66. doi: 10.1038/cr.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Luo J, Mosley YYC, Hedrick VE, Paul LN, Chang J, Zhang G, Wang YK, Banko MR, Brunet A, et al. AMP-Activated Protein Kinase Directly Phosphorylates and Destabilizes Hedgehog Pathway Transcription Factor GLI1 in Medulloblastoma. CellReports. 2015b;12:599–609. doi: 10.1016/j.celrep.2015.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu S, Mihaylova MM, Zheng Bin, Hou X, Jiang B, Park O, Luo Z, Lefai E, Shyy JYJ, et al. AMPK Phosphorylates and Inhibits SREBP Activity to Attenuate Hepatic Steatosis and Atherosclerosis in Diet-Induced Insulin-Resistant Mice. Cell Metabolism. 2011;13:376–388. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Xu ZX, Ding Z, Lu Y, Yu Q, Werle KD, Zhou G, Park YY, Peng G, Gambello MJ, et al. Myristoylation confers noncanonical AMPK functions in autophagy selectivity and mitochondrial surveillance. Nature Communications. 2015;6:7926. doi: 10.1038/ncomms8926. [DOI] [PubMed] [Google Scholar]
- Mack HID, Zheng B, Asara JM, Thomas SM. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy. 2012;8:1197–1214. doi: 10.4161/auto.20586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madiraju AK, Erion DM, Rahimi Y, Zhang XM, Braddock DT, Albright RA, Prigaro BJ, Wood JL, Bhanot S, MacDonald MJ, et al. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature. 2015;510:542–546. doi: 10.1038/nature13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansueto G, Armani A, Viscomi C, D’Orsi L, De Cegli R, Polishchuk EV, Lamperti C, Di Meo I, Romanello V, Marchet S, et al. Transcription Factor EB Controls Metabolic Flexibility during Exercise. Cell Metabolism. 2017;25:182–196. doi: 10.1016/j.cmet.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride A, Ghilagaber S, Nikolaev A, Hardie DG. The Glycogen-Binding Domain on the AMPK β Subunit Allows the Kinase to Act as a Glycogen Sensor. Cell Metabolism. 2009;9:23–34. doi: 10.1016/j.cmet.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, Shaw RJ. Class IIa Histone Deacetylases Are Hormone-Activated Regulators of FOXO and Mammalian Glucose Homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RA, Chu Q, Xie J, Foretz M, Viollet B, Birnbaum MJ. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature. 2013;494:256–260. doi: 10.1038/nature11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Rho E, Sample V, Akano H, Magari M, Ueno T, Gorshkov K, Chen M, Tokumitsu H, Zhang J, et al. Compartmentalized AMPK Signaling Illuminated by Genetically Encoded Molecular Sensors and Actuators. CellReports. 2015;11:657–670. doi: 10.1016/j.celrep.2015.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo JS, Meng Z, Kim YC, Park HW, Hansen CG, Kim S, Lim DS, Guan KL. Cellular energy stress induces AMPK-mediated regulation of YAP and the Hippo pathway. Nat Cell Biol. 2015;17:500–510. doi: 10.1038/ncb3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L, Eichner LJ, Shaw RJ, Auwerx J. Transcriptional Coregulators: Fine-Tuning Metabolism. Cell Metabolism. 2014;20:26–40. doi: 10.1016/j.cmet.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakhill JS, Steel R, Chen ZP, Scott JW, Ling N, Tam S, Kemp BE. AMPK Is a Direct Adenylate Charge-Regulated Protein Kinase. Science. 2011;332:1433–1435. doi: 10.1126/science.1200094. [DOI] [PubMed] [Google Scholar]
- Oakhill JS, Chen ZP, Scott JW, Steel R, Castelli LA, Ling N, Macaulay SL, Kemp BE. β-Subunit myristoylation is the gatekeeper for initiating metabolic stress sensing by AMP-activated protein kinase (AMPK) Proceedings of the National Academy of Sciences. 2010;107:19237–19241. doi: 10.1073/pnas.1009705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J, Parakhia RA, Ochs RS. Metformin activates AMP kinase through inhibition of AMP deaminase. J Biol Chem. 2011;286:1–11. doi: 10.1074/jbc.M110.121806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda CT, Ramanathan S, Tacer KF, Weon JL, Potts MB, Ou YH, White MA, Potts PR. Degradation of AMPK by a Cancer-Specific Ubiquitin Ligase. Cell. 2015;160:715–728. doi: 10.1016/j.cell.2015.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirkmajer S, Kulkarni SS, Tom RZ, Ross FA, Hawley SA, Hardie DG, Zierath JR, Chibalin AV. Methotrexate promotes glucose uptake and lipid oxidation in skeletal muscle via AMPK activation. Diabetes. 2015;64:360–369. doi: 10.2337/db14-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaideau C, Lai YC, Kviklyte S, Zanou N, Löfgren L, Andersén H, Vertommen D, Gailly P, Hue L, Bohlooly-Y M, et al. Effects of Pharmacological AMP Deaminase Inhibition and Ampd1 Deletion on Nucleotide Levels and AMPK Activation in Contracting Skeletal Muscle. Chbiol. 2014;21:1497–1510. doi: 10.1016/j.chembiol.2014.09.013. [DOI] [PubMed] [Google Scholar]
