Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
Whole-genome sequencing of 11 262 Icelanders reveals that clonal hematopoiesis is very common in the elderly.
Somatic mutation of some genes is strongly associated with clonal hematopoiesis, but in most cases, no driver mutations were evident.
Abstract
Clonal hematopoiesis (CH) arises when a substantial proportion of mature blood cells is derived from a single dominant hematopoietic stem cell lineage. Somatic mutations in candidate driver (CD) genes are thought to be responsible for at least some cases of CH. Using whole-genome sequencing of 11 262 Icelanders, we found 1403 cases of CH by using barcodes of mosaic somatic mutations in peripheral blood, whether or not they have a mutation in a CD gene. We find that CH is very common in the elderly, trending toward inevitability. We show that somatic mutations in TET2, DNMT3A, ASXL1, and PPM1D are associated with CH at high significance. However, known CD mutations were evident in only a fraction of CH cases. Nevertheless, the highly prevalent CH we detect associates with increased mortality rates, risk for hematological malignancy, smoking behavior, telomere length, Y-chromosome loss, and other phenotypic characteristics. Modeling suggests some CH cases could arise in the absence of CD mutations as a result of neutral drift acting on a small population of active hematopoietic stem cells. Finally, we find a germline deletion in intron 3 of the telomerase reverse transcriptase (TERT) gene that predisposes to CH (rs34002450; P = 7.4 × 10−12; odds ratio, 1.37).
Introduction
Hematopoietic stem cells (HSC) are responsible for the generation of all mature blood cells throughout life. Clonal hematopoiesis (CH) arises when a single HSC clonal lineage contributes disproportionately to the population of mature blood cells. Early indications of this phenomenon came from observations that the ratio of maternal to paternal X-chromosome inactivation is skewed in the blood of some otherwise healthy individuals, especially among the elderly.1-4 Skewing can be seen in all hematopoietic lineages, consistent with an origin in HSCs, but is most easily seen in nucleated cells of the myeloid lineage because they are short lived and require continuous replenishment from HSCs.5,6 Age-related CH does not arise from a simple depletion of HSCs, as the abundance of HSCs in human bone marrow actually increases in older people.7
Skewed X-inactivation also occurs in myeloid neoplasias, including acute myelogenous leukemia (AML), myelodysplastic syndromes, and myeloproliferative disorders.8-11 In AML, genes encoding epigenetic regulators such as DNMT3A, ASXL1, IDH2, and TET2 tend to mutate early in the development of disease.12 Mutations of these so-called “early genes” can persist in HSCs of patients in remission, creating reservoirs of pre-leukemic clones that can engender a relapse.13-16 Early gene mutations are also detectable in mature blood cells of patients with AML and some subjects with skewed X-inactivation, but ostensibly normal hematopoiesis.13,16,17 The frequencies of the mutant alleles in these cases indicate the mutant cells must have undergone clonal expansions, despite retaining a capacity to differentiate normally. This suggests early myeloid neoplasia-associated mutations in HSCs can drive CH without an obvious phenotypic effect on mature blood cells. In this article, we refer to genes and mutations that are suspected of promoting clonal expansion in CH as candidate drivers (CDs). A provisional list of CD genes, composed primarily of genes showing the characteristics of the early genes mentioned here, has been compiled by Steensma et al.18 Whole-exome sequencing (WES) and candidate gene analyses have shown that low-variant allele fraction (VAF) CD mutations indicative of CH are associated with increased all-cause mortality rates and risks for subsequent hematological malignancy.19,20 This prompted some investigators to propose that the presence of a CD mutation with a VAF above 2% constitutes an at-risk clinical entity, and to consider how it might be managed appropriately.18
In most studies based on DNA sequencing, the detection of CH per se has been inexorably bound up with the detection of mutations in genes previously known to be involved in hematological malignancies.19-23 Until now, this has precluded formal tests of association between CD mutations and CH. In this study we use the observation that HSCs accumulate somatic mutations during their life history, most of which have no apparent effect on cellular phenotype.24 Each HSC and its clonal descendants are therefore “barcoded” with a unique spectrum of mutations. If a particular clone becomes a substantial contributor to hematopoiesis, its unique spectrum of mutations should be evident in the sequence of peripheral blood DNA. The proportion of mature blood cells derived from a particular barcoded HSC clone will (for heterozygous mutant loci) be equivalent to about twice the VAF. Using this method to detect CH does not rely on the detection of a CD mutation, but it does require an extensive amount of sequence data. Genovese et al used a method like this on whole-exome sequence data to show that CH can occur in elderly people without CD mutations being detected.19 Similarly, Holstege et al used this method to detect extreme CH without CD mutations in a 115-year-old woman.25 We have sequenced the whole genomes of a substantial number of Icelanders.26 Here we use the whole-genome sequence (WGS) data to search for CH events by counting the number of low-VAF mutations present in peripheral blood. We find that CH is far more common in the elderly than has previously been demonstrated. In the majority of CH cases, no CD mutation was evident.
Methods
Full details of the methods used are given in the supplemental Data, available on the Blood Web site.
Subject recruitment and phenotyping
The study is based on WGS data from whole blood samples from 11 262 Icelanders participating in various disease projects at deCODE genetics. The study was authorized by the Icelandic National Bioethics Committee and the Data Protection Authority. All individuals gave informed consent. Patients were excluded if they had a diagnosis of hematological malignancy in the Icelandic Cancer Registry before or within 6 months after blood draw.
WGS
DNA samples were isolated from whole blood and prepared for sequencing using TruSeq Nano or TruSeq PCR-free library kits (Illumina). Libraries were sequenced on Illumina GAIIx, HiSeq2000/2500 or HiSeq X instruments. Single nucleotide polymorphisms (SNPs) and indels were called using GATK and GenotypeGVCFs27 and then annotated using Ensembl Variant Effect Predictor.28
Identification of mosaic somatic mutations
To differentiate mosaic somatic mutations from germline ones, we first restricted analysis to mutations that occurred only once in individuals of proven Icelandic ancestry. Because of the large size of our sample and the population structure of Iceland, germline variants are most likely to be observed more than once in our sample. We then imposed VAF restrictions to identify mosaic somatic mutations. Germline mutations can by chance have an observed VAF considerably less than 0.5. To control the false-discovery rate stemming from germline mutations to less than 1%, we considered only singleton mutations with a VAF less than 0.2 as somatic and mosaic. The lower frequency limit of detected somatic mutations was 0.1, as determined by the sensitivity of the variant caller. At least 3 independent reads containing a variant allele were required to call a somatic mutation. The spectrum of mosaic somatic mutations called was similar to what has been observed previously in AML24,29 (supplemental Figure 1).