- Racanelli AC, Rothbart SB, Heyer CL, Moran RG. Therapeutics by cytotoxic metabolite accumulation: pemetrexed causes ZMP accumulation, AMPK activation, and mammalian target of rapamycin inhibition. Cancer Research. 2009;69:5467–5474. doi: 10.1158/0008-5472.CAN-08-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross FA, MacKintosh C, Hardie DG. AMP-activated protein kinase: a cellular energy sensor that comes in 12 flavours. Febs J. 2016;283:2987–3001. doi: 10.1111/febs.13698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders MJ, Ali ZS, Hegarty BD, Heath R, Snowden MA, Carling D. Defining the mechanism of activation of AMP-activated protein kinase by the small molecule A-769662, a member of the thienopyridone family. J Biol Chem. 2007;282:32539–32548. doi: 10.1074/jbc.M706543200. [DOI] [PubMed] [Google Scholar]
- Schaffer BE, Levin RS, Hertz NT, Maures TJ, Schoof ML, Hollstein PE, Benayoun BA, Banko MR, Shaw RJ, Shokat KM, et al. Identification of AMPK Phosphorylation Sites Reveals a Network of Proteins Involved in Cell Invasion and Facilitates Large-Scale Substrate Prediction. Cell Metabolism. 2015;22:907–921. doi: 10.1016/j.cmet.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JW, Ling N, Issa SMA, Dite TA, O’Brien MT, Chen Z-P, Galic S, Langendorf CG, Steinberg GR, Kemp BE, et al. Small Molecule Drug A-769662 and AMP Synergistically Activate Naive AMPK Independent of Upstream Kinase Signaling. Chbiol. 2014;21:619–627. doi: 10.1016/j.chembiol.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Shao D, Oka SI, Liu T, Zhai P, Ago T, Sciarretta S, Li H, Sadoshima J. A Redox-Dependent Mechanism for Regulation of AMPK Activation by Thioredoxin1 during Energy Starvation. Cell Metabolism. 2014;19:232–245. doi: 10.1016/j.cmet.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Pnas. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Lamia KA, Vasquez D, Koo SH, Bardeesy N, DePinho RA, Montminy M, Cantley LC. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HJR, Kim H, Oh S, Lee JG, Kee M, Ko HJ, Kweon MN, Won KJ, Baek SH. AMPK–SKP2–CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. 2016;534:553–557. doi: 10.1038/nature18014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujobert P, Poulain L, Paubelle E, Zylbersztejn F, Grenier A, Lambert M, Townsend EC, Brusq JM, Nicodeme E, Decrooqc J, et al. Co-activation of AMPK and mTORC1 Induces Cytotoxicity in Acute Myeloid Leukemia. CellReports. 2015;11:1446–1457. doi: 10.1016/j.celrep.2015.04.063. [DOI] [PubMed] [Google Scholar]
- Suter M, Riek U, Tuerk R, Schlattner U, Wallimann T, Neumann D. Dissecting the role of 5′-AMP for allosteric stimulation, activation, and deactivation of AMP-activated protein kinase. J Biol Chem. 2006;281:32207–32216. doi: 10.1074/jbc.M606357200. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Bridges D, Nakada D, Skiniotis G, Morrison SJ, Lin JD, Saltiel AR, Inoki K. Inhibition of AMPK Catabolic Action by GSK3. Molecular Cell. 2013;50:407–419. doi: 10.1016/j.molcel.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama EQ, Herzig S, Courchet J, Lewis TL, Losón OC, Hellberg K, Young NP, Chen H, Polleux F, Chan DC, et al. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress Science. 2016;351:275–281. doi: 10.1126/science.aab4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um JH, Yang S, Yamazaki S, Kang H, Viollet B, Foretz M, Chung JH. Activation of 5′-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2. J Biol Chem. 2007;282:20794–20798. doi: 10.1074/jbc.C700070200. [DOI] [PubMed] [Google Scholar]