Definition of clonal hematopoiesis cases
We counted the number of mosaic somatic mutations in each subject. The 99.5% quantile of the distribution of counts in subjects younger than 35 years was 20 mutations. Subjects with more than 20 mutations were defined as WGS-outliers. We took outlier status as evidence that the subject had CH.
Criteria for detection of CD mutations
For 18 CD genes on a list from Steensma et al,18 we included all high-impact mutations and any missense mutation that had been reported in the Catalogue of Somatic Mutations in Cancer (COSMIC) for hematopoietic or lymphoid tissue. For the less stringent “COSMIC5” criterion, we extended this list to include any high- or moderate-impact mutation (in any gene) that had been reported 5 times or more in COSMIC for hematopoietic or lymphoid tissue. Germline mutations were excluded, but somatic mutations were unconstrained by their VAF and were allowed to occur in more than 1 individual. For genome-wide burden testing of RefSeq genes, similar criteria were used for identification of somatic mutations. Mutations were graded by Variant Effect Predictor impact and then binned by gene for association testing against WGS-outlier status.
Panel resequencing
We used an Illumina TruSight Myeloid panel for resequencing 54 genes known to be mutated in myeloid neoplasia (to >5000× on Illumina MiSeq instruments). This was supplemented with a custom assay for the last 2 exons of PPM1D, sequenced to more than 600×. We called somatic mosaic variants down to a VAF of 0.01. Modeling revealed that the probability for a given CD mutation to explain an observed WGS-barcode clone drops precipitously when the VAF of the CD mutation drops < 0.05. At a VAF of 0.01, the probability that a causal CD mutation could generate a clone with a detectable WGS-outlier barcode is only 2.1 × 10−12. We note in passing that under this model, none of the low-VAF CD mutations reported by Young et al23 could have generated a case that meets our criteria for CH.
WGS-based genome-wide association for inherited variants
Long-range haplotype phasing and genotype imputation was performed as described previously.30-32 This permitted us to reliably test for association with germline variants down to a minor allele frequency of about 0.05%.
Telomere length assay
Estimates of telomere length were obtained from WGS data using TelSeq software.33
Statistical analysis
Analyses were performed using R packages, including packages survival and ggplot2. For modeling of clonal hematopoiesis, the model of Dingli et al34 was adapted to our data. Details of all analyses are presented in the supplemental Data.
Results
Clonal hematopoiesis is very common in the elderly
We generated WGS from 11 262 people to an average sequencing depth of 35.6 (median, 34.8; range, 20.4-119.2). We identified 3 300 768 singleton SNPs, of which 146 389 were classified as mosaic somatic mutations. The Icelandic genealogy was used to ensure none of the mosaic somatic mutations were transmitted in the germline (supplemental Data; supplemental Figure 2). The mean VAF of the mutations was 0.17, with a range of 0.11 to 0.20. The upper boundary was set to control the false-discovery rate because of interference from constitutional mutations to a level of less than 1%, whereas the lower boundary was determined by the sensitivity of the variant caller. As shown in Figure 1A, younger subjects showed a distribution with a median of around 3 mosaic somatic mutations. Above an age of 35 to 45 years, the number of people with a high count of mosaic somatic mutations climbed rapidly. We applied a cutoff of more than 20 mosaic somatic mutations (corresponding to the 99.5% quantile of the distribution for ages younger than 35 years) to classify individuals as WGS-outliers (supplemental Table 2). We took WGS-outlier status as evidence that the person had CH. By this criterion, 1403 of 11 262 participants had CH, a prevalence of 12.5% over all age groups. The frequency of WGS-outliers increased from 0.5% (by definition) in subjects younger than 35 years to more than 50% for subjects older than 85 years (Figure 1A-B). This peak prevalence is more than twice that of previous estimates made using WES data.19,20 Hence, it appears that CH is far more common among the elderly than has previously been shown. This confirms a prediction by McKerrell et al that CH would prove to be an almost inevitable consequence of advanced aging.21
Figure 1.
Age distribution of clonal hematopoiesis detected by WGS-outlier status. (A) Histograms showing the number of mosaic somatic mutations per person stratified by their age at blood sampling (adjusted as described in the supplemental Data). The vertical line shows the cutoff at 20 mosaic somatic mutations (corresponding to the 99.5% quantile of the distribution for ages younger than 35 years) that was used to classify individuals as WGS-outliers. (B) Prevalence of clonal hematopoiesis and CD mutations stratified by age class. Lavender bar, the fraction of samples classified as WGS-outliers; red bar, the fraction of samples with detected CD mutations from the 18-gene candidate list18; green bar, the fraction of samples detected as outliers, using exon-restricted analysis; blue bar, combined fraction of samples detected with CD mutation or exon-restricted analysis. Error bars indicate 95% confidence intervals.
To align our analysis with what might be detected by WES, we restricted the scope to mosaic somatic mutations that occurred in exons. Only 176 subjects were then detected as outliers (supplemental Data), 174 of whom had already been identified as WGS-outliers. The exome-restricted detection rate corresponded to 12.4% of the 1403 CH cases detected as WGS-outliers (Figure 1B). These 174 people had a 2.65-fold higher number of mosaic somatic mutations, as measured by WGS (mean number, 138 vs 52; P = 2.8 × 10−81), and were on average 7.8 years older than other WGS-outliers (P = 1.2 × 10−7). The age-specific prevalence of outliers detected by exon-restricted analysis was similar to published estimates using WES (Figure 1B).19,20 Thus, the WGS-outlier method offers much greater sensitivity for detection of CH than exome-restricted methods.
Association of driver mutations with clonal hematopoiesis
We investigated what proportion of our CH cases carries detectable mosaic somatic mutations in CD genes. As candidates, we used a list of 18 CD genes that have previously been seen mutated in myeloid neoplasia.18 The list also includes PPM1D, which was never implicated in hematological malignancy, but shows somatic mutation in blood in association with breast cancer, ovarian cancer, and prior chemotherapy.35-38 For detection of CD mutations, we used a different strategy from the one used to detect barcode mutations. We permitted somatic mutations that were present in more than 1 individual and at a wider range of VAFs (supplemental Data). We found 286 CD mutations in 16 of the genes in 246 of 11 262 people (supplemental Table 2). For 196 individuals, their CD mutation occurred in DNMT3A (n = 93), TET2 (n = 76), ASXL1 (n = 25), or PPM1D (n = 18). The probability of detection of a CD mutation was strongly dependent on age (Figure 1B). Subjects with CD mutations were on average 18 years older than those without (P = 2 × 10−46). The age-specific prevalence of CD mutations was in line with what has been observed previously using WES.19,20 However, mutations in the 18 CD genes were detected in only a small proportion of CH cases 12.6%, 177/1403; Figure 1B). Employing a less stringent criterion to define CD mutations (“COSMIC5”; supplemental Data) explained a further 9 CH cases (supplemental Table 2). Analysis of sample subsets for copy number variations (CNV) resulting in deletions of CD genes, recurrent AML fusion genes, or for mutations detected by high-depth WGS did not yield substantial numbers of additionally explained CH cases (supplemental Data; supplemental Figure 3; supplemental Table 4). We then employed deep resequencing of a panel of amplicons from 54 genes frequently mutated in myeloid malignancies, as well as PPM1D, on 76 CH cases selected by stratified random sampling. The method allowed us to detect mutations reliably down to a VAF of 0.01 (ie, approximately 10-fold lower than the lower limit of WGS-outlier barcode mutations). We found CD mutations in 30 (39.5%) of the 76 cases who were resequenced (supplemental Table 3). Adding the estimated 1.2% of cases who have a large deletion of a CD gene (supplemental Data; supplemental Figure 3), this corresponds to 40.7% of WGS-outliers who might plausibly be accounted for by a CD mutation detectable by our methods. This is likely to be an overestimate, however, because some of the observed CD mutations may not have a sufficiently high VAF to account for the clone generating the associated WGS-outlier barcode (supplemental Data). In any case, the majority of our CH cases remain in need of a satisfactory mechanistic explanation.
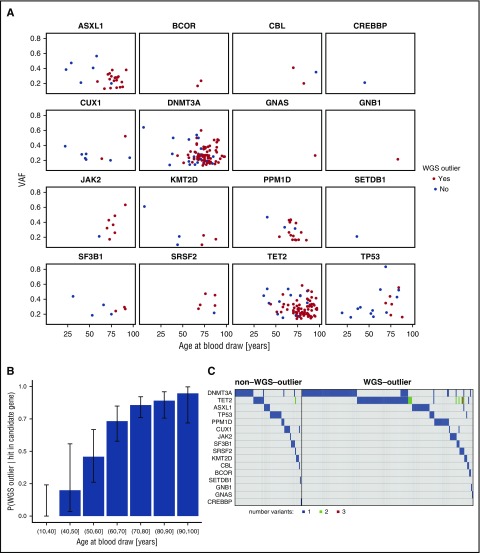
Overall, 177 (72%) of the 246 subjects with WGS-detected CD mutations were also WGS-outliers. For younger individuals in particular, CD mutations sometimes occurred without WGS-outlier status being detected (Figure 2A; supplemental Tables 2 and 3). We speculated that some younger people who have CH with CD mutations might not have been detected as WGS-outliers because they had not accumulated enough mosaic somatic mutations to qualify. Indeed, the fraction of subjects with a CD mutation who were also WGS-outliers increased significantly with age (P = 1.9 × 10−12; Figure 2B). DNMT3A, TET2, and ASXL1 were the genes most commonly mutated in both WGS-outliers and nonoutliers (Figure 2C). So, despite its high positivity rate in older people, the WGS-outlier method is still likely to underreport CH among the young.
Figure 2.
Presence of candidate driver (CD) mutations by age. (A) VAF vs age at blood draw for the 16 CD genes where mutations were detected. The 177 subjects who were classified as WGS-outliers are plotted as blue points, and the 69 subjects who were not outliers are plotted as red points. (B) Conditional probability of being identified as a WGS-outlier given that a CD mutation was detected, stratified by age bins. P = 1.9 × 10−12; β = 0.10, assessed by logistic regression. Error bars indicate 95% confidence intervals. (C) Co-mutation plot for WGS-outliers and nonoutliers in whom CD mutations were detected. Each column represents a subject, each row a candidate pre-leukemic driver gene. Cells are shaded if a mutation was detected, and the color of the shading indicates the number of mutations detected for the particular gene. The vertical black line separates non–WGS-outlier from WGS-outlier subjects.
Unlike previous studies, our WGS-outlier method defines CH cases irrespective of whether or not they have a mutation in a CD gene. Therefore, we could carry out unbiased tests of whether the presence of a mutant CD gene is associated with CH status. As shown in Table 1, 11 of the 18 genes nominated by Steensma et al18 demonstrated an association with CH at a significance of P < .05 in burden tests. The less stringent COSMIC5 criterion added some plausible candidate genes for which mutations were found in WGS-outliers: ATM, IDH2, KIT, MPL, MYD88, and NRAS. Of these, IDH2 and MYD88 reached P < .05 significance.
Table 1.