- Vila IK, Yao Y, Kim G, Xia W, Kim H, Kim SJ, Park MK, Hwang JP, González-Billalabeitia E, Hung MC, et al. A UBE2O-AMPKα2 Axis that Promotes Tumor Initiation and Progression Offers Opportunities for Therapy. Cancer Cell. 2017;31:208–224. doi: 10.1016/j.ccell.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Neinast M, Jang C, Ibrahim YH, Lee G, Babu A, Li J, Hoshino A, Rowe GC, Rhee J, et al. The tumor suppressor FLCN mediates an alternate mTOR pathway to regulate browning of adipose tissue. Genes & Development. 2016;30:2551–2564. doi: 10.1101/gad.287953.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xiao ZD, Li X, Aziz KE, Gan B, Johnson RL, Chen J. AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat Cell Biol. 2015;17:490–499. doi: 10.1038/ncb3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerasekara VK, Panek DJ, Broadbent DG, Mortenson JB, Mathis AD, Logan GN, Prince JT, Thomson DM, Thompson JW, Andersen JL. Metabolic-stress-induced rearrangement of the 14-3-3ζ interactome promotes autophagy via a ULK1- and AMPK-regulated 14-3-3ζ interaction with phosphorylated Atg9. Molecular and Cellular Biology. 2014;34:4379–4388. doi: 10.1128/MCB.00740-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Dickerson K, Heath R, Hong SP, Momcilovic M, Johnstone SR, Carlson M, Carling D. Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metabolism. 2005;2:21–33. doi: 10.1016/j.cmet.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LGD, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D. LKB1 Is the Upstream Kinase in the AMP-Activated Protein Kinase Cascade. Current Biology. 2003;13:2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- Wu N, Zheng Bin, Shaywitz A, Dagon Y, Tower C, Bellinger G, Shen C-H, Wen J, Asara J, McGraw TE, et al. AMPK-Dependent Degradation of TXNIP upon Energy Stress Leads to Enhanced Glucose Uptake via GLUT1. Molecular Cell. 2013;49:1167–1175. doi: 10.1016/j.molcel.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Sanders MJ, Carmena D, Bright NJ, Haire LF, Underwood E, Patel BR, Heath RB, Walker PA, Hallen S, et al. Structural basis of AMPK regulation by small molecule activators. Nature Communications. 2013;4:3017. doi: 10.1038/ncomms4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Sanders MJ, Underwood E, Heath R, Mayer FV, Carmena D, Jing C, Walker PA, Eccleston JF, Haire LF, et al. Structure of mammalian AMPK and its regulation by ADP. Nature. 2011;472:230–233. doi: 10.1038/nature09932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin FJ, Wang J, Zhao RQ, Wang ZX, Wu JW. Coordinated regulation of AMPK activity by multiple elements in the α-subunit. Cell Res. 2013;23:1237–1240. doi: 10.1038/cr.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Liang X, Sun X, Zhang L, Fu X, Rogers CJ, Berim A, Zhang S, Wang S, Wang B, et al. AMPK/α-Ketoglutarate Axis Dynamically Mediates DNA Demethylation in the Prdm16 Promoter and Brown Adipogenesis. Cell Metabolism. 2016;24:542–554. doi: 10.1016/j.cmet.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NP, Kamireddy A, Van Nostrand JL, Eichner LJ, Shokhirev MN, Dayn Y, Shaw RJ. AMPK governs lineage specification through Tfeb-dependent regulation of lysosomes. Genes & Development. 2016;30:535–552. doi: 10.1101/gad.274142.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CS, Jiang B, Li M, Zhu M, Peng Y, Zhang YL, Wu YQ, Li TY, Liang Y, Lu Z, et al. The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metabolism. 2014;20:526–540. doi: 10.1016/j.cmet.2014.06.014. [DOI] [PubMed] [Google Scholar]
- Zhang YL, Guo H, Zhang CS, Lin SY, Yin Z, Peng Y, Luo H, Shi Y, Lian G, Zhang C, et al. AMP as a Low-Energy Charge Signal Autonomously Initiates Assembly of AXIN-AMPK-LKB1 Complex for AMPK Activation. Cell Metabolism. 2013;18:546–555. doi: 10.1016/j.cmet.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Zhou C, Ma K, Gao R, Mu C, Chen L, Liu Q, Luo Q, Feng D, Zhu Y, Chen Q. Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res. 2017;27:184–201. doi: 10.1038/cr.2016.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmijewski JW, Banerjee S, Bae H, Friggeri A, Lazarowski ER, Abraham E. Exposure to hydrogen peroxide induces oxidation and activation of AMP-activated protein kinase. J Biol Chem. 2010;285:33154–33164. doi: 10.1074/jbc.M110.143685. [DOI] [PMC free article] [PubMed] [Google Scholar]