Association of mutant candidate pre-leukemic driver genes with clonal hematopoiesis defined by WGS-outlier status
| Gene symbol | Number of nonoutliers with mutation in gene* | Number of WGS outliers with mutation in gene† | P value‡ | OR | Candidate gene source | Significant? |
|---|---|---|---|---|---|---|
| TET2 | 12 | 64 | 2.5 × 10−46 | 39.2 | Steensma et al18 | * |
| DNMT3A | 22 | 71 | 9.5 × 10−46 | 23.8 | Steensma et al18 | * |
| ASXL1 | 6 | 19 | 4.9 × 10−13 | 22.5 | Steensma et al18 | * |
| PPM1D | 3 | 15 | 1.4 × 10−11 | 35.5 | Steensma et al18 | * |
| JAK2 | 1 | 7 | 3.3 × 10−6 | 49.5 | Steensma et al18 | * |
| SRSF2 | 1 | 5 | 1.6 × 10−4 | 35.2 | Steensma et al18 | * |
| BCOR | 0 | 2 | .016 | Inf | Steensma et al18 | * |
| IDH2 | 0 | 2 | .016 | Inf | COSMIC5 | * |
| MYD88 | 0 | 2 | .016 | Inf | COSMIC5 | * |
| TP53 | 12 | 6 | .018 | 3.5 | Steensma et al18 | * |
| KMT2D | 3 | 3 | .029 | 7 | Steensma et al18 | * |
| CBL | 1 | 2 | .043 | 14.1 | Steensma et al18 | * |
| SF3B1 | 4 | 3 | .046 | 5.3 | Steensma et al18 | * |
| ATM | 0 | 1 | .12 | Inf | COSMIC5 | |
| GNAS | 0 | 1 | .12 | Inf | Steensma et al18 | |
| GNB1 | 0 | 1 | .12 | Inf | Steensma et al18 | |
| KIT | 0 | 1 | .12 | Inf | COSMIC5 | |
| MPL | 0 | 1 | .12 | Inf | COSMIC5 | |
| STAT3 | 0 | 1 | .12 | Inf | COSMIC5 | |
| NRAS | 1 | 1 | .23 | 7 | COSMIC5 | |
| CUX1 | 7 | 2 | .31 | 2 | Steensma et al18 | |
| AKAP17A | 2 | 1 | .33 | 3.5 | COSMIC5 | |
| ACSS2 | 3 | 1 | .41 | 2.3 | COSMIC5 | |
| HRC | 5 | 1 | .55 | 1.4 | COSMIC5 | |
| GOLGA8B | 27 | 2 | .57 | 0.5 | COSMIC5 | |
| CEP112 | 1 | 0 | 1 | 0 | COSMIC5 | |
| CREBBP | 1 | 0 | 1 | 0 | Steensma et al18 | |
| ROBO1 | 1 | 0 | 1 | 0 | COSMIC5 | |
| SETDB1 | 1 | 0 | 1 | 0 | Steensma et al18 | |
| SYNE1 | 3 | 0 | 1 | 0 | COSMIC5 | |
| UMODL1 | 2 | 0 | 1 | 0 | COSMIC5 |
Total number of non–WGS-outliers: n = 9859.
Total number of WGS-outliers: n = 1403.
Fisher's exact test.
To extend the search for mutant genes associated with CH to a genome-wide scale, we carried out burden tests of 17 933 RefSeq genes with high- or moderate-impact mosaic somatic mutations for association with WGS-outlier status. After Bonferroni correction, TET2, DNMT3A, ASXL1, and PPM1D demonstrated highly significant associations with CH in burden tests based on high-impact mutations only, or both high- and moderate-impact mutations (Table 2; supplemental Table 5). DNMT3A and TET2 were also significant when moderate mutations alone were considered. These associations directly implicate those genes as drivers of CH. Other genes reached suggestive levels of significance, notably moderate-impact mutations in ATM (P = 3.9 × 10−6), SRSF2 (P = 3.0 × 10−5), and MTA2 (P = 9.6 × 10−5). The latter gene, Metastasis-Associated Protein 2, encodes a component of the nucleosome remodeling and histone deacetylation (NuRD) complex. It is widely expressed, including in leukocytes and bone marrow, but has no known involvement in myeloid disease. Further investigation of MTA2 is warranted.
Table 2.
Genome-wide burden test for association of mutant genes with clonal hematopoiesis
| Mutation impact and gene symbol | Number of nonoutliers with mutation in gene* | Number of WGS outliers with mutation in gene† | P value‡ | OR |
|---|---|---|---|---|
| High | ||||
| TET2 | 10 | 57 | 7.1 × 10−42 | 41.68 |
| DNMT3A | 6 | 38 | 9.0 × 10−29 | 45.65 |
| ASXL1 | 2 | 18 | 7.0 × 10−15 | 64.04 |
| PPM1D | 3 | 15 | 1.4 × 10−11 | 35.48 |
| ZNF318 | 5 | 8 | 4.1 × 10−5 | 11.30 |
| Moderate | ||||
| DNMT3A | 33 | 68 | 5.1 × 10−38 | 15.16 |
| TET2 | 12 | 33 | 6.4 × 10−21 | 19.76 |
| ATM | 45 | 23 | 3.9 × 10−6 | 3.63 |
| SRSF2 | 0 | 5 | 3.0 × 10−5 | Inf |
| KPNA7 | 106 | 34 | 9.2 × 10−5 | 2.29 |
| MTA2 | 4 | 7 | 9.6 × 10−5 | 12.35 |
| High and moderate | ||||
| DNMT3A | 39 | 105 | 6.6 × 10−64 | 20.36 |
| TET2 | 21 | 85 | 7.8 × 10−58 | 30.18 |
| ASXL1 | 27 | 23 | 5.0 × 10−9 | 6.07 |
| PPM1D | 16 | 16 | 2.6 × 10−7 | 7.09 |
| SRSF2 | 0 | 5 | 3.0 × 10−5 | Inf |
| PRKCG | 5 | 8 | 4.1 × 10−5 | 11.30 |
| ATM | 59 | 24 | 6.3 × 10−5 | 2.89 |
| MTA2 | 4 | 7 | 9.6 × 10−5 | 12.35 |
Data down to P = .01 are shown in supplemental Table 5.
Total number of non–WGS-outliers n = 9859.
Total number of WGS-outliers n = 1403.
Fisher's exact test.
CH is associated with higher death rates and increased risks for hematological malignancy
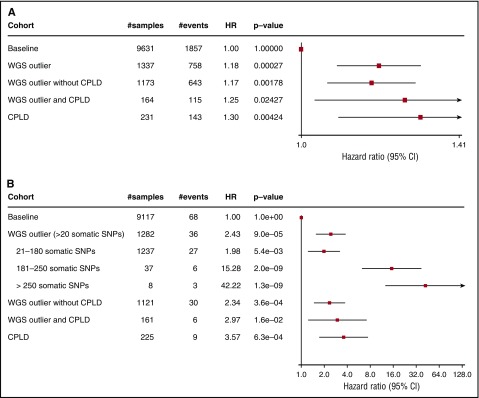
Subjects with CH defined by WGS-outlier status had significantly higher rates of all-cause mortality (hazard ratio [HR], 1.18; P = 2.7 × 10−4; Figure 3A). WGS-outliers with or without detected CD mutations had similar risks. Subjects who had CD mutations were also at increased risk, irrespective of WGS-outlier status (HR, 1.30; P = .0042). To set the increased mortality rates in perspective, we found that smoking (ever) had an HR of 1.19 (P = 2.6 × 10−5). Therefore, the effect of clonal hematopoiesis on mortality rate is similar to ever smoking.
Figure 3.
Survival analysis using Cox proportional hazard model. Baseline was defined as subjects who were neither WGS-outliers nor carriers of a mosaic somatic CD mutation. Plots show HRs with 95% confidence intervals. (A) HRs for all-cause mortality adjusted for age at blood draw, year of birth, sex, previous diagnoses of cancer, and smoking. (B) HRs for subsequent hematological malignancy adjusted for age at blood draw and year of birth. Details of the subjects who developed hematological malignancies are shown in supplemental Table 6.
We also assessed the risk for a hematological malignancy arising 6 months or more after sampling (Figure 3B). CH defined by WGS-outlier status substantially increased the risk for a subsequent hematological malignancy (HR, 2.43; P = 9.0 × 10−5). Again, WGS-outliers with or without detected CD mutations had similar risks, and all subjects with CD mutations were at increased risk. We noted that the risk increased with count of mosaic somatic mutations to an HR of 42.2 (P = 1.3 × 10−9) in subjects with more than 250 mutations. This might be indicative of disease risk associated with the age (in divisions) of the HSC clone when it began to expand, its rate of expansion, its degree of predominance in supporting hematopoiesis (as the number of detected mutations is correlated with VAF), or the probability that a leukemic driver gene had been hit by a mutation. Alternatively, it might indicate that individuals with very high mutation counts have an undiagnosed, non-HSC hematological malignancy.
Association of CH with other phenotypes
We tested for association between the WGS-outlier status and 1482 case–control phenotypes from the deCODE database (supplemental Table 7). After consideration of a Bonferroni correction threshold (P < 3.4 × 10−5), significant associations were found with smoking (P = 6.0 × 10−13), treatment of addiction (P = 4.2 × 10−8), psychiatric disease (P = 9.5 × 10−6), smoking-related diseases (P = 1.1 × 10−5), and chronic pulmonary disease (P = 1.4 × 10−5). In analyses limited to those for whom smoking information was available, addiction and psychiatric disease remained significant (P < .05) after adjustment for smoking. However, these traits do have known correlations with smoking behavior, including metrics of smoking quantity, which were not taken into account. Smoking was also associated with the number of mosaic somatic mutations detected, irrespective of WGS-outlier status (P = 1.3 × 10−11). No cancer phenotypes reached Bonferroni corrected significance, the lowest P value being for lung adenocarcinoma (P = .001). Testing association with 4078 quantitative traits revealed significant increases in counts of white blood cells (P = 5.0 × 10−11), monocytes (P = 2.2 × 10−10), platelets (P = 1.2 × 10−9), lymphocytes (P = 6.4 × 10−8), granulocytes (P = 6.8 × 10−6), and an increase in total platelet volume (P = 7.8 × 10−10; supplemental Table 8).
Mosaic loss of the Y chromosome is reportedly similar to CH in its age and phenotypic associations.39-41 Mosaic loss-of-Y showed a highly significant association with CH in our WGS data (P = 5.02 × 10−110) and had a similar age distribution (supplemental Data; supplemental Figure 4). This suggests that mosaic loss-of-Y and CH are related phenomena.
Modeling clonal hematopoiesis caused by neutral drift
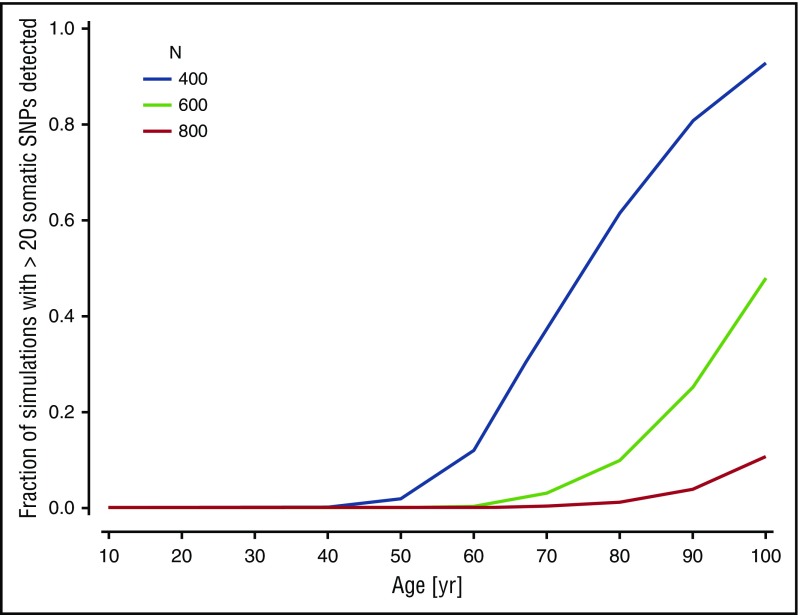
The lack of identified CD mutations in most of the WGS-outliers led us to question whether such mutations are essential for CH to arise. Modeling studies show that in a stem cell pool of constrained size, and in the absence of any clonal selective advantage, a large fraction of the stem cells will eventually derive from a single clone as a result of neutral drift.42 The question is, therefore, not whether CH will manifest itself, but over what timescale in relation to the human life span. To examine whether neutral drift could possibly account for some of our CH cases, we considered a simple model of stem cell dynamics34 adapted to our data (supplemental Data). Simulations with a plausible set of parameters indicated that clonal expansion could occur so rapidly by neutral drift that a substantial proportion of the WGS-outliers without known CD mutations would be explained (Figure 4). Neutral drift should therefore be considered as one possible mechanism underlying CH.
Figure 4.
Computer simulation of clonal hematopoiesis arising under neutral drift. The graph shows the proportion of simulations producing more than 20 observable mosaic somatic mutations with a VAF less than 0.2 as a function of subject age, for different choices of N, the size of the active HSC compartment. The value of p, the probability that an HSC division will produce 2 daughter stem cells, was set at 0.25.51 Other parameters were fixed at λ = 1 division per 40 weeks,43 mutation rate µ = 6.4 × 10−10 per base pair per division.52
We noted that the simulations were very sensitive to N, the assumed size of the active HSC pool (Figure 4). Estimates for the size of this pool vary widely.43-45 The model was also sensitive to the value of p, the probability that a given cell division will produce 2 daughter stem cells. Although a high pi can give a particular HSC clone i a competitive advantage, a high p value could apply equally to all members of the HSC pool and still promote rapid development of CH (supplemental Figure 5). In other words, a person with an endogenously high p could be predisposed to develop CH early through neutral drift.
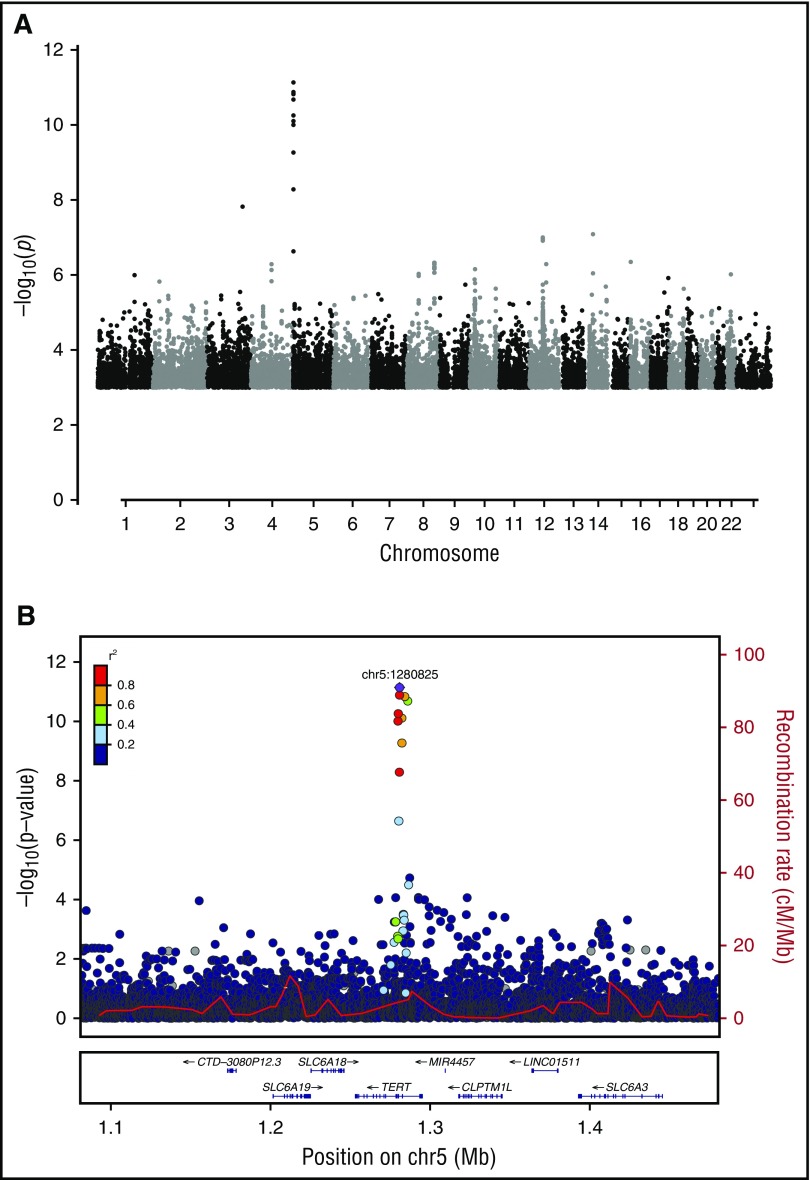
Association of germline TERT variants with CH
To examine further the concept of constitutive predisposition to CH, we carried out a WGS-based genome-wide association study for germ-line SNPs and small indels,26 using WGS-outlier status as the query phenotype. The strongest association came from an 8-bp deletion in intron 3 of the telomerase reverse transcriptase (TERT) gene (rs34002450, g.1280826_1280833delAGCCCACC; P = 7.4 × 10−12; odds ratio [OR], 1.37; allele frequency, 40.6% in Iceland; Figure 5). Previously, we reported an association between myeloproliferative neoplasms and a common variant in TERT (rs2736100, r2 = 0.28 vs rs34002450).46 Conditional analysis showed that the CH association mapped preferentially to rs34002450 (Padj = 3.6 × 10−8), whereas the myeloproliferative neoplasms association mapped preferentially to rs2736100 (Punadj = 2.2 × 10−8 , Padj = 6.4 × 10−4). Rare coding mutations in TERT have been implicated in marrow failure and HSC dysfunction.47,48 In our study, no high- or moderate-impact germline variants (with CH-association P values of <.05 and <2.5 × 10−3, respectively) were seen in TERT or any of the 18 CD genes.
Figure 5.
Genome-wide association for germline variants associated with clonal hematopoiesis detected by WGS-outlier status. (A) Manhattan plot of association [expressed as –log10(P)] with WGS-outlier status, determined using logistic regression. (B) Locus zoom of the signal in the TERT gene on chromosome 5. The location of the 8-bp indel rs34002450 (chr5:1280825) giving the strongest signal is indicated by a purple diamond. Other variants are plotted in colors corresponding to their r2 values relative to rs34002450, as indicated in the legend. Recombination rates, in cM/Mb and based on Icelandic data, are plotted as a red line. The lower panel shows the locations of RefSeq genes and the chromosomal position (GRCh38/hg38).
The TERT association suggested telomerase activity might have a role in CH. To explore this further, we estimated telomere lengths for 2703 samples based on the occurrence of the telomeric TTAGGG motif in WGS reads (supplemental Data). Telomere length estimates were significantly lower in people with CH defined by WGS-outlier status (β, −0.19 standard deviations; P = 1.0 × 10−3). The presence of CD mutations had no effect in addition to WGS-outlier status. Moreover, we could detect no effect of the rs34002450 deletion on telomere length (P = .22, linear regression adjusted for sex, age at blood draw, chronic obstructive pulmonary disease, cancer diagnoses, and smoking). Both telomere length and rs34002450 genotype were significant independent predictors of WGS-outlier status in a multivariate regression (P = .008 and .003, respectively, adjusted for age at blood draw and smoking). The involvement of TERT and telomere length with CH merits further investigation. Moreover, studies that ascribe phenotypic effects to variations in telomere length should consider a possible role of CH.
Discussion
Using WGS data from peripheral blood of 11 262 people, we have developed a method to identify individuals with CH based on the accumulation of somatic mutations in the dominant HSC clone. Unlike previous studies of CH, this method does not depend on prior knowledge of CD genes or mutations. We detected a high prevalence of CH among the elderly, trending toward inevitability.
Holstege et al25 studied WGS of a 115-year-old woman with extreme CH and found no CD mutations (nor indeed any mutations present in the COSMIC catalog). This suggested that CH might occur in the absence of detectable CD mutations. Similarly, Genovese et al,19 using WES, did not detect CD mutations in a substantial fraction of their patients with CH. In our study, we did not find obvious CD mutations in the majority of subjects with WGS-outlier status indicative of CH. Conceivably, the lack of identified CD mutations, first, could be the result of a simple inability to detect them with the methods applied. Our sensitivity to detecting a driver mutation might be rather limited for clones with low degrees of predominance and high mutation burdens. Second, because of genetic heterogeneity, each individual driver mutation might occur at too low a frequency in the CH population to be recognized as such, or be located in a nonexonic region. Third, CH could be driven by variation in clonally inherited epigenetic states that affect the self-renewal and proliferative capacities of HSCs.49 Fourth, some CH may be a simple (or indeed, inevitable) consequence of neutral drift operating on the small, aging population of active HSCs.42 If this latter scenario is true, it invites the question of why CH without defined CD mutations still carries high hazards of hematological malignancy and all-cause mortality, as we and others19 have seen. Perhaps some instances of CH are indications the HSC compartment is in a permissive state that allows clonal expansions to occur over short timescales. The situation may be akin to melanoma, in which a high number of benign nevi is a strong risk factor even though few nevi progress to melanoma.50
The ability to identify CH cases presents opportunities for monitoring and intervention. A number of authors have recommended the development of strategies to target and eliminate clonal reservoirs of HSCs containing pre-leukemic mutations.13,15,16,18 Such strategies must take into account that CH appears to be quite common in elderly asymptomatic individuals and that absolute risks for progression to hematological malignancy are low. On the basis of the observations described here, targeting known pre-leukemic driver mutations may address only a fraction of the at-risk individuals. Moreover, some people may be reliant on surviving mutant HSC clones to support their normal hematopoiesis, and this may be associated with a general frailty. Clearly a deeper understanding of the nature and associated risks for clonal hematopoiesis would be valuable.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
This work was supported, in part, by National Institutes of Health, National Institute on Drug Abuse grants R01-DA017932 and R01-DA034076.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: The study was designed and the results interpreted by F.Z., S.N.S., G.L.N., A.K., and K.S.; subject ascertainment and recruitment was carried out by S.N.S., I.J., T.E.T., J.G., J.G.J., L.T., T.J., T.R., and U.T.; sequencing and genotyping was done by F.Z., S.N.S., O.T.M., A.S., and G.M.; statistical and bioinformatics analysis was done by F.Z., S.N.S., G.L.N., M.L.F., S.A.G., A.H., A.G., P.S., D.F.G., G.M., and A.K.; the manuscript was drafted by F.Z., S.N.S., G.L.N, A.K., and K.S.; and all authors contributed to the final version of the paper.
Conflict-of-interest disclosure: All deCODE authors are employees of the biotechnology company deCODE genetics/AMGEN.
Correspondence: Augustine Kong, deCODE genetics/AMGEN, Sturlugata 8, 101 Reykjavik, Iceland; e-mail: augustine.kong@decode.is; and Kari Stefansson, deCODE genetics/AMGEN, Sturlugata 8, 101 Reykjavik, Iceland; e-mail: kari.stefansson@decode.is.
References
- 1.Busque L, Mio R, Mattioli J, et al. Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood. 1996;88(1):59-65. [PubMed] [Google Scholar]
- 2.Busque L, Paquette Y, Provost S, et al. Skewing of X-inactivation ratios in blood cells of aging women is confirmed by independent methodologies. Blood. 2009;113(15):3472-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fey MF, Liechti-Gallati S, von Rohr A, et al. Clonality and X-inactivation patterns in hematopoietic cell populations detected by the highly informative M27 beta DNA probe. Blood. 1994;83(4):931-938. [PubMed] [Google Scholar]
- 4.Gale RE, Wheadon H, Linch DC. X-chromosome inactivation patterns using HPRT and PGK polymorphisms in haematologically normal and post-chemotherapy females. Br J Haematol. 1991;79(2):193-197. [DOI] [PubMed] [Google Scholar]
- 5.Champion KM, Gilbert JG, Asimakopoulos FA, Hinshelwood S, Green AR. Clonal haemopoiesis in normal elderly women: implications for the myeloproliferative disorders and myelodysplastic syndromes. Br J Haematol. 1997;97(4):920-926. [DOI] [PubMed] [Google Scholar]
- 6.Sawai CM, Babovic S, Upadhaya S, et al. Hematopoietic stem cells are the major source of multilineage hematopoiesis in adult animals. Immunity. 2016;45(3):597-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pang WW, Price EA, Sahoo D, et al. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc Natl Acad Sci USA. 2011;108(50):20012-20017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anger B, Janssen JW, Schrezenmeier H, Hehlmann R, Heimpel H, Bartram CR. Clonal analysis of chronic myeloproliferative disorders using X-linked DNA polymorphisms. Leukemia. 1990;4(4):258-261. [PubMed] [Google Scholar]
- 9.Fialkow PJ, Faguet GB, Jacobson RJ, Vaidya K, Murphy S. Evidence that essential thrombocythemia is a clonal disorder with origin in a multipotent stem cell. Blood. 1981;58(5):916-919. [PubMed] [Google Scholar]
- 10.Raskind WH, Tirumali N, Jacobson R, Singer J, Fialkow PJ. Evidence for a multistep pathogenesis of a myelodysplastic syndrome. Blood. 1984;63(6):1318-1323. [PubMed] [Google Scholar]
- 11.Wiggans RG, Jacobson RJ, Fialkow PJ, Woolley PV III, Macdonald JS, Schein PS. Probable clonal origin of acute myeloblastic leukemia following radiation and chemotherapy of colon cancer. Blood. 1978;52(4):659-663. [PubMed] [Google Scholar]
- 12.Grove CS, Vassiliou GS. Acute myeloid leukaemia: a paradigm for the clonal evolution of cancer? Dis Model Mech. 2014;7(8):941-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corces-Zimmerman MR, Hong W-J, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci USA. 2014;111(7):2548-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jan M, Snyder TM, Corces-Zimmerman MR, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med. 2012;4(149):149ra118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krönke J, Bullinger L, Teleanu V, et al. Clonal evolution in relapsed NPM1-mutated acute myeloid leukemia. Blood. 2013;122(1):100-108. [DOI] [PubMed] [Google Scholar]
- 16.Shlush LI, Zandi S, Mitchell A, et al. ; HALT Pan-Leukemia Gene Panel Consortium. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Busque L, Patel JP, Figueroa ME, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44(11):1179-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steensma DP, Bejar R, Jaiswal S, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKerrell T, Park N, Moreno T, et al. ; Understanding Society Scientific Group. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Reports. 2015;10(8):1239-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun. 2016;7:12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welch JS, Ley TJ, Link DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holstege H, Pfeiffer W, Sie D, et al. Somatic mutations found in the healthy blood compartment of a 115-yr-old woman demonstrate oligoclonal hematopoiesis. Genome Res. 2014;24(5):733-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gudbjartsson DF, Helgason H, Gudjonsson SA, et al. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet. 2015;47(5):435-444. [DOI] [PubMed] [Google Scholar]
- 27.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26(16):2069-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ley TJ, Miller C, Ding L, et al. ; Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong A, Masson G, Frigge ML, et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat Genet. 2008;40(9):1068-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong A, Steinthorsdottir V, Masson G, et al. ; DIAGRAM Consortium. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462(7275):868-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinthorsdottir V, Thorleifsson G, Sulem P, et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet. 2014;46(3):294-298. [DOI] [PubMed] [Google Scholar]
- 33.Ding Z, Mangino M, Aviv A, Spector T, Durbin R; UK10K Consortium. Estimating telomere length from whole genome sequence data. Nucleic Acids Res. 2014;42(9):e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dingli D, Traulsen A, Michor F. (A)symmetric stem cell replication and cancer. PLOS Comput Biol. 2007;3(3):e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akbari MR, Lepage P, Rosen B, et al. PPM1D mutations in circulating white blood cells and the risk for ovarian cancer. J Natl Cancer Inst. 2014;106(1):djt323. [DOI] [PubMed] [Google Scholar]
- 36.Pharoah PDP, Song H, Dicks E, et al. ; Australian Ovarian Cancer Study Group; Ovarian Cancer Association Consortium. PPM1D mosaic truncating variants in ovarian cancer cases may be treatment-related somatic mutations. J Natl Cancer Inst. 2016;108(3):djv347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruark E, Snape K, Humburg P, et al. ; Breast and Ovarian Cancer Susceptibility Collaboration; Wellcome Trust Case Control Consortium. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature. 2013;493(7432):406-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swisher EM, Harrell MI, Norquist BM, et al. Somatic mosaic mutations in PPM1D and TP53 in the blood of women with ovarian carcinoma. JAMA Oncol. 2016;2(3):370-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forsberg LA, Rasi C, Malmqvist N, et al. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat Genet. 2014;46(6):624-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacobs PA, Brunton M, Court Brown WM, Doll R, Goldstein H. Change of human chromosome count distribution with age: evidence for a sex differences. Nature. 1963;197:1080-1081. [DOI] [PubMed] [Google Scholar]
- 41.Pierre RV, Hoagland HC. Age-associated aneuploidy: loss of Y chromosome from human bone marrow cells with aging. Cancer. 1972;30(4):889-894. [DOI] [PubMed] [Google Scholar]
- 42.Klein AM, Simons BD. Universal patterns of stem cell fate in cycling adult tissues. Development. 2011;138(15):3103-3111. [DOI] [PubMed] [Google Scholar]
- 43.Catlin SN, Busque L, Gale RE, Guttorp P, Abkowitz JL. The replication rate of human hematopoietic stem cells in vivo. Blood. 2011;117(17):4460-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dingli D, Pacheco JM. Allometric scaling of the active hematopoietic stem cell pool across mammals. PLoS One. 2006;1:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rozhok AI, Salstrom JL, DeGregori J. Stochastic modeling reveals an evolutionary mechanism underlying elevated rates of childhood leukemia. Proc Natl Acad Sci USA. 2016;113(4):1050-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oddsson A, Kristinsson SY, Helgason H, et al. The germline sequence variant rs2736100_C in TERT associates with myeloproliferative neoplasms. Leukemia. 2014;28(6):1371-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gramatges MM, Bertuch AA. Short telomeres: from dyskeratosis congenita to sporadic aplastic anemia and malignancy. Transl Res. 2013;162(6):353-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Townsley DM, Dumitriu B, Young NS. Bone marrow failure and the telomeropathies. Blood. 2014;124(18):2775-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schroeder T. Hematopoietic stem cell heterogeneity: subtypes, not unpredictable behavior. Cell Stem Cell. 2010;6(3):203-207. [DOI] [PubMed] [Google Scholar]
- 50.Holman CD, Armstrong BK. Pigmentary traits, ethnic origin, benign nevi, and family history as risk factors for cutaneous malignant melanoma. J Natl Cancer Inst. 1984;72(2):257-266. [PubMed] [Google Scholar]
- 51.Werner B, Beier F, Hummel S, et al. Reconstructing the in vivo dynamics of hematopoietic stem cells from telomere length distributions. eLife. 2015;4:e08687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomasetti C, Vogelstein B, Parmigiani G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc Natl Acad Sci USA. 2013;110(6):1999-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.